Abstract
Female and male mice of the BTBR T+Itpr3tf/J (BTBR) strain have behaviors that resemble autism spectrum disorder. In comparison to C57BL/6 (B6) mice, BTBR mice have elevated humoral immunity, in that they have naturally high serum IgG levels and generate high levels of IgG antibodies, including autoantibodies to brain antigens. This study focused on the specificities of autoantibodies and the immune cells and their transcription factors that might be responsible for the autoantibodies. BTBR IgG autoantibodies bind to neurons better than microglia and with highest titer to nuclear antigens. Two of the antigens identified were alpha-enolase (ENO1) and dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial (DLST). Surprisingly based on IgG levels, the blood and spleens of BTBR mice have more CD4+ and CD8+ T cells, but fewer B cells than B6 mice. The high levels of autoantibodies in BTBR relates to their splenic T follicular helper (Tfh) cell levels, which likely are responsible for the higher number of plasma cells in BTBR mice than B6 mice. BTBR mice have increased gene expression of interleukin-21 receptor (Il-21r) and Paired Box 5 (Pax5), which are known to aid B cell differentiation to plasma cells, and an increased Lysine Demethylase 6B (Kdm6b)/DNA Methyltransferase 1 (Dnmt1) ratio, which increases gene expression. Identification of gene expression and immune activities of BTBR mice may aid understanding of mechanisms associated with autism since neuroimmune network interactions have been posited and induction of autoantibodies may drive the neuroinflammation associated with autism.
Keywords: Autism, BTBR, Autoantibody, Plasma cell, T follicular helper cell, Pax5, Kdm6b, Dnmt1, Alpha-enolase, IL-21r
Abbreviations: ASD, autism spectrum disorder; Ab, antibody; Ag, antigen; BM, bone marrow; Tfh, T follicular helper cell; Eno1, alpha-enolase; IL21R, interleukin-21 receptor; Dlst, dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial; Kdm6b, Lysine Demethylase 6B; Pax5, Paired Box 5; Dnmt1, DNA Methyltransferase 1
Highlights
-
•
BTBR mice have more autoantibodies, e.g., anti-α-enolase and anti-DLST than B6 mice.
-
•
T follicular helper cells are increased in BTBR mice compared to B6 mice.
-
•
BTBR mice have more plasma cells with Pax5 and IL-21r than B6 mice.
-
•
A high Kdm6b to Dmnt1 ratio influences the BTBR’s increase in plasma cells.
1. Introduction
Autism spectrum disorder (ASD) is a developmental disorder categorized by deficits in social communication and interaction, stereotypic repetitive patterns of activities, and restricted interests and behaviors (Healy et al., 2018; Sharma et al., 2018; Zwaigenbaum and Penner, 2018). A recent study estimated that 1 in 60 US children are diagnosed with ASD (Baio et al., 2018). ASD prevalence is increasing throughout the world, but the exact pathogenesis of ASD remains unclear. Several environmental toxicants such as air pollutants, heavy metals, pesticides and organic pollutants (Bolte et al., 2019) and mutation of genes such as Foxp1, Grin2brin2brin, Scnia, Lamc3, Chd8 and several others have been posited to contribute to the development of ASD (Bi et al., 2012; Cotney et al., 2015; O’Roak et al., 2011). Most ASD associated genes are brain development related factors, including neuron and glia proliferation and differentiation, cellular synapses, and connections and signal transductions between neuron-neuron and neuron-glia. However, many brain functions and immune functions are centrally and peripherally interconnected; the peripheral associations have been termed the “Neuroimmune Interactome” (Jain et al., 2020). Nervous system abnormalities in ASD can be found in the frontal and temporal lobes of the cortex, regions for body emotions, social behavior communication, and learning language.
Many children with autism have immune dysfunction, including microglial activation as well as elevated pro-inflammatory cytokine and chemokine production (Ashwood et al., 2006), which might modulate neuron-neuron and neuron-glia interactions. Immunological disproportions are considered a key etiological element in ASD based on the elevated proinflammatory cytokine levels observed in the postmortem brain (Sweeten et al., 2003; Vargas et al., 2005). Immune system abnormalities have been suggested to play a major part in the development and progression of ASD (Bock, 2002). Several studies have demonstrated changes in the cytokine levels in the cerebrospinal fluid, blood, and brain of ASD patients (Ahmad et al., 2018; Xu et al., 2015). Pro-inflammatory mediators such as cytokines, chemokines, and their receptors are associated with the progression and development of autism (Garbett et al., 2008). Furthermore, it has been recently demonstrated that the children with autism displayed decreased level of Forkhead Box P3 (FOXP3) expressing T regulatory (Treg) cells and increased level of helper T (Th) cell subsets expressing Retinoic acid-related orphan receptor gamma t (RORγt), Signal transducer and activator of transcription 3 (STAT-3), T-box transcription factor (T-bet), or GATA binding protein 3 (GATA-3) (Ahmad et al., 2017).
The BTBR mouse model has been considered a useful animal model for autism studies with comparison to B6 mice, which have normal behaviors; this strain comparison has been increasingly used to study the underlying mechanisms for ASD behaviors (Heo et al., 2011; McFarlane et al., 2008; Silverman et al., 2010; Zhang et al., 2013). BTBR mice exhibit several behavioral and immune abnormalities that are also observed in children with autism (Li et al., 2009). BTBR mice showed highly replicable impairments in social interactions, including high levels of repetitive self-grooming and minimal vocalization in social settings (McFarlane et al., 2008; Silverman et al., 2010). BTBR mice have been shown to produce interleukin (IL)-6, IL-17, and tumor necrosis factor-alpha (TNF-α) in greater amounts than B6 mice (Schwartzer et al., 2013). Studies also have reported that a distinct immune profile with higher levels of chemokine expression and alterations in Th type 1 (Th1), type 2 (Th2), type 17 (Th17), and Treg cells, which preferentially express transcription factor T-bet, GATA-3, RORγt and FOXP3 signaling, respectively, in the BTBR mice (Bakheet et al., 2016, 2017).
In our previous studies, we examined the expression of many inflammatory mediators in the whole brain and multiple brain regions of BTBR mice (Heo et al., 2011). BTBR mice had significantly higher levels of serum IgG and anti-brain antibodies (Abs), elevated cytokines expression, especially IL-33, IL-18 and IL-1β, and an increased percentage of MHC class II-expressing microglia compared to B6 mice (Heo et al., 2011). The BTBR’s maternal environment, which includes maternal anti-brain autoantibodies (autoAbs) as well as other maternal conditions are critical for the development of ASD-like behavior in offspring (Zhang et al., 2013). The role of maternal environment was further suggested to explain the impaired social behavior of B6 offspring that developed in BTBR dams and the improved social behavior of BTBR offspring that developed in B6 dams (Zhang et al., 2013).
Herein, we further assess the immune profile leading to high autoAb production and the specificities of the autoAbs suggested to be contributing to the ASD development. An autoimmune profile with Tfh cells promoting elevated production of plasma cells making autoAbs to brain antigens is described for the aberrant behavior of BTBR mice (Ahmad et al., 2020); our results confirm and extend this report with involvement of specific gene expression.
2. Material and methods
2.1. Animals
BTBR T+ Itpr3tf/J (BTBR) and C57BL/6J (B6) mice used in this study were 8–12 weeks old and were purchased from Jackson Laboratories (Bar Harbor, ME, USA). BTBR mice are an inbred strain with autism-like behaviors; they were not genetically modified for any factors known to influence ASD. Male and female mice were used. Mice were housed in the AAALAC-approved Wadsworth Center Animal Facility under standard conditions of temperature and humidity in a 12h light-dark cycle (lights on at 7AM). The study was approved by Wadsworth Center’s IACUC.
2.2. Cells
Spleens and bone marrow (BM) were harvested to make single cell suspensions. Blood, spleens, and BM cells were used immediately after collection for flow cytometry analyses and/or molecular analyses. Sera were collected by centrifugation at 12,000×g for 10 min and frozen at -20 °C until use.
2.3. IgG ELISA
The level of total IgG in serum was determined with a sandwich ELISA. Easywash 96 well plates (ThermoFisher.com) were coated with goat anti-mouse IgG (Fab Specific) (Sigma-Aldrich, St. Louis, MO) and HRP-conjugated goat anti-mouse IgG Fc (Novex by Life Technologies) was used as the detection Ab; 3,3′,5,5′-tetramethylbenzidine (TMB) was used as substrate (Sigma). Plates were washed with a BioTek ELx405Select CW (Winooski, VT) and read at OD450 with an ELISA analyzer (BioTek EL808). Sera were diluted 1/100,000 or 1/200,000. To detect the brain-specific IgG Abs, sera were diluted 1/100 or 1/200 and measured as previously described (Mondal et al., 2008; Heo et al., 2011). Briefly, SCID mouse whole brain lysates (10 μg/well) were used to coat the 96-well plate overnight at 4 °C with brain antigens (Ags), the plate was blocked with 2.5% bovine serum albumin in phosphate buffer saline containing 0.05% Tween 20 (BSA-PBST) for 2 h. After 3X washes, sera from B6 and BTBR mice were added into the wells and incubated for 2 h at room temperature. After 6X washes, HRP-goat anti-mouse IgG detection Ab was added and incubated for another 2 h at room temperature. The plates were washed 6X, and the TMB (T444, Sigma) substrate solution was added for color development. Absorbance was measured at 450 nm.
2.4. Cell ELISA
Six different mouse cell lines N1E-115 (ATCC® CRL-2263™ tyrosine hydroxylase (TH) expressing neuroblastoma from A/J mouse), CATH.a (ATCC® CRL-11179™ TH+ expressing D2 brain neuronal tumor from B6 mouse), NE-GFP-4C (ATCC® CRL-2926™ neuroectodermal stem cell from an embryonic day E9 (E9) B6/129Svp53−/− mouse), MN9D (dopaminergic mesencephalic tegmentum and neuroblastoma from an E14 B6 mouse), C8-B4 (ATCC® CRL-2540™ cerebellar macrophage/microglia from a postnatal day 8 (PND8) B6 mouse), and N9 (microglia) were used. Sera were diluted to 1:100-1/1000 from untreated adult BTBR and B6 mice. The IgG bound to the cells was quantified as described earlier. MN9D cells were from Dr. Alfred Heller, Department of Pharmacological and Physiological Sciences, University of Chicago; N9 cells were from Dr. P. Ricciadi-Castagnoli, Department of Biotechnology and Bioscience, University of Milano-Bicocca.
