Abstract
Introduction
DNA repair capacity, as exemplified by BRCA1 gene expression, is related with outcome to EGFR tyrosine kinase inhibitors in patients with EGFR-mutant NSCLC. Olaparib, a PARP inhibitor, reduces BRCA1 expression. Olaparib was tested in combination with gefitinib versus gefitinib single agent, as a first-line therapy for patients with EGFR-mutant NSCLC in the GOAL study (trial registration: NCT01513174). Here, we report the results of the biomarker-related prespecified secondary objectives of the GOAL study.
Methods
We evaluated the impact of BRCA1 mRNA expression in 91 patients with EGFR-mutant NSCLC. Of those 91 patients, 51 were randomized to treatment with gefitinib and 40 were randomized to treatment with gefitinib plus olaparib. We explored in vitro whether BRCA1 mRNA levels are related with outcome to gefitinib plus olaparib. The expression levels of 53BP1, CtIP, and AXL were also explored and correlated with the treatment outcome.
Results
Overall, as what happened in the GOAL study, no statistically significant difference was observed in median progression-free survival (PFS) between the two treatment arms, for the 91 patients of the present study (p = 0.2419). For patients with high BRCA1 mRNA expression (BRCA1-high group), median PFS was 12.9 months in the gefitinib plus olaparib arm, compared with 9.2 months in the gefitinib arm (p = 0.0449). In the gefitinib arm, median PFS was 9.1 months for the BRCA1-high group and 10.2 months for the BRCA1-low group (p = 0.0193). We observed a more pronounced synergism of gefitinib plus olaparib in cells with higher BRCA1 compared with those with low BRCA1 mRNA expression.
Conclusions
High BRCA1 mRNA expression identified patients with NSCLC who benefited from gefitinib plus olaparib in the GOAL phase 2 clinical trial.
Keywords: EGFR, Lung cancer, Gefitinib, Olaparib, GOAL study
Introduction
We identified that low BRCA1 gene mRNA levels were an independent favorable predictive marker to erlotinib in patients with EGFR-mutant advanced NSCLC.1,2 On the basis of the findings, we proposed a model for a BRCA1-dependent DNA repair of erlotinib-induced DNA damage through an H2AX-independent pathway. We speculate that DNA breakage caused by erlotinib is different from that caused by radiotherapy or platinum-based chemotherapy. In addition, poly(ADP)-ribosylation of proteins by PARP1 is a rapid response to DNA lesions. PARP1 inhibitors down-regulate BRCA1 expression.3 We posit that BRCA1 by itself could be a predictive biomarker, and studies are warranted to use PARP inhibitors in combination with erlotinib in patients with elevated BRCA1 gene expression.2
The GOAL study, performed by the Spanish Lung Cancer Group, was a phase 1B and 2B study to evaluate the efficacy and tolerability of gefitinib plus olaparib versus gefitinib alone as first-line therapy in patients with metastatic EGFR-mutant NSCLC (NCT01513174). In the phase 1B dose escalation part of the study, tolerance in the absence of pharmacokinetic interactions and the activity of gefitinib plus olaparib were confirmed in 22 patients with EGFR-mutant NSCLC.4 The recommended phase 2 dose was 250 mg of gefitinib once daily plus 200 mg of olaparib three times daily.4 In the phase 2B part of the GOAL study, between July 2013 and July 2016, a total of 186 patients with previously untreated metastatic EGFR-mutant NSCLC were included in 34 centers in Spain and one in Mexico.5 The intent-to-treat (ITT) analysis included 91 patients with EGFR-mutant NSCLC in the gefitinib arm and 91 patients with EGFR-mutant NSCLC in the gefitinib plus olaparib arm.5 Progression-free survival (PFS) and overall survival (OS) were evaluated at the final data cutoff point on July 2017. The median follow-up time was 26.2 months (95% confidence interval [CI]: 20.3–27.8) for gefitinib and 21.2 months (95% CI: 17.5–28.3) for gefitinib plus olaparib (p = 0.2858).5
The primary end point of the phase 2B GOAL study, which was to determine whether the addition of olaparib to gefitinib improved PFS in previously untreated patients with metastatic EGFR-mutant NSCLC, was not met.5 Median PFS was 10.9 months (95% CI: 9.3–13.3) for gefitinib versus 12.8 months (95% CI: 9.1–14.7) for gefitinib plus olaparib (p = 0.1242; hazard ratio [HR] = 0.75, 95% CI: 0.52–1.08).5 Among 174 patients assessable for response, the objective response rate was 68% in the gefitinib arm versus 71% in the gefitinib plus olaparib arm (p = 0.4873).5 No statistically significant differences were found between the two treatment arms in median OS.5 As far as safety concerns, there was an increase in hematological and gastrointestinal toxicities for gefitinib plus olaparib, compared with gefitinib alone.5
Here, we report the results of the biomarker-related prespecified secondary objectives of the GOAL study. A secondary objective of the GOAL study was to evaluate whether the mRNA expression levels of BRCA1 may affect PFS in the two treatment arms.2 The correlation of the mRNA expression levels of other biomarkers, including 53BP1, CtIP,6 and AXL,7 with PFS, is also explored. All patients have provided written informed consent before being enrolled in the study. Preclinical in vitro evidence of the potential effect of BRCA1 mRNA expression levels on the outcome to gefitinib plus olaparib is finally provided.
