Abstract
Introduction
Despite initial benefit, virtually all patients suffering from EGFR-mutant NSCLC experience acquired resistance to tyrosine kinase inhibitors (TKIs), driven by multiple mechanisms. Recent reports have identified oncogenic kinase fusions as off-target resistance mechanisms; however, these alterations have been rarely investigated at EGFR TKIs progression.
Methods
Patients with EGFR-mutated metastatic NSCLC (N = 62) with tissue and plasma biopsies at EGFR TKI progression between January 2015 and June 2019, at a French hospital and optionally before progression, were identified from the prospective MATCH-R study (NCT02517892). Postprogression biopsy samples were analyzed for gene fusions using targeted gene panel sequencing, whole-exome sequencing, RNA sequencing, and comparative genomic hybridization array.
Results
Six gene fusions were detected in tumor progression biopsies under an EGFR TKI from 62 consecutive patients (9.7%) with EGFR-mutated advanced NSCLC. Among 31 patients progressing to first- or second-generation EGFR TKIs, one (3%) had an Eukaryotic translation initiation factor 4 gamma 2–GRB2 associated binding protein 1 (EIF4G2-GAB1) fusion. Among 31 patients progressing to the third-generation osimertinib, five (16%) presented oncogene fusions of fibroblast growth factor receptor 3–transforming acidic coiled-coil containing protein 3 (FGFR3-TACC3) (n = 2), kinesin family member 5B–Ret proto-oncogene (KIF5B-RET) (n = 1), striatin–anaplastic lymphoma kinase (STRN-ALK) (n = 1), and zinc finger DHHC-Type palmitoyltransferase 20–Thr790Met (ZDHHC20-BRAF) (n = 1) transcripts. Out of two patients that received osimertinib at first-line, one acquired an FGFR3-TACC3 fusion at progression. In all patients, fusions co-occurred with the original activating EGFR mutation; however, among four patients with an acquired T790M mutation, three (75%) lost the T790M mutation.
Conclusions
Oncogenic fusions at the time of EGFR TKI resistance were identified at a relatively high frequency, mainly after the third-generation TKI osimertinib. Patients progressing to EGFR TKIs may have a new opportunity for targeted therapy when oncogenic fusions are identified.
Keywords: EGFR resistance, Oncogene fusions, NSCLC, EGFR tyrosine kinase inhibitors
Introduction
During the past decade, tyrosine kinase inhibitors (TKIs) have displayed a substantial clinical benefit for patients with EGFR-mutant NSCLC.1 Despite tremendous advances, the long-term effectiveness of these targeted therapies has been limited by the unavoidable development of acquired resistance, leading to clinical disease progression. Several resistance mechanisms have been studied recently, schematically dichotomized between on-target (molecular alterations involving the target itself) and off-target (alterations involving other molecular elements). Gene fusions that activate tyrosine kinase receptors, such as ALK, ROS1, and RET, which occur in 1% to 5% of NSCLC, are usually mutually exclusive with EGFR mutations and represent meaningful therapeutic targets.2 Recent studies have also documented the emergence of oncogenic fusions as an off-target resistance mechanism to EGFR TKIs; however, limited cases have been reported and the estimated frequency remains unclear.3,4
Through tumor genotyping of tissue and plasma biopsies, we analyzed the presence of fusions and concurrent genetic alterations at biopsy progression under EGFR TKIs in patients with advanced NSCLC.
