Abstract
Introduction
EGFR G724S has been described to mediate resistance to first- and third-generation EGFR tyrosine kinase inhibitors (TKIs). In vitro experiments have provided compelling evidence that G724S retains sensitivity for afatinib. Nevertheless, limited data have reported the clinical efficacy of afatinib in patients with NSCLC harboring G724S mutation.
Methods
We identified 52 patients with NSCLC with EGFR G724S from an inhouse database and comprehensively profiled their concurrent mutation statuses. Treatments and clinical outcomes were also collected.
Results
Of 52 G724S-positive patients, 39 harbored concomitant EGFR exon 19 deletion (19del), and all 37 of the 39 patients who had available clinical data were detected with a G724S mutation after receiving EGFR TKIs. A rare variant of 19del E746_S752delinsV co-occurred with G724S the most frequently (n = 29), whereas 7 of 10 patients with concomitant EGFR exon 20 mutation were TKI treatment naive. S768I was the most common mutation in exon 20 (n = 7). One patient harbored a concomitant EGFR exon 21 mutation, and two lacked co-occurring EGFR mutations. A total of 23 patients provided valid clinical outcome data, of whom eight were treated with afatinib after the emergence of G724S, whereas 15 received non-afatinib treatment (alternative EGFR TKI, chemotherapy, or best supportive care). The disease control rate in afatinib-treated patients (n = 8) reached 100% with a median progression-free survival of 4.5 months, significantly longer than that of non–afatinib-treated (n = 15, 1.7 mo, hazard ratio [HR] = 0.32, p = 0.037) and alternative EGFR TKI-treated (n = 11, 1.8 mo, HR = 0.28, p = 0.042) patients. In the subset who had progressed on osimertinib, afatinib also yielded a superior progression-free survival (6.2 mo) than non-afatinib therapies (1.0 mo, HR = 0.04, p = 0.005) and alternative EGFR TKIs (1.8 mo, HR = 0.06, p = 0.033). Analysis of acquired mutations at afatinib progression revealed re-emergence of EGFR T790M or MET amplification as the potential mechanism of afatinib resistance.
Conclusions
EGFR G724S emerges as a resistant mutation against EGFR TKI preferentially in the context of a rare variant of 19del, whereas it might mediate differential mechanisms in the context of exon 20 mutation. We also found that afatinib could be a potential therapeutic option for patients with NSCLC with G724S.
Keywords: EGFR G724S, EGFR 19 deletion, Non–small Cell Lung Cancer, Afatinib
Introduction
The use of EGFR tyrosine kinase inhibitors (TKIs) has dramatically altered the therapeutic routine for patients with NSCLC and substantially improved the prognosis of EGFR-mutant population.1,2 First-generation EGFR TKI, such as erlotinib or gefitinib, was designed to target EGFR-activating mutations in the tyrosine kinase domain (exon19 deletion (del) or exon21 L858R). Afatinib, a second-generation EGFR TKI selectively and irreversibly blocking ErbB family (including EGFR and HER2), also has clinical activity in patients with NSCLC with uncommon EGFR mutation.3,4 Nevertheless, a secondary EGFR T790M gatekeeper mutation often emerges, leading to acquired resistance toward EGFR TKIs.5,6 To overcome the resistance induced by T790M, third-generation EGFR TKIs, such as osimertinib, have been developed. Osimertinib has recently been approved as first-line therapy for EGFR-mutated metastatic NSCLC.7 Unfortunately, patients treated with osimertinib often inevitably acquire resistance with the emergence of EGFR C797S as the most well-described mechanism.8,9
EGFR exon 18 G724S is a rare mutation and has recently been described in some case reports mediating the resistance to third-generation EGFR TKIs.10, 11, 12 Both in vitro and in vivo studies have revealed that G724S limits the activity of third-generation EGFR TKIs.13 Li et al.14 reported G724S arising in 0.43% (5 of 1170) of osimertinib treatment-naive patients with NSCLC and revealed its association with the resistance to first-generation EGFR TKIs. In vitro experiments have provided compelling evidence that EGFR G724S retains its sensitivity for second-generation inhibitors, including afatinib.13,15 Nevertheless, owing to its rarity, comprehensive characterization of EGFR G724S mutation is still lacking and limited data have reported the clinical efficacy of afatinib in patients with NSCLC harboring this rare mutation.
In this study, we identified 52 patients with lung cancer harboring EGFR G724S mutation from our inhouse database, aiming to comprehensively profile their concurrent mutation statuses in EGFR and other genes and explore potential mechanisms mediated by G724S. We also investigated patients’ treatment responses and survival outcomes and explored the mechanism of afatinib resistance in G724S-positive patients.