2.5. Isolation of brain fractions for ELISA and Western blot analysis
Whole brains were collected from BTBR (n = 6) and B6 (n = 6) mice following intracardial perfusion with warm phosphate-buffered saline (PBS; 50 ml/mouse). The subcellular protein fractions used as Ags were isolated as previously described (Cox and Emili, 2006). Briefly, minced tissue was dounced and centrifuged to obtain supernatant cyto-1 containing mitochondria, cytosol and microsomes and the pellet (nuclei-I). The nuclei-I was further homogenized and extracted to obtain Nuc-S, which contains soluble proteins and proteins loosely bound to DNA, and Nuc-T, which contains nuclear membrane proteins and other proteins that are tightly bound to DNA, such as histones. Cyto-I and cyto-II are combined to pellet out mitochondria, which is extracted and centrifuged to produce Mito-S, which is the supernatant fraction and Mito-M, which contains mitochondrial membrane proteins. The supernatant is Cyto-III, and it is centrifuged, and the pellet is extracted to give the microsomal protein fraction. Six different fractions were used to coat a 96-well ELISA plate with 10 μg/well. Total protein concentration was detected by BCA assay using bovine serum albumin (BSA; Sigma) as the standard. The assay assessing IgG bound to each fraction was performed as described in 2.2.
2.6. Western blot
Proteins from the neuronal cell lines (CATH.a, MN9D, NE-GFP-4C and N1E-115), proteins from BTBR and B6 brain fractions as described in 2.4, and protein from human brain regions (Bioreclamation, Hicksville, NY) were used to assess IgG Ab activity in BTBR vs B6 sera by Western blot analysis. Equivalent amounts of proteins were loaded into each lane (approximately 10 μg/lane), separated by 10–12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 20 mM Tris base, pH 7.6, 140 mM NaCl, 0.05% v/v Tween-20 (TBST) containing 5% (v/v) fish gelatin (Sigma) and were incubated on a rocker 2 h at room temperature to prevent nonspecific binding of Abs. After blocking, the membranes were incubated with equivalent amounts of diluted (1:100) sera (BTBR or B6). IgG was detected with HRP conjugated goat anti-mouse IgG (H + L) (Invitrogen). The blot was developed by incubating with super signal/chemiluminescent substrate (Pierce Biotechnology, Inc., Rockford, IL) for 5 min and was analyzed with a LAS-1000plus (Fuji Film, USA).
2.7. Protein identification by mass spectrometry (MS)
Protein bands excised from a Coomassie stained SDS-PAGE gel were identified by MS as previously described (https://www.ncbi.nlm.nih.gov/pubmed/26871944). Briefly, the gel bands were manually excised from the gel and cut into pieces. The gel pieces were dehydrated with acetonitrile followed by reduction and alkylation using Tris (2-carboxyethyl) phosphine hydrochloride and iodoacetamide, respectively. In gel digestion was performed using a sequencing grade modified trypsin (12.5 ng/μl) (Promega, Madison, WI, USA) in 25 mM ammonium bicarbonate, pH 8.5, at 37 °C for overnight. Following the digestion, tryptic peptides were extracted three times with 50% acetonitrile containing 5% formic acid for 15 min each time with moderate sonication. The extracted solutions were pooled and evaporated to dryness under vacuum. LC-MS/MS analysis was performed on an integrated QSTAR XL nanoLC-MS/MS system (ABSCIEX, ON, Canada) comprising three micro pumps with an autosampler, a stream select module configured for precolumn plus analytical capillary column, and a QSTAR XL mass spectrometer fitted with nano sprayer III, operated under both Analyst 1.1 and MassLynx 4.0 control with a contact closure. Injected samples were first trapped and desalted isocratic ally on an Everest C18 precolumn (5 μm, 500 μm ID X 15 mm, Grace, Deerfield, IL) for 6 min with 0.1% formic acid delivered by an auxillary pump at 40 μl/min. The peptides were eluted off the precolumn and separated on an analytical C18 capillary column (15 cm × 100 μm I.D. packed with 3 μm Jupitor C18 particles, Phenomenex, Torrance, CA, USA) at 300 nl/min using a 50 min fast gradient of 5%–60% acetonitrile in 0.1% formic acid and 0.0075%TFA. The eluted peptides were electrosprayed into QSTAR XL mass spectrometer and the three most intense peptide ions (+2, +3, or +4) from each survey scan (MS scan) were selected for fragmentation by collision-induced dissociation and detection (MS/MS scan). The peak lists of the MS/MS data were generated using an Analyst “script” mascot.dll from Matrixscience. Then the file was submitted to Mascot V2.5 (Matrix Science, London, United Kingdom) for protein identification by searching against NCBI non-redundant mouse database. The allowed mass search tolerances were 0.3 Da for precursor m/z and 0.3 Da for MS/MS fragment m/z, respectively. In addition, two missed cleavages were allowed. Variable modifications considered were methionine oxidation, cysteine carboxyamidomethylation, and deamidation. The peptides identified at the 95% confidence level by Mascot were selected for a positive identification. The protein name was inferred by a group of positively identified peptides.
2.8. Antibodies to alpha-enolase (ENO1) and dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial (DLST)
Rabbit polyclonal Abs anti-ENO1 and anti-DLST (ABclonal, Woburn, MA) were used to coat wells (1 μg/well) to capture ENO1 or DLST in BTBR brain homogenates (10 μg). BTBR and B6 serum was diluted (1:10) and assayed for Abs to these Ags with HRP-anti-IgG Fc, which was used at 1:1000 dilution.
2.9. Flow cytometry
Blood was collected with heart puncture and transferred into a Na EDTA-containing tube. Single cell suspensions were prepared from spleens and BM as described (Heo, 2005). Whole blood (50 μl) was stained and erythrocytes were lysed with FACSlyse (BD Biosciences, San Jose) before analysis. Splenocytes and BM cells were counted, and 1 × 106 cells were used for staining followed by FACSlyse and analysis by six-color flow cytometry with a FACSCanto flow cytometer (BD Biosciences), and the following fluorochrome conjugated Abs: PerCP Cy5.5-CD45, APC Cy7-CD45, PerCP Cy5.5-CD4, PE-CD4, FITC-CD4, FITC-CD8a, PE-CD8a, PerCP-CD8a, FITC-CD3, APC-CD3e, PerCP-CD3e, PE Cy7-CD24, FITC-CD43, PerCP-CD249- APC Cy7-B220, PE -IgD, APC-IgM, APC Cy7-CD3, PE Cy7-CD19, PerCP cy5.5-CD93, APC-CD138, PE-CXCR4, FITC-MHC-II, PE-CD19, FITC-CD19, PE Cy7-CD19, APC-CXCR5, PE Cy7-PD1, PerCP Cy5.5-CD138 and anti-CD16/32 (Fc block). The Abs were purchased from BD Pharmingen (San Diego, CA), Biolegend (San Diego, CA), or eBiosciences (Thermo Fisher Scientific, MA, USA). Frequencies and numbers of populations in the blood and spleens of B6 and BTBR mice were gated based on FSC-A and SSC-A, followed by gating out the doublets and finally gated on CD45+ cell. Data were analyzed by Flow Jo-V10.
2.10. RNA isolation and cDNA preparation
Fresh mouse spleen (100 mg) was added 700 μl QIAzol Lysis Reagent (QIAGEN). After homogenization and incubation for 5 min at room temperature (RT), 140 μl chloroform was added and centrifuged for 15 min at 12,000×g at 4 °C. Upper aqueous phase was transferred to a new collection tube and washed with 100% ethanol, 700 μl RWT buffer and 500 μl RPE buffer twice. After RNA isolation, the absorbance measurements OD260/OD280 and OD260/OD230 using a NanoDrop Spectrophotometer, together with RNA electrophoresis were utilized to detect the RNA purity. Collected RNA (1 μg) was used for cDNA reverse transcription.
2.11. Real-time PCR for gene detections
Five μl of PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) was used for 10 μl Real-time PCR system, and 1 μl primer was used for each gene detection. Relative expression data were calculated using 2-ΔΔCt, and Gapdh was used as reference (Lech et al., 2012). The forward (F) and reverse (R) gene sequences used were:
Il21r(F:CTCACGGTCACTTGCTTGTCT,R:CAAGTCCAGAATCAGGGGAGTC);
Pax5(F:AGGCTGGATACCTAGGGAACTT,R:CATTCAGGGAGGCTCCTTTCTT);
Xbp1(F:TAAGAACACGCTTGGGAATGGA,R:CTTGTCCAGAATGCCCAAAAGG);
Irf4 (F:GGGACTTGGTTCTTAGGTCCAG,R:AGAAGAGGGAGCAAACAGGATG);
Kdm6b(F:ATGTCAGCCTCATAGCAGGACC,R:CCTTGTCTCCGCCTCAGTAAA);
Gapdh(F:CATCACTGCCACCCAGAAGACTG,R:ATGCCAGTGAGCTTCCCGC).
2.12. Statistical analysis
Data reported as mean ± SEM, and p values determined by unpaired two-tailed Student’s t-test with p < 0.05 considered as significantly different.
3. Results
3.1. Higher levels of serum IgG and anti-brain autoAbs in BTBR than B6 mice
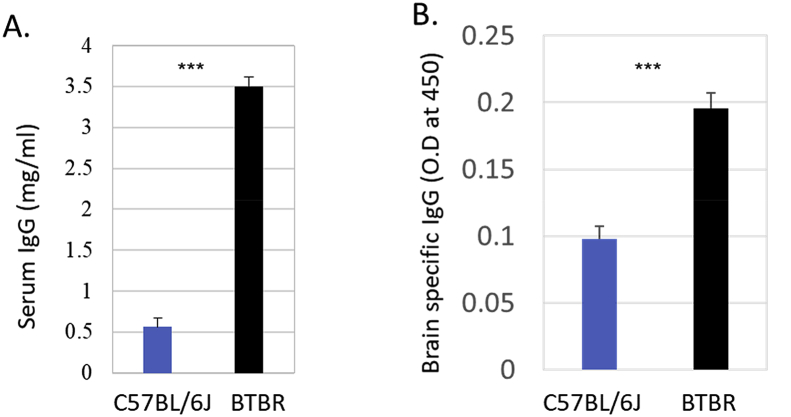
The amounts of total serum IgG (Fig. 1A) and serum IgG to brain Ags (Fig. 1B) from BTBR and B6 mice (10–12 weeks of age) were quantified. Since we did not find any male and female differences in our measures, results were combined across sex. The average value of total sera IgG levels from BTBR mice was 3.5 ± 0.39 mg/ml, which was six times higher than that from B6 mice (0.56 ± 0.36 mg/ml); the differences were significant (p < 0.001). Whole brain homogenates from SCID mice were used as Ags to detect the brain-reactive IgG. The sera levels of brain-reactive Abs are presented as optical density (OD) values, and BTBR sera had at least two times greater OD values than B6 sera (p < 0.001).
