Abstract
The gut microbiome comprised of microbes from multiple kingdoms, including bacteria, fungi, and viruses. Emerging evidence suggests that the intestinal fungi (the gut “mycobiome”) play an important role in host immunity and inflammation. Advances in next generation sequencing methods to study the fungi in fecal samples and mucosa tissues have expanded our understanding of gut fungi in intestinal homeostasis and systemic immunity in health and their contribution to different human diseases. In this review, the current status of gut mycobiome in health, early life, and different diseases including inflammatory bowel disease, colorectal cancer, and metabolic diseases were summarized.
Keywords: Gut fungi, Mycobiome, Early life, Health, Disease
Introduction
Most studies of the human gut microbiome have focused on the bacterial component of the microbiome, but the fungal microbiome (i.e. the mycobiome) has recently gained recognition as a fundamental part of the human microbiome. Fungi inhabit the mucosal surface to maintain intestinal homeostasis and systemic immunity.1 Until recently, there has been a lack of research focusing on the interactions of the fungal kingdom of microorganisms with other constituents of the gut microbiome and their contribution to health and diseases. To date, less than 3% of the microbiome literature accounts for the presence of fungi in microbial communities. Recent advances in sequencing technology have provided comprehensive tools to profile the fungal component of the gut microbiome2 and emerging data suggest that the gut mycobiome may act as a reservoir for potentially opportunistic pathogens in inflammatory bowel disease (IBD),3,4 graft-versus-host disease (GVHD),5 gastrointestinal cancer,6–8 and other diseases. Here, we discuss the importance of developing standardized methodology to define the gut mycobiome in early life, health, and diseases. We also highlight future directions of studies to delineate the role of gut mycobiome and the potential of therapeutic intervention targeting the gut mycobiome.
Methods to characterize the gut mycobiome
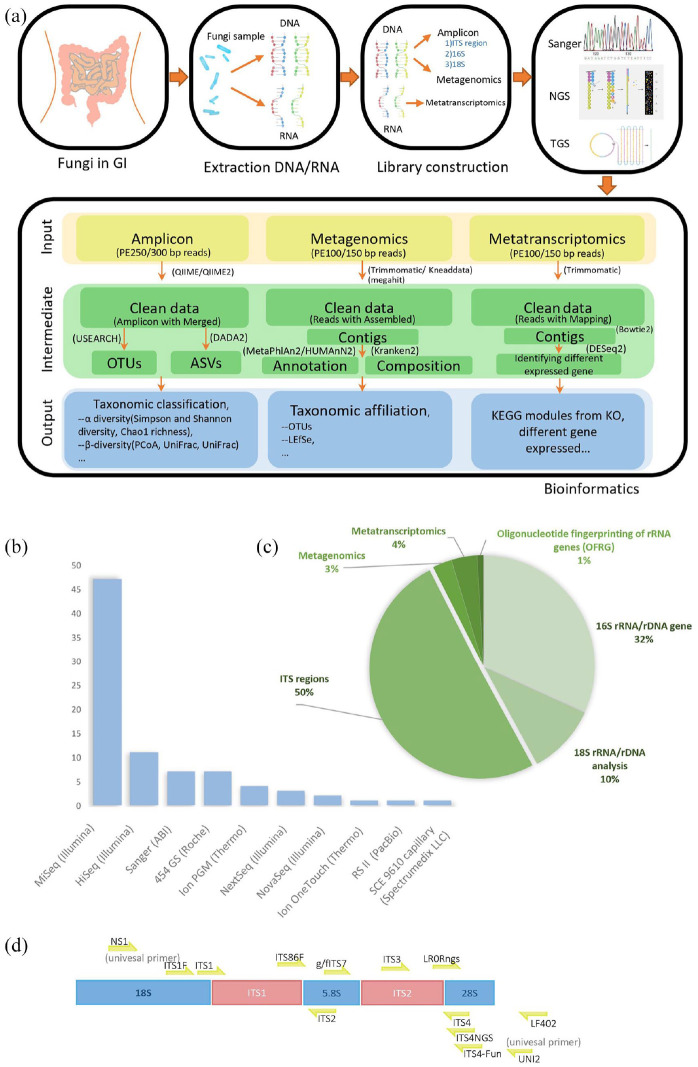
Characterization of the gut mycobiome has been complicated by the lack of comprehensive, well-curated, and high-resolution taxonomic annotation within fungal databases. Development of standardized methods for mycobiome analyses is critical to minimize variations across studies. A comprehensive literature search using broad searching criteria was conducted in the PubMed and Google Scholar databases. In short, 103 relevant literatures related to gut mycobiome are carefully selected based on the key words including “gut mycobiome”, “fungi”, “fungus”, “eukaryotes”, “fungal community”, “yeast,” and “sequencing”. The fungal study workflow contains extraction, library construction, and sequencing, and bioinformatic analysis was shown in Figure 1(a). There were 33 different commercial extraction kits used for fungi studies2,5,6,8–74 with 13 customized protocols.5,9,21,26,34,35,39,47,53,58,73–75 QIAamp DNA Mini Kit (Qiagen) was the most commonly used kit in 16 papers.2,6,8,16,22,23,33,43–47,49,54,61,67 Fast DNA Spin Kit (MP Biomedicals) was a common extraction kit for fungi.11,12,17,27,36,65,66,70
Figure 1.