Materials and Methods
Patient Information
We retrospectively reviewed the genomic profiling data of 42,316 patients with lung cancer from an inhouse database (BR) and identified 52 patients harboring EGFR G724S mutation (Supplementary Fig. 1). Formalin-fixed, paraffin-embedded tissue, plasma, cerebrospinal fluid, or pleural fluid samples of these patients were sequenced using a capture-based targeted panel including 520 cancer-related genes or a panel consisting 168 lung cancer genes (Burning Rock Biotech, Guangzhou, People’s Republic of China). Clinical characteristics and treatment histories of patients were also retrospectively collected. The study was approved by the institutional review board of Sun Yat-sen University Cancer Center (B2020-323-01). Owing to the retrospective nature of the study, patient’s informed consent was waived.
DNA Library Preparation and Sequencing
DNA was extracted using a QIAamp DNA formalin-fixed, paraffin-embedded tissue kit (Qiagen, Venlo, The Netherlands) or a QIAamp circulating nucleic acid kit (Qiagen) accordingly. DNA was sheared using Covaris M220. End repair and A tailing were followed by adaptor ligation. Ligated fragments of 200 to 400 base pairs were selected by beads (Agencourt AMPure XP kit; Beckman Coulter, Brea, CA), hybridized with RNA probe, purified by magnetic beads, and amplified by polymerase chain reaction. Indexed libraries were sequenced on a NextSeq500 (Illumina, San Diego, CA) with pair-end reads.
Sequencing Data analysis
All reads were trimmed for adapters and mapped to the reference human genome (hg19) using the Burrows-Wheeler Aligner version (v.)0.7.10.16 Local alignment optimization, duplication marking, and variant calling were performed using the Genome Analysis Tool Kit v.3.2,17 Picard and VarScan v.2.4.3.18 Variants were filtered using the VarScan fpfilter pipeline, and loci with depth less than 100 were filtered out. Variants with population frequency more than 0.1% in the ExAC, 1000 Genomes, dbSNP, or ESP6500SI-V2 databases were grouped as single-nucleotide polymorphisms and excluded from further analysis. The remaining variants were annotated with ANNOVAR (2016-02-01 release)19 and SnpEff v.3.6.20
Clinical Data Collection and Evaluation of Clinical Outcomes
Clinicopathologic data and treatment histories of EGFR G724S-positive patients were retrospectively collected. Tumor response was evaluated according to the Response Evaluation Criteria in Solid Tumors version 1.1. Investigator-assessed objective response and survival outcomes were analyzed. Progression-free survival (PFS) and overall survival were defined from the start of afatinib therapy until the date of progression and date of death or last follow-up, respectively. A total of 23 patients had clinical outcomes available, and eight of them were treated with afatinib. Acquired resistance mechanisms to afatinib therapy were also described for three patients who had rebiopsy at afatinib progression (Supplementary Fig. 1).
Statistical Analysis
Statistical analysis was performed using R version 3.3.3 software. Pearson’s chi-square test was performed to compare the prevalence difference in groups. Kaplan-Meier analysis was used to estimate survival functions, and a log-rank test was used to determine the difference in the survival curves between groups. In addition, p value less than 0.05 was considered to be statistically significant.
Results
G724S Preferentially Co-Occurring With a Rare Variant of EGFR Exon 19 Del Mediates Resistance to EGFR TKI
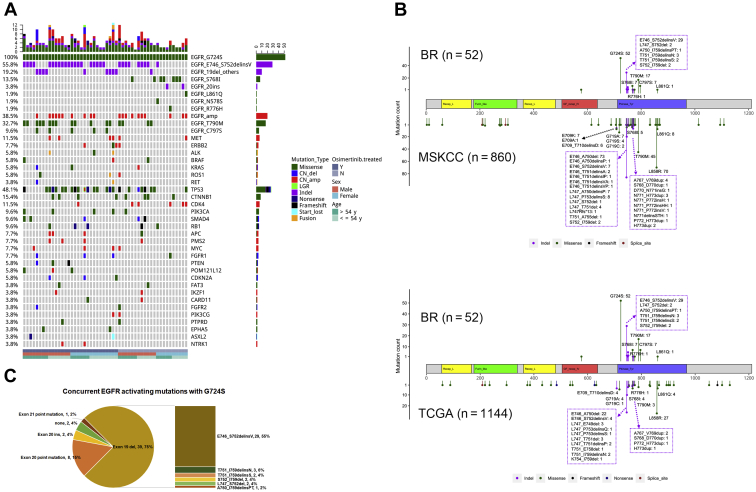