Materials and Methods
Patients with EGFR-mutant advanced NSCLC with tissue and plasma biopsies at the time of TKI progression (and optionally before starting targeted therapy) were selected from the prospective MATCH-R study (NCT02517892) (Supplementary methods). Postprogression samples were obtained from formalin-fixed, paraffin-embedded pathology blocks or fresh biopsies, if available. Blood samples were collected longitudinally during treatment and at progression for circulating tumor DNA (ctDNA) sequencing. Targeted gene panel sequencing was performed with an Ion Torrent PGM (ThermoFisher Scientific) sequencer using a customized panel (Mosc3 or 4) covering 75 to 82 critical oncogenes or tumor suppressor genes developed with Ion AmpliSeq custom design.5 Whole-exome sequencing, RNA sequencing (RNA-seq), and Affymetrix CytoScan HD comparative genomic hybridization array were performed as previously reported (see Supplementary Methods).5 CtDNA samples were analyzed by next-generation sequencing (50-gene panel) (Supplementary Methods). All molecular oncogenic alterations were respectively classified in either definitive (or potential) resistance or concomitant genetic alterations according to OncoKB and Cancer GenomeInterpreter.6,7 Patients were analyzed according to first- or second-generation TKIs (erlotinib, gefitinib, or afatinib) and the third-generation TKI osimertinib. The Kaplan-Meier method was used to estimate progression-free survival 2 (time from initiation of subsequent line therapy after osimertinib progression to the first documented disease progression or death) and overall survival in the post-osimertinib cohort (Supplementary methods).
Results
Between January 2015 and June 2019, 62 consecutive patients with EGFR-mutated advanced NSCLC underwent genotyping of tumor tissue and ctDNA samples collected at the time of EGFR TKIs progression and were analyzed according to TKI-generation (Supplementary Fig. 1). A total of 60 patients (97%) had adenocarcinomas, 37 (60%) were nonsmokers, and the mean age was 58 years (± SD 10.7). An exclusive thoracic progression was more frequent at osimertinib recurrence, and extrathoracic progression patterns were more frequent after first- or second-generation EGFR TKI (p = 0.03) (Table 1).
Table 1.
Patient Clinical Characteristics
| Characteristics | First- or Second-Generation EGFR TKI Cohort, n (%) | Third-Generation EGFR TKI Cohort, n (%) | p value |
|---|---|---|---|
| Total | 31 | 31 | |
| Median age (range), y | 60 (37–89) | 58 (40–72) | 0.40 |
| Sex, n (%) | |||
| Male | 7 (23) | 8 (26) | 0.77 |
| Female | 24 (77) | 23 (74) | |
| Smoking history, n (%) | |||
| Never | 21 (68) | 16 (52) | 0.24 |
| Current and former | 9 (29) | 13 (42) | |
| NS | 1 (3) | 2 (6) | |
| Baseline driver alteration | |||
| Exon 19, deletion | 20 (65) | 24 (77) | 0.40 |
| Exon 21, L858R | 10 (32) | 7 (23) | |
| Exon 18, G719A | 1 (3) | 0 | |
| First- or second-generation EGFR TKI before resistance biopsy | |||
| Erlotinib or Gefitinib | 26 (84) | 20 (65) | 0.17 |
| Afatinib | 5 (16) | 9 (29) | |
| Third-generation EGFR TKI before resistance biopsy | |||
| Osimertinib | 0 | 31 (100) | |
| Response to TKI | |||
| CR/PR | 24 (77) | 22 (71) | 0.71 |
| SD/PD | 7 (23) | 8 (26) | |
| NS | 0 | 1 (3) | |
| Progression pattern at TKI resistance | |||
| Solitary | 19 (61) | 24 (77) | 0.23 |
| Multiple | 11 (36) | 7 (23) | |
| NS | 1 (3) | 0 | |
| Site of progression | |||
| Thoracic | 10 (32) | 19 (61) | 0.03 |
| Extrathoracic | 20 (65) | 12 (39) | |
| NS | 1 (3) | 0 | |
Missing data were excluded from the statistical analysis.
NS, not specified; TKI, tyrosine kinase inhibitor; PD, progressive disease; CR, complete response, PR, partial response.