Materials and Methods
Real-Time Polymerase Chain Reaction Analyses
Paraffin-embedded samples and slides and cell lines were processed as previously reported for gene mRNA expression.8,9 Tumor tissue was available from 91 patients of the GOAL study, 51 of whom were randomized to treatment with gefitinib and 40 were randomized to treatment with gefitinib plus olaparib.5 The present gene expression study was conducted in the ISO 15189-certified Pangaea Oncology laboratory located in Hospital Universitari Dexeus—Grupo Quirónsalud (Barcelona, Spain). Pangaea Oncology was the central laboratory for the GOAL study. The primer and probe sequences for BRCA1, 53BP1, CtIP, and AXL were designed using Primer Express 3.0 Software (Applied Biosystems) according to their reference sequence (http://www.ncbi.nlm.nih.gov/LocusLink). β-actin was used as the endogenous gene. Gene expression analyses were performed as previously described with quantitative real-time polymerase chain reaction.8,9
Cell Culture and Reagents
PC9 (EGFR exon 19 deletion) cells were provided by F. Hoffmann-La Roche Ltd. The brain metastatic lung cancer cell line, PC9-BrM3, was generously provided by Professor Joan Massagué (Cancer Biology and Genetics Program, Memorial Sloan Kettering Cancer Center, New York, NY).10 Gefitinib was purchased from Tocris Bioscience Company (Bristol, United Kingdom). Olaparib was purchased from Selleckchem (Houston, TX). Drugs were prepared in DMSO at a concentration of 10 to 100 mmol/liter stock solutions and stored at −20°C. Further dilutions were made in culture medium to final concentration before use, as previously described.7
Cell Viability Assay
Cells were seeded on 96-well plates at the following densities: 2 × 103, 3 × 103, and 4 × 103 and incubated for 24 hours.7 Cells were treated with serial dilutions of the drugs administered at indicated doses. After 72 hours of incubation, 0.5 mg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reagent (Sigma-Aldrich, St. Louis, MO) was added to the medium in the wells for 2 hours at 37°C. Formazan crystals in viable cells were solubilized with 100 μL DMSO and spectrophotometrically quantified using a microplate reader (Varioskan Flash; Thermo Fisher Scientific, Waltham, MA) at 550 nm of absorbance. Fractional survival was calculated as percentage to control cells. Data of combined drug effects were analyzed by the Bliss method.11
Statistical Analyses
The primary end point of this study was to examine the potential effects of gene mRNA expression levels on survival. PFS and OS were estimated by means of the Kaplan-Meier method and compared with a nonparametric log-rank test. Biomarker expression was assessed as a dichotomous estimate (low versus high using the median as the cutoff).7 A Cox proportional hazard model was applied with potential risk factors as covariates, obtaining HR and their 95% CI. Each analysis was performed with the use of a two-sided 5% significance level and a 95% CI. The statistical analyses were performed using SAS version 9.4. In vitro data were analyzed using unpaired t test (GraphPad Prism, GraphPad Software, Inc.). Values of p less than 0.05 were considered statistically significant. Proportion hazard regression analyses were generated using GraphPad Prism.
Results
Patients
From the 182 patients of the ITT analysis of the GOAL study,5 91 had sufficient tumor tissue for the gene expression analysis of BRCA1, CtIP, 53BP1, and AXL. The characteristics of the 91 patients, enrolled at 30 centers in Spain and one in Mexico, are illustrated in Table 1. Of those 91 patients, 51 were randomized to treatment with gefitinib and 40 were randomized to treatment with gefitinib plus olaparib. Patients allocated to the two arms were well balanced for baseline characteristics including age, sex, smoking history, Eastern Cooperative Oncology Group performance status, and type of EGFR mutation (Table 1). The median age of all patients was 67.5 years, and 69% of them were female. Most of the patients had Eastern Cooperative Oncology Group performance status 1 (68%). In both arms, almost two-thirds of the patients had EGFR exon 19 deletions and one-third had EGFR exon 21 L858R substitutions.
Table 1.
Baseline Characteristics
| Characteristic | Gefitinib (N = 51) | Gefitinib + Olaparib (N = 40) | p Value Test |
|---|---|---|---|
| Sex, N (%) | |||
| Male | 18 (35) | 10 (25) | Chi-square: 0.2910 |
| Female | 33 (65) | 30 (75) | |
| Age, y | |||
| Median (range) | 70 (36–85) | 65 (39–85) | Wilcoxon: 0.1500 |
| Smoking history, N (%) | |||
| Never smoker | 33 (65) | 25 (63) | Fisher: 0.9369 |
| Ex-smoker | 15 (29) | 13 (32) | |
| Current smoker | 3 (6) | 2 (5) | |
| ECOG PS, N (%) | |||
| 0 | 14 (27) | 9 (22) | Fisher: 0.7242 |
| 1 | 33 (65) | 29 (73) | |
| ≥2 | 4 (8) | 2 (5) | |
| Bone metastases, N (%) | |||
| Yes | 13 (25) | 12 (30) | NA |
| No | 38 (75) | 28 (70) | |
| Brain metastases, N (%) | |||
| Yes | 6 (12) | 4 (10) | NA |
| No | 45 (88) | 36 (90) | |
| Type of EGFR mutation, N (%) | |||
| Exon 19 deletion | 30 (59) | 22 (55) | Fisher: 0.3958 |
| L858R | 20 (39) | 14 (35) | |
| Exon 18 | 1 (2) | 2 (5) | |
| Exon 20 | 0 (0) | 2 (5) |
ECOG PS, Eastern Cooperative Oncology Group performance status; NA, not applicable.
