Abstract
Introduction
Molecular profiling is considered a standard of care in advanced NSCLC. A comprehensive next-generation sequencing panel can discover somatic or germline BRCA1/2 mutations that are new druggable molecular alterations. However, the phenotypic and potential therapeutic relevance of BRCA1/2 mutation in NSCLC remains poorly defined.
Methods
From April 2014 to March 2017, 600 newly diagnosed, EGFR/ALK negative patients with advanced NSCLC were enrolled in the SAFIR02-Lung trial. Molecular profiling was done at study entry on archival tissue or frozen tissue collected from a new biopsy specimen before the third cycle of platinum-based chemotherapy. The prevalence of BRCA1/2 variants and its biological relevance were assessed. A homologous recombinant deficiency (HRD) score was based on the copy number variation data, and the germline status was determined by blood analysis. The BRCA Share database and the French CGG consortium were the references for the variant classification.
Results
Of 379 patients with a molecular profile discussed in a tumor molecular board, BRCA1/2 variants were identified in 20 patients (5.3%), including eight patients (2.1%) with a confirmed pathogenic BRCA mutation. Two patients (0.5%) harbored a germline BRCA2 mutation, and for six others, a somatic BRCA mutation was identified (1.6%). All were men and mainly smokers (88%). The overall response rate to chemotherapy was 13%. BRCA variants of unknown significance were detected in 12 patients (3.2%), achieving an 8.3% overall response rate with chemotherapy. One-third of tumors carrying pathogenic BRCA mutations or variants of unknown significance had biallelic inactivation and high HRD score. Overall survival of this cohort was 12.8 months.
Conclusions
Pathogenic BRCA1/2 mutations occur in 2.1% of patients with advanced NSCLC. The predictive role of BRCA mutation for making treatment decisions in NSCLC seems limited based on clinical response (low platinum sensitivity) and molecular features (discrepancy between biallelic inactivation and high HRD score).
Keywords: BRCA mutation, Advanced non–small cell lung cancer, PARP inhibitors, Next-generation sequencing, SAFIR trial
Introduction
Since the discovery of driver oncogenic alterations in advanced NSCLC, mainly in adenocarcinoma, the therapeutic treatment landscape has grown exponentially. Specific tyrosine kinase inhibitors targeting different genomic alterations, such as EGFR, BRAF mutations, and ALK or ROS11, 2, 3, 4 fusions, have impressively improved the outcomes of patients with advanced NSCLC,5 and genomic profiling is proposed as the standard of care in this population.6 The emergence of additional genomic alterations, such as NTRK fusions, found in a small subset of patients with cancer, including NSCLC, is also a strong predictor of response to targeted agents irrespective of tumor type, called agnostic agents.7,8 Genomic profiling has led to an increase in the percentage of patients with NSCLC suitable for matched therapies and the number of patients with molecular screening is increasing over time. It is speculated that the broader and earlier testing for molecular alterations that have not yet been recognized as standard-of-care predictive biomarkers of drug response in specific malignancies could accelerate the development and approval of targeted agents that could result in improved clinical outcomes,9 increasing the percentage of patients suitable for personalized treatment.
The breast and ovarian cancer genes 1 and 2 (BRCA1/2) are multifunctional tumor suppressor genes mainly involved in DNA double-strand break repair by homologous recombination (HR). Recently, overall BRCA1/2 alterations (pathogenic or variants of unknown significance [VUS]) were observed in 4.7% of cancers, especially in BRCA1/2-associated cancers (ovarian, breast, prostate, pancreatic). Across non–BRCA1/2-associated cancers, BRCA1/2 alterations were observed at a 3.0% frequency overall and greater than 1% frequency in each individual cancer type assessed.10 Germline and somatic BRCA1/2 mutation occur in 2.7% and 1.8% of advanced-stage cancers, respectively.11 The most common germline BRCA1/2 tumor carriers are 5% to 10% of breast cancers,12 10% to 15% of ovarian cancers,13,14 4% to 7% of pancreatic cancers,15 and 6% of patients with metastatic prostate cancer;16 cancer in BRCA1/2 germline carriers occurs at a younger age than in patients with germline wild-type cancer.11 Contrary to this, somatic BRCA mutations are reported in 2% to 5% and 3% of ovarian and breast cancers, respectively.15,17 The BRCA families have not exhibited considerable increase in the risk of NSCLC.17 Moreover, BRCA1/2-mutant tumors are often deficient in the repair of double-stranded DNA breaks by HR and consequently exhibit increased therapeutic sensitivity to platinum-based chemotherapy, and to inhibitors of poly(ADP-ribose) polymerase (PARP)11 in different tumor types, such as olaparib (in patients with ovarian,18 pancreatic,15 and breast cancer19 with BRCA mutation and metastatic castration–resistant prostate cancer with HR gene alterations20), niraparib (reporting higher efficacy in homologous recombinant deficiency [HRD]–positive patients with ovarian cancer21), rucaparib (in patients with BRCA-mutant ovarian cancer22), and talazoparib (in patients with BRCA-mutant breast cancer).23
However, the phenotypic and therapeutic relevance of mutations in BRCA1/2 remains poorly defined in most other cancer types. Both, BRCA mutations9,11 and BRCAness phenotype (deficiency of HR with or without BRCA mutation)24 have been reported in patients with NSCLC. However, the clinicopathologic characteristics of these patients and the pathogenic relevance of these mutations have not been clearly described. As in other tumors, preclinical work revealed that defects in BRCA and in other tumor suppressors that control DNA repair processes, such as ERCC1 defects in NSCLC models, cause PARP inhibitor sensitivity,25,26 initially suggesting the biological relevance of these alterations. However, for lung cancer with germline BRCA mutations, a higher frequency of loss of heterozygosis (LOH) of the mutated allele was reported, suggesting that it was not implicated in the tumorigenesis of lung cancer.11 Therefore, it is relevant to assess this response with all the molecular features in tumors harboring a BRCA mutation.