The fungal study workflow contains extraction, libraries construction, sequencing, and bioinformatic analysis. (a) Initially DNA/RNA will be isolated from fungal samples. Then DNA or RNA extraction was performed following the protocol of amplicon, metagenomics, and metatranscriptomics according to strategy. Depending on strategy of research, different sequencer is selected from first-generation sequencing to third-generation sequencing. Generally, next generation sequencing (NGS) platform is chosen by higher output like Illumina sequencing by synthesis (SBS) sequencing technology, while-third generation sequencing has also become competitive for sequence full-length small genomes without assembly like Pacbio Single-molecule real-time (SMRT) sequencing. Analyze sequencing data using amplicon, metagenomics, and metatranscriptomics bioinformatics pipeline to analysis 16S rRNA/rDNA, DNA and RNA, respectively. (b) Among the studies involving sequencing methods, there are 10 sequencing platforms are used, in which NGS platform is used frequently and Illumina MiSeq platform is used the most in NGS method. (c) The fungal sequencing strategies are the study of ITS region, analysis of 16S rRNA/rDNA and 18S rRNA/rDNA gene. (d) ITS sequences and commonly and locatable primers used in fungal target region.
About 90% of users selected next generation sequencing (NGS) platform for higher throughput and faster turnaround time. Among all the sequencing platforms, Illumina MiSeq platform is the most widely utilized sequencer for 16S amplicon (Figure 1(b)). Widely used platform of NGS is Illumina which has high throughput, high quality, and high speed for large number of samples. Compared with Nanopore which belongs to one of the third-generation sequencing (TGS) technique, Illumina sequencers involve the following features: (1) it has implemented hundreds of metabarcoding of complex samples per run through dual-index method compared with less than 24 sequences in a Nanopore single run.76 (2) It has high-quality rate with 99.99% accuracy higher than approximately 90% of Nanopore.77 (3) For each sequencing run of massive fungal specimen, the MiSeq generates 24Gbp with PE300 and the NovaSeq 6000 generates 2190Gbp with PE150, respectively, but 10Gbp was generated by MinION in general research lab.78 With the continuous development of sequencing technology, new technologies have been used in research to identify fungi, especially Nanopore,79 and TGS sequencers have two significant advantages compared with Illumina. Initially, as an advantage of long reads sequencing in Nanopore, 0–200 kbp DNA could be covered, while 400–600 bp reads are sequenced in illumina sequencing. Long fragment could be reconstructed and identified as species precisely, decreasing the number of mislabeled species identified in illumine.80 Secondarily, researches use Nanopore to detect fungi for the advantage of amplicon-based and PCR-free metagenomics technology.77
For sequencing strategy, as indicated in Figure 1(c), 50% of articles sequenced the internal transcribed spacer (ITS) region, including ITS1 and ITS2, while 32% articles focused on 16s rRNA/rDNA gene and 10% papers on 18s rRNA/rDNA regions. Only 3% of the papers have performed metagenomics and 4% with metatranscriptomic fungal analysis. The fungal ribosomal region contains the internal transcribed spacers (ITS1 and ITS2) and the 5.8s, 18s, and 28s rRNA (Figure 1(d)). Currently, ITS1 and ITS211–16,51,58,65,66,73,75,81 are two major optional primers for human gut mycobiome study and the primer ITS2 is longer than primer ITS1 which has lower species resolution but fewer amplification and sequencing errors.82 Besides, the primer ITS1 is biased toward the amplification of Basidiomycetes and the primer ITS2 for the Ascomycetes.83 There were also reported ITS target regions outperformed compared with other subunit rRNA region in the human microbiome.28 Because ITS regions show not only fewer number of ribosomal gene copies but also less relative abundance of variants and breakpoints in various regions per unit in human rDNA.84 A preview results nonetheless support that the result of 18s rDNA PCR, which compares with one of ITS PCR, shows significant diagnostic and accurate identification of fungal pathogens in clinical sample from fresh biopsies and punctate and deep wound secretions.85 Compared with target sequence, metagenomics reads might be decreased biases from primer choices and increased community taxonomic characterization providing information of gene composition and the function.86
Overall, although significant advancements have been made on bioinformatics methodologies for gut mycobiome analyses, further development of pipelines and databases is required to define and characterize fungal communities more accurately.