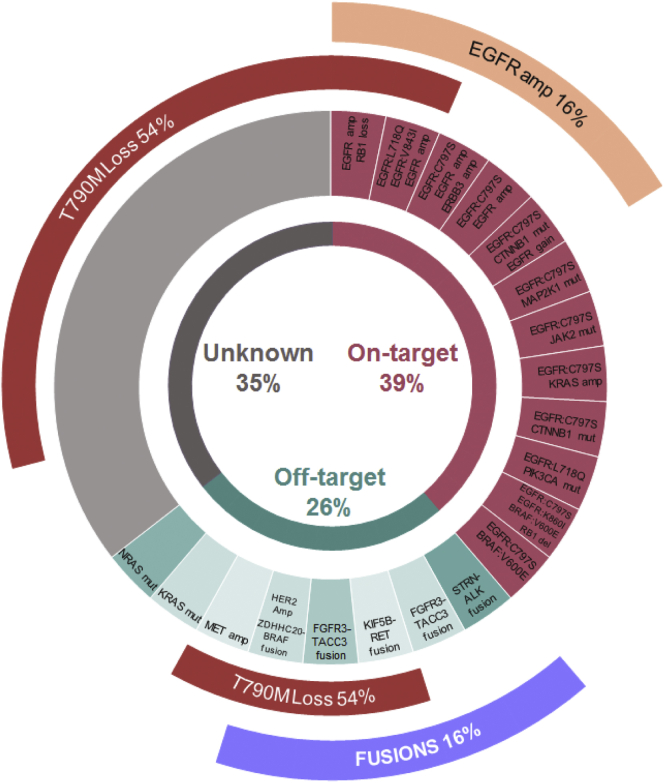
In six patients (9.7%), fusions were detected by RNA-seq analyses on tissue samples (Table 2). In the post–first- or second-generation EGFR TKIs cohort (n = 31), one case (3%) had a transcript fusion involving Eukaryotic translation initiation factor 4 gamma 2 (EIF4G2) and GRB2-associated binding protein 1 (GAB1) after gefitinib treatment. In the post-osimertinib cohort (n = 31, two and 29 receiving the drug in the first and subsequent lines, respectively), the resistance alteration landscapes at progression biopsy are described in Figure 1. Five patients (16%) presented oncogenic fusions including fibroblast growth factor receptor 3–transforming acidic coiled-coil containing protein 3 (FGFR3-TACC3) (n = 2), kinesin family member 5B–Ret proto-oncogene (KIF5B-RET) (n = 1), striatin–anaplastic lymphoma kinase (STRN-ALK) (n = 1), and zinc finger DHHC-type palmitoyltransferase 20 (ZDHHC20)-BRAF (n = 1) (Table 2). One of the FGFR3-TACC3 fusions was acquired after osimertinib first-line treatment, and the remaining in the subsequent lines of treatment. In terms of EGFR mutations identified at the time of fusion occurrence, all tumors retained the original activating EGFR mutation, but three of four patients (75%) lost the acquired Thr790Met (T790M) mutation. Median progression-free survival 2 in patients that presented fusions at osimertinib progression was longer than patients with other known resistance alteration, but not statistically significant (6 months [95% confidence interval [CI]: 0.7–16.8] versus 3 months [95% CI: 2.5–3.5], respectively; hazard ratio, 3.31 (95% CI: 0.7–16.7); p = 0.09) (Supplementary Fig. 2). No difference was observed either in the same analysis on overall survival (p = 0.95) (Supplementary Fig. 3).
Table 2.