BRCA1 mRNA Expression and Treatment Outcome
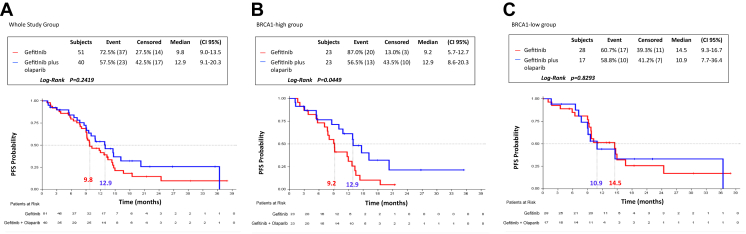
Similarly to the whole population of the GOAL study,5 in this analysis of the 91 patients, there was no statistically significant difference in median PFS between the two treatment arms (p = 0.2419) (Fig. 1A). However, when we dichotomized the 91 patients into two groups, on the basis of the median mRNA expression of BRCA1 (BRCA1-high group [N = 46] and BRCA1-low group [N = 45]), in the BRCA1-high group, a statistically significant longer median PFS was found with gefitinib plus olaparib compared with gefitinib single agent. Specifically, as illustrated in Figure 1B, in the BRCA1-high group, median PFS was 12.9 months (95% CI: 8.6–20.3) in the gefitinib plus olaparib arm, compared with 9.2 months (95% CI: 5.7–12.7) in the gefitinib arm, p = 0.0449 (HR for gefitinib versus gefitinib plus olaparib = 2.04, 95% CI: 1.00–4.13, p = 0.0492). In the BRCA1-low group, a longer median PFS of 14.5 months (95% CI: 9.3–16.7), which did not reach the statistical significance, was found in the gefitinib arm compared with the gefitinib plus olaparib arm, in which patients experienced a shorter median PFS of 10.9 months (95% CI: 7.7–36.4, p = 0.8293) (Fig. 1C).
Figure 1.
PFS by treatment arm and by BRCA1 mRNA expression in 91 patients with EGFR-mutant NSCLC from the GOAL study. (A) mPFS was 9.1 months (95% CI: 9.0–13.5) for the 51 patients in the gefitinib arm and 12.9 months (95% CI: 9.1–20.3) for the 40 patients in the gefitinib plus olaparib arm; p = 0.2419. (B) In the BRCA1-high group, mPFS was 9.2 months (95% CI: 5.7–12.7) for the 23 patients in the gefitinib arm and 12.9 months (95% CI: 8.6–20.3) for the 23 patients in the gefitinib plus olaparib arm; p = 0.0449. (C) In the BRCA1-low group, mPFS was 14.5 months (95% CI: 9.3–16.7) for the 28 patients in the gefitinib arm and 10.9 months (95% CI: 7.7–36.4) for the 17 patients in the gefitinib plus olaparib arm; p = 0.8293. CI, confidence interval; mPFS, median PFS; PFS, progression-free survival.
Furthermore, in the gefitinib treatment arm, the BRCA1-high group had a significantly shorter median PFS of 9.1 months (95% CI: 7.1–12.0) compared with 10.2 months (95% CI: 9.2–16.7) for the BRCA1-low group (p = 0.0193) (HR = 2.08, 95% CI: 1.11–3.91, p = 0.0223) (Table 2). In the gefitinib plus olaparib arm, the BRCA1-high group had a longer median PFS of 14.6 months (95% CI: 8.6–20.3) compared with 10.9 months (95% CI: 7.2–36.4) for the BRCA1-low group, although this increase did not reach statistical significance (p = 0.8755) (Table 2). No statistically significant differences were observed in the responses in the BRCA1-high and BRCA1-low groups according to the treatment arm or in the two treatment arms according to the BRCA1 mRNA expression levels (Table 3).
Table 2.