Fig. 1.
Total IgG and brain reactive IgG in mouse serum. BTBR mice have significantly higher serum IgG levels (a) and serum anti-brain antibodies than B6(C57BL/6) mice. Sera were obtained from 8-12 weeks old mice. To detect anti-brain antibodies sera were added to wells coated with SCID whole brain proteins (10 μg/well) and the level of brain-reactive IgG was measured by using the HRP conjugated goat anti-mouse IgG. Data are representative of three (A) and two (B) independent experiments with 3 or 4 pairs of mice in each experiment. The p values were determined by unpaired two-tailed Student’s t-test, and p < 0.05 is considered as significantly different. Error bar indicates mean ± SEM. ∗ indicates a significant difference between the two strains.∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
3.2. BTBR mice have higher amounts of serum IgG Abs reactive with neural cell lines and brain fractions than B6 mice
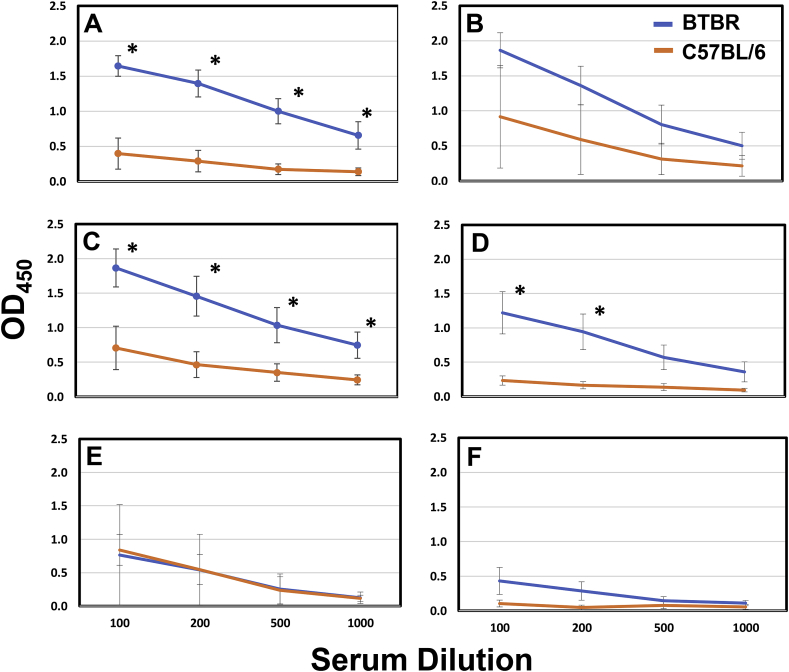
Sera were diluted 1:100-1/1000 for the cellular ELISA with the MN9D, CATH.a, NIE-115 and NE-GPA-4C neuronal cell lines. There was more Ab to all four neuronal cell lines in the sera from BTBR mice than B6 mice. The titer was greatest with MN9D and CATH.a cells. At the 1/1000 dilution, there was significantly more binding with the BTBR sera (Fig. 2). With two macrophage/microglial cell lines (C8-B4 and N9), there were no differences with BTBR and B6 sera, and the binding was lower than with the neuronal cells. At the 1/100 dilution, the ODs were 0.75 and < 0.5 for C8-B4 and N9 cells, respectively.
Fig. 2.
Serum autoAbs to neuronal cell line. Neural cellular ELISA with BTBR and B6 sera demonstrating that BTBR have higher titers to neural cell lines than B6. CATH.a (A), N1E-115 (B), MN9D (C), NE-GFP-4C (D), C8-B4 (E) and N9 (F) cell lines were growth to confluency, fixed, and assayed with male BTBR or B6 sera diluted 1/100 to 1/1000; sera were from 3-11 mice. The ∗ indicates a significant difference at the indicated dilution.
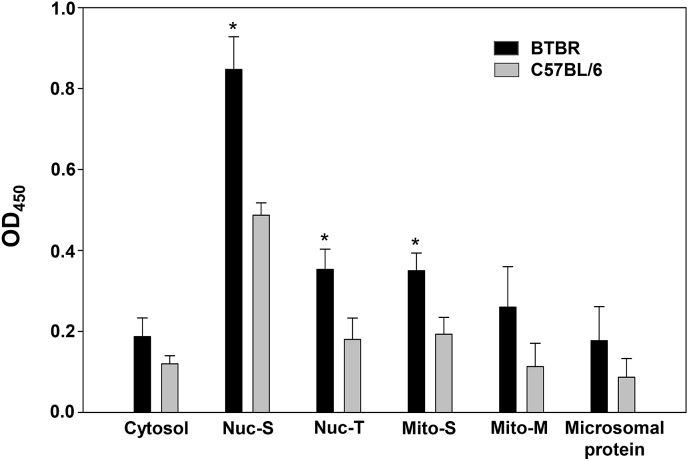
Whole brains were collected from BTBR and B6 mice for the isolation of subcellular protein fractions. Six different protein fractions (cytosol, Nuc-S, Nuc-T, Mito-S, Mito-M, and the microsomal protein fraction) were produced as described earlier and were coated to wells at 10 μg/well. Interestingly, the IgG from the BTBR mice bound significantly better to Nuc-S, Nuc-T, and Mito-S fractions from BTBR brains than from B6 brains, which suggests either more Ag or different Ags and antigenic epitopes (Fig. 3).
Fig. 3.
Serum autoAbs to subcellular protein fractions of brain tissue. Subcellular protein fractions from whole BTBR (B6) and C57BL/6 (B6) brains were used to detect brain-reactive IgG antibodies from BTBR mice by ELISA. ELISA plates were coated with 10 μg of each protein fraction per well and 100 μl of BTBR sera (n = 3, mean ± SD) diluted 1/100 were assayed as described. The ∗ indicates a significant difference between BTBR and B6 brain protein fractions.
3.3. Western blot analyses of serum IgG from BTBR and B6 mice
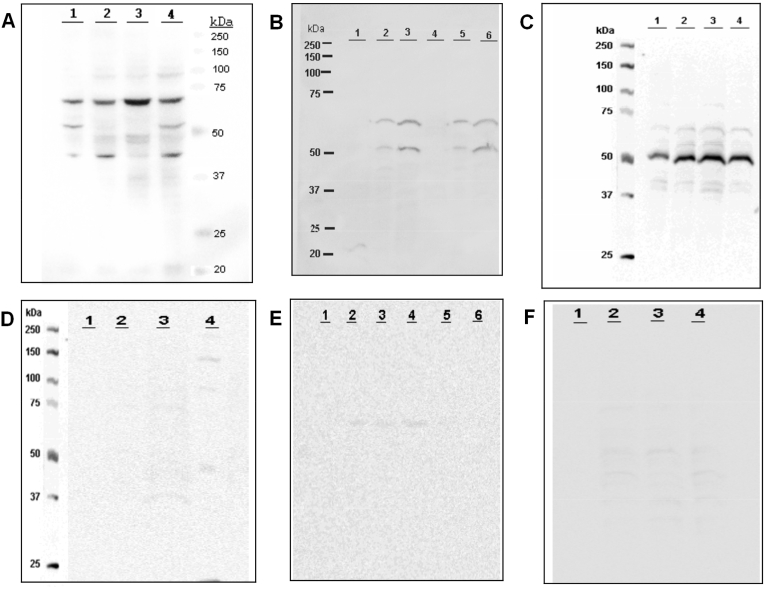
We further investigated the IgG Abs from BTBR mice reactive to different neuronal cell lysates, fractionated BTBR and B6 subcellular brain proteins, and proteins isolated from different regions of human brain were analyzed with Western blots. We used the four different neuronal cell lines (CATH.a, MN9D, NE-GFP-4C and N1E-115). BTBR IgG Abs reacted with the cell lysates from all neuronal cell lines (Fig. 4A), but with different intensities to different Ags. The highest reactivity was with a ~70 kDa band. For IgG binding to the fractions of mouse brains, the proteins in the Mito-S and Mito-M fractions from both B6 and BTBR brains strongly reacted with IgG in BTBR serum (Fig. 4 B). The IgG reactive bands at ~65 and ~50 kDa were those with the greatest binding. Interestingly, the higher titer of BTBR serum IgG to Nuc-S (Fig. 3) was lost in the Western blot analysis suggesting that the SDS treatment destroys the epitope(s). To examine the reactivity of BTBR IgG to human brain proteins, we isolated protein from different regions (frontal cortex, parietal cortex, hippocampus and cerebellum) of a deidentified cadaver human brain. Western blot analysis indicated that serum IgG from BTBR mice also bound to human brain Ags. All four homogenates of the human brain regions strongly interacted with BTBR IgG; the intense protein band observed at ~50 kDa in L3 (hippocampus homogenate) was extracted from a Coomassie stained gel and was tentatively identified as alpha-enolase (Eno1) or dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial (Dlst) by mass spectrometry protein sequencing analysis. This ~50 kDa band was reactive with rabbit polyconal Abs to ENO1 (A1033) and DLST (A6901) from ABclonal (Woburn, MA). For the Western analyses of BTBR IgG of mouse brain Ags (Fig. 4B), and for all B6 IgG (Fig. 4D–F), the contrast had to be increased to observe any hint of IgG binding, which prevents quantitative comparisons of BTBR and B6 IgG specificities to specific bands.
Fig. 4.
