Abstract
Background
Substance use disorder emerges from a complex interaction between genetic predisposition, life experiences, exposure, and subsequent adaptation of biological systems to the repeated use of drugs. Recently, investigators have proposed that the human microbiota may play a role in brain health and disease. In particular, the human oral microbiome is a distinct and diverse ecological niche with its composition influenced by external factors such as lifestyle, diet, and oral hygiene. This investigation examined whether individuals with substance use disorder (SU) show a different oral microbiome pattern and whether this pattern is sufficient to delineate the SU group from healthy comparison (HC) subjects.
Methods
Participants were a sub-sample (N = 177) of the Tulsa 1000 (T-1000) project. We analyzed 123 SU and 54 HC subjects using 16S rRNA marker gene sequencing to characterize the oral microbiome.
Results
The groups differed significantly based on the UniFrac distance, a phylogenetic-based measure of beta diversity, but did not differ in alpha diversity. Using a machine learning approach, microbiome features combined with socio-demographic variables successfully categorized group membership with 87%–92% accuracy, even after controlling for external factors such as smoking or alcohol consumption. SU individuals with relatively lower diversity also reported higher levels of negative reinforcement experiences associated with their primary substance of abuse.
Conclusions
Oral microbiome features are useful to sufficiently differentiate SU from HC subjects. There is some evidence that subjects whose drug use is driven by negative reinforcement show an impoverished oral microbiome. Taken together, the oral microbiome may help to understand the dysfunctional biological processes that promote substance use or may be pragmatically useful as a risk or severity biological marker.
Keywords: Substance use disorders, Alcohol use disorder, Microbiome, Genomics, Machine learning
Highlights
-
•
Oral microbiome features differentiate substance use disorder and healthy subjects.
-
•
Machine learning with microbiome and socio-demographic variables categorizes groups.
-
•
Substance use individuals have lower microbiome diversity.
-
•
Substance use individuals have higher levels of negative reinforcement.
-
•
Oral microbiome may be useful as a risk or severity biological marker.
1. Introduction
Substance use disorder is a complex disease that affects multiple body systems including the adaptive immunity in the peripheral and central nervous system (Kohno et al., 2019; Wang and Roy, 2017). The characteristics of an adaptive immune system are important determinants for interactions between the hosts and micro-organisms. However, the contributions of neuroimmune interactions to the formation and maintenance of addictive behaviors have only recently garnered appreciation (Hofford et al., 2019). The characteristics of the adaptive immune system are also important determinants of the interactions between the human host and their microbiome, the trillions of microbes residing on and within the body (Turnbaugh et al., 2007). The human microbiome consists of thousands of microbial species forming distinct ecological niches across the body that contribute to site-specific functions which are essential to the host's health (Consortium, 2012; IntegrativeP (iP) Re, 2019; Qin et al., 2010). Not only is the health of the microbiome linked with normal development and function of the host, but lasting perturbations to these microbial communities, termed dysbiosis, have been associated with a variety of diseases such as inflammatory bowel disease, colorectal cancer (McDonald et al., 2018; Yatsunenko et al., 2012; Vázquez-Baeza et al., 2018), and most recently, mental health disorders, such as depression, anxiety, schizophrenia and bipolar disorder (Nguyen et al., 2018; Collins et al., 2012; Cryan and Dinan, 2012). These mental health disorders are associated with significant medical comorbidities, including substance use. To date, the role of the human microbiome in substance use disorders remains largely unknown (Roy-Byrne et al., 2008).
The human oral microbiome is a distinct ecological niche from the highly-studied gut microbiome and is formed early in development (Koenig et al., 2011; Costello et al., 2009; Demmit et al., 2017). Its composition is influenced by environmental factors including diet, substance use, oral health, and overall health and disease. Studies of twins have revealed that more than 50% of microbial phenotypes are heritable (Demmitt et al., 2017; Stahringer et al., 2012). The human oral cavity contains a number of different habitats, including the teeth, gingival sulcus, tongue, cheeks, hard and soft palates, and tonsils, which are colonized by bacteria, and comprises over 600 prevalent taxa at the species level (Dewhirst et al., 2010). Sequencing results indicate that distinct sites host microbial communities that are not only distinguishable, but to a meaningful degree, are composed of entirely different microbes and that most oral microbes are site specialists (Dewhirst et al., 2010; Welch et al., 2019). The spatial organization of complex natural microbiomes is critical to understanding the interactions of the individual taxa that comprise a community. Within the structure, individual taxa are localized at the micron scale in ways that suggest their functional niche in the consortium (Mark Welch et al., 2016). Thus, assessing the oral microbiome provides a unique opportunity to examine whether drugs of abuse affect the colonization of the oral cavity and how this information could be used to differentiate individuals with substance use disorder from healthy comparison subjects.
The current investigation aimed to determine whether the oral microbiome of recently abstinent individuals with substance use disorder differed from that of comparable healthy subjects. We selected individuals with a diagnosed amphetamine or opioid substance use disorder and healthy comparison subjects who participated in the Tulsa 1000 (T-1000) project. The T-1000 project is a longitudinal observational study of treatment-seeking individuals with mental health problems across the categories of mood, anxiety, eating, and substance use disorders and those without mental health conditions (Victor et al., 2018). In addition, we carefully examined both current alcohol and nicotine use as covariates of interest to examine whether or not amphetamine use disorder or opioid use disorder independently contributed to differences between substance using individuals and healthy comparison subjects.