Summary Table of Univariate PFS Analysis for BRCA1 mRNA Expression Level in the Two Treatment Arms
| Variable | N | Stratified Kaplan-Meier Model |
Cox Regression |
|||
|---|---|---|---|---|---|---|
| PFS, Median (95% CI) | p Value | Contrast | HR (95% CI) | p Value | ||
| Gefitinib | ||||||
| BRCA1-low | 29 | 10.2 (9.2–16.7) | 0.0193 | BRCA1-high vs. BRCA1-low | 2.08 (1.11–3.91) | 0.0223 |
| BRCA1-high | 27 | 9.1 (7.1–12.0) | ||||
| Gefitinib plus olaparib | ||||||
| BRCA1-low | 22 | 10.9 (7.2–36.4) | 0.8755 | BRCA1-high vs. BRCA1-low | 0.94 (0.41–2.14) | 0.8749 |
| BRCA1-high | 25 | 14.6 (8.6–20.3) | ||||
CI, confidence interval; HR, hazard ratio; PFS, progression-free survival.
Table 3.
Summary Table of Objective Response According to BRCA1 mRNA Expression Level in the Two Treatment Arms or According to the Two Treatment Arms in the Two BRCA1 mRNA Expression Level Groups
| Variable | BRCA1-High Group |
BRCA1-Low Group |
Gefitinib |
Gefitinib + Olaparib |
||||
|---|---|---|---|---|---|---|---|---|
| Gefitinib + Olaparib (n = 23) | Gefitinib (n = 23) | Gefitinib + Olaparib (n = 17) | Gefitinib (n = 28) | BRCA1-High (n = 23) | BRCA1-Low (n = 28) | BRCA1-High (n = 23) | BRCA1-Low (n = 17) | |
| Objective response | ||||||||
| Complete response, N (%) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (3.57) | 0 (0.00) | 1 (3.57) | 0 (0.00) | 0 (0.00) |
| Partial response, N (%) | 17 (73.91) | 14 (60.87) | 13 (76.47) | 17 (60.71) | 14 (60.87) | 17 (60.71) | 17 (73.91) | 13 (76.47) |
| Stable disease, N (%) | 3 (13.04) | 7 (30.43) | 3 (17.65) | 8 (28.57) | 7 (30.43) | 8 (28.57) | 3 (13.04) | 3 (17.65) |
| Progressive disease, N (%) | 2 (8.70) | 2 (8.70) | 1 (5.88) | 1 (3.57) | 2 (8.70) | 1 (3.57) | 2 (8.70) | 1 (5.88) |
| Not assessable, N (%) | 1 (4.35) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (3.57) | 1 (4.35) | 0 (0.00) |
| p value test (Fisher) | 0.4012 | 0.8663 | 0.9705 | 1.0000 | ||||
| Objective response rate, N (%) | 17 (73.91) | 14 (60.87) | 13 (76.47) | 18 (64.29) | 14 (60.87) | 18 (64.29) | 17 (73.91) | 13 (76.47) |
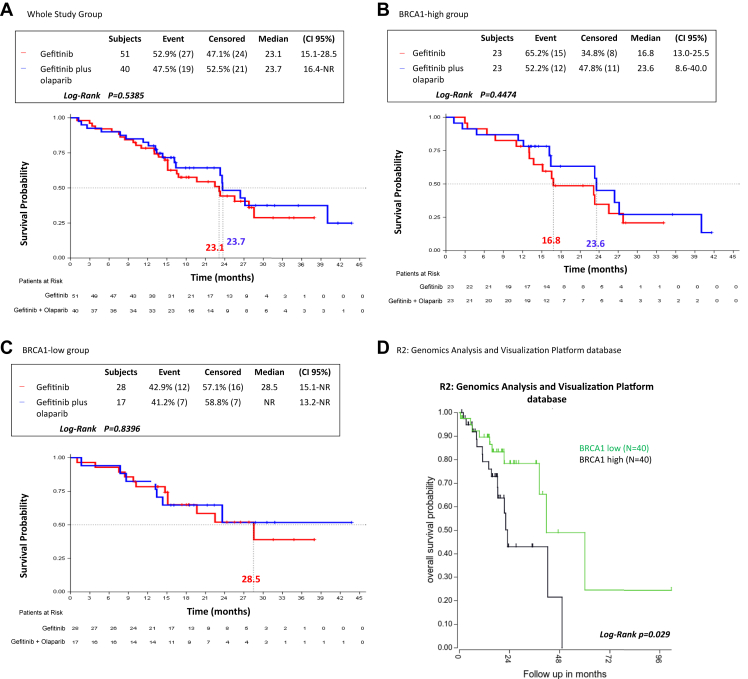
Overall, for the 91 patients, similar median OS was observed between the two treatment arms (23.1 mo, 95% CI: 15.1–28.5 for gefitinib versus 23.7 mo, 95% CI: 16.4–not reached for gefitinib plus olaparib, p = 0.5385). No differences in median OS were found between the two treatment arms, neither in the BRCA1-high group nor in the BRCA1-low group, although the patients in the BRCA1-high group experienced numerically shorter OS compared with those in the BRCA1-low group, independently of the treatment arm (Fig. 2A–C). This observation was reinforced by data obtained from the R2: Genomics Analysis and Visualization Platform database (http://r2.amc.nl, Data Set Tumor Lung-Bild-114-MAS5.0-u133p2, EGFR-mutant). As presented in Figure 2D, patients with EGFR-mutant NSCLC with high BRCA1 mRNA expression have a significantly worse prognosis than those with low BRCA1 mRNA expression (p = 0.029).