We aimed to prospectively investigate in a clinical setting the prevalence of germline and somatic BRCA mutations in patients with advanced NSCLC enrolled in the phase II UNICANCER SAFIR02-Lung/IFCT1301 trial (NCT02117167). In addition, we sought to evaluate the biological relevance of these mutations by determining whether these mutations were associated with biallelic inactivation, a high HRD score, or platinum sensitivity. We also analyzed the clinicopathologic characteristics of this subgroup of patients with NSCLC.
Materials and Methods
Patients
UNICANCER SAFIR02-Lung/IFCT1301 (NCT02117167) is an open-label, ongoing French multicentric phase II randomized trial. Biopsy specimens were obtained from treatment-naive and EGFR-activating mutation or ALK-rearranged–negative patients with advanced NSCLC at baseline or within the two initial platinum-based chemotherapy cycles. A single nucleotide polymorphism (SNP) array and next-generation sequencing tests are performed and the results analyzed during the two subsequent cycles as a therapeutic decision tool. If fresh frozen tissue was not available, the test was carried out on paraffin-embedded tissue, and in case of failure, on circulating tumor DNA. Only patients with objective response or stable disease (SD) after four cycles of platinum-based chemotherapy, as assessed by the investigator according to the Response Evaluation Criteria in Solid Tumors (RECIST) 1.1 criteria, are randomized. The study aims to evaluate, in the maintenance setting, the improvement in progression-free survival in the targeted drug arm guided by the molecular anomalies of the tumor compared with the standard-of-care maintenance treatment as per guidelines. Patients with no molecular profile available are randomized to durvalumab versus the standard of care. All the patients signed the informed consent document of the SAFIR02-Lung trial.
On the basis of the molecular profile of this population in the SAFIR02-Lung trial, the study aimed to evaluate the following: (1) the frequency of pathogenic BRCA mutations in treatment-naive patients with advanced NSCLC; (2) the response rate to platinum-based chemotherapy as assessed by the investigator according to the RECIST 1.1 criteria; and (3) the clinicopathologic characteristics of this subgroup of patients. We also report the overall survival (OS) of patients with BRCA mutation reported in this cohort.
Outcomes
OS (mo) was defined as the time from platinum-based treatment initiation to death because of any reason or last follow-up. Median follow-up was computed using the reverse Kaplan-Meier method. The outcomes were described separately for BRCA VUS and pathogenic mutation.
Tumor Tissue DNA Analysis
As a primary quality control measure, tumor cellularity was assessed by a senior pathologist on a hematoxylin and eosin slide from the same biopsy core as the one used for nucleic acid extraction and molecular analysis. When tumor cellularity was greater than or equal to 30%, both an SNP array test and targeted sequencing were performed. When the tumor cellularity ranged from 10% to 30% for formalin-fixed, paraffin-embedded samples, both techniques were engaged, or priority was given to next-generation sequencing when a low DNA quantity was extracted. Targeted sequencing was performed using either Personal Genome Machine (Ion Torrent PGM, ThermoFisher Scientific) or NextSeq (Illumina) with Ion AmpliSeq multiple gene panels, based on the multiplex polymerase chain reaction. The biallelic inactivation was assessed either by demonstrating the presence of LOH by the SNP array or by the absence of the SNP array data, which determined the imbalance of the allelic fraction.
The targeted gene panel consisted of the Ion AmpliSeq Cancer Panel, which covers 65 critical oncogenes or tumor suppressor genes (Supplementary Table 1) using Ion AmpliSeq custom design. The library preparation and the sequencing analysis were performed according to the manufacturer’s recommendation for Ion AmpliSeq Workflow. The bioinformatic analysis was performed with Torrent Suite software variantCaller (ThermoFisher Scientific), completed by homemade filtering and annotation pipeline. Each retained variant was reviewed by a molecular geneticist and classified as pathogenic variant, variant of unknown pathogenicity, or probably nonpathogenic variant. The median coverage depth on retained variants was over 700 reads, offering sensitivity down to 5% of allelic frequency.
The SNP array scanning was performed on the Affymetrix platform with CytoScan HD assays for DNA extracted from frozen samples and OncoScan copy number variation (CNV) assays for formalin-fixed, paraffin-embedded DNA. Default parameters and quantification from the devices were used. Scans were then analyzed and annotated with our own bioinformatic pipeline. The SNP array analysis was mainly used to detect tumor gene amplifications as well as deletions (amplifications > ×0.7 log2 ratio and deletions <0.5 log2 ratio) and was discussed during the tumor board review.