2. Materials & methods
2.1. Participants
The current study focused on the first 500 participants of the Tulsa 1000 (T-1000) project (Victor et al., 2018) (recruited January 5, 2015 to February 22, 2017). Complete data were available from 177 participants, aged 18–55 years, with substance use disorders (SU, n = 123) and healthy controls (HC, n = 54). Full inclusion/exclusion criteria are described in (Victor et al., 2018). Subjects were seeking treatment for substance use disorders and mainly recruited from local substance use treatment facilities, including 12&12, Inc. (n = 57, 49 male), the Women in Recovery program through Family and Children's Services (n = 52, 51 female), but also the Laureate Psychiatric Clinic and Hospital and the Tulsa metropolitan area. The mean age of regular substance use for the SUD group was 20.3 years (SD = 6.4). Healthy control subjects were recruited from the Tulsa metropolitan area through online, radio, and informational flyer advertisements. Additional details on subject characteristics are provided in Table 1.
Table 1.
Characteristics for 177 combined SU and HC subjects. BMI: Body Mass Index, PHQ-9: Patient Health Questionnaire, OASIS: Overall Anxiety Severity and Impairment Scale, DAST: Drug Abuse Screening Test, PROMIS: Patient-Reported Outcomes Measurement Information System.
| Sociodemographics (Mean/SD) | Healthy Control (n = 54) | Substance Use Disorder (n = 123) | p-value |
|---|---|---|---|
| Age | 32.09 (10.87) | 34.19 (8.93) | 0.18 |
| Gender = Male (%) | 27 (50.0) | 57 (46.0) | 0.74 |
| Weight [kg] | 81.77 (17.95) | 84.16 (16.33) | 0.39 |
| Height [cm] | 171.27 (8.30) | 172.06 (10.21) | 0.63 |
| BMI [kg/m2] | 27.81 (5.37) | 28.35 (4.56) | 0.46 |
| Percent Body Fat | 30.16 (10.81) | 31.19 (10.03) | 0.54 |
| Race/Ethnicity (%) | 0.05 | ||
| White | 40 (74.1) | 67 (54.5) | |
| Asian | 2 (3.7) | 1 (0.8) | |
| Black | 2 (3.7) | 8 (6.5) | |
| Hispanic | 3 (5.6) | 5 (4.1) | |
| Native American | 6 (11.1) | 36 (29.3) | |
| Other | 1 (1.9) | 6 (4.9) | |
| Education (highest level obtained) | <0.001 | ||
| Less than High School | 0 (0.0) | 28 (22.8) | |
| High School | 7 (13.0) | 44 (35.8) | |
| Some College | 22 (40.7) | 31 (25.2) | |
| College or Higher | 25 (46.3) | 20 (16.3) | |
| Symptom Scores (Mean/SD) | |||
| PHQ-9 | 0.80 (1.19) | 6.33 (5.91) | <0.001 |
| OASIS | 1.07 (1.36) | 5.71 (4.77) | <0.001 |
| DAST | 0.13 (0.39) | 7.74 (2.01) | <0.001 |
| PROMIS: Alcohol Use T-score | 44.14 (6.76) | 50.02 (3.55) | <0.001 |
| PROMIS: Nicotine Dependence T-score | 25.57 (7.46) | 41.98 (13.66) | <0.001 |
| SU Psychiatric Medication Status | |||
| Medicated (%) | 48 (39.02) | NA | |
| SU Psychiatric Comorbidities | |||
| Comorbidities (%) | 58 (47.15) | NA | |
| Anxiety (%) | 47 (38.21) | ||
| Depression (%) | 38 (30.89) |
2.2. Self-report questionnaires and clinician-based assessments
Participants completed self-report assessments for demographic and clinical and psychiatric features, with a primary focus on measurements of mood, anxiety, childhood and adult traumatic experiences, history and recent drug use, personality, social and physical functioning, cognition, sleep, fatigue, pain, impulsivity, and eating dimensions. A full report on assessments obtained as part of the Tulsa 1000 study is described in Victor et al., 2018) (Victor et al., 2018). Pertinent measurements related to substance use disorders for the current study are included in Table 1 and Supplementary Tables 1-2. These included: The Patient Health Questionnaire (PHQ-9) (Kroenke and Spitzer, 2002), Overall Anxiety Severity and Impairment Scale (OASIS) (Campbell-Sills et al., 2009; Norman et al., 2006), Drug Abuse Screening Test (DAST-10) (Cocco and Carey, 1998), Customary Drinking and Drug Use Record (CDDR with Michigan Negative Reinforcement Questionnaire (MNRQ) (Brown et al., 1998; Pomerleau et al., 2003), PROMIS® (Patient Reported Outcome Measurement Information System) measures (Cella et al., 2010; Gershon et al., 2010), Wide Range Achievement Test (WRAT-4 reading) (Wilkinson, 2006) and Mini International Neuropsychiatric Interview (MINI Version 6.0) (Lecrubier et al., 1997).
2.3. Ethical approval
Ethical approval was obtained from Western Institutional Review Board T1000 protocol #20142082. All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained by trained members of the research team. ClinicalTrials.gov identifier: #NCT02450240.
2.4. Microbiome
2.4.1. Sample collection and sequencing
Participants were provided with home oral collection kits (BD SWUBE Dual Swab Collection System; BD Worldwide). Participants were instructed to collect the oral sample a minimum of 2 h after brushing their teeth, to swab inside the mouth for 20 s and cover all sides of the swab by rubbing back and forth. Swabs were placed in labeled tubes without touching the sides, secured, and returned to an assessment team member at the Laureate Institute for Brain Research within 24 h of collection. Returned samples were transported to the University of Oklahoma Integrative Immunology Center (IIC) Laboratories for DNA extraction and long-term storage in secure freezers at −80 °C.