Figure 2.
OS by treatment arm and by BRCA1 mRNA expression in 91 patients with EGFR-mutant NSCLC from the GOAL study and in 80 patients with EGFR-mutant NSCLC from the R2: Genomics Analysis and Visualization Platform database. (A) mOS was 23.1 months (95% CI: 15.1–28.5) for the 51 patients in the gefitinib arm and 23.7 months (95% CI: 16.4–NR) for the 40 patients in the gefitinib plus olaparib arm; p = 0.5385. (B) In the BRCA1-high group, mOS was 16.8 months (95% CI: 13.0–25.5) for the 23 patients in the gefitinib arm and 23.6 months (95% CI: 8.6–40-0) for the 23 patients in the gefitinib plus olaparib arm; p = 0.4474. (C) In the BRCA1-low group, mOS was 28.5 months (95% CI: 15.1–NR) for the 28 patients in the gefitinib arm and NR (95% CI: 13.2–NR) for the 17 patients in the gefitinib plus olaparib arm; p = 0.8396. (D) Correlation between OS and BRCA1 mRNA expression levels in patients with EGFR-mutant NSCLC as determined by means of Kaplan-Meier analysis obtained from the R2: Genomics Analysis and Visualization Platform database. CI, confidence interval; mOS, median OS; NR, not reached; OS, overall survival.
Our data indicate that patients with EGFR-mutant NSCLC with high BRCA1 mRNA expression may derive a better outcome with gefitinib plus olaparib compared with gefitinib single agent. High BRCA1 mRNA expression appears to be a poor theranostic factor for patients with EGFR-mutant NSCLC treated with EGFR tyrosine kinase inhibitors (TKIs), as we have previously reported.2
BRCA1 mRNA Expression and Sensitivity to Gefitinib or Gefitinib Plus Olaparib In Vitro
To figure out whether the effect of gefitinib plus olaparib in EGFR-mutant NSCLC is related with BRCA1 mRNA expression, we performed in vitro experiments in two EGFR-mutant NSCLC cell lines, the PC9 lung adenocarcinoma cell line with the EGFR exon 19 deletion and the PC9-BrM3 cell line, a brain metastatic variant derived from the PC9 cell line. It has been previously reported that PARP1 knockdown in PC9-BrM3 cells attenuates anchorage-independent colony formation in soft agar and decreases invasion and transendothelial migration.12
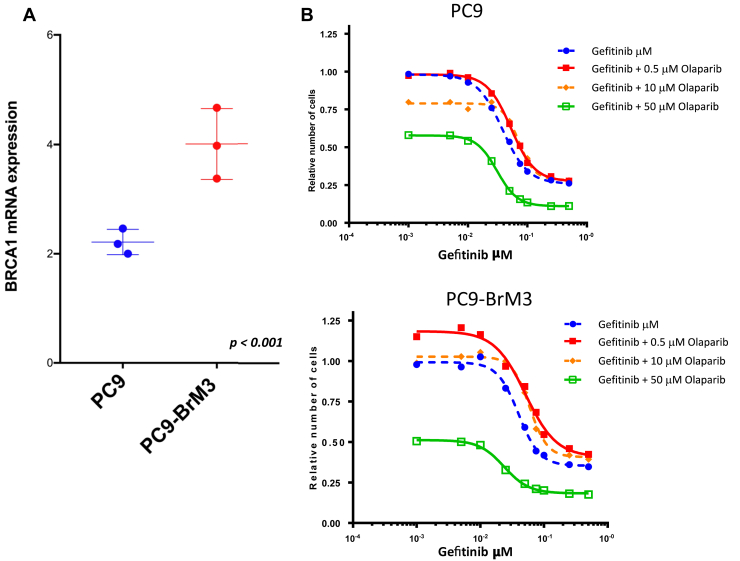
We found that PC9-BrM3 cells have significantly higher BRCA1 mRNA expression levels compared with parental PC9 cells (Fig. 3A). However, this difference in BRCA1 mRNA expression was not translated to a differential sensitivity to gefitinib single agent, as the half maximal inhibitory concentration of gefitinib was the same for parental PC9 and PC9-BrM3 cells (0.11 μM). This is probably because of the fact that both parental PC9 and PC9-BrM3 cell lines are dependent on EGFR signaling for survival but PC9-BrM3 cells have a marked increase in the capacity to invade and colonize distant organs.10
Figure 3.
Quantitative RT-PCR analysis of BRCA1 and the effects of the combination of gefitinib plus olaparib in the PC9 and PC9-BrM3 cell lines. (A) BRCA1 mRNA expression in the PC9 and PC9-BrM3 cell lines. Data are means ± SD of three independent experiments. Data were analyzed using unpaired t test (GraphPad Prism, GraphPad Software, Inc.). (B) PC9 cells were treated with serial dilutions of gefitinib alone or in combination with 50 μM of olaparib for 72 hours. Cell viability was measured by MTT, and the synergy between the drugs was determined using the Chou and Talalay method (Chou and Talalay plot or fraction affected plot). (C) PC9-BrM3 cells were treated with serial dilutions of gefitinib alone or in combination with 50 μM of olaparib for 72 hours. Cell viability was measured by MTT, and the synergy between the drugs was determined using the Chou and Talalay method as in (B). The results in (B) and in (C) represent the means of at least three independent experiments. MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; RT-PCR, real-time polymerase chain reaction.