Characteristics and Genomic Alterations in Pre- and Post-EGFR TKIs Samples in Patients With a Fusion at EGFR TKI Progression
| Case | Age/Sex | EGFR TKI/Line Before Resistance Biopsy | Genomic Alterations Pre-EGFR TKI (TGPS on Tissue) | Genomic Alterations on Tissue at EGFR TKI Progression (TGPS, CGH Array, and WES-RNAseq on Tissue) |
|
|---|---|---|---|---|---|
| Fusion | Other Alterations | ||||
| MR 04 | 62/F | Gefitinib/2 | EGFR: L858R EGFR exon 18, I706Y CTNNB1: S33C NOTCH4: G1821E |
EIF4G2-GAB1a | EGFR: L858Ra,b EGFR: I706Ta,b MET: R988Cc EGFR: T790Mc NOTCH4: G1821Eb CTNNB1: G34Va FGFR3: S804Ac BRD4: R1329Qa GATA2: D184Aa WT1: S255Aa TSC1: T1020Pa PDPK1 ampd |
| MR 211 | 57/F | Osimertinib/4 | EGFR exon 19 del EGFR: T790M TP53 lossd CDK4 ampd HMGA2 ampd MDM2 ampd |
FGFR3-TACC3a | EGFR exon 19 dela,b CDK4 ampa,d HMGA2 ampd MDM2 ampa,d TP53 lossd MAP3K7: p.Asp211Hisa GATA3: p.Thr156Proa |
| MR 393 | 64/F | Osimertinib/1 | EGFR: L858R | FGFR3-TACC3a | EGFR exon 21, L858Ra PIK3R1 muta MYCN: Pro345Thra FANCA: p.Pro1220Arga |
| MR 48 | 51/F | Osimertinib/8 | EGFR exon 19 del EGFR: T790M TP53: C238F |
KIF5B-RETa | EGFR exon 19 dela,b TP53: C238Fa,b |
| MR 240 | 65/F | Osimertinib/2 | EGFR exon 19 del EGFR: T790Mc CDKN2A muta,c FGFR2: S252Sc TP53 mutc |
STRN-ALKa,d | EGFR exon 19 dela,b EGFR: T790Ma,b TP53: p.Lys132fa,b PTEN: F278Lb TERT ampd |
| MR 01 | 54/M | Osimertinib/3 | EGFR exon 19 del EGFR: T790M TP53 mut |
DHHC20-BRAFa | EGFR exon 19 dela,b TP53: Cys135Serfsa,b ERBB2 ampa,d CDK12 ampa |
CGH, comparative genomic hybridization; ctDNA circulating tumor DNA; F, female; M, male; RNA-seq, RNA sequencing; TGPS, targeted gene panel sequencing; TKI, tyrosine kinase inhibitor; WES, whole-exome sequencing; Amp, amplification; MR, patient number.
Genomic alterations found in WES-RNAseq analyses.
Genomic alterations found in TGPS analyses.
Genomic alterations found only in the ctDNA analyses.
Genomic alterations found in cytoscan HD CGH array.
Figure 1.
Distribution of resistance alterations to third-generation EGFR tyrosine kinase inhibitor (n = 31). A total of 28 patients tested positive for T790M pre-osimertinib (two received osimertinib as first-line and T790M was not investigated at baseline in one patient). Among them, 15 (54%) lost this mutation at osimertinib progression biopsy. T790M, Thr790Met mutation of the epidermal growth factor receptor (EGFR); ERBB3, Erb-b2 receptor tyrosine kinase; CTNNB1, catenin beta 1; MAP2K1, mitogen-activated protein kinase kinase 1; JAK2, Janus kinase 2; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; RB1, retinoblastoma gene; STRN, striatin gene; ALK, anaplastic lymphoma kinase; FGFR3, fibroblast growth factor receptor 3; TACC3, transforming acidic coiled-coil containing protein 3; KIF5B, kinesin family member 5B; RET, ret proto-oncogene; HER2 Amp, human epidermal growth factor receptor 2 amplification; ZDHHC2, zinc finger DHHC-type palmitoyltransferase 20; MET; met proto-oncogene (hepatocyte growth factor receptor); NRAS, N-ras proto-oncogene (neuroblastoma RAS viral oncogene homolog).
Only two tumors, in particular, had other well-established resistance alterations to EGFR TKI (receptor tyrosine-protein kinase erbB-2 [ERBB2] amplification and T790M mutation) concomitantly with the fusions. When the putative resistance mechanisms differed from fusions, 37% (23 of 62) had more than one concomitant resistance alterations in the progression biopsy.