Of the 52 EGFR G724S-positive patients, 48 were diagnosed with adenocarcinomas, one with adenosquamous carcinoma, and one with small cell carcinoma. The histopathology classification of the remaining two patients was unclear. A variant of EGFR exon 19 del E746_S752delinsV co-occurred most frequently with G724S (n = 29, 55.8%), which was only observed in 7 of 860 (0.8%) of Memorial Sloan Kettering Cancer Center (MSKCC) NSCLCs (p < 0.001) and 4 of 1144 (0.3%) of The Cancer Genome Atlas (TCGA) lung cancers (p < 0.001) (Fig. 1A and B). Other co-occurring exon 19 del variants accounted for 19.2%, including T751_I759delinsN/S (n = 5), S752_I759del (n = 2), L747_S752del (n = 2), and A750_I759delinsPT (n = 1), which were also rarely found in MSKCC and TCGA (Fig. 1B and C). The mutation S768I in exon 20 occurred in 13.5% of G724S-positive lung cancers (n = 7), compared with 0.3% to 0.5% in unselected lung cancers in TCGA and MSKCC (Fig. 1A and B). We also observed two 20 exon insertion variants N771dup (n = 1) and V769_D770insGT (n = 1) and two missense mutations in exon 20 (R776H, n = 1) and exon 21 (L861Q, n = 1), co-occurring with G724S (Fig. 2A and B). Two patients lacked a concurrent EGFR mutation (Fig. 1A). Collectively, 75% (n = 39) of G724S-positive patients harbored a concurrent mutation of exon 19 del and insertions (delins) and 15% (n = 8) carried a concomitant exon 20 point mutation. The exon 20 insertion and exon 21 point mutation were present in 4% (n = 2) and 2% (n = 1) of patients, respectively (Fig. 1C). Of note, none of the G724S-positive patients harbored the common mutation EGFR L858R. In comparison, the exon19 del and exon21 point mutation comprised 44% and 43% of all EGFR mutations, respectively, in our whole cohort without selection (Supplementary Fig. 2). E746_S752delinsV, the most enriched exon19 del variant in the G724S-positive cohort, only accounted for 3.2% of all exon19 dels with a prevalence of 0.5% in the total cohort of 42,316 patients, which is similar to that of 0.3% to 0.5% in MSKCC and TCGA. The distribution of the G724S among each of the different EGFR mutations and variants was also evaluated (Supplementary Table 1). We observed a significantly higher frequency of G724S in patients harboring exon 20 point mutation than those with other types of EGFR mutations (2.77% versus 0.26%, p < 0.001). Among different exon19 del variants, G724S occurred the most frequently in the context of A750_I759delinsPT (16.67%) and E746_S752delinsV (12.83%). The prevalence of G724S was significantly higher in the context of E746_S752delinsV than other exon19 del variants (12.83% versus 4.5%, p = 0.003).
Figure 1.
The genomic profiles of EGFR G724S-positive lung cancers. (A) Oncoprint of EGFR and concurrent mutations (n = 52). (B) Comparisons of EGFR mutational spectra in G724S-positive lung cancers from an inhouse (BR) database (n = 52) versus NSCLCs from MSKCC (n = 860) and pan-lung cancer from TCGA (n = 1144). (C) The distribution of EGFR-activating mutations co-occurring with G724S. amp, amplification; del, deletion; Indel, insertion and deletion; ins, insertion; MSKCC, Memorial Sloan Kettering Cancer Center; N, no; TCGA, The Cancer Genome Atlas; Y, yes.
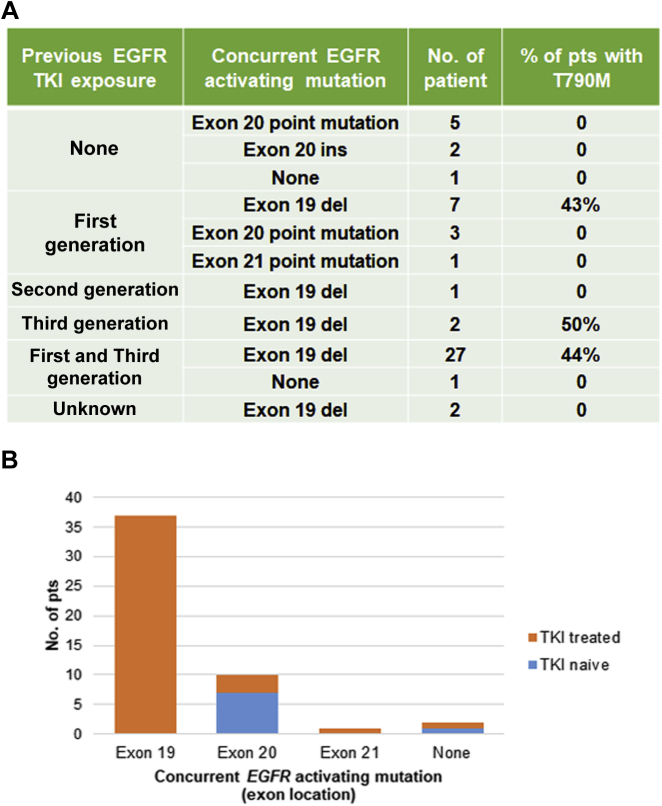
Figure 2.
The association between previous EGFR TKI exposure and the type of concurrent EGFR-activating mutation. (A) List of concurrent EGFR mutations in Pts treated with different TKIs. (B) The proportions of EGFR TKI exposure in Pts with EGFR-activating mutation in different exons. del, deletion; ins, insertion; Pts, patients; TKI, tyrosine kinase inhibitor.