DNA extraction was performed at the University of Oklahoma IIC Laboratories and shipped for further processing and sequencing. Amplification and sequencing were performed at the University of California San Diego using standard Earth Microbiome Project (EMP) protocols (Caporaso et al., 2012; Minich et al., 2018). Whole DNA was extracted from samples using the EMP 16S rRNA amplicon extraction protocol (Earth Microbiome Project, 2016). PCR amplification and library preparation were performed similarly to the protocol described by Caporaso et al. (2012). Illumina primers with unique reverse primer barcodes were used to target the V4 region of the 16S ribosomal RNA gene. V4 amplicons were sequenced on the Illumina HiSeq 2000 platform, yielding paired-end, 150-base-pair reads. Sequencing was performed at the UCSD IGM Genomics Center. Feature tables along with sample and preparation information may be accessed in Qiita (qiita.ucsd.edu (Gonzalez et al., 2018);) as study ID 10424. Raw sequencing data was deposited in EBI-ENA under accession number EBI: ERP123404.
2.4.2. Bioinformatic and statistical analyses
All sequence processing was performed using QIIME 2 version 2019.1 (qiime2.org (Bolyen et al., 2019);). Raw sequencing results were demultiplexed and microbial Amplicon Sequence Variants (ASVs) were identified using the Deblur algorithm (Amir et al., 2017). Rarefied to 5000 sequences/sample. The output feature table was characterized using QIIME 2: feature table summary, alpha and beta diversity. Measures of alpha diversity were: Shannon, observed OTUs (richness), and Faith's phylogenetic diversity. Measures of beta diversity used were: Bray-Curtis distance, weighted, and unweighted UniFrac. Principal coordinate (PCoA) plots were generated using Emperor (Vázquez-Baeza et al., 2013). The significance of alpha diversity results was measured using the Kruskal-Wallis H test and the significance of beta diversity results was measured using PERMANOVA with 999 iterations, both tests as implemented in QIIME 2.
2.4.3. Machine learning
The machine learning analysis of the results was performed using scikit-learn Python package and calour (Xu et al., 2019; https://github.com/biocore/calour), similarly to previously proposed approaches (Belk et al., 2018; Teng et al., 2015; Xu et al., 2014). Microbial features were filtered out at 5000 and then normalized to 10,000 counts per sample. Random forest algorithm was used to predict classes of interest using a grid search optimization of the parameter space. For each classifier a 5-fold stratified cross validation was used and 20% of samples were left out as a separate test set. Depending on the specific classification problem, different scoring functions were used (highlighted in the Results section). In any case where non-microbiome variables were used in model training, they were 1-hot encoded as features. Each ROC curve was generated from cross-validation results. The confusion matrices were generated from the test set.
2.4.4. Taxonomic assignment
Taxonomy was assigned to ASVs using the q2-feature-classifier plugin (Bokulich et al., 2018). We trained a classifier specific to the 16S region covered by our primers, and additionally incorporated environment-specific taxonomic abundance information acquired from the readytowear taxonomic weights repository (https://github.com/BenKaehler/readytowear). This weighted bespoke method has been shown to significantly improve classification accuracy over common Naive Bayes classification methods without taxonomic weights (Kaehler et al., 2019).
3. Results
177 microbiome samples were collected from SU and HC subjects recruited through the Tulsa-1000 project (Table 1).
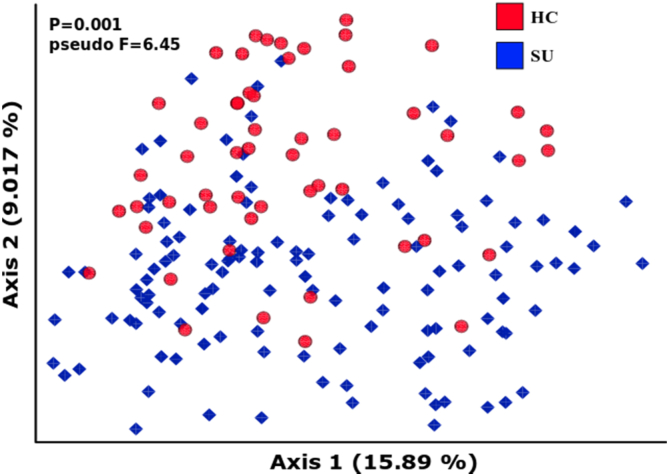
As a first approximation, we examined community-level differences to evaluate whether healthy subjects (HC) and the substance use cohort (SU) exhibit microbial differences. Indeed, using beta diversity, or between-subject microbial diversity, we observed such differences (Fig. 1).
Fig. 1.
Community-level differences. Unweighted UniFrac Principal Coordinate Analysis (PCoA) visualization of differences between groups.
Although the differences were not visually striking using both weighted (not shown) and unweighted UniFrac measures (Lozupone and Knight, 2005), they were significant in both cases as determined by a PERMANOVA test (p-values = 0.001). UniFrac is a phylogenetic-based beta diversity distance metric that quantifies the unique fraction of the phylogenetic tree occupied by microbes belonging to one of the two compared samples. The visualization in Fig. 1 is a dimensionally reduced (using principal coordinate analysis; PCoA) representation of an all-vs-all UniFrac distance matrix. The relative effect size (expressed as pseudo-F) was higher in weighted UniFrac (7.83), as compared to unweighted UniFrac (5.65). Weighted UniFrac takes into account the relative abundance of particular microbes, while the unweighted version considers only presence/absence.
Comparing subsets of the SU cohort (stimulant users, SU-S and opioid + users, SU-O, Supplementary Table 2) we did not observe beta diversity differences in UniFrac measures (PERMANOVA unweighted UniFrac p-value = 0.503; weighted UniFrac p-value = 0.152). There were no significant alpha diversity (within-subject diversity) differences observed between HC vs. SU or SU-O vs. SU-S (all p-values > 0.1).