In addition, for this study, an HRD score was calculated from the CNV data. This genomic instability score was calculated as the number of altered segments superior to 15 Mbp and inferior to chromosome arm, and samples were classified into three categories using unsupervised machine learning (K-means python scikit). The scores have been validated retrospectively in an unpublished ovarian cancer cohort and in a published endometrial cancer cohort reports.27
The germline BRCA1/2 status was based on blood analysis for all variants. The BRCA Share database was the reference for the variant classification and experience of the French Cancer Genetic Group consortium, which is the curator of this national database.28,29 All variants were reported from class 3 (of unknown significance) to class 5 (pathogenic mutation).
Results
From April 2014 to data cutoff for this analysis in March 2017, 600 advanced, newly diagnosed, patients with NSCLC were enrolled in the SAFIR02-Lung trial. Among the enrolled patients, tissue biopsy analysis failed as a consequence of low tumor cellularity in 177 patients (30%), and in 44 additional patients (7%), the analysis was ongoing without results being declared at the time of our analysis. A molecular profile was successfully performed and discussed in a tumor molecular board for 379 patients (63%).
BRCA variants were reported in 20 of 379 patients (5.3%), all observed on a genomic profile of the tumor tissue. Baseline patient characteristics are provided in Table 1, and the specific information about the BRCA variants is reported in Supplementary Table 2.
Table 1.
Characteristics of Patients With Advanced NSCLC With BRCA Alterations at the Time of Enrollment in SAFIR02-Lung Trial
| Patient | Sex M/F | Age | Smoking (packs/y) | Histology | Cancer Family History | BRCA origin | Mutation | Status | Metastatic Sites | Best RRa |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 67 | CS (40) | ADC | N | S | BRCA1 | D | Bone, lymph nodes | PD |
| 2 | M | 60 | CS (40) | ADC | UK | S | BRCA1 | D | Muscle, lung | SD |
| 3 | M | 65 | CS (47) | ADC | N | S | BRCA2 | D | Suprarenal, bone, CNS, liver, lymph nodes | SD |
| 4 | M | 48 | CS (30) | ADC | UK | S | BRCA2 | D | Suprarenal, bone, liver, lymph nodes | SD |
| 5 | M | 62 | CS (88) | SCC | UK | S | BRCA2 | D | Bone, carcinomatous lymphangitis, lung, lymph nodes, muscle, pleura | PR |
| 6 | M | 65 | CS (45) | ADC | Y Colon Breast |
S | BRCA2 | D | Bone, lung, lymph nodes, pleural | SD |
| 7 | M | 60 | CS (30) | ADC | UK | G | BRCA2 | D | Bone, liver, lung, lymph nodes | SD |
| 8 | M | 68 | NS | ADC | Y Pancreas Breast |
G | BRCA2 | D | Suprarenal, CNS, lung | SD |
| 9 | F | 47 | FS (12) | ADC | N | U |
BRCA1 BRCA2 BRCA2 |
VUS | Bone, lymph nodes, muscle | SD |
| 10 | F | 52 | CS (25) | ADC | N | S | BRCA1 | VUS | Suprarenal, lung, lymph nodes | SD |
| 11 | M | 68 | CS (60) | ADC | Y Prostate |
G | BRCA1 | VUS | Pleural | SD |
| 12 | F | 73 | FS (7) | LCC | N | G | BRCA1 | VUS | Bone | PD |
| 13 | F | 67 | NS | SCC | UK | G | BRCA2 | VUS | NR | PR |
| 14 | M | 68 | CS (53) | ADC | N | G | BRCA2 | VUS | Lung | PD |
| 15 | F | 61 | FS (17) | ADC | Y Breast |
G | BRCA2 | VUS | Lung | SD |
| 16 | M | 61 | CS (45) | LCC | N | S | BRCA2 | VUS | Pleural effusion | SD |
| 17 | M | 56 | CS (40) | ADC | Y Prostate |
S | BRCA2 | VUS | Lung | SD |
| 18 | F | 71 | CS (42) | LCC | N | G | BRCA2 | VUS | Suprarenal, CNS, lymph nodes | PD |
| 19 | M | 62 | CS (43) | SCC | N | S | BRCA2 | VUS | Bone, lung, lymph nodes, pleural | PD |
| 20 | M | 63 | CS (50) | ADC | Y MPM |
S | BRCA2 | VUS | Adrenal, Bone, liver, lymph nodes | SD |
ADC, adenocarcinoma; CS, current smoker; D, deleterious; FS, former smoker; G, germline; LCC, large cell carcinoma; M/F, male/female; MPM, malignant pleural mesothelioma; NS, never smoker; PD, progressive disease; PR, partial response; RECIST, Response Evaluation Criteria in Solid Tumors, RR, response rate; S somatic; SCC, squamous cell carcinoma; SD, stable disease; U, undetermined; UK, unknown; VUS, variants of unknown significance; Y/N: yes/no.
Best RR to chemotherapy is according to the RECIST 1.1 criteria by the investigator. BRCA origin: S, G, U, MPM.