Discussion
We found a higher rate of fusions at resistance after a third-generation EGFR TKI (five of 31, 16%) compared with first- or second-generation TKIs (one of 31, 3%). Recent reports have described fusions as off-target resistance mechanisms to EGFR TKIs, but at lower frequencies. Among 3873 patients with EGFR-positive NSCLC, Xu et al.8 found 16 fusions (0.4%) at progression to EGFR TKI, including RET (n = 6), ALK (n = 5), neurotrophic receptor tyrosine kinase 1 (NTRK1) (n = 4), ROS1 (n = 1), and FGFR3 (n = 1).8 Analyzing ctDNA and tumor tissue samples at osimertinib progression from three lung cancer studies, fusions were found in 3% (one of 91) to 7% (three of 41) of cases.10, 11, 9 Interestingly, all our fusions were detected on tissue but not on ctDNA analysis, which may explain our higher incidence, because most previous studies have used targeted gene panels sequencing DNA extracted from blood rather than tissue biopsies.
FGFR3-TACC3 rearrangement has been identified as a driver alteration in several solid tumors and could lead to EGFR TKI resistance by promoting sustained activation of the ERK pathway.12 Fusions involving RET were also described at EGFR TKI resistance and were successfully reversed by a combination of EGFR and RET TKIs.13
The ALK partner STRN, found in our study, has rarely been established in the EGFR TKI resistance setting. Our patients did not respond to the ALK inhibitor crizotinib but achieved a stable disease of 6 months when both EGFR and ALK were inhibited with brigatinib. A published case report offers conflicting results about the effectiveness of ALK inhibitors on STRN-ALK translocated tumors.14
Although BRAF alterations in NSCLC are represented mainly by mutations (2%–4%), fusions display another mechanism of BRAF activation, which has also been suggested as a resistance mechanism to EGFR TKIs at a low frequency (∼2%).4 The ZDHHC20 fusion partner has not been previously reported. It is anticipated to be an oncogenic fusion because the loss of the N-terminal inhibitory domain permits the constitutive dimerization of RAF proteins with consequent activation of the downstream pathways.
The EIF4G2-GAB1 rearrangement found at gefitinib progression is also a novel reported fusion. GAB1 activity is relevant for some cellular functions such as the regulation of proliferation, migration, and survival by associating with TKI receptors such as met proto-oncogene (hepatocyte growth factor receptor) (cMET). Aberrant GAB1 activity has been associated with resistance mechanisms in BRAF-mutant melanomas owing to altered feedback regulation of cMET signaling.15 Despite the well-established oncogenic property of this fusion, its role in EGFR resistance needs preclinical validations.
Interestingly, at osimertinib resistance, tumors with fusion emergence retained the original activating EGFR mutation, but most of them (75%) lost the resistant T790M mutation. In agreement with these findings, Xu et al.8 reported a 50% T790M loss from 10 patients who presented fusions after osimertinib treatment.8 Furthermore, Oxnard et al.10 found similar results in three tumors with fusions and without T790M mutations at osimertinib progression (cell division cycle 6 [CDC6]-RET, FGFR3-TACC3, and extended synaptotagmin 2 [ESYT2]-BRAF).10 Together, these findings suggest that tumors drive the resistance and growth through these oncogenic fusions over the EGFR-dependent pathway.
Limitations
Our study has limitations. The sample size is limited and comes from a single center. Nevertheless, it is the first study using systematic RNA-seq at resistance to EGFR TKI. In addition, the lack of whole-exome sequencing–RNA-seq analyses in the TKI-naive biopsies does not allow us to define these oncogenic fusions as confirmed acquired resistance to EGFR TKIs. However, it should be noted that oncogenic fusions have been reported at a very low frequency at diagnosis.2
Conclusions
In our cohort, oncogenic fusions identified at the time of EGFR TKI resistance were more frequent than expected, in particular after treatment with a third-generation TKI. A significant proportion of these fusions can be targeted so their identification could influence treatment selection and overall survival of patients failing EGFR TKIs.