Serum autoAbs to subcellular protein fractions of neuronal cell line and human brain regions. Western blot analyses of BTBR and B6 IgG specificities. An equal amount of protein from CATH.a (L1), MN9D (L2), NE-GFP-4C (L3) and N1E-115 (L4) cell homogenates were separated with 10–12% SDS-PAGE gel and blotted with BTBR (A) or B6 (D) sera as primary antibodies. This experiment was done more than three times. For (B and E), equivalent amounts of protein from BTBR brain subcellular fractions (Nuc-s,L1; Mito-s, L2; Mito-M; L-3) and B6 brain sub-cellular fractions (Nuc-s, L4; Mito-s, L5; Mito-m, L6) were loaded, separated by 10–12% SDS-PAGE, and probed with diluted BTBR (B) or B6 (E) sera as primary antibody. This experiment was done more than six times. For C and F, equivalent amounts of protein from different human brain sections (frontal cortex-L1, Parietal cortex-L2, Hippocampus-L3 and cerebellum-L4) were loaded, separated by 10–12% SDS-PAGE, and blotted with diluted BTBR (C) or B6 (F) sera as a primary antibody. Data are representative of minimum three independent experiments with equivalent results.
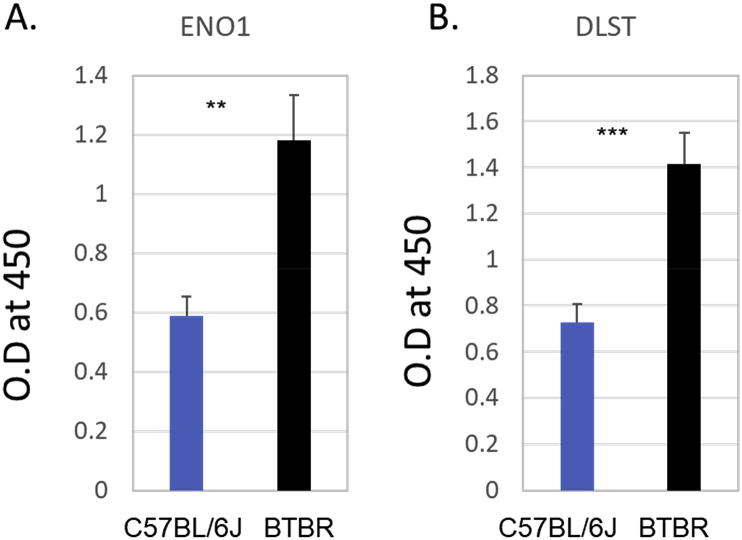
3.4. ELISA evaluation of Abs to ENO1 and DLST
Since the ~50 kDa band observed in the Western blot (Fig. 4C) was suggested to be ENO1 or DLST with MS analysis, we assayed BTBR and B6 sera for IgG to these Ags; BTBR sera had significantly more IgG anti-Eno1 and IgG anti-Dlst than B6 sera (Fig. 5).
Fig. 5.
Serum autoAbs to ENO1 and DLST ELISA of BTBR and B6 serum Abs to Eno1 (A) and Dlst (B). A 96-well plate was coated with 1 μg rabbit polyclonal Ab to ENO1 or DLST, blocked and used to capture Ag from 10 μg SCID brain homogenate for assessment of anti-ENO1 or anti-DLST IgG Ab in BTBR or B6 sera, which was diluted 1:10, and detected with HRP conjugated goat ant-mouse IgG Fc. The ∗ indicates a significant difference between BTBR and B6 Ab levels. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
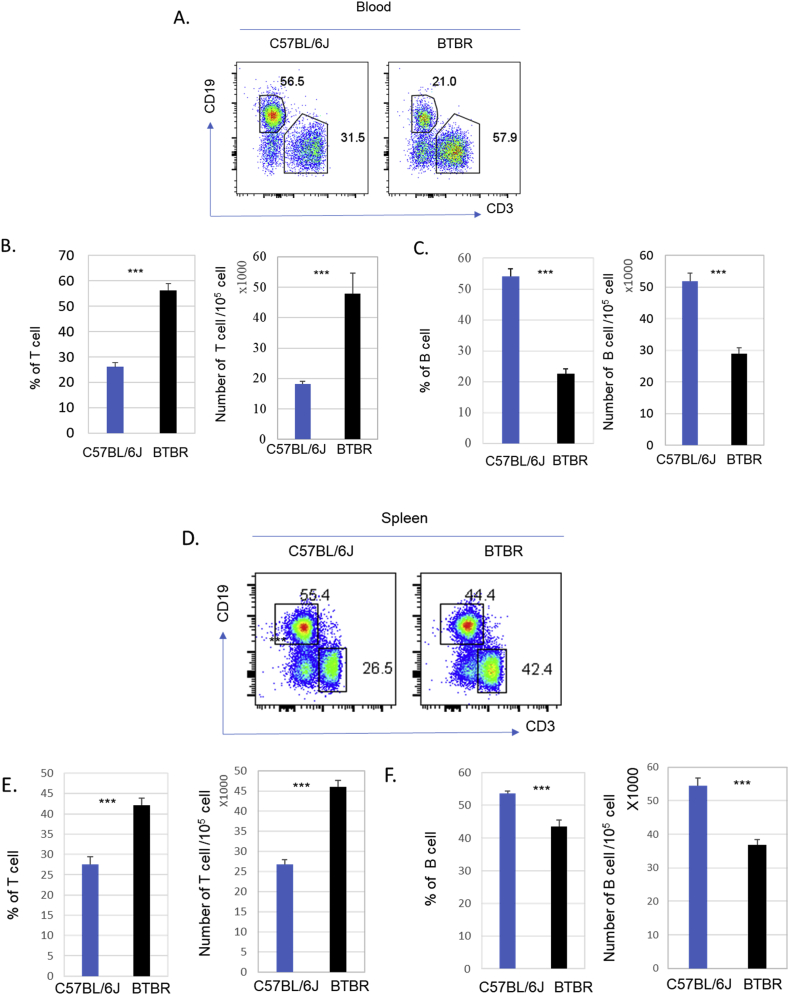
3.5. BTBR mice have more T cells but fewer B cells than B6 mice
Since some immune cell types have been linked with development of neuroinflammatory disorders, including autism (Ashwood et al., 2006; Bilbo and Schwarz, 2009; Vargas et al., 2005), we assessed the frequency and numbers of key immune cell types in the blood, spleen and BM of BTBR mice with comparison to B6 mice. Both the frequencies and numbers of CD3+ T cells were higher in the blood of BTBR mice compared to B6 mice, and the differences were statistically significant (Fig. 6A and B; p < 0.001). Similarly, in spleens, the frequencies and numbers of CD3+ T cells were significantly higher (Fig. 6D and E; p < 0.001) in BTBR mice compared to B6 mice. In contrast to T cells, blood (Fig. 6C) and spleens (Fig. 6 F) had fewer B cells in BTBR than B6 mice.
Fig. 6.
CD3+ T and CD19+ B cell populations in BTBR and B6 mice. BTBR mice have significantly more T cells and fewer B cells than B6 mice. Representative flow cytometric analysis of B cells and T cells are shown for blood (A) and spleen (D) of B6 and BTBR mice. Frequency and number of CD3+ T cells and CD19+ B cells in the blood (B, C) and spleens (E, F) of B6 and BTBR mice (mean ± SEM) are shown. Data are representative of three or four independent experiments with 4 pairs of mice in each experiment; significant differences between the two strains are indicated by ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
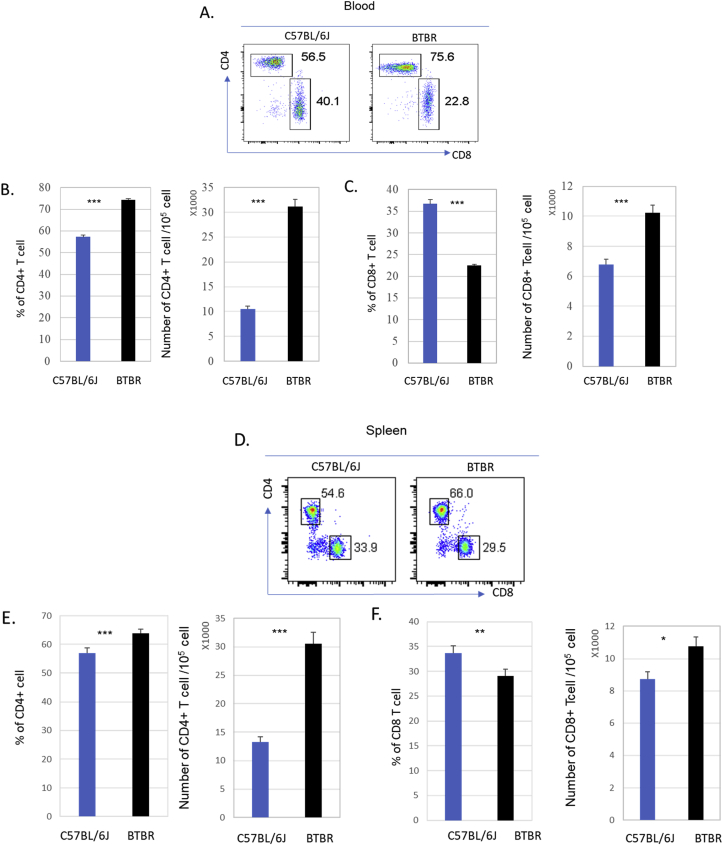
3.6. Higher numbers of CD4+ and CD8+ T cells in peripheral blood and spleens of BTBR mice than B6 mice
Since the numbers of T cells appeared higher in BTBR mice compared to B6 mice, we assessed the frequencies and numbers of CD4+ and CD8+ T cells in blood and spleens. Both the frequencies and numbers of CD4+ T cell were significantly higher in the blood (Fig. 7A and B, p < 0.001) and spleens (Fig. 7D and E, p < 0.001) of BTBR mice compared to B6 mice. The frequencies of CD8+ T cells were lower in blood (Fig. 7A and C, p < 0.001) and spleens (Fig. 7D and F, p = 0.008) of BTBR mice compared to B6 mice, but the numbers were significantly higher both in blood (Fig. 7A and C, p < 0.001) and spleens (Fig. 7 D and F, p = 0.019) of BTBR mice compared to B6 mice.
Fig. 7.