We then combined gefitinib with three different concentrations of olaparib (0.5, 10, and 50 μM) in both PC9 and PC9-BrM3 cells, and cell proliferation was investigated after 72 hours of treatment. Gefitinib plus 50 μM of olaparib was able to inhibit cell proliferation more potently than gefitinib alone in both cell lines (Fig. 3B and C). A more pronounced synergistic interaction of gefitinib with olaparib (at the highest concentration of 50 μM) with a median combination index of 0.48 (range: 0.42–0.58) was found in the PC9-BrM3 cell line, compared with the parental PC9 cell line with median combination index of 0.52 (range: 0.29–0.86).
These preliminary and exploratory preclinical findings reinforce our clinical observations in the GOAL study, in which in the BRCA1-high group the combination of gefitinib plus olaparib conferred a significantly longer PFS compared with gefitinib alone (p = 0.0449), whereas no differences between the two treatment arms were found in the BRCA1-low group.
Correlation of CtIP, 53BP1, and AXL mRNA Expression and Treatment Outcome
We then explored whether the mRNA expression of other than BRCA1 biomarkers may have an impact on the treatment outcome, as prespecified in the protocol of the GOAL study. Similarly, to what we performed for the BRCA1 mRNA expression, the 91 patients were dichotomized into two groups, on the basis of the median mRNA expression of each of the three biomarkers, CtIP, 53BP1, and AXL.
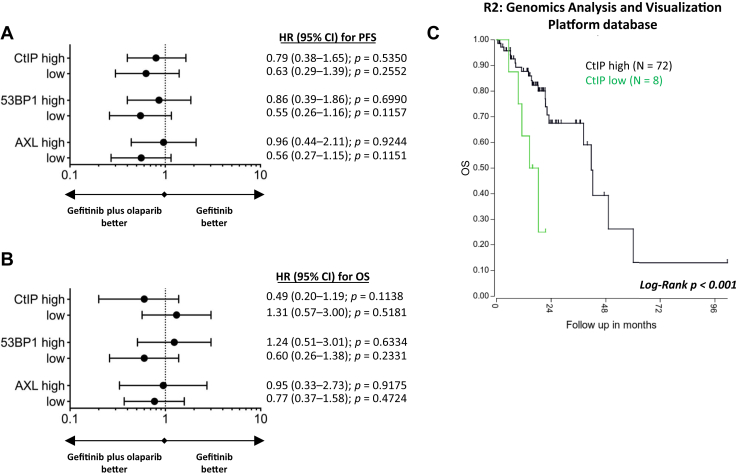
In the case of CtIP mRNA expression, we obtained statistically significant associations with gefitinib treatment, and we were able to make some interesting observations that merit further investigation. First, in the CtIP-low group, median PFS was numerically longer with gefitinib plus olaparib (12.5 mo, 95% CI: 5.4–not reached) compared with gefitinib alone (9.2 mo, 95% CI: 6.6–10.2) (p = 0.2479) (Fig. 4A). Second, in the gefitinib treatment arm, the CtIP-low group had a statistically significant (Fig. 4A) shorter median PFS of 9.0 months (95% CI: 5.4–9.3) compared with 13.3 months (95% CI: 9.7–14.9) for the CtIP-high group (p = 0.0071; HR: 2.52, 95% CI: 1.26–5.07, p = 0.0093). No statistically significant or clinically relevant correlations were found on the basis of the expression levels of 53BP1 or AXL (Fig. 4A).
Figure 4.
Progression-free and OS by CtIP, 53BP1, and AXL mRNA expression in 91 patients with EGFR-mutant NSCLC from the GOAL study and OS by CtIP mRNA expression in 80 patients with EGFR-mutant NSCLC from the R2: Genomics Analysis and Visualization Platform database. (A) Forest plots of the predictive value of the biomarkers for PFS to gefitinib compared with gefitinib plus olaparib. (B) Forest plots of the predictive value of the biomarkers for OS to gefitinib compared with gefitinib plus olaparib. (C) Correlation between OS and CtIP mRNA expression levels in patients with EGFR-mutant NSCLC as determined by means of Kaplan-Meier analysis obtained from the R2: Genomics Analysis and Visualization Platform database. CI, confidence interval; HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
In our study, no significant or relevant findings were found for OS on the basis of the mRNA expression levels of the three biomarkers or the treatment arm (Fig. 4B). In the R2: Genomics Analysis and Visualization Platform database (http://r2.amc.nl, Data Set Tumor Lung-Bild-114-MAS5.0-u133p2, EGFR-mutant), patients with EGFR-mutant NSCLC with low CtIP mRNA expression have a significantly worse prognosis than those with high CtIP mRNA expression (p < 0.001) (Fig. 4C). In our previous study, we were not able to find any significant correlation between CtIP mRNA expression and outcome to erlotinib.6
Collectively, these data point out that besides high BRCA1 mRNA expression, low mRNA expression of CtIP may be an indicator of worse outcome to single therapy with EGFR TKIs. Whether patients with EGFR-mutant NSCLC who are CtIP-low expressers may derive benefit from the combination of EGFR TKIs with PARP inhibitors requires further research.