The gastrointestinal tract mycobiome
Fecal samples are the most commonly used specimens to represent the human gut mycobiome. According to Human Microbiome Project, Saccharomyces, Malassezia, Candida, Cyberlindnera, Penicillium, Cladosporium, Aspergillus, Debaryomyces, Pichia, Clavispora, and Galactomyces are the most prevalent fungal genera in the human gut based on ITS2 and 18S rRNA sequencing.87 Cultivable gut mycobiota including Candida albicans, Candida glabrata, Candida deformans, Aspergillus glaucus, Cryptococcus saitoi, Cryptococcus neoformans, Lichtheimia ramosa, Mucor circinelloides, Pleurostomophora richardsiae, Rhodotorula mucilaginosa, Trichosporon asahii and Yarrowia lipolytica are frequently isolated from human feces.88,89 By pyrosequencing, Hoffmann et al.90 have found 12 fungal genera in fecal samples and Saccharomyces (present in 89% of the samples) was the most dominant. In another study of 45 healthy individuals, a total of 72 distinct operational taxonomic units (OTU) of gut fungi were found using ITS sequencing. Candida tropicalis, Geotrichum gigas, C. sake, and Phichia jadinii were the most commonly detected fungal microbiota from the fecal samples.91 Moreover, edible mushrooms (Agaricus bisporus) and plant pathogen (Epicoccum nigrum and Alternaria spp.) were also identified in the gastrointestinal tract of vegetarian individuals.43 Fecal-associated mycobiome are less persistent and varied over time and is easily influenced by food intake with “passing through” fungi.91,92 For instance, Saccharomyces cerevisiae and C. albicans were the two abundant fungal species in healthy individuals’ gut.92,93 However, in a controlled-diet experiment, Saccharomyces declined below the limit of detection in stool when the volunteer consumed a S. cerevisiae-free diet, and the levels of C. albicans in stool were dramatically reduced when the volunteer cleaned their teeth more frequently.71
Mucosa is also colonized by fungi. Compared with luminal-associated gut mycobiota, a more stable fungal community was found in the intestinal mucosa.6,94 There is limited research reporting the fungal composition of mucosa tissues as specimens are less accessible compared with fecal samples.95 A study applied 18s rDNA sequencing to study the mucosa-associated fungi of colonic biopsy tissues from 47 healthy individuals. They found that R. mucilaginosa, Galactomyces geotrichum, C. albicans, Septoria epambrosiae, Cryptococcus carnescens, Bullera crocea, C. dubliensi, Cladosporium cladospoirioides, Raciborskiomyces longisetosum, and Penicillium ialicum were the most detected in mucosal samples.96 Future studies comparing fecal and mucosa mycobiota and the contribution of their individual role in intestinal homeostasis and pathogenesis of disease are warranted.
Factors affecting the gut mycobiome
Despite emerging research on the gut mycobiome, a consensus healthy mycobiome has yet to be established. Several factors have been shown to be associated with alterations in mycobiota community composition including host genetics, gender, age, comorbidities, drugs, lifestyle factors including hygiene, socioeconomic status, diet, occupation, and the immune system.81 Interestingly, unlike the bacteriome, the mycobiome was shown a higher diversity in highly acidic stomach environment in piglet.97 This suggested that the imbalance of gut mycobiota may be related to stomach disease. Indeed, recent study reveals a perturbation of fungal compositional and ecological changes in gastric cancer development and C. albicans was characterized as a biomarker for gastric cancer.98 Given that early life factors are known to influence host microbiome status, data on early life gut mycobiome and factors influencing their development were reviewed.