CD4+ and CD8+ T cells in BTBR and B6 mice. BTBR mice have significantly more CD4+ and CD8+ T cells than B6 mice. Representative flow cytometric analyses are shown for CD4+ T cells and CD8+ T cells in the blood (A) and spleen (D) of B6 and BTBR mice. To observe the CD4+ and CD8+ populations the cells were first gated on the CD3+ population; B cells were gated out based on CD19+ cells. Frequencies and numbers of CD3+CD8+ T cells and CD3+CD4+ cells and populations in the blood (B &C) and spleens (E&F) of B6 and BTBR mice are shown. Data are representative of three or four independent experiments with 4 pairs of mice in each experiment. P values determined by unpaired two-tailed Student’s t-test are indicated and p < 0.05 is considered as significantly different. Error bars indicate mean ± SEM. ∗ indicates a significant difference between the two strains. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
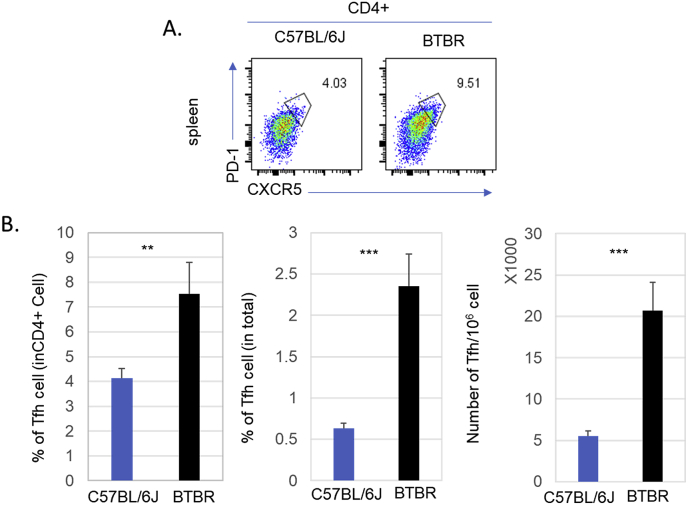
3.7. Higher numbers of T follicular helper (Tfh) cells in BTBR mice than B6 mice
To become an antibody-producing plasma cell, B cells require help through interacting with a specialized CD4+ T cell population called follicular T helper cell (Tfh). We quantified Tfh cells in the spleen by flow cytometry, which were identified by PD1 and CXCR5 expression on CD4+ T cells. The frequencies and numbers of Tfh cells were significantly higher in BTBR mice than B6 mice (Fig. 8); the frequencies of Tfh cells were significantly different (p = 0.0025, Fig. 8B). When compare for the total CD45+ population, the differences were even more significant (p < 0.001, Fig. 8B), because the total numbers of CD4+ T cells are higher in BTBR mice. When we compared the total numbers of Tfh cells, BTBR mice had 4-5 times more splenic Tfh cells compared to B6 mice.
Fig. 8.
Splenic T follicular helper (Tfh) cells of BTBR and B6 mice. BTBR mice have significantly higher Tfh cell population than B6 mice. Representative flow cytometric analyses are shown for Tfh cells (CD4+PD1+CXCR5+) in the spleens of B6 and BTBR mice (A). The frequencies and numbers of splenic Tfh cells of B6 and BTBR mice are in (B). Tfh cell population were identified based on PD1+CXCR5+ cell in CD3+CD4+ population. Data are representative of two independent experiments with 4 pairs of mice in each experiment. ∗ indicates a significant difference between the two strains. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
3.8. BTBR mice have more plasma cells than B6 mice
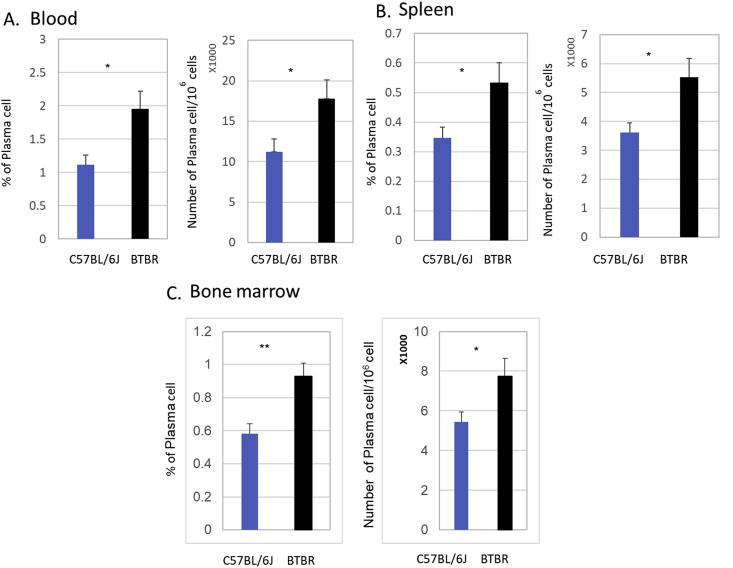
Given our result showing relatively high levels of IgG in BTBR mice and low numbers of B cells, we investigated the numbers of plasma cells in the blood, spleen and BM using CD138 expression in the CD3-/CD19- population by flow cytometry. The frequencies and numbers of plasma cells were higher in BTBR mice compared to B6 mice (Fig. 9). The frequencies of plasma cells in blood were statistically significant (p = 0.025, Fig. 9 A), and the number within the CD3-/CD19- population was also significant (p = 0.035 Fig. 9A). In spleens, the frequencies of plasma cells were higher in BTBR mice compared to B6 mice based on CD138+ cells in the CD45+ population, and the difference is statistically significant (p = 0.029 Fig. 9B). The numbers of plasma cells were also significantly (p = 0.023) higher in the spleens of BTBR mice compared to B6 mice. In BM, both the frequencies and numbers were significantly higher in BTBR mice compared to B6 mice (Fig. 9C, p = 004 and p = 0.04). The higher frequencies of plasma cells are the suggested contributing factor for the higher levels of IgG in the BTBR sera.
Fig. 9.
Plasma cells in the periphery and lymphoid organs of BTBR and B6 mice. BTBR mice have more plasma cells than B6(C57BL/6) mice. Frequencies and numbers of CD138+ plasma cells in the blood (A), spleen (B) and bone marrow (C) are reported for B6 and BTBR mice. Plasma cells were identified based on CD138+ cell in CD19-CD3- population. Data are representative of three or four independent experiments with 4 pairs of mice in each experiment. ∗ indicates a significant difference between the two strains. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
3.9. B cell lineage populations in spleen and bone marrow
We observed fewer B cells and higher numbers of plasma cells in BTBR mice than in B6 mice. To determine a difference in the B cell lineage between BTBR and B6 mice, we investigated different developmental stages of the B cell population in spleen and BM. Higher frequencies of Pro-B cells (B220lowCD19+CD24+CD43+CD249-) were detected in spleen and BM (Table 1) of BTBR mice compared to B6 mice, and at least in BM, the difference was statistically significant. Immature B cells (B220+CD19+CD24+CD43-IgM+IgD-) and Transitional B cells (B220+CD19+CD24+CD43-IgM+IgDlow) were significantly reduced in spleen (Table 1). Both in spleen and BM, the frequencies of Mature B cells (B220+ CD19+CD24+CD43-IgM+IgD+) were significantly reduced (Table 1). Increased numbers of Pro-B cells and lower numbers of Immature and Transitional B cells do not adequately explain the reduced numbers of Mature B cell (B220+ CD19+CD24+CD43-IgM+IgD+) in the spleens and BM of BTBR mice. However, the increased numbers of plasma cells may explain the low numbers of B cells. In the spleen, Ab-producing plasmablasts (B220lowCD138+CXCR4+MHCII+), plasma cells (CD19-CD138+CXCR4+) and long-lived plasma cells (CD19-CD138+CXCR4+CD93-) were increased in BTBR mice compared to B6 mice, and the differences in plasma cells were statistically significant. Regarding BM plasmablasts, plasma cells and long lived-plasma cells, all were significantly higher in BTBR mice compared to B6 mice. The increased populations of these highly efficient Ab-producing cells relate to the higher Ab levels in BTBR mice.
Table 1.
B cell lineage phenotype (percentage).
| Cell Stage (surface immunophenotype)a |
Spleen |
Bone marrow |
||
|---|---|---|---|---|
| B6 | BTBR | B6 | BTBR | |
| PrePro B cell (B220lowCD24-CD43+IgM-IgD-) |
1.77 ± 0.98 | 2.01 ± 1.23 | 1.83 ± 0.24 | 1.84 ± 0.29 |
| Pro B cell (B220lowCD19+CD24+CD43+CD249- |
2.78 ± 0.52 | 3.99 ± 1.41 | 45.65 ± 2.07 | 48.64 ± 2.59b |
| PreB cell (B220lowCD19+CD24+CD43lowCD249+) |
0.25 ± 0.05 | 0.24 ± 0.06 | 1.26 ± 0.45 | 0.98 ± 0.49 |
| Immature B cell (B220+CD19+CD24+CD43- IgM+IgD-) |
1.2 ± 0.47 | 0.41 ± 0.16b | 1.53 ± 0.46 | 1.23 ± 0.45 |
| Transitional B cell (B220+CD19+CD24+CD43- IgM+IgDlow) |
7.5 ± 0.49 | 4.89 ± 0.88b | 3.07 ± 0.46 | 2.75 ± 0.91 |
| Mature B cell (B220+ CD19+CD24+CD43- IgM+ IgD+) |
45.8 ± 2.05 | 37.24 ± 3.23b | 10.72 ± 1.67 | 7.53 ± 2.17b |
| Plasmablast (B220lowCD138+CXCR4+ MHCII+) |
0.41 ± 0.06 | 1.18 ± 1.92 | 0.13 ± 0.04 | 0.21 ± 0.03b |
| Plasma cell (CD19-CD138+CXCR4+) |
0.26 ± 0.06 | 0.47 ± 0.18b | 0.58 ± 0.15 | 0.93 ± 0.2b |
| Plasma cell -Short lived (CD19-CD138+CXCR4+CD93+) |
0.005 ± 0.004 | 0.030 ± 0.057 | 0.06 ± 0.04 | 0.06 ± 0.04 |
| Plasma cell-Long lived (CD19-CD138+CXCR4+CD93-) |
0.24 ± 0.07 | 0.37 ± 0.36 | 0.52 ± 0.15 | 0.85 ± 0.16b |
The immunophenotyping of B cell and plasma cell development was based on references (Allman and Pillai, 2008; Shapiro-Shelef and Calame, 2005).
Significant difference between B6 and BTBR.