In addition to the EGFR, other driver genes co-altered in G724S-postive lung cancers consisted of MET (n = 6), ERBB2 (n = 4), ALK (n = 3), BRAF (n = 3), ROS (n = 3), and RET (n = 2) (Fig. 1A). TP53 alterations co-occurred the most frequently (48.1%), followed by CTNNB1 (15.4%).
We also investigated the time point when EGFR G724S emerged and its association with the type of concurrent EGFR activating mutation. G724S was detected in 11 patients after progression on first-generation TKI, with seven of them harboring concomitant exon 19 delins, three harboring exon 20 point mutation, and one with exon 21 point mutation (Fig. 2A). G724S was detected in one patient after progression on second-generation TKI afatinib accompanied by an exon 19 delins. A total of 30 patients had G724S identified after progression on third-generation TKI, 28 of whom also had received first-generation TKI. All 30 of them carried concurrent exon 19 delins except for one. Eight patients (five with exon 20 point mutation, two with exon 20 insertion, and one without concurrent EGFR mutation) were TKI treatment naive before the emergence of G724S. T790M was detected in 40% to 50% of patients harboring co-occurring exon 19 delins who had received TKI treatment. Two patients with exon 19 delins had unknown TKI exposure status. Interestingly, all patients harboring EGFR 19 exon delins with known TKI exposure status (n = 37) received EGFR TKI treatment before G724S arising, whereas 7 of 10 patients harboring exon 20 mutations were TKI treatment naive (p < 0.001, Fig. 2B). This phenomenon suggests that the emergence of EGFR G724S mediates the TKI resistance preferentially in the context of EGFR-activating mutation exon 19 del, whereas the G724S co-occurring with a mutation in exon 20 is more likely a primary mutation.
The Clinical Outcomes of EGFR G724S-Positive Patients
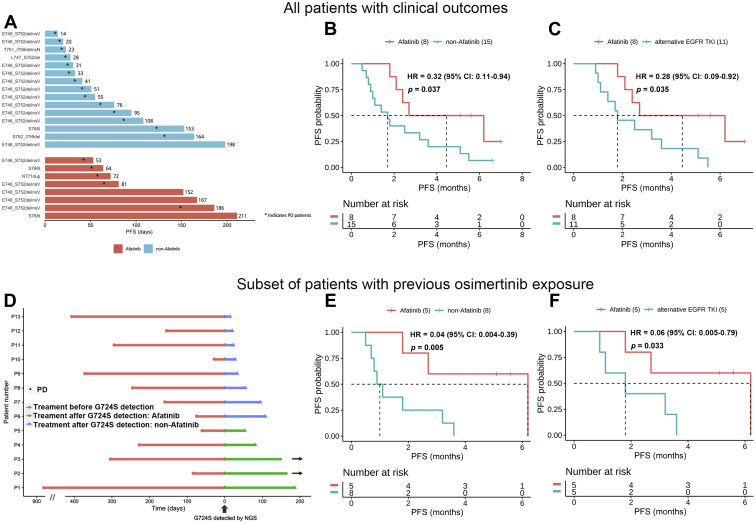
Of the 52 G724S-positive patients, 23 provided valid data for clinical outcomes with eight of them treated with afatinib after the emergence of EGFR G724S (Fig. 3A). The remaining received non-afatinib treatment, including alternative EGFR TKI, chemotherapy, or best supportive care (Supplementary Table 2). The survival curves revealed that afatinib treatment resulted in a significantly longer PFS in G724S-positive patients than non-afatinib therapy (4.5 versus 1.7 mo, hazard ratio [HR] = 0.32, p = 0.037) (Fig. 3B). All the eight afatinib-treated patients achieved stable disease, resulting in a disease control rate of 100% (Table 1). Among the 15 non-afatinib-treated patients, 11 received alternative EGFR TKI (Supplementary Table 2). A longer median PFS (mPFS) was also observed in the afatinib group compared with the alternative EGFR TKI group (4.5 versus 1.8 mo, HR = 0.28, p = 0.035, Fig. 3C). Among the eight afatinib-treated patients, G724S was identified from two treatment-naive patients (baseline), one patient after gefitinib failure and five patients after osimertinib failure (Table 1), in concurrence with other rare EGFR mutations, including EGFR exon 20 insertion (n = 1), EGFR S768I (n = 2), and E746_S752delinsV (19delins; n = 5) (Fig. 3A). Interestingly, all the five patients who acquired G724S at osimertinib progression had co-occurring EGFR exon 19delins (Table 1). Patient 24 harboring baseline G724S in concurrence with EGFR S768I had the longest PFS of 7.0 months (ongoing) after the first-line treatment of afatinib. Nevertheless, the other patient harboring baseline G724S (patient 2) only obtained a PFS of 2.4 months after afatinib treatment who carried a concurrent EGFR exon 20 insertion (Table 1, Fig. 3D).
Figure 3.