Acknowledgments
MATCH-R trial (NCT02517892) is supported by a Natixis Foundation grant (https://clinicaltrials.gov/ct2/show/NCT02517892). The authors thank Sarah MacKenzie for the English language editing.
Footnotes
Disclosure: Dr. Loriot reported receiving honoraria (self) from Roche, MSD, Astellas, Janssen, AstraZeneca, BMS, Seattle Genetics, Sanofi, Clovis, and Pfizer; honoraria (institution) from Roche, MSD, Astellas, Janssen, AstraZeneca, BMS, Seattle Genetics, Sanofi, Clovis, Incyte, and Pfizer; research grant or funding (institution) from Roche, MSD, and Sanofi; travel or accommodation expenses from Roche, MSD, Janssen, AstraZeneca, and Seattle Genetics; and licensing royalties from a pending patent (USA 62/455211, Europe 17209098.7). Dr. Planchard reported receiving advisory or consultancy fees from AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Celgene, Daiichi Sankyo, Eli Lilly, Merck, MedImmune, Novartis, Pfizer, prIME Oncology, Peer CME, and Roche; honoraria (self) from AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Celgene, Eli Lilly, Merck, Novartis, Pfizer, prIME Oncology, Peer CME, and Roche; research grant or funding (institution) from AstraZeneca, Bristol-Myers Squibb, AbbVie, Boehringer Ingelheim, Eli Lilly, Merck, Novartis, Pfizer, Roche, Medimmune, Sanofi-Aventis, Taiho Pharma, Novocure, and Daiichi Sankyo; and travel or accommodation expenses from AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Roche, Merck, Novartis, prIME Oncology, and Pfizer. Dr. Mezquita reported receiving speakers’ bureau honoraria from Bristol-Myers Squibb and Tecnofarma; and is a consultant or advisory board member for Roche. Dr. Michiels reported giving advisory or consultancy services and statistical advice to Janssen Cilag France; and serving as advisor, consultant, or data and safety monitoring member of Hexal, Johnson & Johnson, Ipsen, Neovacs, Genticel, Mabxience, Steba, IQVIA, Roche, Sensorion, and Biophytis. Dr. Massard reported having a leadership role in Aduro Biotech, Agios Pharmaceuticals, Amgen, Argen-X Bvba, Arno Therapeutics, Astex Pharmaceuticals, AstraZeneca, Aveo, Bayer Healthcare AG, Bbb Technologies BV, BeiGene, Bioalliance Pharma, BioNTech AG, Blueprint Medicines, Boehringer Ingelheim, and Bristol; receiving research grant or funding (institution) from AstraZeneca, BMS, Boehringer Ingelheim, Janssen Cilag, Merck, Novartis, Pfizer, Roche, and Sanofi; participating in nonremunerated activities of AstraZeneca, Bayer, BMS, Boringher Ingelheim, Johnson & Johnson, Lilly, MedImmune, Merck, NH TherAguix, Pfizer, and Roche; and giving advisory or consultancy services to Amgen, Astellas, AstraZeneca, Bayer, BeiGene, Bristol-Myers Squibb, Celgene, Debiopharm, Genentech, Ipsen, Janssen, Lilly, MedImmune, Novartis, Pfizer, Roche, Sanofi, and Orion. Dr. Remon reported receiving advisory fees from Merck Sharp and Dohme, Boehringer Ingelheim, Bristol-Myers Squibb, and AstraZeneca; speaker fees from Pfizer; and travel fees from OSE Immunotherapeutics, Bristol-Myers Squibb, AstraZeneca, and Roche. Dr. Soria reported giving advisory or consultancy services to AstraZeneca, Astex, Clovis, GSK, GamaMabs, Lilly, MSD, Mission Therapeutics, Merus, Pfizer, PharmaMar, Pierre Fabre, Roche/Genentech, Sanofi, Servier, Symphogen, and Takeda; serving as full- or part-time employee of AstraZeneca; and is a shareholder, stockholder or having stock options at AstraZeneca. Dr. André reported receiving research grant or funding (institution) from Novartis, AstraZeneca, Roche, Daiichi, Lilly, and Pfizer. Dr. Vassal reported serving as advisory board member or unpaid consultant for Pfizer. Dr. Besse reported receiving research grant or funding (institution) from AbbVie, Amgen, AstraZeneca, Biogen, Blueprint Medicines, BMS, Celgene, Eli Lilly, GSK, Ignyta, Ipsen, Merck KGaA, MSD, Nektar, Onxeo, Pfizer, Pharma Mar, Sanofi, Spectrum Pharmaceuticals, Takeda, and Tiziana Pharma. Other authors declare no conflicts of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2020.100023.