3.10. Factors influencing differentiation and proliferation of B cells and plasma cells
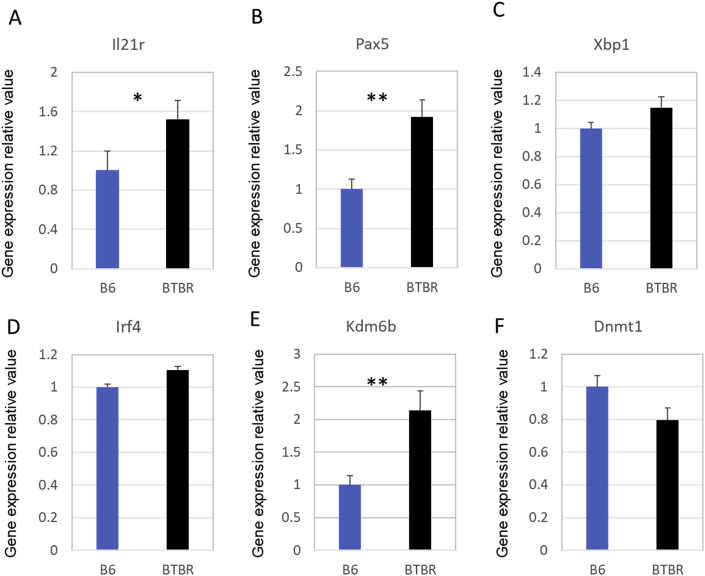
Biomarkers known to affect gene expression and B cell proliferation and differentiation into plasma cells were investigated. We selected for analysis the transcription factors and receptors known to influence B cell activities. As shown in Fig. 10, gene expression of Il-21r (A) and Pax5 (B) in BTBR mice were significantly increased compared with B6 mice (set as 1), other transcriptional factors such as Xbp1 (C) and Irf4 (D), involved in B cell activation and differentiation into plasma cells showed only a modest increase. To better understand the up-regulation of these factors, we assayed expression of factors associated with methylation processes. Kdm6b (E), a lysine-specific demethylase, was found greatly increased in BTBR spleens while Dnmt1 (F), which is a DNA methyltransferase, was slightly decreased; the Kdm6b/Dnmt1 ratio of BTBR and B6 spleens were 2.6514 ± 0.333 and 1.0 ± 0.168 (p = 0.0013), respectively. Thus, epitranscriptomic mechanisms may be responsible for the increased level of plasma cells.
Fig. 10.
Factors influencing differentiation and proliferation of B cells and plasma cells. Six adult B6 and BTBR mice spleen (3 male and 3 female mice in each group) were used for gene detection. Value in B6 group was set as 1 and value in BTBR group were calculated by comparison with B6 group. Data are representative of three independent experiments with 6 pairs of mice in each experiment and expressed as Mean ± S.E.M. P values are determined by two-tailed Student’s t-test. ∗ indicates a significant difference between the two strains. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
4. Discussion
This study investigated the immunological profile of the BTBR mice regarding their autoAb specificities to brain Ags and associated immune cell levels. BTBR mice have been described to have atypical behavior including lack of sociability and restricted repetitive behavior that resemble autism (McFarlane et al., 2008; Wohr et al., 2011). Several recent studies have demonstrated that there is a strong association between immune dysfunction observed in ASD and irregular behavior (Masi et al., 2017; Onore et al., 2012). ASD patients have higher levels of total IgG and brain Ag-specific autoAbs (Dalton et al., 2003; Wills et al., 2007). The generation of autoAbs is one of the key features in many autoimmune neurologic disorders with IgG specificity for various neuronal Ags (Moscavitch et al., 2009; Steinman, 2004). Human serum samples have been used for assessment of brain Ag-specific autoAbs, and specific neuronal proteins were used as brain Ags (Croen et al., 2008; Singer et al., 2008). Transferring Abs from mothers of ASD children (Martin et al., 2008; Singer et al., 2009) or BTBR IgG (Zhang et al., 2013) into animals with typical development induced ASD-like behavior, but the responsible Ab specificities have not been fully delineated. BTBR mice have higher levels of serum IgG and IgG anti-brain antibodies at postnatal day 21 than B6 mice (Heo et al., 2011), and these early levels of autoAbs are maintained throughout adulthood. The autoAbs of BTBR mice were specifically reactive to brain fractions and neuronal cell lines but had lower binding capacity to microglia cells, further demonstrating the specificity to neuronal Ags. The Abs of BTBR mice also interacted with human Ags from various brain parts and different fractions of brain Ags. Interestingly, autoAbs to alpha-enolase, as reported herein, have also been reported to be associated with other aberrant immune responses including Hashimoto’s encephalopathy (Yoneda et al., 2007; Churilov et al., 2019) and asthma (Nahm et al., 2006). DLST polymorphism has been connected to Alzheimer’s disease (Sheu et al., 1999), and the autoAb to DLST may disrupt its function. It is increasingly becoming more apparent that autoAbs to brain Ags play an important role in the development of autism, but like autoimmune diseases, the etiology for the development and expansion of T and B cells with receptors for self-constituents and production of autoAbs remain unclear. A key question is why mothers with normal behaviors develop Abs that gestationally affect aberrant behaviors in offspring.
Like the multitude of genes that have been implicated in the development of ASD (Ramaswami and Geschwind, 2018), the autoAb specificities appear to be numerous. The BTBR IgG autoAbs bind to neuronal Ags with highest interaction to nuclear antigens like the anti-nuclear Abs diagnostic for lupus patients (Kavanaugh et al., 2000). As for many autoimmune diseases, certain gene expression influences prevalence, but environmental exposures also have an effect. There is a strong hereditary component in the etiology of ASD, but its not 100% (Campisi et al., 2018; Tick et al., 2016), which suggests a role for environmental factors. As a dynamic epigenetic modification, DNA methylation plays a critical role in immune development and activities. Myeloid and lymphoid cell differentiation and function are controlled by DNA methylation with modulation of immune cell types and stimulus transcriptional factors as well as affecting host defense and organ homeostasis. Dysregulation of DNA methylation will result in immune system related diseases such as infections, autoimmune diseases, blood malignancies, and other solid tumors. Epitranscriptomics (Siu and Weksberg, 2017) have been implicated in ASD, and high on the list of factors affecting gene expression is DNA methylation and modulation of histone structure. Herein, we have shown that BTBR mice have a high gene expression ratio of Kdm6b to Dnmt1. As a lysine-specific demethylase, KDM6B specifically demethylates di- or tri-methylated lysine 27 of histone H3, whose trimethylation is a repressive epigenetic mark as chromatin controlling and gene silencing (Wiles and Selker, 2017). KDM6B plays a critical role in cellular proliferation, differentiation and body development, inflammatory diseases including cancers, and neurodegeneration (Nakamura et al., 2010; Wei et al., 2018). KDM6B (or JMJD3) is known to affect activation of germinal center B cells and their differentiation into plasma cells (Barwick et al., 2018), which was shown to occur in this study. This demethylase activity has also been reported for development of rheumatoid arthritis and lupus (Jia et al., 2018; Yin et al., 2017). In contrast, DNMT1 promotes DNA methylation by transferring methyl groups to DNA specific CpG structures and is important for T-cell homeostasis (Lee et al., 2001) and regulating B-cell activation (Lai et al., 2013). Plasma cells are the source of the majority of the autoAbs.
During B lymphocyte activation, some transcription factors, such as PAX5, and receptors, such as IL-21R, play a critical role for transition. After activation, B cells undergo immunoglobulin class-switch recombination (CSR) and then differentiated into immature plasma cells. Immature B cells are regulated and proliferated into antibody-secreting plasma cells by expression of numerous factors. The expressions of Pax5 and Il-21r, which were significantly increased in BTBR spleens, are potentially connected with the differential activities of KDM6B and DMNT1. PAX5 is a transcription factor that belongs to the paired box (PAX) family. It encodes the B-cell lineage specific activator protein (BSAP) in early B-cell activation, differentiation, proliferation and maturation (Thevenin et al., 1998). Pax5 has been identified as one of 64 genes reported to influence autism (O’Roak et al., 2014). PAX5 is necessary for GABAergic neurons affecting normal lateral ventricular development (Ohtsuka et al., 2013). Herein, Pax5 is increased in BTBR spleens; its gene regulation linkage with more immune and nervous system functions is needed for future investigation. IL-21R is the receptor of IL21, which is critical for the activation, differentiation, proliferation and maturation of B cells (Konforte et al., 2009). IL21 ligand and receptor lead to the activation of downstream signaling molecules, such as JAK1, JAK3, STAT1, and STAT3 (Berglund et al., 2013). Xbp1 and Irf4 were also were slightly increased. XBP1 aids B cells becoming plasmablasts and colonization of BM with plasma cells that have sustained Ab production (Hu et al., 2009). As a transcription factor, IRF4 is essential for Th2 cells, Th17 cells, and Th9 cells development (Staudt et al., 2010). IRF4 is also required for the plasma cell generation (Klein et al., 2006). Conditional deletion of Irf4 in germinal center B cells causes a reduction of post-germinal center plasma cells development as well as memory B cells failing to differentiate into plasma cells (Willis et al., 2014).
IL-21 enhances germinal center activities by enhancing B cell and Tfh cell effects needed for plasma cell differentiation (Alinikula and Lassila, 2011). Tfh cells are increased in BTBR mice. Tfh cells are linked with a wide range of autoimmune diseases, and activated Tfh cells are observed in children with current onset of type 1 diabetes (T1D) or at danger of T1D (Crotty, 2019; Viisanen et al., 2017). In systemic lupus erythematosus (SLE), there is a clear correlation between Tfh cells and an autoAb-mediated autoimmune disease. Tfh cells have also been involved in a number of autoimmune diseases of mice (Gensous et al., 2018; Linterman et al., 2009; Quinn et al., 2018). The higher number of Ab-secreting plasma cells in BTBR mice corresponds to the combination of more Tfh cells, IL-21, and transcriptional factors, such as PAX5, which enhance the generation of plasma cells.
Surprisingly, the population of B lymphocytes was reduced in the blood, spleens and BM of BTBR mice. In contrast, the numbers of plasma cells are increased in blood, spleens and BM; however, the increase does not seem to account for the lower numbers of B cells. This suggests that there may be more sites not measured such as brain, meninges or peritoneal fluid. The Increased production of plasma cells has been reported to be involved in some autoimmune diseases (Nutt et al., 2015).