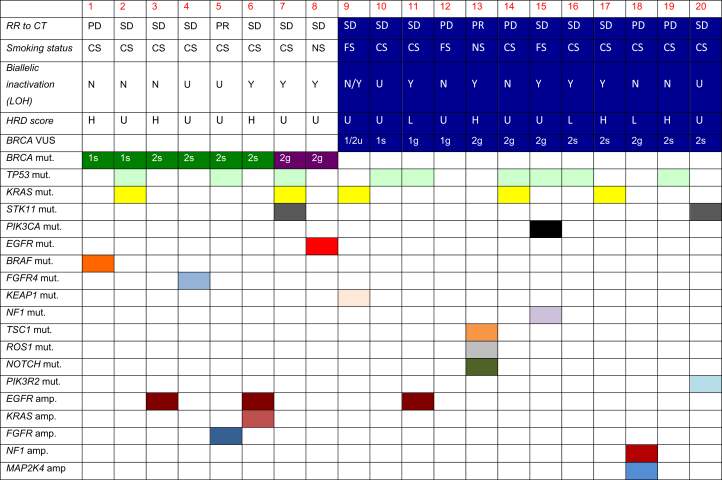
Eight of the 379 patients (2.1%) were diagnosed as having a pathogenic BRCA mutation, with six and two patients having somatic and germline BRCA mutations, respectively. BRCA mutations occurred in BRCA2 genes in 75% of cases (six of eight). Median age was 61.8 years (range, 48–68 y); all patients were men, mainly smokers (88% smokers) with a median of 48 packs/year. All but one tumor were adenocarcinomas, and bone and brain metastases were reported in 63% and 25% of the patients, respectively. All cases had additional oncogenic alterations (Fig. 1 and Supplementary Table 2), with 38% (N = 3 of 8) and 25% (N = 2 of 8) of concurrent TP53 and KRAS mutations, respectively.
Figure 1.
Concurrence genomic alterations in patients with advanced NSCLC with BRCA alterations (for the specific subtype mutations, see Supplementary Table 2). BRCA mutation (in green somatic/in purple germline mutation). 1/2, mutation in BRCA1 or BRCA2; Amp, amplification; CS, current smoker; CT, chemotherapy; FS, former smoker; G, germline; H, high; LOH, loss of heterozygosis; Mut, mutation; N, no; NS, never smoker; PD, progressive disease; PR, partial response; RECIST, Response Evaluation Criteria in Solid Tumors; RR, response rate; S, somatic; SD, stable disease; U, undetermined; Wt, wild type; Y, yes. aRR to CT is according to RECIST 1.1 criteria by the investigator.
Two of the 379 patients (0.5%) harbored a germline, pathogenic BRCA2 mutation (one with breast and pancreas cancer familial history). All had biallelic inactivation on tumor analysis (Table 2); however, the HRD score was unknown for these two patients as a consequence of low DNA quality. Both patients also had co-occurrence of other oncogenic mutations (one STK11 and KRAS mutation, and one EGFR exon 20 insertion) and achieved SD as the best response with platinum chemotherapy (Fig. 1).
Table 2.
HRD Signature and LOH Status Relevant to the Inactivation of the HR Pathway Related to the Mutation
| Status |
BRCA VUS Mutation N = 12/20 |
BRCA Pathogenic Mutation N = 8/20 |
Total | ||
|---|---|---|---|---|---|
| Somatic/unknowna N = 6/12 | Germline N = 6/12 | Somatic N = 6/8 | Germline N = 2/8 | ||
| HRD | |||||
| Cases with HRD score | 3/6 | 3/6 | 3/6 | 0/2 | 9/20 |
| High | 2/3 | 1/3 | 3/3 | — | 6/9 |
| Low | 1/3 | 2/3 | 0/3 | — | 3/9 |
| LOH | |||||
| Cases with LOH status | 4/6 | 6/6 | 4/6 | 2/2 | 16/20 |
| Biallelic inactivation | 3/4a | 3/6 | 1/4 | 2/2 | 9/16 |
| No biallelic inactivation | 1/4 | 3/6 | 3/4 | 0/2 | 7/16 |
HR, homologous recombination; HRD, homologous recombinant deficiency; LOH, loss of heterozygosis; VUS, variants of unknown significance.
Case 9: biallelic inactivation was considered for variants in the BRCA2 gene.
Somatic pathogenic BRCA mutation was identified in six of the 379 patients (1.6%, 2 BRCA1 mutations and 4 BRCA2 mutations, respectively). Biallelic inactivation was successfully assessed in four of the six cases, and only one case among the evaluable cases had a biallelic inactivation (Table 2). Somatic BRCA mutations were accompanied with co-occurring genomic alterations, such as BRAF non-V600 and KRAS mutations and amplifications (Fig. 1 and Supplementary Table 2). Patients’ responses to chemotherapy were as follows: SD in four patients, partial response (PR) in one patient, and one progressive disease. Three of six tumors with somatic BRCA mutations had SNP array data for CNV profiling and were evaluable for HRD score analysis; all three had a high HRD score (Table 2). The tumor with known biallelic inactivation had also high HRD, but response to platinum-based chemotherapy was SD (Fig. 1).
BRCA VUS were detected in 12 patients (3.2%), with a median age of 62.4 years (range, 47–73 y); 50% were women, mainly current smokers (67%) with different histologic subtypes (N = 7 adenocarcinoma, 58%; N = 3 large cell carcinoma, 25%; and N = 2 squamous cell carcinoma, 17%). Four patients (33%) had familial cancer history (two prostate cancer, one breast cancer, and one malignant pleural mesothelioma). Concurrent genomic alterations were reported in all except one patient (Fig. 1 and Supplementary Table 2), with 50% (N = 6) and 25% (N = 3) of cases with concurrent TP53 and KRAS mutations, respectively. Patients’ responses to chemotherapy were as follows: seven SD, one PR, and four progressive disease. LOH status was successfully assessed in 10 samples, with biallelic inactivation in 50% of the cases (five of 10). CNV profile was evaluable for HRD in six samples, and 50% (three of six) had a high score (Table 2). Two BRCA VUS carrier patients had matched biallelic inactivation and high HRD with PR and SD to platinum-based chemotherapy, respectively (Fig. 1).