Since the majority of the SU subjects were recruited from two different treatment facilities based on their sex (see Methods section), we investigated whether there were systematic community-level differences between them. We compared sex differences within the HC group and within the SU group. All alpha- and beta diversity differences for the HC group were not significant for alpha-diversity: Shannon diversity (p-value = 0.441), Faith's PD (p-value = 0.586); beta diversity: unweighted UniFrac (p-value = 0.942), Bray-Curtis (p-value = 0.581). For the SU group we did not find significant differences for alpha-diversity measures: Shannon diversity (p-value = 0.917), Faith's PD (p-value = 0.424). However, we found borderline significant values for beta diversity measures: unweighted UniFrac (p-value = 0.051; pseudo-F = 1.886), Bray-Curtis (p-value = 0.018; pseudo-F = 3.119). Since most of the females were recruited from a single center, it was impossible to disambiguate whether the observed effect resulted from true microbiome sex differences in SU cohort, or whether it was a center effect and a consequence of varying therapeutic strategies. Nevertheless, the relative effect size of those differences (expressed as pseudo-F) was close to 3 times smaller than the differences between HC and SU groups (unweighted UniFrac HC vs. SU pseudo-F = 5.65; SU males vs. SU females pseudo-F = 1.89).
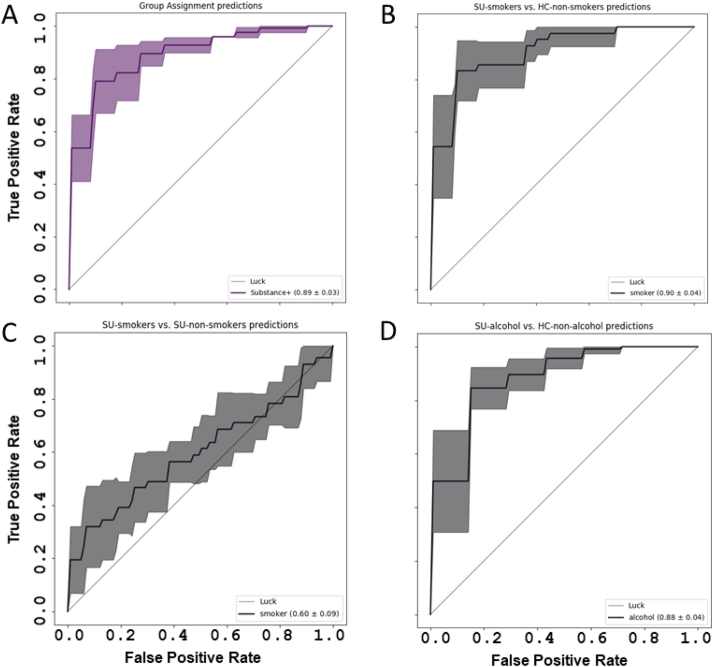
A series of random forest classifiers were conducted using the bacterial abundance table to determine the discriminative utility of the oral microbiome, i.e. to generate individual-level group membership predictions. First, we addressed whether on the basis of microbiome and socio-demographic features it was possible to predict group membership of participants (Fig. 2, Table 2). Aside from microbiome features (represented as amplicon sequence variants; ASVs), we used basic socio-demographic variables (sex, body mass index (BMI), age, ethnicity), as well as PROMIS nicotine dependence T-score and PROMIS alcohol use T-score as input variables for the model. On a test set of 35 samples (20% of subjects), we were able to achieve 83% classification accuracy (Table 2). Correctly assigning HC status was easier (92% accuracy) than assigning the SU label (73% accuracy).
Fig. 2.
Random Forest classifier ROC curves. The results of a random forest classifier. (A) Group assignment (Substance Use (SU) vs. Healthy Control (HC)) predictions controlled for gender, BMI, age, ethnicity, PROMIS Nicotine Dependence T-score, PROMIS Alcohol Use T-score. (B) SU-smokers vs. HC non-smokers predictions controlled for gender, BMI, age, ethnicity. (C) smokers vs. non-smokers predictions only within the SU cohort. (D) SU-alcohol consumers vs. HC non-alcohol consumers controlled for gender, BMI, age, ethnicity.
Table 2.
Random Forest classifier statistics.
| classifier | Accuracy | TPR | FPR | PPV |
|---|---|---|---|---|
| Group Assignment | 0.83 | 0.73 | 0.08 | 0.90 |
| Substance Use: smokers vs. non-smokers | 0.58 | 0.82 | 0.67 | 0.55 |
| Substance Use smokers vs. Healthy Control non-smokers | 0.81 | 0.73 | 0.11 | 0.87 |
| Substance Use alcohol vs. Healthy Control non-alcohol | 0.75 | 0.92 | 0.43 | 0.68 |
TPR - true positive rate.
FPR - false positive rate.
PPV - positive predictive value.
PPV - positive predictive value.
We suspected that current nicotine and alcohol use might substantially alter the oral microbiome composition, therefore contributing to our classification accuracy. To evaluate, we created models to predict SU-smoker vs. HC-non-smoker status (taking only smokers from the SU cohort and non-smokers from the healthy control cohort; Fig. 2B) and, analogously, SU-alcohol vs. HC-non-alcohol (Fig. 2D), as well as a within group assessment of smoking predictions, i.e. SU-smokers vs. SU-non-smokers (Fig. 2C). A score of 50 was used as a threshold to separate positive and negative groups (PROMIS alcohol use T-score > 50 = alcohol group; PROMIS nicotine dependence T-score > 50 = smoker group). Both classifiers include socio-demographic variables (sex, BMI, age, and ethnicity).
By creating classifiers for smoking and alcohol use groups, we observed similar accuracy (81% and 75%, respectively) to the result achieved by the HC vs. SU classifier (Table 2). However, creating a similar classifier for differentiating smokers vs. non-smokers within the SU cohort yielded a close to random accuracy of 58% and PPV of 55% (Fig. 2C; Table 2), suggesting that the dominant effect was driven by substances of abuse and not from smoking status, or other confounding factors. For other within-group comparisons, e.g. alcohol vs. non-alcohol within HC or SU, the group sizes were too small to create a reliable classifier.