The clinical outcomes of patients with EGFR G724S-positive lung cancers. (A) PFS according to the type of co-occurring EGFR-activating mutations. (B) Kaplan-Meier curves for PFS in patients treated with afatinib versus those with non-afatinib treatments. (C) Kaplan-Meier curves for PFS in patients treated with afatinib versus those with other TKIs. (D) Treatment time of the patients with previous osimertinib exposure. (E) Kaplan-Meier curves for PFS in patients treated with afatinib versus non-afatinib therapy post-osimertinib progression. (F) Kaplan-Meier curves for PFS in patients treated with afatinib versus other TKIs post-osimertinib progression. p value was adjusted by sex and age. CI, confidence interval; HR, hazard ratio; NGS, next-generation sequencing; PFS, progression-free survival; TKI, tyrosine kinase inhibitor.
Table 1.
Characteristics of Eight Cases With Lung Adenocarcinoma Who Received Afatinib Treatment After the Emergence of EGFR G724S
| P | Sex | Age, y | Clinical Stage | Concurrent EGFR Mutation | Line of Afatinib | Previous TKI | Best Response | PFS (mo) | PD Status | OS (mo) | OS Status |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P2 | Female | 44 | IVb | N771dup+amp | 1 | NA | Stable disease | 2.4 | Yes | 2.4 | No |
| P24 | Male | 63 | IVa | S768I | 1 | NA | Stable disease | 7.0 | No | 7.0 | No |
| P10 | Female | 52 | IV | S768I | 2 | Gefitinib | Stable disease | 2.1 | Yes | 2.7 | Yes |
| P3 | Male | 49 | IVa | E746_S752delinsV | 3 | Erlotinib, osimertinib | Stable disease | 5.1 | No | 5.1 | No |
| P31 | Female | 62 | IV | E746_S752delinsV | 3 | Erlotinib, osimertinib | Stable disease | 6.2 | Yes | 6.2 | No |
| P32 | Female | 55 | IVb | E746_S752delinsV | 4 | Erlotinib, osimertinib | Stable disease | 5.6 | No | 5.6 | No |
| P38 | Male | 48 | IVb | E746_S752delinsV | 3 | Erlotinib, osimertinib | Stable disease | 2.7 | Yes | 3.6 | Yes |
| P39 | Male | 50 | IV | E746_S752delinsV | 5 | Gefitinib, osimertinib | Stable disease | 1.8 | Yes | 21.4 | No |
amp, amplification; delins, deletion and insertion; NA, not applicable; OS, overall survival; P, patient; PD, progressive disease; PFS, progression-free survival; TKI, tyrosine kinase inhibitor.
In the subset of 13 patients who had received and progressed on osimertinib treatment, afatinib also yielded a superior PFS (n = 5, 6.2 mo) than non-afatinib therapies (n = 8, 1.0 mo, HR = 0.04, p = 0.005, Fig. 3E) and alternative EGFR TKIs (n = 5, 1.8 mo, HR = 0.06, p = 0.033, Fig. 3F). Of note, two of the five afatinib-treated patients (P3 and P32) remained on afatinib with a PFS of 5.1 and 5.7 months (and counting), respectively (Table 1, Fig. 3D).
Mechanisms of Afatinib Resistance in G724S-Positive Patients
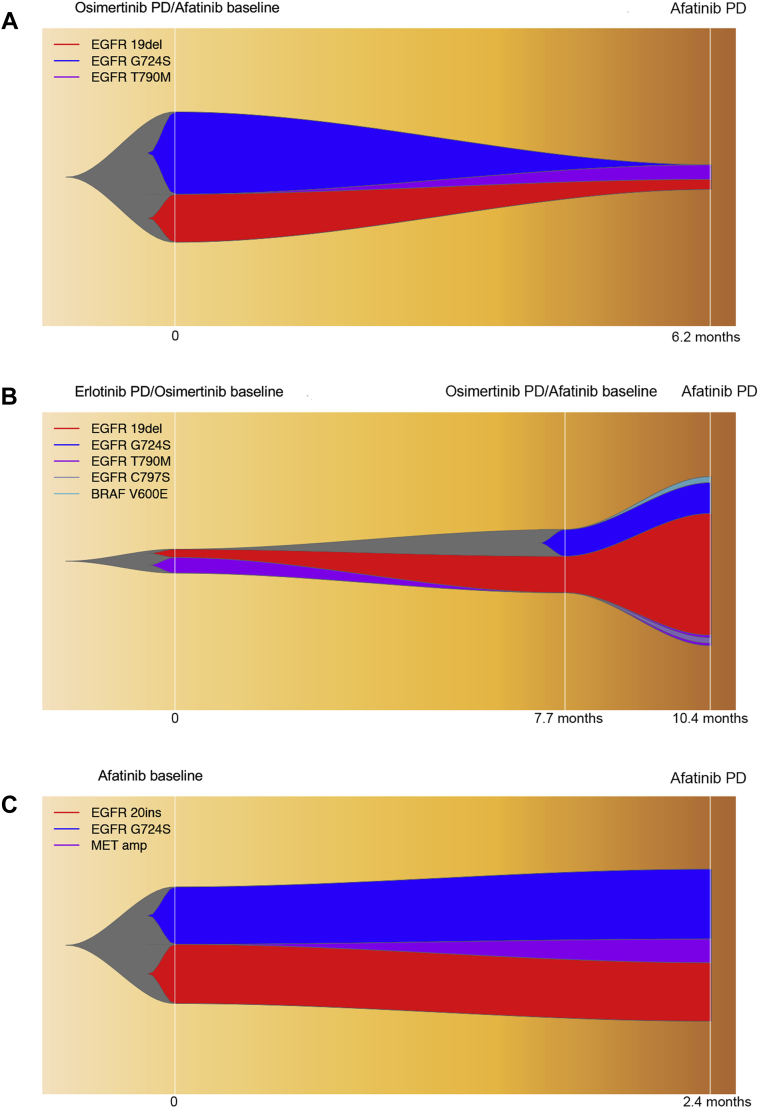
Analysis of acquired mutation profile at afatinib progression revealed re-emergence of EGFR T790M as the mechanism of afatinib resistance in two of the three patients who had available rebiopsy samples. Patient 31, a female patient with baseline EGFR exon 19delins who progressed on osimertinib after 28.4 months, lost EGFR T790M and acquired G724S at osimertinib progression. After receiving afatinib for 6.2 months, EGFR G724S was undetectable but EGFR T790M re-emerged which mediated her resistance to afatinib (Fig. 4A). Furthermore, patient 38, a male patient with baseline EGFR exon 19delins who acquired G724S and lost T790M at osimertinib progression, eventually