We suggest this is the first report of the types of immune cells and their characteristics that are implicated in the higher level of IgG and brain-specific autoAbs of BTBR mice, which lead to their aberrant behavior. The molecular mechanisms for the spontaneously higher amounts of IgG and autoAbs in BTBR mice need further investigation along with the Ags and their role in brain functions and behavior. The elevated numbers of T cells, especially Tfh cells, and their influences on the B cell lineage are partially described regarding the epitranscriptomic influences on autoAb production; however, the dominant initiating influences have not been identified.
Declaration of competing interest
None of the authors have any conflicts of interest.
Acknowledgements
The work reported in this manuscript was supported by an NIH grant (R01 ES025584) to DAL.
References
- Ahmad S.F., Ansari M.A., Nadeem A., Bakheet S.A., Alsanea S., Al-Hosaini K.A., Mahmood H.M., Alzahrani M.Z., Attia S.M. Inhibition of tyrosine kinase signaling by tyrphostin AG126 downregulates the IL-21/IL-21R and JAK/STAT pathway in the BTBR mouse model of autism. Neurotoxicology. 2020;77:1–11. doi: 10.1016/j.neuro.2019.12.003. [DOI] [PubMed] [Google Scholar]
- Ahmad S.F., Nadeem A., Ansari M.A., Bakheet S.A., Al-Ayadhi L.Y., Attia S.M. Downregulation in Helios transcription factor signaling is associated with immune dysfunction in blood leukocytes of autistic children. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2018;85:98–104. doi: 10.1016/j.pnpbp.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Ahmad S.F., Zoheir K.M.A., Ansari M.A., Nadeem A., Bakheet S.A., Al-Ayadhi L.Y., Alzahrani M.Z., Al-Shabanah O.A., Al-Harbi M.M., Attia S.M. Dysregulation of Th1, Th2, Th17, and T regulatory cell-related transcription factor signaling in children with autism. Mol. Neurobiol. 2017;54:4390–4400. doi: 10.1007/s12035-016-9977-0. [DOI] [PubMed] [Google Scholar]
- Alinikula J., Lassila O. Gene interaction network regulates plasma cell differentiation. Scand. J. Immunol. 2011;73:512–519. doi: 10.1111/j.1365-3083.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- Allman D., Pillai S. Peripheral B cell subsets. Curr. Opin. Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwood P., Wills S., Van de Water J. The immune response in autism: a new frontier for autism research. J. Leukoc. Biol. 2006;80:1–15. doi: 10.1189/jlb.1205707. [DOI] [PubMed] [Google Scholar]
- Baio J., Wiggins L., Christensen D.L., Maenner M.J., Daniels J., Warren Z., Kurzius-Spencer M., Zahorodny W., Robinson Rosenberg C., White T., Durkin M.S., Imm P., Nikolaou L., Yeargin-Allsopp M., Lee L.C., Harrington R., Lopez M., Fitzgerald R.T., Hewitt A., Pettygrove S., Constantino J.N., Vehorn A., Shenouda J., Hall-Lande J., Van Naarden Braun K., Dowling N.F. Prevalence of autism spectrum disorder among children aged 8 Years - autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR Surveill. Summ. 2018;67:1–23. doi: 10.15585/mmwr.ss6706a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet S.A., Alzahrani M.Z., Nadeem A., Ansari M.A., Zoheir K.M.A., Attia S.M., Al-Ayadhi L.Y., Ahmad S.F. Resveratrol treatment attenuates chemokine receptor expression in the BTBR T+tf/J mouse model of autism. Mol. Cell. Neurosci. 2016;77:1–10. doi: 10.1016/j.mcn.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Bakheet S.A., Alzahrani M.Z., Ansari M.A., Nadeem A., Zoheir K.M.A., Attia S.M., Al-Ayadhi L.Y., Ahmad S.F. Resveratrol ameliorates dysregulation of Th1, Th2, Th17, and T regulatory cell-related transcription factor signaling in a BTBR T + tf/J mouse model of autism. Mol. Neurobiol. 2017;54:5201–5212. doi: 10.1007/s12035-016-0066-1. [DOI] [PubMed] [Google Scholar]
- Barwick B.G., Scharer C.D., Martinez R.J., Price M.J., Wein A.N., Haines R.R., Bally A.P.R., Kohlmeier J.E., Boss J.M. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat. Commun. 2018;9:1900. doi: 10.1038/s41467-018-04234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund L.J., Avery D.T., Ma C.S., Moens L., Deenick E.K., Bustamante J., Boisson-Dupuis S., Wong M., Adelstein S., Arkwright P.D., Bacchetta R., Bezrodnik L., Dadi H., Roifman C.M., Fulcher D.A., Ziegler J.B., Smart J.M., Kobayashi M., Picard C., Durandy A., Cook M.C., Casanova J.L., Uzel G., Tangye S.G. IL-21 signalling via STAT3 primes human naive B cells to respond to IL-2 to enhance their differentiation into plasmablasts. Blood. 2013;122:3940–3950. doi: 10.1182/blood-2013-06-506865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi C., Wu J., Jiang T., Liu Q., Cai W., Yu P., Cai T., Zhao M., Jiang Y.H., Sun Z.S. Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility. Hum. Mutat. 2012;33:1635–1638. doi: 10.1002/humu.22174. [DOI] [PubMed] [Google Scholar]
- Bilbo S.D., Schwarz J.M. Early-life programming of later-life brain and behavior: a critical role for the immune system. Front. Behav. Neurosci. 2009;3:14. doi: 10.3389/neuro.08.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölte S., Girdler S., Marschik P.B. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell Mol Life Sci. 2019;76:1275–1297. doi: 10.1007/s00018-018-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock K.A. In: DAN! (Defeat Autism Now!) Spring Conference Practitioner Training. San Diego, CA. Rimland B., editor. 2002. Integrative approach to autism spectrum disorders. [Google Scholar]
- Campisi L., Imran N., Nazeer A., Skokauskas N., Azeem M.W. Autism spectrum disorder. Br. Med. Bull. 2018;127:91–100. doi: 10.1093/bmb/ldy026. [DOI] [PubMed] [Google Scholar]
- Churilov L.P., Sobolevskaia P.A., Stroev Y.I. Thyroid gland and brain: enigma of Hashimoto’s encephalopathy. Best Pract. Res. Clin. Endocrinol. Metabol. 2019;33:101364. doi: 10.1016/j.beem.2019.101364. [DOI] [PubMed] [Google Scholar]
- Cotney J., Muhle R.A., Sanders S.J., Liu L., Willsey A.J., Niu W., Liu W., Klei L., Lei J., Yin J., Reilly S.K., Tebbenkamp A.T., Bichsel C., Pletikos M., Sestan N., Roeder K., State M.W., Devlin B., Noonan J.P. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B., Emili A. Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nat Protoc. 2006;1:1872–1878. doi: 10.1038/nprot.2006.273. [DOI] [PubMed] [Google Scholar]
- Croen L.A., Braunschweig D., Haapanen L., Yoshida C.K., Fireman B., Grether J.K., Kharrazi M., Hansen R.L., Ashwood P., Van de Water J. Maternal mid-pregnancy autoantibodies to fetal brain protein: the early markers for autism study. Biol. Psychiatr. 2008;64:583–588. doi: 10.1016/j.biopsych.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton P., Deacon R., Blamire A., Pike M., McKinlay I., Stein J., Styles P., Vincent A. Maternal neuronal antibodies associated with autism and a language disorder. Ann. Neurol. 2003;53:533–537. doi: 10.1002/ana.10557. [DOI] [PubMed] [Google Scholar]
- Garbett K., Ebert P.J., Mitchell A., Lintas C., Manzi B., Mirnics K., Persico A.M. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol. Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensous N., Charrier M., Duluc D., Contin-Bordes C., Truchetet M.E., Lazaro E., Duffau P., Blanco P., Richez C. T follicular helper cells in autoimmune disorders. Front. Immunol. 2018;9:1637. doi: 10.3389/fimmu.2018.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy S., Nacario A., Braithwaite R.E., Hopper C. The effect of physical activity interventions on youth with autism spectrum disorder: a meta-analysis. Autism Res. 2018;11:818–833. doi: 10.1002/aur.1955. [DOI] [PubMed] [Google Scholar]
- Heo Y. In vitro model for modulation of helper T cell differentiation and activation. Curr. Protoc. Toxicol. 2005;18 doi: 10.1002/0471140856.tx1809s24. Unit18 19. [DOI] [PubMed] [Google Scholar]
- Heo Y., Zhang Y., Gao D., Miller V.M., Lawrence D.A. Aberrant immune responses in a mouse with behavioral disorders. PloS One. 2011;6 doi: 10.1371/journal.pone.0020912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.C., Dougan S.K., McGehee A.M., Love J.C., Ploegh H.L. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28:1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Hakim S., Woolf C.J. Unraveling the plastic peripheral neuroimmune interactome. J. Immunol. 2020;204(2):257–263. doi: 10.4049/jimmunol.1900818. [DOI] [PubMed] [Google Scholar]
- Jia W., Wu W., Yang D., Xiao C., Su Z., Huang Z., Li Z., Qin M., Huang M., Liu S., Long F., Mao J., Liu X., Zhu Y.Z. Histone demethylase JMJD3 regulates fibroblast-like synoviocyte-mediated proliferation and joint destruction in rheumatoid arthritis. Faseb. J. 2018;32:4031–4042. doi: 10.1096/fj.201701483R. [DOI] [PubMed] [Google Scholar]
- Kavanaugh A., Tomar R., Reveille J., Solomon D.H., Homburger H.A. Guidelines for clinical use of the antinuclear antibody test and tests for specific autoantibodies to nuclear antigens. American College of Pathologists. Arch. Pathol. Lab Med. 2000;124:71–81. doi: 10.5858/2000-124-0071-GFCUOT. [DOI] [PubMed] [Google Scholar]
- Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Konforte D., Simard N., Paige C.J. IL-21: an executor of B cell fate. J. Immunol. 2009;182:1781–1787. doi: 10.4049/jimmunol.0803009. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., Mav D., Shah R., Grimm S.A., Phadke D., Hatzi K., Melnick A., Geigerman C., Sobol S.E., Jaye D.L., Wade P.A. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 2013;23:2030–2041. doi: 10.1101/gr.155473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lech M., Susanti H.E., Römmele C., Gröbmayr R., Günthner R., Anders H.J. Quantitative expression of C-type lectin receptors in humans and mice. Int. J. Mol. Sci. 2012;13:10113–10131. doi: 10.3390/ijms130810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Perez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., Cherry S.R., Tsai J.H., Tucker S.M., Weaver W.M., Kelso A., Jaenisch R., Wilson C.B. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Li X., Chauhan A., Sheikh A.M., Patil S., Chauhan V., Li X.M., Ji L., Brown T., Malik M. Elevated immune response in the brain of autistic patients. J. Neuroimmunol. 2009;207:111–116. doi: 10.1016/j.jneuroim.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Rigby R.J., Wong R.K., Yu D., Brink R., Cannons J.L., Schwartzberg P.L., Cook M.C., Walters G.D., Vinuesa C.G. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L.A., Ashwood P., Braunschweig D., Cabanlit M., Van de Water J., Amaral D.G. Stereotypies and hyperactivity in rhesus monkeys exposed to IgG from mothers of children with autism. Brain Behav. Immun. 2008;22:806–816. doi: 10.1016/j.bbi.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi A., Glozier N., Dale R., Guastella A.J. The immune system, cytokines, and biomarkers in autism spectrum disorder. Neurosci. Bull. 2017;33:194–204. doi: 10.1007/s12264-017-0103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H.G., Kusek G.K., Yang M., Phoenix J.L., Bolivar V.J., Crawley J.N. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Gene Brain Behav. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- Mondal T.K., Saha S.K., Miller V.M., Seegal R.F., Lawrence D.A. Autoantibody-mediated neuroinflammation: pathogenesis of neuropsychiatric systemic lupus erythematosus in the NZM88 murine model. Brain Behav Immun. 2008;22:949–959. doi: 10.1016/j.bbi.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Moscavitch S.D., Szyper-Kravitz M., Shoenfeld Y. Autoimmune pathology accounts for common manifestations in a wide range of neuro-psychiatric disorders: the olfactory and immune system interrelationship. Clin. Immunol. 2009;130:235–243. doi: 10.1016/j.clim.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Nahm D.H., Lee K.H., Shin J.Y., Ye Y.M., Kang Y., Park H.S. Identification of alpha-enolase as an autoantigen associated with severe asthma. J. Allergy Clin. Immunol. 2006;118:376–381. doi: 10.1016/j.jaci.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Nakamura A., Osonoi T., Terauchi Y. Relationship between urinary sodium excretion and pioglitazone-induced edema. J. Diabetes Investig. 2010;1:208–211. doi: 10.1111/j.2040-1124.2010.00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- Ohtsuka N., Badurek S., Busslinger M., Benes F.M., Minichiello L., Rudolph U. GABAergic neurons regulate lateral ventricular development via transcription factor Pax5. Genesis. 2013;51:234–245. doi: 10.1002/dvg.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A., Bernier R., Shendure J., Eichler E.E. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B.J., Deriziotis P., Lee C., Vives L., Schwartz J.J., Girirajan S., Karakoc E., Mackenzie A.P., Ng S.B., Baker C., Rieder M.J., Nickerson D.A., Bernier R., Fisher S.E., Shendure J., Eichler E.E. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onore C., Careaga M., Ashwood P. The role of immune dysfunction in the pathophysiology of autism. Brain Behav. Immun. 2012;26:383–392. doi: 10.1016/j.bbi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.L., Kumar G., Agasing A., Ko R.M., Axtell R.C. Role of TFH cells in promoting T helper 17-induced neuroinflammation. Front. Immunol. 2018;9:382. doi: 10.3389/fimmu.2018.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Geschwind D.H. Genetics of autism spectrum disorder. Handb. Clin. Neurol. 2018;147:321–329. doi: 10.1016/B978-0-444-63233-3.00021-X. [DOI] [PubMed] [Google Scholar]
- Schwartzer J.J., Careaga M., Onore C.E., Rushakoff J.A., Berman R.F., Ashwood P. Maternal immune activation and strain specific interactions in the development of autism-like behaviors in mice. Transl. Psychiatry. 2013;3 doi: 10.1038/tp.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Calame K. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- Sharma S.R., Gonda X., Tarazi F.I. Autism spectrum disorder: classification, diagnosis and therapy. Pharmacol. Ther. 2018;190:91–104. doi: 10.1016/j.pharmthera.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Sheu K.F., Brown A.M., Haroutunian V., Kristal B.S., Thaler H., Lesser M., Kalaria R.N., Relkin N.R., Mohs R.C., Lilius L., Lannfelt L., Blass J.P. Modulation by DLST of the genetic risk of Alzheimer’s disease in a very elderly population. Ann. Neurol. 1999;45:48–53. [PubMed] [Google Scholar]
- Silverman J.L., Tolu S.S., Barkan C.L., Crawley J.N. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology. 2010;35:976–989. doi: 10.1038/npp.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer H.S., Morris C., Gause C., Pollard M., Zimmerman A.W., Pletnikov M. Prenatal exposure to antibodies from mothers of children with autism produces neurobehavioral alterations: a pregnant dam mouse model. J. Neuroimmunol. 2009;211:39–48. doi: 10.1016/j.jneuroim.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Singer H.S., Morris C.M., Gause C.D., Gillin P.K., Crawford S., Zimmerman A.W. Antibodies against fetal brain in sera of mothers with autistic children. J. Neuroimmunol. 2008;194:165–172. doi: 10.1016/j.jneuroim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Siu M.T., Weksberg R. Epigenetics of autism spectrum disorder. Adv. Exp. Med. Biol. 2017;978:63–90. doi: 10.1007/978-3-319-53889-1_4. [DOI] [PubMed] [Google Scholar]
- Staudt V., Bothur E., Klein M., Lingnau K., Reuter S., Grebe N., Gerlitzki B., Hoffmann M., Ulges A., Taube C., Dehzad N., Becker M., Stassen M., Steinborn A., Lohoff M., Schild H., Schmitt E., Bopp T. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Steinman L. Elaborate interactions between the immune and nervous systems. Nat. Immunol. 2004;5:575–581. doi: 10.1038/ni1078. [DOI] [PubMed] [Google Scholar]
- Sweeten T.L., Posey D.J., McDougle C.J. High blood monocyte counts and neopterin levels in children with autistic disorder. Am. J. Psychiatr. 2003;160:1691–1693. doi: 10.1176/appi.ajp.160.9.1691. [DOI] [PubMed] [Google Scholar]
- Thevenin C., Nutt S.L., Busslinger M. Early function of Pax5 (BSAP) before the pre-B cell receptor stage of B lymphopoiesis. J. Exp. Med. 1998;188:735–744. doi: 10.1084/jem.188.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tick B., Bolton P., Happe F., Rutter M., Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. JCPP (J. Child Psychol. Psychiatry) 2016;57:585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas D.L., Nascimbene C., Krishnan C., Zimmerman A.W., Pardo C.A. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- Viisanen T., Ihantola E.L., Nanto-Salonen K., Hyoty H., Nurminen N., Selvenius J., Juutilainen A., Moilanen L., Pihlajamaki J., Veijola R., Toppari J., Knip M., Ilonen J., Kinnunen T. Circulating CXCR5+PD-1+ICOS+ follicular T helper cells are increased close to the diagnosis of type 1 diabetes in children with multiple autoantibodies. Diabetes. 2017;66:437–447. doi: 10.2337/db16-0714. [DOI] [PubMed] [Google Scholar]
- Wei Y., Zheng H., Bao N., Jiang S., Bueso-Ramos C.E., Khoury J., Class C., Lu Y., Lin K., Yang H., Ganan-Gomez I., Starczynowski D.T., Do K.A., Colla S., Garcia-Manero G. KDM6B overexpression activates innate immune signaling and impairs hematopoiesis in mice. Blood Adv. 2018;2:2491–2504. doi: 10.1182/bloodadvances.2018024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles E.T., Selker E.U. H3K27 methylation: a promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2017;43:31–37. doi: 10.1016/j.gde.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis S.N., Good-Jacobson K.L., Curtis J., Light A., Tellier J., Shi W., Smyth G.K., Tarlinton D.M., Belz G.T., Corcoran L.M., Kallies A., Nutt S.L. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J. Immunol. 2014;192:3200–3206. doi: 10.4049/jimmunol.1303216. [DOI] [PubMed] [Google Scholar]
- Wills S., Cabanlit M., Bennett J., Ashwood P., Amaral D., Van de Water J. Autoantibodies in autism spectrum disorders (ASD) Ann. N. Y. Acad. Sci. 2007;1107:79–91. doi: 10.1196/annals.1381.009. [DOI] [PubMed] [Google Scholar]
- Wohr M., Roullet F.I., Crawley J.N. Reduced scent marking and ultrasonic vocalizations in the BTBR T+tf/J mouse model of autism. Gene Brain Behav. 2011;10:35–43. doi: 10.1111/j.1601-183X.2010.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Li X., Zhong Y. Inflammatory cytokines: potential biomarkers of immunologic dysfunction in autism spectrum disorders. Mediat. Inflamm. 2015;2015:531518. doi: 10.1155/2015/531518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Wu H., Zhao M., Zhang Q., Long H., Fu S., Lu Q. Histone demethylase JMJD3 regulates CD11a expression through changes in histone H3K27 tri-methylation levels in CD4+ T cells of patients with systemic lupus erythematosus. Oncotarget. 2017;8:48938–48947. doi: 10.18632/oncotarget.16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M., Fujii A., Ito A., Yokoyama H., Nakagawa H., Kuriyama M. High prevalence of serum autoantibodies against the amino terminal of alpha-enolase in Hashimoto’s encephalopathy. J. Neuroimmunol. 2007;185:195–200. doi: 10.1016/j.jneuroim.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao D., Kluetzman K., Mendoza A., Bolivar V.J., Reilly A., Jolly J.K., Lawrence D.A. The maternal autoimmune environment affects the social behavior of offspring. J. Neuroimmunol. 2013;258:51–60. doi: 10.1016/j.jneuroim.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Zwaigenbaum L., Penner M. Autism spectrum disorder: advances in diagnosis and evaluation. BMJ. 2018;361:k1674. doi: 10.1136/bmj.k1674. [DOI] [PubMed] [Google Scholar]