Discussion
The biomarker-related prespecified secondary objective of the GOAL study was achieved. The combination of gefitinib plus olaparib significantly improved PFS in the BRCA1-high group, as was predicted in our model (p = 0.0449).2 In addition, the BRCA1-low group had significantly longer PFS when treated with gefitinib alone compared with the BRCA1-high group (p = 0.0193). Nevertheless, no significant benefit of the combination of gefitinib plus olaparib was observed in the whole population of EGFR-mutant patients in the GOAL study.5 In our preclinical experiments, there was no difference in sensitivity to gefitinib between cells with higher BRCA1 mRNA expression (PC9-BrM3 cells) compared with those with lower BRCA1 mRNA expression (parental PC9 cells).
BRCA1 protein contains two BRCT (BRCA1 C-terminal) repeats, which form exclusive complexes with Abraxas, BACH1 and CtIP. These complexes are defined as BRCA1 A complex (RAP80 and Abraxas), B complex (BACH1), and C complex (CtIP and RAP80). The BRCA1 A complex is involved in DNA damage response; however, depletion of only BRCA1 has stronger effect on the various DNA damage response assays compared with depletion of either Abraxas or RAP80.13 Both the A and C complexes are required for the G2-M checkpoint. They are also implicated in transcription.13
In this study, we observed that low CtIP mRNA levels predicted longer PFS in EGFR-mutant patients treated with gefitinib plus olaparib, in comparison with those treated with gefitinib single agent. Noteworthy was the fact that, in the gefitinib treatment arm, the CtIP-low group had significantly shorter median PFS in comparison with the CtIP-high group (HR = 2.52, p = 0.0093). In the case of 53BP1 mRNA expression,14 we were not able to find any statistically significant correlation with the outcome to gefitinib or gefitinib plus olaparib. These findings are in concordance with our previous model that BRCA1-dependent repair of erlotinib-induced DNA damage occurs independently of the homologous recombination pathway.2 Although AXL overexpression is associated with intrinsic and acquired resistance to EGFR TKIs,7,15 in the current study, we did not find any significant association between AXL mRNA expression and treatment outcome. AXL has been noted to be associated to many components of the homologous recombination pathway.16
In our previous study, BRCA1 mRNA expression predicted outcome to gefitinib or erlotinib in patients with EGFR-mutant NSCLC opening opportunities for alternative therapies, including PARP inhibitors or chemotherapy customization.2 BRCA1 regulation is associated with resistance to cisplatin17 and differential expression of BRCA1 supports the use of taxanes.18,19 In fact, intercalated combination of chemotherapy and erlotinib leads to a PFS of 16.8 months in patients with EGFR-mutant NSCLC, in comparison with 6.9 months in the chemotherapy group. These differences also translate to significant improvement in median OS in the chemotherapy plus erlotinib group.20
Finally, we noted that in the PC9-BrM3 cell line, a brain metastatic variant derived from the PC9 cell line, highly metastatic to the bones and brain, BRCA1 mRNA expression was higher, and a stronger synergistic effect was observed when gefitinib was combined with olaparib, compared with the parental PC9 EGFR-mutant cell line. Importantly, PC9 cells are sensitive to chemotherapy with etoposide or camptothecin through cyclic GMP-AMP synthase (cGAS) overexpression, which disrupts the formation of the PARP1-timeless complex for DNA repair by homologous recombination.21,22 Aberrant up-regulation of cGAS transcripts is often noted in NSCLC and could represent a biomarker to predict response to chemotherapy. This mechanism of action is in contrast with the effect of PARP inhibitors, which prevent the translocation of cGAS from the cytoplasm to the nucleus.22
The study has some shortcomings. Pretreatment tumor specimens for genetic analyses were available from only 91 of the 182 patients of the ITT analysis of the GOAL study. The relatively small sample size may have limited the statistical power of our results. The expression of PARP1 and its effect on transcription regulation were not examined.23 PARP1 up-regulation has been found in models of castration-resistant prostate cancer and promotes cancer progression by DNA repair and transcriptional regulation.23 PARP1 interacts with and poly-ADP-ribosylates BRCA1.24 Finally, the fact that PARP1 enhances lung adenocarcinoma metastases implicates that PARP1 has a tumor progression effect independently of its role in DNA repair.12
In conclusion, BRCA1 mRNA expression may potentially predict the benefit of combining gefitinib plus olaparib and usher the investigation of chemotherapy plus EGFR TKIs in patients with EGFR-mutant NSCLC. BRCA1 mRNA expression levels may be used for customizing therapy. The role of CtIP as a biomarker to predict the outcome to EGFR TKIs warrants further research.
Acknowledgments
This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 765492. Work in Dr. Rosell’s laboratory is partially supported by a grant from la Caixa Foundation, an Instituto de Salud Carlos III grant (RESPONSE, PIE16/ 00011), and a Spanish Association Against Cancer (AECC) grant (PROYE18012ROSE).