acquired resistance to afatinib within 2.7 months of treatment. He had the re-emergence of T790M and acquisition of EGFR C797S and BRAF V600E while retaining EGFR G724S and 19delins (Fig. 4B). In contrast, patient 2, a female patient with stage IV recurrent lung adenocarcinoma, received afatinib as first-line treatment, who had baseline EGFR 20 exon insertion (N771dup) in concomitant with G724S. She acquired MET amplification at afatinib progression and retained the EGFR G724S and N771dup (Fig. 4C), revealing a differential afatinib resistance mechanism in treatment-naive compared with osimertinib-treated patients with EGFR G724S.
Figure 4.
The dynamic changes of plasma mutational profile in three cases treated with afatinib. (A) P31; (B) P38; and (C) P2. amp, amplification; del, deletion; ins, insertion; P, patient; PD, progressive disease.
Discussion
Our study revealed EGFR G724S emerging primarily after the progression of third-generation TKI (n = 30) and partially after the failure of first-generation TKI (n = 11). Recent case reports have revealed the potential role of G724S in acquired resistance to osimertinib10, 11, 12,21 and first-generation TKI14 in lung adenocarcinoma. Fassunke et al.13 revealed the emergence or persistence of EGFR G724S in osimertinib-resistant clones and described increasing G724S frequency accompanied by declining EGFR T790M under third-generation TKI treatment. This phenomenon was also observed in our study (Fig. 4B) and in other case reports.12,21 Moreover, in vitro study revealed that EGFR G724S limits the activity of erlotinib, and both in vitro and in vivo experiments suggested that G724S confers resistance against third-generation TKI by inducing a conformational change in the glycine-rich loop, which reduces the binding affinity of third-generation TKI.13,15
EGFR exon 19 del and L858R are the two activating mutations that most often occur in NSCLCs with approximately equal prevalence.22 Intriguingly, our data revealed that G724S emerges as a resistant mutation against TKI (especially third-generation) preferentially in the context of exon 19 del but not in concurrence with L858R, which is concordant with previous reports.10, 11, 12,21 Furthermore, the most frequent exon 19 del co-occurring with G724S was a rare variant E746_S752delinsV (55.8%, Fig. 1B), which only accounts for less than 2% of exon 19 cases,22,23 whereas E746_A750del, the most common 19 exon del (~67%), was not identified from our G724S-positive cohort. Studies have revealed that G724S reduces the binding affinity of osimertinib selectively in the context of exon 19 del rather than L868R.13,15 More interestingly, it has been suggested that E746_S752delinsV/G724S double mutant enhances the dimerization-dependent αC-helix inward conformation compared with E746_S752delinsV, whereas E746_A750del/G724S reduces the dimerization-dependent activation versus E746_A750del, which might explain the unexpected enrichment of E746_S752delinsV/G724S double mutant.15
G724S also occurred in the context of EGFR exon 20 mutation in approximately 20% of cases (Fig. 1C). Unlike exon 19 del, from the current but limited data, it seemed that most patients harboring exon 20 mutation lacked exposure to TKI before G724S arising (Fig. 2B). This observation highlights a distinct underlying mechanism mediated by G724S in the context of exon 20 mutation, which merits further elucidation.