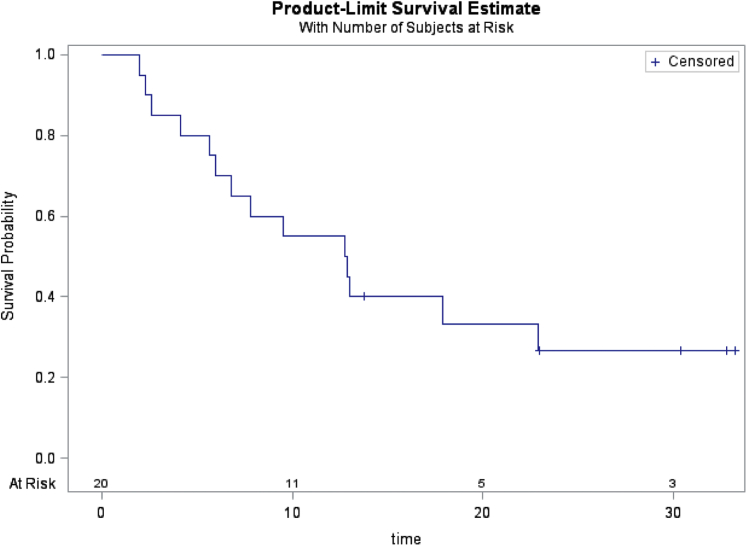
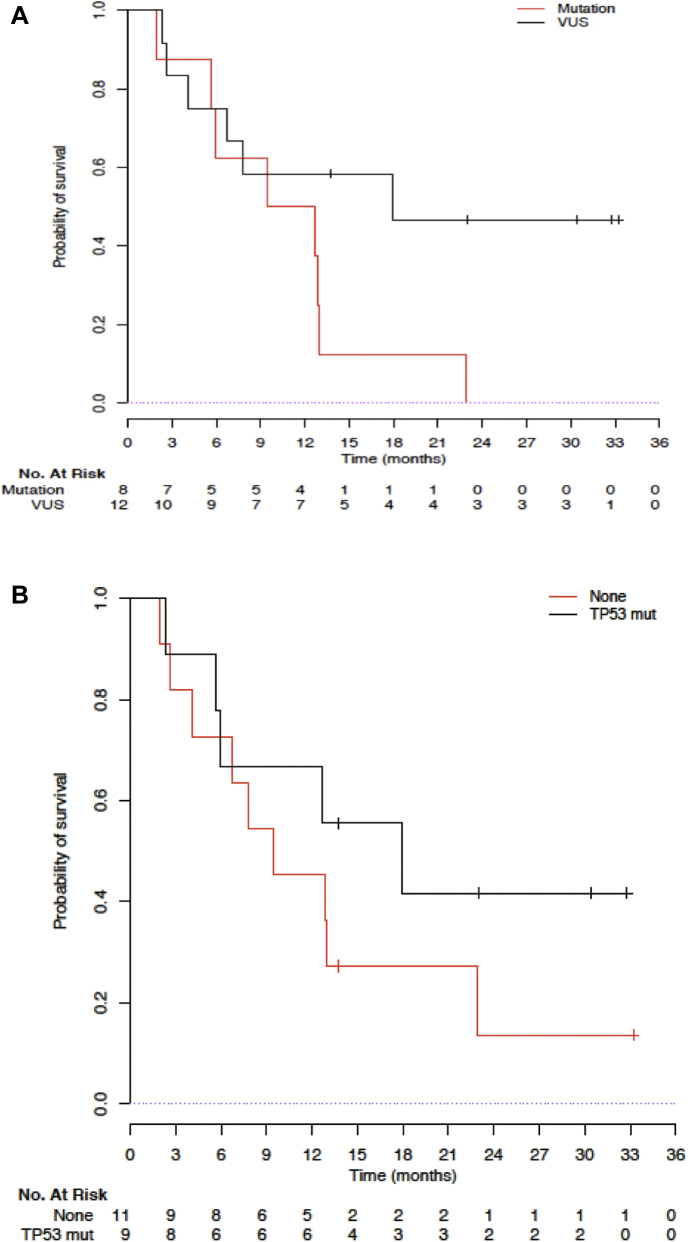
With a median follow-up of 30.4 months (95% confidence interval: 13.7–33.2), 14 of the 20 patients died, and the median OS was 12.8 months (95% confidence interval: 5.6–22.9, Fig. 2). Pathogenic BRCA-mutant tumors seem to have a shorter OS compared with BRCA VUS tumors (11.1 mo versus 17.9 mo, log-rank p = 0.07; Fig. 3A). TP53 comutation in nine patients seemed not to confer a poorer prognosis, with a median OS of 17.9 months and 9.5 months for TP53 mutation and no TP53 mutation (p = 0.3), respectively (Fig. 3B).
Figure 2.
OS of BRCA-positive patients with NSCLC. OS, overall survival.
Figure 3.
(A) OS of BRCA-positive patients with NSCLC according to pathogenic BRCA mutation or VUS. (B) OS of BRCA-positive (deleterious plus VUS) patients with NSCLC according to TP53 comutation present or absent. OS, overall survival; VUS, variant of unknown significance.