The microbiome features that contributed to the classifiers collectively showed low feature importance (<5%) and high variability. This suggests that there is not a single feature or combination of several features (i.e. socio-demographic variables and the microbiome) to robustly predict the phenotype. The effects observed are likely on the community-level, and therefore require information about the entire microbiome in order to be effective. This also provides further confirmation that neither PROMIS nicotine dependence score, nor PROMIS alcohol use score are significant contributors for classifying subjects into healthy or substance use groups.
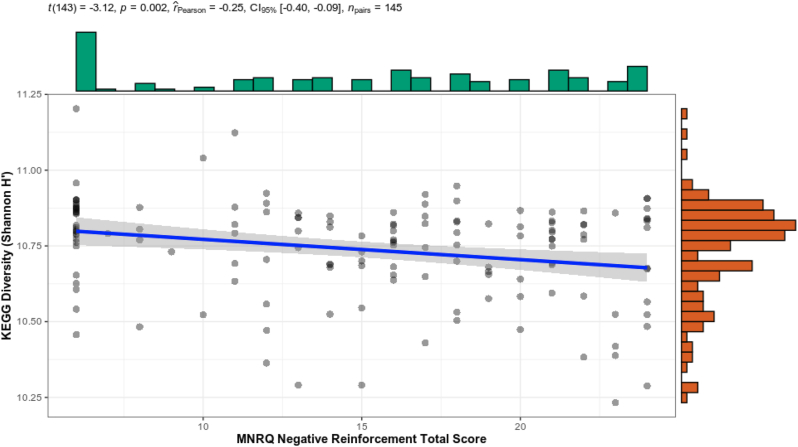
To gain a better understanding of the potential underlying mechanisms contributing to the observed differences, we performed a PICRUSt2 analysis which aimed to reconstruct the functional potential of microbial communities within each sample using 16S rRNA data (Langille et al., 2013; Douglas et al., 1101). PICRUSt2 may predict, among others, KEGG ontologies classifying functions within each sample. We previously argued that negative reinforcement processes, i.e. “relief use”, the use of a substance to prevent the occurrence of an aversive future state, is central to the maintenance of the use disorder and is modulated by (Kohno et al., 2019) internal body sensations (Wang and Roy, 2017), aversive interoception, and (Hofford et al., 2019) conditioned stimuli that are associated with heightened negative emotions, drug craving, withdrawal, or stress (May et al., 2020). As part of the T-1000 project assessments, we modified the Customary Drinking and Drug Use Record (CDDR) to include the Michigan Negative Reinforcement Questionnaire (MNRQ) to evaluate subject beliefs concerning the positive and negative consequences of the subject's drug use in reference to their primary drug of abuse. With an analysis approach similar to microbe-level ones, we found that Shannon alpha-diversity decreased as negative reinforcement scores increased (Fig. 3), i.e. those individuals with relatively lower diversity report higher levels of negative reinforcement experiences associated with their primary substance of abuse. The relatively small correlation (Spearman rho = −0.23; p = 0.005) included all subjects with CDDR Negative Reinforcement measured (146 subjects). This suggests that the process of addiction decreases the functional potential of the microbiome.
Fig. 3.
Alpha correlation (Shannon) between KEGG ontology pathways and CDDR Negative Reinforcement score at timepoint 0 for all subjects.
4. Discussion
This investigation aimed to determine pragmatically whether oral microbiome differed across individuals with substance use disorder relative to healthy comparison subjects. A secondary aim was to determine whether there was evidence for a particular process or a particular microbiota that differentiated these groups. There were four main results. First, the oral microbiome differed significantly between individuals with SU relative to HC subjects, even after adjusting for BMI, age, ethnicity, current nicotine dependence, and current alcohol use. In combination, the microbiome characteristics were able to produce a prediction accuracy in a test set of 83% (Table 2). Second, a similar accuracy was observed for both smokers and individuals with alcohol use (Table 2). Third, the detailed characterization of the microbiota contribution to the differentiation between the groups did not yield evidence for a particular microbe that differed between the groups, even though the beta diversity (between sample) differences at the whole microbial community level as measured by unweighted UniFrac were significant (Fig. 1). It is not surprising, as the oral microbiome is primarily impacted by differences in individual's oral hygiene or dietary habits, which we expect to be the major contributors to microbial diversity changes (57). Fourth, there was some evidence for a more strongly reduced functional diversity of microbiota in those individuals who also reported greater levels of negative reinforcement symptoms, i.e. admitted to stronger relief use (Fig. 3). Taken together, these results support the hypothesis that individuals with substance use disorder have a significantly altered microbiome and that this difference is sufficiently distinct to accurately classify this group. However, future investigations will need to determine whether these differences contribute directly to the pathophysiological process of addiction.