Footnotes
Disclosure: The authors declare no conflict of interest.
Dr. Karachaliou is now with Global Clinical Development, Merck Healthcare KGaA, Darmstadt, Germany.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2020.100113.
Supplementary Data
References
- 1.Rosell R., Moran T., Queralt C. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R., Molina M.A., Costa C. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17:1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- 3.Hegan D.C., Lu Y., Stachelek G.C., Crosby M.E., Bindra R.S., Glazer P.M. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc Natl Acad Sci U S A. 2010;107:2201–2206. doi: 10.1073/pnas.0904783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campelo R.G., Felip E., Massuti B. Phase IB study to evaluate efficacy and tolerability of olaparib (AZD2281) plus gefitinib in patients (P) with epidermal growth factor receptor (EGFR) mutation positive advanced non-small cell lung cancer (NSCLC) (NCT=1513174/GECP-GOAL) J Clin Oncol. 2014;32(suppl 15) 8079–8079. [Google Scholar]
- 5.García-Campelo R., Arrieta O., Massuti B. Combination of gefitinib and olaparib versus gefitinib alone in EGFR mutant non-small-cell lung cancer (NSCLC): a multicenter, randomized phase II study (GOAL) Lung Cancer. 2020;150:62–69. doi: 10.1016/j.lungcan.2020.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Karachaliou N., Costa C., Gimenez-Capitan A. BRCA1, LMO4, and CtIP mRNA expression in erlotinib-treated non-small-cell lung cancer patients with EGFR mutations. J Thorac Oncol. 2013;8:295–300. doi: 10.1097/JTO.0b013e31827db621. [DOI] [PubMed] [Google Scholar]
- 7.Karachaliou N., Chaib I., Cardona A.F. Common co-activation of AXL and CDCP1 in EGFR-mutation-positive non-smallcell lung cancer associated with poor prognosis. EBioMedicine. 2018;29:112–127. doi: 10.1016/j.ebiom.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karachaliou N., Bracht J.W.P., Fernandez Bruno M. Association of PALB2 messenger RNA expression with platinum-docetaxel efficacy in advanced non-small cell lung cancer. J Thorac Oncol. 2019;14:304–310. doi: 10.1016/j.jtho.2018.10.168. [DOI] [PubMed] [Google Scholar]
- 9.Karachaliou N., Cardona A.F., Bracht J.W.P. Integrin-linked kinase (ILK) and Src homology 2 domain-containing phosphatase 2 (SHP2): novel targets in EGFR-mutation positive non-small cell lung cancer (NSCLC) EBioMedicine. 2019;39:207–214. doi: 10.1016/j.ebiom.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen D.X., Chiang A.C., Zhang X.H. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bliss C.I. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 12.Choi E.B., Yang A.Y., Kim S.C. PARP1 enhances lung adenocarcinoma metastasis by novel mechanisms independent of DNA repair. Oncogene. 2016;35:4569–4579. doi: 10.1038/onc.2016.3. [DOI] [PubMed] [Google Scholar]
- 13.Wang B., Matsuoka S., Ballif B.A. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunting S.F., Callen E., Wong N. 53BP1 inhibits homologous recombination in BRCA1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Lee J.C., Lin L. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen T., Tong P., Diao L. Targeting AXL and mTOR pathway overcomes primary and acquired resistance to WEE1 inhibition in small-cell lung cancer. Clin Cancer Res. 2017;23:6239–6253. doi: 10.1158/1078-0432.CCR-17-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husain A., He G., Venkatraman E.S., Spriggs D.R. BRCA1 up-regulation is associated with repair-mediated resistance to cis-diamminedichloroplatinum(II) Cancer Res. 1998;58:1120–1123. [PubMed] [Google Scholar]
- 18.Quinn J.E., Kennedy R.D., Mullan P.B. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63:6221–6228. [PubMed] [Google Scholar]
- 19.Karachaliou N., Moreno M.D., Sosa A.E. Using genetics to predict patient response to platinum-based chemotherapy. Expert Rev Precis Med Drug Dev. 2017;2:21–32. [Google Scholar]
- 20.Wu Y.L., Lee J.S., Thongprasert S. Intercalated combination of chemotherapy and erlotinib for patients with advanced stage non-small-cell lung cancer (FASTACT-2): a randomised, double-blind trial. Lancet Oncol. 2013;14:777–786. doi: 10.1016/S1470-2045(13)70254-7. [DOI] [PubMed] [Google Scholar]
- 21.Xie S., Mortusewicz O., Ma H.T. Timeless interacts with PARP-1 to promote homologous recombination repair [published correction appears in Mol Cell. 2016;61:181. Poon, Randy R Y [Corrected to Poon, Randy Y C]] Mol Cell. 2015;60:163–176. doi: 10.1016/j.molcel.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Liu H., Zhang H., Wu X. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 23.Schiewer M.J., Goodwin J.F., Han S. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y., Petit S.A., Ficarro S.B. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014;4:1430–1447. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.