Discussion
Pathogenic BRCA1/2 mutations occurred in 2.1% of the 379 patients with advanced NSCLC included in the SAFIR02-Lung trial, of which 75% were of somatic origin (six of eight) and 75% targeted BRCA2 genes. However, the proportion of BRCA1/2-mutant NSCLC, including mutants with inactivation of the second allele, high HRD score, and platinum sensitivity, is low, calling into the question the real relevance of this potential druggable genomic alteration in NSCLC.
The prevalence of BRCA1/2 mutations in our cohort is comparable with the 1.2% previously reported in a cohort of 860 lung adenocarcinomas; in that study, however, clinical data were not reported, and most BRCA mutations, especially BRCA2, were somatic missense mutations of uncertain significance.9 In our NSCLC cohort, pathogenic germline BRCA mutations are a minority (0.5%), and this prevalence seems slightly lower than reported by other authors (∼1%), but ethnicity differences cannot be excluded.30 Finally, our data confirm higher incidence of BRCA2 than BRCA1 mutation in the NSCLC population, concordant with previous data, especially among germline BRCA mutations.11,30
Contrary to other NSCLC genomic alterations in which female predominance exists as a consequence of higher correlation with nonsmoking habits,5 in our cohort, all patients with pathogenic BRCA mutations were men and mainly heavy smokers, and up to one-third of the patients presented with baseline brain metastases. Although BRCA-deficient tumors are described as more sensitive to platinum salts,31 only one patient with BRCA mutation from our cohort achieved PR as the best response to platinum-based chemotherapy. Of note, according to the most recent studies, the expected RR to platinum-based chemotherapy ranges from 32% to 38%.32,33 However, caution should be exercised, given the low numbers that do not allow obtaining robust conclusions. We also report 12 patients whose tumors harbored 13 VUS, and again just one patient achieved PR with platinum-based chemotherapy. In breast or ovarian cancer, BRCA mutations are associated with better prognosis. In our BRCA-positive NSCLC cohort, median OS was 12.8 months, concordant with the median OS of patients with advanced NSCLC treated with platinum-based chemotherapy,34 probably implying that BRCA status does not reflect higher platinum sensitivity in NSCLC.
In the era of cancer-type agnostic approvals, it is beneficial to assess the biological relevance of germline or somatic BRCA1/2 mutations as tumor-agnostic biomarkers in advanced cancers. In germline BRCA-associated cancer types (breast, ovary, prostate, and pancreas), PARP inhibitors have been reported to have clinical benefit regardless of tumor origin.15,18, 19, 20 However, data from our NSCLC cohort revealed that pathogenic BRCA mutation was more common in men and smokers and without increased platinum sensitivity questioning the predictive value of BRCA1/2 status. The HRD score and/or the biallelic inactivation can be a point about the putative actionability of BRCA mutation in NSCLC and other tumors. Recently, it has been reported that biallelic inactivation is higher in BRCA1/2-associated cancers than in non–BRCA1/2-associated cancers (8.9% versus 1.3%), and up to 90% of germline or somatic BRCA1/2 mutations are biallelic in BRCA1/2-associated cancers, whereas only 46% and 25%, respectively, of germline and somatic BRCA1/2 mutations in non–BRCA1/2-associated cancers are biallelic.10 In our cohort, a high HRD score was present in three somatic pathogenic variants (only one with biallelic inactivation) achieving SD as the best response to platinum-based chemotherapy. The biallelic inactivation occurred in three pathogenic variants, all with SD response to chemotherapy. Globally, we identified a percentage of discordance between the biallelic status and HRD score, suggesting in some cases the mechanism of inactivation might be caused by events other than LOH, such as methylation or mutation. Probably the genomic instability observed is not related to BRCA but to other mechanisms of tumorigenesis, such as those induced by tobacco.
The analyses of the VUS were also concordant with the result observed in the pathogenic variants. Of the two BRCA VUS with both biallelic inactivation and high HRD score, the BRCA2 c.5634C>G p.Asn1878Lys is now considered as benign; this case is the only one achieving a PR, which can be because of an undetected variant in the same allele. The BRCA2 c.8893G>C p.Asp2965His is a very conserved interspecies, and algorithms are in favor of pathogenicity. The other two cases with biallelic inactivation and low HRD score (BRCA1 p.Glu1576Lys and BRCA2 p.Val2610Leu, and p.Asp189Asn) were probably not implied in the tumorigenesis of those lung cancers.
Altogether, these data suggest that in most patients with NSCLC, the pathogenic BRCA mutation could be considered an incidental event not related to tumorigenesis and not suitable for consideration as an agnostic biomarker for making treatment decisions. This observation is concordant with recent data suggesting that the true phenotypic dependency on mutant BRCA in non–BRCA1/2-associated cancers would be for tumors with biallelic mutations; therefore, it is ultimately necessary to incorporate methods to discriminate biallelic from monoallelic BRCA 1/2 alterations,10,11 before proposing a basket trial for non–BRCA1/2–associated tumors. Although in lung cancer, preclinical evidence exists for the efficacy of PARP inhibitors in deficient BRCA lung cancers,25 the clinical efficacy remains unknown. In patients with advanced NSCLC, the addition of PARP inhibitors with chemotherapy35 as maintenance treatment36 failed to reveal a sufficient level of efficacy, even in the subset of squamous NSCLC with homologous recombinant repair deficiency.36 No patient from our cohort enrolled in SAFIR02-Lung trial received PARP inhibitor drugs, as olaparib was not yet offered in the personalized treatment arm during the enrollment of our first 600 patients.