Supplementary Data
References
- 1.Recondo G., Facchinetti F., Olaussen K.A., Besse B., Friboulet L. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol. 2018;15:694–708. doi: 10.1038/s41571-018-0081-4. [DOI] [PubMed] [Google Scholar]
- 2.Gkolfinopoulos S., Mountzios G. Beyond EGFR and ALK: targeting rare mutations in advanced non-small cell lung cancer. Ann Transl Med. 2018;6 doi: 10.21037/atm.2018.04.28. 142–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klempner S.J., Bazhenova L.A., Braiteh F.S. Emergence of RET rearrangement co-existing with activated EGFR mutation in EGFR-mutated NSCLC patients who had progressed on first- or second-generation EGFR TKI. Lung Cancer. 2015;89:357–359. doi: 10.1016/j.lungcan.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Vojnic M., Kubota D., Kurzatkowski C. Acquired BRAF Rearrangements Induce Secondary Resistance to EGFR therapy in EGFR-Mutated Lung Cancers. J Thorac Oncol. 2019;14:802–815. doi: 10.1016/j.jtho.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massard C., Michiels S., Ferté C. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7:586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 6.Chakravarty D., Gao J., Phillips S.M. OncoKB: a precision oncology knowledge base. JCO Precision Oncology. 2017;1:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamborero D., Rubio-Perez C., Deu-Pons J. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018;10:25. doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H., Shen J., Xiang J. Characterization of acquired receptor tyrosine-kinase fusions as mechanisms of resistance to EGFR tyrosine-kinase inhibitors. Cancer Manag Res. 2019;11:6343–6351. doi: 10.2147/CMAR.S197337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadimitrakopoulou VA, Wu Y, Han J, et al. Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Paper presented at: ESMO 2018 Congress. October 19, 2018.
- 10.Oxnard G.R., Hu Y., Mileham K.F. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramalingam S.S., Cheng Y., Zhou C. Mechanisms of acquired resistance to first-line osimertinib: preliminary data from the phase III FLAURA study. Ann Oncol. 2018;29(suppl 8):viii740. [Google Scholar]
- 12.Daly C., Castanaro C., Zhang W. FGFR3-TACC3 fusion proteins act as naturally occurring drivers of tumor resistance by functionally substituting for EGFR/ERK signaling. Oncogene. 2017;36:471–481. doi: 10.1038/onc.2016.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piotrowska Z., Isozaki H., Lennerz J.K. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi Y., Masuda S., Iida Y., Takahashi N., Hashimoto S. Case report of non-small cell lung cancer with STRN-ALK translocation: a nonresponder to alectinib. J Thorac. 2017;12:e202–e204. doi: 10.1016/j.jtho.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Caenepeel S., Cooke K., Wadsworth S. MAPK pathway inhibition induces MET and GAB1 levels, priming BRAF mutant melanoma for rescue by hepatocyte growth factor. Oncotarget. 2017;8:17795–17809. doi: 10.18632/oncotarget.14855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.