We also identified a single G724S in two patients without other EGFR-activating mutations (Fig. 1A). G724S has been suggested as an independent oncogenic mutation potentially. G724S single mutant also reveals sensitivity to erlotinib, afatinib, and osimertinib. Furthermore, unlike exon 19Del/G724S double mutant, G724S single mutant does not mediate osimertinib resistance.15 In our study, one of the single G724S was identified in a treatment-naive patient with resected adenocarcinoma, which supports the oncogenic role of single G724S. The other case with recurrent adenocarcinoma received gefitinib for 1 year followed by 1 year of chemotherapy, and osimertinib was administrated subsequently but immediately failed. Although the single EGFR G724S was detected after the progression on osimertinib, it is still uncertain when the G724S first emerged owing to the lack of genomic profile before osimertinib failure. Therefore, the role of single EGFR G724S in tumorigenesis and TKI resistance remains elusive.
Both in vitro and in vivo studies have revealed that Ex19Del/G724S retains sensitivity to afatinib.13,15 A case report of a patient with lung adenocarcinoma with EGFR 19 del/G724S achieved stable disease to the combination of osimertinib and afatinib after osimertinib failure but experienced progressive disease within 2 months.21 A recent study also described a case with acquired G724S in the context of EGFR E746_S752delinsV, who achieved PR after afatinib monotherapy with a PFS of more than 3.8 months.11 Oztan et al.12 reported two cases of stage IV lung adenocarcinomas harboring EGFR G724S concomitantly with exon9 del. One patient received carboplatin and pemetrexed and the other was treated with nivolumab, but neither of the regimens revealed efficacy. Complementing the limited clinical evidence, our study further supports a better survival after afatinib than other treatments including alternative TKIs in EGFR G724S-positive patients with lung cancers (HR = 0.33, p = 0.04, Fig. 3B), and the survival advantage seems more significant in the osimertinib-resistant subset (HR = 0.04, p = 0.006, Fig. 3E). The disease control rate in afatinib-treated patients reached 100% regardless of the genotype of pre-existing EGFR mutation (Table 1).
In addition to the well-described EGFR 19 del, our data revealed that G724S-positive patients in the context of S768I or 20 exon insertion also achieved stable disease to afatinib. Notably, of the two patients who received afatinib as the first-line treatment, patient 24 with concurrent S768I had the longest PFS of 7.0 months and patient 2 with 20 exon insertion progressed rapidly with a PFS of 2.4 months (Table 1). Concordantly, the LUX-Lung study reported an inferior response to afatinib in patients with 20 exon insertion (objective response rate = 8.7%, mPFS = 2.7 mo) than those with other EGFR mutation, whereas patients with S768I revealed an objective response rate of 100% and mPFS of 14.7 months.4 Nevertheless, patient 10 experienced rapid disease progression who also harbored S768I but received afatinib treatment after progressing on gefitinib (Table 1). Collectively, our results suggest a potentially better response to afatinib in TKI-naive patients with EGFR S768I/G724S.
T790M has been revealed as major resistance mechanism on afatinib treatment in EGFR 19del- or L858R-mutant patients.5,24 Our study also revealed that EGFR T790M re-emerged as the mechanism of afatinib resistance in two osimertinib-treated patients, accompanied by the disappearance of G724S or the acquisition of other resistant mutations (EGFR C797S and BRAF V600E) (Fig. 4A and B), whereas MET amplification might mediate the acquired resistance in the patients with afatinib as first-line treatment (Fig. 4C). Peled et al.21 reported a decline of C724S clone accompanied with the afatinib and osimertinib combinational treatment and a slight increase of C724S plus emergence of C797S on progression on the combination. Diverse mechanisms observed might be partially attributable to the different regimens administered but also indicate the complexity of acquiring resistance to afatinib in G724S-positive patients.
Our study also has limitations. Despite the large screening cohort, we only identified 52 EGFR G724S-positive patients owing to the rarity of this mutation. In addition, the treatment information and clinical outcomes were retrospectively obtained from half of the patients, with only eight treated with afatinib. The small number of patients and the retrospective nature of the study weaken the strength of our finding on the efficacy of afatinib. Furthermore, we were unable to statistically identity genomic modifiers (such as co-occurring EGFR-activating variants or other gene alterations) that might be associated with clinical outcomes because of the limited number of patients with therapeutic information. Further well-designed studies with large cohorts are warranted to evaluate the efficacy of afatinib and explore predictive and prognostic biomarkers for patients with advanced NSCLC harboring EGFR G724S.
In conclusion, our study reveals that G724S emerges as a resistant mutation against TKI preferentially in the context of a rare variant of EGFR exon 19 del and provides clinical evidence that afatinib monotherapy could be a potential therapeutic option for patients with NSCLC with EGFR G724S.
Availability of Data and Materials
The data sets used or analyzed during the current study are available from the corresponding author on reasonable request. The data supporting our trial will be found at the online Research Data Deposit website (http://www.researchdata.org.cn) (Research Data Deposit number: RDDA2020001827).
CRediT Authorship Contribution Statement
Yang Wei: Conceptualization, Investigation, Writing—review and editing.
Shulin Liu: Investigation, Resources, Writing—review and editing.
Benyuan Jiang: Investigation, Resources, Writing—review and editing.
Zhonghan Zhang: Investigation, Data curation, Writing—original draft, Writing—review and editing.