It has been reported that BRCA1 and BRCA2 alterations and POLE are significantly enriched with high tumor mutational burden,37 and most pathogenic BRCA-mutant patients in our cohort were smokers. Both factors have been correlated as potential predictive markers for the efficacy of immune checkpoint inhibitors (ICIs).38,39 Indeed, preclinical models have provided the rationale for a potential synergism with a combination of a PARP inhibitor and ICIs in NSCLC.40 Several clinical trials are currently assessing this combination in patients with NSCLC (NCT03308942 in first line, NCT02484404 in previously treated, and ORION trial NCT03775486, as maintenance treatment), but inclusion criteria for patients in all these trials include the BRCA status. However, recently it has been reported that a monoallelic event such as a BRCA mutation, in the context of high TMB in a smoker patient, is probably a passenger event and unlikely to be relevant for predicting the efficacy of ICIs.11
Our study has some limitations. The population was selected and might not reflect the all-comer population in the routine setting (cases of NSCLC with EGFR mutations or ALK fusions were excluded as were patients with history of cancer within 5 y). Another limitation of our study is the tissue sample availability for all patients, which limits the assessment of HRD score validation in this population. Tumor response assessment to chemotherapy according to the RECIST 1.1 criteria in the screening phase of the SAFIR02-Lung trial was not centralized. Finally, no patients were treated with PARP inhibitors. However, genomic analysis was performed with a centralized technique, providing the same gene coverage for molecular profiling.
In conclusion, our data provide prospective evidence that pathogenic BRCA1/2 mutations occur in 2.1% of patients with advanced NSCLC with mainly somatic mutations. However, clinical data and biological characteristics endorsing the agnostic biomarker value of this mutation in NSCLC are scarce.
Footnotes
Disclosure: Dr. Rouleau reports grants, personal fees, and other fees from AstraZeneca; grants and other fees from Bristol-Myers Squibb; grants from Roche during the conduct of the study. Dr. Masip reports other fees from Merck Sharp & Dohme, Boehringer Ingelhim, Pfizer, Ose Immunotherapeutics, Bristol-Myers Squibb, AstraZeneca, and Roche outside of the submitted work. Dr. Michels reports other fees from the International Drug Development Institute, Janssen, Hexal, Johnson & Johnson, Mabxiene, Roche, Steba, Iqvia, Sensorion, Biophytis, Servier, and Yuhan outside of the submitted work. Dr. Leary reports other fees from Clovis Oncology, Pharma Mar, AstraZeneca, Tesaro, Merck Sharp & Dohme, Merck-Serono, Pfizer, Seattle Genetics, and Roche; grants from Sanofi, Inivata, Merus, and GammaMabs during the conduct of the study. Dr. Besse reports grants from AbbVie, Amgen, AstraZeneca, Biogen, Blueprint Medicines, Bristol-Myers Squibb, Celgène, Eli Lilly, GSK, Ignyta, Ipsen, Merck KGaA, Merck Sharp & Dohme, Nektar, Onxeo, Pfizer, Pharma Mar, Sanofi, Spectrum Pharmaceuticals, Takeda, and Tiziana Pharma outside of the submitted work. Dr. Barlesi reports grants and personal fees from AstraZeneca, Bristol-Myers Squibb, Roche, Boehringer Ingelheim, Eli Lilly, Novartis, Merck, Merck Sharp & Dohme, Pierre Fabre, Takeda, and Pfizer; personal fees from Bayer; grants from AbbVie, ACEA, Amgen, Ipsen, Ignyta, Innate Pharma, Loxo Oncology, and Medimune outside of of the submitted work. Dr. Audigier-Valette reports other fees from AstraZeneca, AbbVie, Boehringer Ingelheim, Bristol-Myers Squibb, Eli Lilly, Novartis, Merck Sharp & Dohme, Pfizer, and Roche outside of the submitted work. Dr. Madroszyk reports personal fees from AstraZeneca and Roche; nonfinancial support from Pfizer and Eli Lilly outside of the submitted work. Dr. Soria reports personal fees from Astex, AstraZeneca, Bayer, Blend Therapeutics, Boehringer Ingelheim, Clovis Pharmaceuticals, Eli Lilly, GammaMabs, Merus, Mission Therapeutics, Pfizer, Pharma Mar, Pierre Fabre, Roche, Sanofi, Servier, Symphogen, and Tarveda outside of the submitted work; was a full-time employee in AstraZeneca from September 2017 to December 2019; is a stock shareholder of Gritstone. The remaining authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2020.100068.