Early life gut mycobiota
Early life fungal colonization has an impact on health outcomes of infants by training their immune system. Gut mycobiome in early life and factors that influence their development are shown in Figure 2. Infant fecal mycobiome was characterized by a low relative abundance of fungi composition and richness. Mother and infant fecal mycobiome were profiled in 15 mother–infant pairs in the first year of life by 18s rRNA gene amplicon sequencing.99 Phyla Basidiomycota and Ascomycota, with the genus Saccharomyces and the class Exobasidiomycetes, were the most represented fungal component found in meconium. In another study,100 fecal samples from 298 mother–offspring pairs were analyzed by ITS1 amplicon sequencing. S. cerevisiae was the most abundant species in the gut of infants from 1 year of age onwards. Besides, Debaryomyces hansenii prevailed up to 3 months in the infant’s feces. The 10-day, 1-year, and 2-year fecal samples from infants were richer in R. mucilaginosa, whereas the 3-month samples showed a high colonizer of C. parapsilosis. To further investigate early fungal community establishment, Ward et al.101 assessed the skin, oral, and anal mycobiomes of 16 infants over the first month of life and the anal and vaginal mycobiomes of 17 mothers using ITS2 amplicon sequencing. Infant mycobiomes varied by three body sites; skin mycobiome were dominated by C. tropicalis, C. parapsilosis, S. cerevisiae, C. albicans, and C. orthopsilosis; oral mycobiome was enriched with C. parapsilosis, C. tropicalis, S. cerevisiae, C. orthopsilosis, C. albicans, and Cladosporium velox; and anal mycobiome were colonized with C. parapsilosis, C. tropicalis, C. albicans, S. cerevisiae, C. orthopsilosis, and Cryptococcus pseudolongus, respectively.99 However, the infant’s mycobiome did not show a trajectory toward maturity within first 30 days of life, and this could be in part explained by the use of a relatively consistent food source of either breast milk or infant formula. Moreover, Kasper has demonstrated that mycobiota could be transferred from mothers to their offspring. In the study, five infants shared D. hansenii and S. cerevisiae observed in their mothers.100 Notably, the mother-to-child bacterial transmission patterns have been well profiled at strain level,102,103 whereas limited studies on mother-to-child vertical fungal transmission were reported. Overall, these studies all demonstrated that fungi could colonize in the neonatal gut at very early stage; nevertheless, how the mycobiome is shaped and how the succession occurs in the gut of neonates remained to be elucidated in the future.
Figure 2.
The gut mycobiome through life and the influencing factors in early life. Infants receive mycobiota colonization in the gut at birth, and fungal community increased with age in infancy but decreased when they grow up to young adults. 10 days after birth, Rhodotorula mucilaginosa and Debaryomyces hansenii predominated in the gut of infants, while Candida parapsilosis, C. tropicalis, C. albicans, Saccharomyces cerevisiae, C. orthopsilosis, and Cryptococcus pseudolongus enriched in the anus of infants in the first month. In the adult stage, C. albicans, S. cerevisiae, C. tropicalis, C. glabrata, C. deforans, and Aspergillus glaucus occupy the gut. In later years, Penicillium, Candida, Aspergillus, and Saccharomyces were dominant in the gut of elders. The major factors contributing to neonatal gut fungal communities were mode of feeding and mode of delivery. Compared with neonates by cesarean section, a higher level of Candida and Pleosporales, decreased level of Malassezia were shown in the gut of infants born via vaginal delivery. Moreover, infants could directly acquire mycobiota from breast milk feeding by their mothers, which contains abundant R. mucilaginosa and C. parapsilosis.