While there is evidence that immune responses in the periphery and the central nervous system are altered by exposure to drugs of abuse, the contributions of neuroimmune interactions to addictive behaviors are just beginning to be appreciated and brain-immune system interactions in substance use disorders are much more complex and important than previously understood (Hofford et al., 2019). In particular, no systematic programs of research have examined the role of microbiota in drug addiction (Skosnik and Cortes-Briones, 2016). Our results point to small, if any, differences in the diversity of the microbiome, i.e. in alpha or beta diversity. However, we did find that those individuals who reported greater negative reinforcement symptoms, e.g. use of a substance to alleviate an aversive state such as withdrawal, had a less varied microbiome in terms of its metabolic capabilities. Over the course of addiction, substance users switch from using drugs to feel “good” to using drugs to avoid feeling “bad”, and this transition from positive to negative reinforcement parallels important changes in brain function that drive chronic use and impair adaptive decision-making (Koob and Le Moal, 2001; Koob, 2013; Koob and Volkow, 2016; Volkow et al., 2016; Kwako and Koob, 2017). During early stages of use, excessive striatal dopamine release associated with drug highs may be suppressed by neurochemical stress signals from the extended amygdala; these signals are thought to amplify over time, further suppressing dopamine responses to drug reward so that users experience tolerance and no longer feel positive effects (Koob et al., 2014). Drug use then becomes “relief use”: a way to avoid withdrawal symptoms (aversive bodily feeling states) that are accompanied by neurochemical stress signals. Moreover, in a recent morphine-murine model, evidence suggested the role of the gut microbiome and metabolome as a potential mechanism that contributes to the negative consequences associated with opioid use. These investigators found a significant shift in the gut microbiome and metabolome within one day following morphine treatment compared to that observed after placebo (Wang et al., 2018). Thus, the changes in microbiome diversity could be a consequence of late stage addiction. Longitudinal studies would be needed in order to distinguish among these possible causes for the patterns shown here.
The detailed analysis of the features differentiating the substance users from healthy comparison subjects revealed low feature importance scores which suggests that the effects we are observing are due to broad ecological changes within the oral microbiome, rather than specific ASVs or covariates. Substance use is associated with a number of behaviors that can affect the oral microbiome such as poor eating habits, poor oral health behaviors, direct vasoconstrictive effects of the oral mucosa, and other poor health maintenance behaviors. Therefore, this cross-sectional study cannot delineate the predominant process that contributes to the observed differences. Nevertheless, the fact that these differences were sufficient to accurately classify the subjects provides some pragmatic justification to further investigate the role of the microbiome in the pathophysiology of substance use disorder. Interestingly, however, the observed differences were not just simply a consequence of smoking behavior because the microbiome did not differ significantly between substance users who smoked versus those that did not. This is particularly interesting because current smoking had the strongest effect on the overall microbial community structure among the tested lifestyle factors. The abundances of Veillonella and Megasphaera were higher in current-smokers, and increased with the pack-year value and the Fagerstrom Test of Nicotine Dependence (FTND) score (Lim et al., 2016). In contrast, Haemophilus decreased with the pack-year of smoking and the FTND score. Thus, it will be important to examine in future studies how the oral microbiome adjusts as smoking behavior changes.
This study has several limitations. First, the cross-sectional nature of the study and the case-control design limit the inferences that can be drawn from the results. Specifically, there is no possibility to go beyond associative statements, i.e. that there are substantial differences between the groups, yet the causal sequence of events is unclear. Future investigations will need to use both interventional or experimental approaches as well as longitudinal studies to investigate the cause and consequences of these observed differences. Second, the broad changes in the microbiome make it difficult to suggest that substance use promotes or inhibits a particular microbial agent. Third, we cannot differentiate between the differences in hygiene and true ecological differences between SU and HC subjects. Thus, we will need to examine prospectively whether changes in oral hygiene among SU individuals are associated with changes in oral microbiome and whether this change differs from changes observed during prolonged recovery.
This study aimed to examine whether the oral microbiome of substance using individuals is sufficiently different from that of healthy comparison subjects to warrant further investigations into the causal role such differences could contribute to the process of substance use. The results support the notion that there are broad differences in the oral microbiome and that these differences are sufficiently significant to robustly differentiate users from comparison subjects. Moreover, there is some evidence that measures of severity or progression of substance use, as indicated by increased negative reinforcement experiences, is also associated with reduced oral microbiome diversity. However, future investigations will need to examine the degree to which these differences contribute to the persistence of substance use or whether these differences are epiphenomal and not disease relevant. In addition, one will need to examine whether microbiome differences are associated with differential brain processing that is relevant for substance use such as reward-related processes. Nevertheless, the possible effects of substance use disorder on the microbiome could lead to new diagnostic approaches, and the possible contribution of the microbiome to substance use disorder could provide a unique approach to developing new intervention for this condition.
Disclosures
No financial disclosures or conflicts of interest to report: TK, TAV, RK, MR, ME, GA, RK, RA, JB, JF, SSK, JS, JLS.
MPP is an advisor to Spring Care, Inc., a behavioral health startup, he has received royalties for an article about methamphetamine in UpToDate.
Declaration of competing interest
All authors contributed substantially to the scientific process leading up to the writing of the manuscript and contributed to the writing and/or revising of the manuscript for important intellectual content. All authors have approved the final version of the manuscript. None of the authors had any disclosures to report.
Acknowledgements
This work has been supported in part by The William K. Warren Foundation and the National Institute of General Medical Sciences Center Grant Award Number 1P20GM121312. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
TK is funded by the Polish National Agency for Academic Exchange grant PPN/PPO/2018/1/00014.
The ClinicalTrials.gov identifier for the clinical protocol associated with data published in the current paper is NCT02450240, “Latent Structure of Multi-level Assessments and Predictors of Outcomes in Psychiatric Disorders”.