Supplementary Data
References
- 1.Soria J.C., Ohe Y., Vansteenkiste J. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 2.Planchard D., Smit E.F., Groen H.J.M. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 2017;18:1307–1316. doi: 10.1016/S1470-2045(17)30679-4. [DOI] [PubMed] [Google Scholar]
- 3.Peters S., Camidge D.R., Shaw A.T. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med. 2017;377:829–838. doi: 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 4.Shaw A.T., Ou S.H.I., Bang Y.J. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlesi F., Mazieres J., Merlio J.P. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic InterGroup (IFCT) Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 6.Reck M., Rabe K.F. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377:849–861. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 7.Lassen U.N., Albert C.M., Kummar S. Larotrectinib efficacy and safety in TRK fusion cancer: an expanded clinical dataset showing consistency in an age and tumor agnostic approach. Ann Oncol. 2018;29(suppl 9):ix23–ix27. [Google Scholar]
- 8.Drilon A., Laetsch T.W., Kummar S. Efficacy of Larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan E.J., Kim H.R., Arcila M.E. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokol E.S., Pavlick D., Khiabanian H. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol. 2020;4:442–465. doi: 10.1200/PO.19.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonsson P., Bandlamudi C., Cheng M.L. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–579. doi: 10.1038/s41586-019-1382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemel D., Domchek S.M. Breast cancer predisposition syndromes. Hematol Oncol Clin North Am. 2010;24:799–814. doi: 10.1016/j.hoc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Russo A., Calò V., Bruno L., Rizzo S., Bazan V., Di Fede G. Hereditary ovarian cancer. Crit Rev Oncol Hematol. 2009;69:28–44. doi: 10.1016/j.critrevonc.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golan T., Hammel P., Reni M. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard C.C., Mateo J., Walsh M.F. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streff H., Profato J., Ye Y. Cancer incidence in first- and second-degree relatives of BRCA1 and BRCA2 mutation carriers. Oncologist. 2016;21:869–874. doi: 10.1634/theoncologist.2015-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore K., Colombo N., Scambia G. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 19.Robson M., Im S.A., Senkus E. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 20.Hussain M., Mateo J., Fizazi K. LBA12_PR—PROfound: phase III study of olaparib versus enzalutamide or abiraterone for metastatic castration-resistant prostate cancer (mCRPC) with homologous recombination repair (HRR) gene alterations. Ann Oncol. 2019;30(suppl 5):v881–v882. [Google Scholar]
- 21.González-Martín A., Pothuri B., Vergote I. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 22.Coleman R.L., Oza A.M., Lorusso D. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litton J.K., Rugo H.S., Ettl J. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul I., Savage K.I., Blayney J.K. PARP inhibition induces BAX/BAK-independent synthetic lethality of BRCA1-deficient non-small cell lung cancer. J Pathol. 2011;224:564–574. doi: 10.1002/path.2925. [DOI] [PubMed] [Google Scholar]
- 26.Postel-Vinay S., Bajrami I., Friboulet L. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene. 2013;32:5377–5387. doi: 10.1038/onc.2013.311. [DOI] [PubMed] [Google Scholar]
- 27.de Jonge M.M., Auguste A., van Wijk L.M. Frequent homologous recombination deficiency in high-grade endometrial carcinomas. Clin Cancer Res. 2019;25:1087–1097. doi: 10.1158/1078-0432.CCR-18-1443. [DOI] [PubMed] [Google Scholar]
- 28.Caputo S., Benboudjema L., Sinilnikova O. Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res. 2012;40:D992–D1002. doi: 10.1093/nar/gkr1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Béroud C., Letovsky S.I., Braastad C.D. BRCA share: a collection of clinical BRCA gene variants. Hum Mutat. 2016;37:1318–1328. doi: 10.1002/humu.23113. [DOI] [PubMed] [Google Scholar]
- 30.Tian P., Cheng X., Zhao Z. Spectrum of pathogenic germline mutations in Chinese lung cancer patients through next-generation sequencing. Pathol Oncol Res. 2020;26:109–114. doi: 10.1007/s12253-019-00771-5. [DOI] [PubMed] [Google Scholar]
- 31.Sonnenblick A., de Azambuja E., Azim H.A., Piccart M. An update on PARP inhibitors—moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- 32.West H., McCleod M., Hussein M. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019;20:924–937. doi: 10.1016/S1470-2045(19)30167-6. [DOI] [PubMed] [Google Scholar]
- 33.Paz-Ares L., Luft A., Vicente D. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379:2040–2051. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 34.Planchard D., Popat S., Kerr K. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(suppl 4):iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 35.Novello S., Besse B., Felip E. A phase II randomized study evaluating the addition of iniparib to gemcitabine plus cisplatin as first-line therapy for metastatic non-small-cell lung cancer. Ann Oncol. 2014;25:2156–2162. doi: 10.1093/annonc/mdu384. [DOI] [PubMed] [Google Scholar]
- 36.Owonikoko T.K., Redman M.W., Byers L.A. A phase II study of talazoparib (BMN 673) in patients with homologous recombination repair deficiency (HRRD) positive stage IV squamous cell lung cancer (Lung-MAP Sub-Study, S1400G) J Clin Oncol. 2019;37(suppl 15) 9022–9022. [Google Scholar]
- 37.Spigel D.R., Schrock A.B., Fabrizio D. Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J Clin Oncol. 2016;34(suppl 15) 9017–9017. [Google Scholar]
- 38.Rizvi N.A., Hellmann M.D., Snyder A. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J.H., Kim H.S., Kim B.J. Prognostic value of smoking status in non-small-cell lung cancer patients treated with immune checkpoint inhibitors: a meta-analysis. Oncotarget. 2017;8:93149–93155. doi: 10.18632/oncotarget.18703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chabanon R.M., Muirhead G., Krastev D.B. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest. 2019;129:1211–1228. doi: 10.1172/JCI123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.