Wenfeng Fang: Resources, Writing—review and editing.
Yunpeng Yang: Resources, Writing—review and editing.
Xin Li: Resources, Writing—review and editing.
Jingyi Zhao: Resources, Writing—review and editing.
Hongyun Zhao: Conceptualization, Funding acquisition, Supervision, Writing—review and editing.
Acknowledgments
This work was supported by the Natural Science Foundation of Guangdong Province of People’s Republic of China (2018A0303130243) and the China International Medical Foundation (z-2018-32-190013). The authors appreciate the assistance in data analyses and manuscript writing from Drs. Jianxing Xiang, Min Li, and Lin Shao from Burning Rock Biotech.
Footnotes
Cite this article as: Wei Y, Jiang B, Liu S, et al. Afatinib as a potential therapeutic option for patients with NSCLC with EGFR G724S. JTO Clin Res Rep. 2021;2:100193.
Dr. Wei and Dr. Jiang contributed equally to this work.
Disclosure: The authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2021.100193.
Supplementary Data
References
- 1.Mok T.S., Wu Y.L., Thongprasert S. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Wu Y.L., Zhou C., Liam C.K. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015;26:1883–1889. doi: 10.1093/annonc/mdv270. [DOI] [PubMed] [Google Scholar]
- 3.Chiu C.H., Yang C.T., Shih J.Y. Epidermal growth factor receptor tyrosine kinase inhibitor treatment response in advanced lung adenocarcinomas with G719X/L861Q/S768I mutations. J Thorac Oncol. 2015;10:793–799. doi: 10.1097/JTO.0000000000000504. [DOI] [PubMed] [Google Scholar]
- 4.Yang J.C., Sequist L.V., Geater S.L. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 5.Camidge D.R., Pao W., Sequist L.V. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 6.Wu S.G., Shih J.Y. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17:38. doi: 10.1186/s12943-018-0777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soria J.C., Ohe Y., Vansteenkiste J. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 8.Piotrowska Z., Isozaki H., Lennerz J.K. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thress K.S., Paweletz C.P., Felip E. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., He B., Zhou D., Li M., Hu C. Newly emergent acquired EGFR exon 18 G724S mutation after resistance of a T790M specific EGFR inhibitor osimertinib in non-small-cell lung cancer: a case report. Onco Targets Ther. 2019;12:51–56. doi: 10.2147/OTT.S188612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang W., Huang Y., Gan J., Zheng Q., Zhang L. Emergence of EGFR G724S after progression on osimertinib responded to afatinib monotherapy. J Thorac Oncol. 2020;15:e36–e37. doi: 10.1016/j.jtho.2019.09.198. [DOI] [PubMed] [Google Scholar]
- 12.Oztan A., Fischer S., Schrock A.B. Emergence of EGFR G724S mutation in EGFR-mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer. 2017;111:84–87. doi: 10.1016/j.lungcan.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Fassunke J., Müller F., Keul M. Overcoming EGFRG724S-mediated osimertinib resistance through unique binding characteristics of second-generation EGFR inhibitors. Nat Commun. 2018;9:4655. doi: 10.1038/s41467-018-07078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Wang Z., Groen H.J.M. Uncommon EGFR G724S mutations arise in non-small-cell lung cancer patients with acquired resistance to first-generation EGFR-TKIs. Lung Cancer. 2018;118:173–175. doi: 10.1016/j.lungcan.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Brown B.P., Zhang Y.K., Westover D. On-target resistance to the mutant-selective EGFR inhibitor osimertinib can develop in an allele-specific manner dependent on the original EGFR-activating mutation. Clin Cancer Res. 2019;25:3341–3351. doi: 10.1158/1078-0432.CCR-18-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A., Hanna M., Banks E. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koboldt D.C., Zhang Q., Larson D.E. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K., Li M., Hakonarson H., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cingolani P., Platts A., Wang le L. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peled N., Roisman L.C., Miron B. Subclonal therapy by two EGFR TKIs guided by sequential plasma cell-free DNA in EGFR-mutated lung cancer. J Thorac Oncol. 2017;12:e81–e84. doi: 10.1016/j.jtho.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi Y., Mitsudomi T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: perspectives for individualized treatment strategy. Cancer Sci. 2016;107:1179–1186. doi: 10.1111/cas.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su J., Zhong W., Zhang X. Molecular characteristics and clinical outcomes of EGFR exon 19 indel subtypes to EGFR TKIs in NSCLC patients. Oncotarget. 2017;8:111246–111257. doi: 10.18632/oncotarget.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campo M., Gerber D., Gainor J.F. Acquired resistance to first-line afatinib and the challenges of prearranged progression biopsies. J Thorac Oncol. 2016;11:2022–2026. doi: 10.1016/j.jtho.2016.06.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used or analyzed during the current study are available from the corresponding author on reasonable request. The data supporting our trial will be found at the online Research Data Deposit website (http://www.researchdata.org.cn) (Research Data Deposit number: RDDA2020001827).