The Tulsa 1000 Investigators include the following contributors: Robin Aupperle, Ph.D., Jerzy Bodurka, Ph.D., Justin Feinstein, Ph.D., Sahib S. Khalsa, M.D., Ph.D., Jonathan Savitz, Ph.D., Jennifer L. Stewart, Ph.D. The primary contact/principal investigator for the Tulsa 1000 Investigators group is Sahib S. Khalsa, M.D., Ph.D.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2021.100271.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems [Internet] 2017 Mar;2(2) doi: 10.1128/mSystems.00191-16. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belk A., Xu Z.Z., Carter D.O., Lynne A., Bucheli S., Knight R. Microbiome data accurately predicts the postmortem interval using random forest regression models. Genes [Internet] 2018 Feb 16;9(2) doi: 10.3390/genes9020104. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome. 2018 May 17;6(1):90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019 Aug;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.A., Myers M.G., Lippke L., Tapert S.F., Stewart D.G., Vik P.W. Psychometric evaluation of the Customary Drinking and Drug Use Record (CDDR): a measure of adolescent alcohol and drug involvement. J. Stud. Alcohol. 1998 Jul;59(4):427–438. doi: 10.15288/jsa.1998.59.427. [DOI] [PubMed] [Google Scholar]
- Campbell-Sills L., Norman S.B., Craske M.G., Sullivan G., Lang A.J., Chavira D.A. Validation of a brief measure of anxiety-related severity and impairment: the overall anxiety severity and impairment scale (OASIS) J. Affect. Disord. 2009 Jan;112(1–3):92–101. doi: 10.1016/j.jad.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012 Aug;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella D., Riley W., Stone A., Rothrock N., Reeve B., Yount S. The Patient-Reported Outcomes Measurement Information System (PROMIS) developed and tested its first wave of adult self-reported health outcome item banks: 2005-2008. J. Clin. Epidemiol. 2010 Nov;63(11):1179–1194. doi: 10.1016/j.jclinepi.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco K.M., Carey K.B. vol. 10. Psychological Assessment; 1998. (Psychometric Properties of the Drug Abuse Screening Test in Psychiatric Outpatients [Internet]). 408–14. Available from: [DOI] [Google Scholar]
- Collins S.M., Surette M., Bercik P. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol. 2012 Nov;10(11):735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- Consortium T.H.M.P. The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome [Internet] Nature. 2012;486 doi: 10.1038/nature11234. 207–14. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009 Dec 18;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012 Oct;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Demmit B.A., Corley R.P., Huibregtse B.M., Keller M.C., Hewitt J.K., McQuinn M.B., Knight R., McDermott I., Krauter K.S. Genetic influences on the human oral microbiome. BMC Genom. 2017;18(1):659. doi: 10.1186/s12864-017-4008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmitt B.A., Corley R.P., Huibregtse B.M., Keller M.C., Hewitt J.K., McQueen M.B. Genetic influences on the human oral microbiome. BMC Genom. 2017 Aug 24;18(1):659. doi: 10.1186/s12864-017-4008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F.E., Chen T., Izard J., Paster B.J., Tanner A.C.R., Yu W.-H. The human oral microbiome. J. Bacteriol. 2010 Oct;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas GM, Maffei VJ, Zaneveld J, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2: an improved and customizable approach for metagenome inference [Internet]. Available from:: 10.1101/672295. [DOI]
- Gershon R.C., Rothrock N., Hanrahan R., Bass M., Cella D. The use of PROMIS and assessment center to deliver patient-reported outcome measures in clinical research. J. Appl. Meas. 2010;11(3):304–314. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Navas-Molina J.A., Kosciolek T., McDonald D., Vázquez-Baeza Y., Ackermann G. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods. 2018 Oct;15(10):796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofford R.S., Russo S.J., Kiraly D.D. Neuroimmune mechanisms of psychostimulant and opioid use disorders. Eur. J. Neurosci. 2019 Aug;50(3):2562–2573. doi: 10.1111/ejn.14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Integrative HMP (iHMP) Research Network Consortium The integrative human microbiome project. Nature. 2019 May;569(7758):641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehler B.D., Bokulich N.A., McDonald D., Knight R., Caporaso J.G., Huttley G.A. Species abundance information improves sequence taxonomy classification accuracy. Nat. Commun. 2019 Oct 11;10(1):4643. doi: 10.1038/s41467-019-12669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U. S. A. 2011 Mar 15;(108 Suppl. 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno M., Link J., Dennis L.E., McCready H., Huckans M., Hoffman W.F. Neuroinflammation in addiction: a review of neuroimaging studies and potential immunotherapies. Pharmacol. Biochem. Behav. 2019 Apr;179:34–42. doi: 10.1016/j.pbb.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G.F. Addiction is a reward deficit and stress surfeit disorder. Front. Psychiatr. 2013 Aug 1;4:72. doi: 10.3389/fpsyt.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G.F., Le Moal M. Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology. 2001 Feb;24(2):97–129. doi: 10.1016/S0893-133X(00)00195-0. [DOI] [PubMed] [Google Scholar]
- Koob G.F., Volkow N.D. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016 Aug;3(8):760–773. doi: 10.1016/S2215-0366(16)00104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G.F., Buck C.L., Cohen A., Edwards S., Park P.E., Schlosburg J.E. Addiction as a stress surfeit disorder. Neuropharmacology. 2014 Jan;76 Pt B:370–382. doi: 10.1016/j.neuropharm.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke K., Spitzer R.L. vol. 32. Psychiatric Annals; 2002. (The PHQ-9: A New Depression Diagnostic and Severity Measure [Internet]). 509–15. Available from: [DOI] [Google Scholar]
- Kwako L.E., Koob G.F. Neuroclinical framework for the role of stress in addiction. Chronic stress (thousand oaks) [internet] 2017 Feb;1. Available from: [DOI] [PMC free article] [PubMed]
- Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013 Sep;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecrubier Y., Sheehan D.V., Weiller E., Amorim P., Bonora I., Harnett Sheehan K. vol. 12. European Psychiatry; 1997. pp. 224–231. (The Mini International Neuropsychiatric Interview (MINI). A Short Diagnostic Structured Interview: Reliability and Validity According to the CIDI [Internet]). Available from: [DOI] [Google Scholar]
- Lim M.Y., Yoon H.S., Rho M., Sung J., Song Y.-M., Lee K. Analysis of the association between host genetics, smoking, and sputum microbiota in healthy humans. Sci. Rep. 2016 Mar 31;6:23745. doi: 10.1038/srep23745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005 Dec;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch J.L., Rossetti B.J., Rieken C.W., Dewhirst F.E., Borisy G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U. S. A. 2016 Feb 9;113(6):E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A.C., Aupperle R.L., Stewart J.L. Dark times: the role of negative reinforcement in methamphetamine addiction. Front. Psychiatr. 2020 doi: 10.3389/fpsyt.2020.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Hyde E., Debelius J.W., Morton J.T., Gonzalez A., Ackermann G. American gut: an open platform for citizen science microbiome research. mSystems [Internet] 2018 May;3(3) doi: 10.1128/mSystems.00031-18. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minich J.J., Humphrey G., Benitez R.A.S., Sanders J., Swafford A., Allen E.E. High-Throughput miniaturized 16S rRNA amplicon library preparation reduces costs while preserving microbiome integrity. mSystems [Internet] 2018 Nov;3(6) doi: 10.1128/mSystems.00166-18. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.T., Kosciolek T., Eyler L.T., Knight R., Jeste D.V. Overview and systematic review of studies of microbiome in schizophrenia and bipolar disorder. J. Psychiatr. Res. 2018 Apr;99:50–61. doi: 10.1016/j.jpsychires.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman S.B., Cissell S.H., Means-Christensen A.J., Stein M.B. vol. 23. Depression and Anxiety; 2006. pp. 245–249. (Development and Validation of an Overall Anxiety Severity and Impairment Scale (OASIS) [Internet]). Available from: [DOI] [PubMed] [Google Scholar]
- Pomerleau O.F., Fagerström K.-O., Marks J.L., Tate J.C., Pomerleau C.S. Development and validation of a self-rating scale for positive- and negative-reinforcement smoking: the Michigan Nicotine Reinforcement Questionnaire. Nicotine Tob. Res. 2003 Oct;5(5):711–718. doi: 10.1080/1462220031000158627. [DOI] [PubMed] [Google Scholar]
- Qin J., MetaHIT Consortium. Li R., Raes J., Arumugam M., Burgdorf K.S. A human gut microbial gene catalogue established by metagenomic sequencing [Internet] Nature. 2010;464 doi: 10.1038/nature08821. 59–65. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Byrne P.P., Davidson K.W., Kessler R.C., Asmundson G.J.G., Goodwin R.D., Kubzansky L. Anxiety disorders and comorbid medical illness. Gen. Hosp. Psychiatr. 2008 May;30(3):208–225. doi: 10.1016/j.genhosppsych.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Skosnik P.D., Cortes-Briones J.A. Targeting the ecology within: the role of the gut-brain axis and human microbiota in drug addiction. Med. Hypotheses. 2016 Aug;93:77–80. doi: 10.1016/j.mehy.2016.05.021. [DOI] [PubMed] [Google Scholar]
- Stahringer S.S., Clemente J.C., Corley R.P., Hewitt J., Knights D., Walters W.A., Knight R., Krauter K.S. Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res. 2012;22(11):2146. doi: 10.1101/gr.140608.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F., Yang F., Huang S., Bo C., Xu Z.Z., Amir A. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe. 2015 Sep 9;18(3):296–306. doi: 10.1016/j.chom.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007 Oct 18;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y., Pirrung M., Gonzalez A., EMPeror Knight R. vol. 2. GigaScience; 2013. (A Tool for Visualizing High-Throughput Microbial Community Data [Internet]). Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y., Callewaert C., Debelius J., Hyde E., Marotz C., Morton J.T. Impacts of the human gut microbiome on therapeutics. Annu. Rev. Pharmacol. Toxicol. 2018 Jan 6;58:253–270. doi: 10.1146/annurev-pharmtox-042017-031849. [DOI] [PubMed] [Google Scholar]
- Victor T.A., Khalsa S.S., Simmons W.K., Feinstein J.S., Savitz J., Aupperle R.L. Tulsa 1000: a naturalistic study protocol for multilevel assessment and outcome prediction in a large psychiatric sample. BMJ Open. 2018 Jan 24;8(1) doi: 10.1136/bmjopen-2017-016620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N.D., Koob G.F., McLellan A.T. Neurobiologic advances from the brain disease model of addiction. N. Engl. J. Med. 2016 Jan 28;374(4):363–371. doi: 10.1056/NEJMra1511480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Roy S. Gut homeostasis, microbial dysbiosis, and opioids. Toxicol. Pathol. 2017 Jan;45(1):150–156. doi: 10.1177/0192623316679898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Meng J., Zhang L., Johnson T., Chen C., Roy S. Morphine induces changes in the gut microbiome and metabolome in a morphine dependence model. Sci. Rep. 2018 Feb 26;8(1):3596. doi: 10.1038/s41598-018-21915-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J.L.M., Mark Welch J.L., Dewhirst F.E., Borisy G.G. Biogeography of the oral microbiome: the site-specialist hypothesis [internet] Annu. Rev. Microbiol. 2019;73:335–358. doi: 10.1146/annurev-micro-090817-062503. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G.S. fourth ed. Professional Manual; 2006. WRAT 4: Wide Range Achievement Test. [Google Scholar]
- Xu Z., Malmer D., Langille M.G.I., Way S.F., Knight R. Which is more important for classifying microbial communities: who's there or what they can do? ISME J. 2014 Dec;8(12):2357–2359. doi: 10.1038/ismej.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.Z., Amir A., Sanders J., Zhu Q., Morton J.T., Bletz M.C. Calour: an interactive, microbe-centric analysis tool. mSystems [Internet] 2019 Jan;4(1) doi: 10.1128/mSystems.00269-18. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012 May 9;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.