Factors affecting mycobiome in early life
Fungi are ubiquitous in the environment and the infant mycobiome may originate from the mother during birth, from the mother’s breast milk, parental skin, or anywhere else in the hospital or home environment with which the offspring come in contact with.100
Mode of delivery. In newborn babies, initial fungal colonization is largely dependent on delivery mode (Figure 2). Infants born vaginally obtained fungi that colonize the vagina, whereas infants born via Caesarian section acquire fungal species that are related to the skin.104–106 Azevedo et al.107 found that delivery mode might be associated with a higher carriage of oral fungi at a young age. Ward et al.108 highlighted that the mode of birth likely influence the fungal composition of the infant. It was believed that infants born vaginally appeared to have a higher level of Candida in their fecal mycobiome given that Candida was found predominantly in the vagina canal and they also had a more diverse mycobiome compared to those born by cesarean section (C-section) given the exposure of the infant to the mother’s fecal mycobiota. In contrast, infants delivered by C-section were colonized by a relatively higher abundance of Malassezia which originated from their mother’s skin. However, there are limited studies to support these hypotheses. In a separate study, Zhu et al.109 found that relative abundance of the order Pleosporales was higher in infants born by vaginal delivery than in those born by C-section.
Mode of feeding. Breast milk is considered the most ideal nutrition for infants offering protection from neonatal sepsis and facilitating infant growth and development.110 Several studies have shown that the mode of feeding impact on the gut bacterial microbiome.111,112 Boix-Amorós et al.113 characterized mycobiome composition in breast milk from healthy mothers and showed that the fungal composition of human breast milk was dominated by Malassezia (44%), followed by Candida (19%) and Saccharomyces (12%). The most abundant viable fungi detected were R. mucilaginosa and C. parapsilosis (Figure 2).24 Azevedo et al.107 suggested that the feeding mode of the infant (formula-fed or breast-fed infants) did not result in a difference in their oral yeast carriage. In contrast, Ward et al.108 suggested that feeding mode might affect the infant oral mycobiota. Currently, the relationship between infant and maternal gut mycobiome and whether breast milk leads to transmission of mycobiome from mother to infant remain controversial and require future studies. Future studies should focus on longitudinal tracking of the early-life mycobiota using mother–baby pairs while monitoring health outcomes during the first year of life. In addition, given the known interactions between the bacteria and fungi, future studies should also focus on the analysis of both bacteria and fungi to better characterize their structural and functional relationships, as well as their combined effects on health outcomes.
Gut mycobiota in elder subjects
Fungal community increased with age in infancy but decreased when they grow up to young adults (Figure 2).88 However, gut mycobiota alteration in elder subjects was less reported,114 and most of the older adults studied were reported with known diseases including hypertriglyceridemia (HG),115 Alzheimer’s disease (AD),51,116 and type 2 diabetes mellitus (T2DM).19 In Denmark, gut mycobiome analysis was performed in 100 elderly participants (70 with normotriglyceridemia versus 30 with hypertriglyceridemia) aged 65–81 years by ITS 2 amplicon sequencing on their fecal samples. Penicillium, Candida, and Aspergillus were the top three genera among the elderly Danes, and genus Penicillium was strongly correlated with the hypertriglyceridemia.115 Another study on 17 older subjects (11 mild cognitive impairment versus 6 cognitively normal) with average age of 64.6 years showed that Saccharomyces, Candida, and Aspergillus were major fungal genera in their fecal samples through ITS 1 sequencing. Higher abundance of Botrytis, Kazachstania, Phaeoacremonium, and Cladosporium and decreased abundance of Meyerozyma at genus level were found in the gut of patients with mild cognitive impairment compared with controls.51 Besides, Alonso et al.116 suggest that higher proportion of fungi were found in the brain tissue of older patients with AD compared with controls, and the percentages of Aspergillus and Candida were higher in elder controls than that in young controls by ITS 1 amplicon sequencing. However, to date no studies have fully profiled the longitudinal changes of mycobiota in the gut of old adults. Further studies are needed for a more complete picture of microbial development throughout the whole life.
The role of gut mycobiome and disease susceptibilities
Mycobiota and inflammatory bowel disease (IBD)
IBD, including ulcerative colitis (UC) and Crohn’s disease (CD), is a chronic and relapsing, inflammatory disorder of the gastrointestinal tract. Its pathogenesis involves a complex interaction among host genetics, host immunity, microbiome, and environmental exposures.117 The alterations of fecal and mucosal mycobiota in IBD in the pediatric and adult populations were summarized in Figure 3. As early as 1990, anti-Saccharomyces cerevisiae antibodies (ASCA) were found to be significantly higher in sera of patients with CD, which suggests that the gut mycobiota may be involved in the pathogenesis of CD.118 Several studies have also confirmed increased ASCA titers in sera of CD patients and considered it as a potential biomarker for disease diagnosis.119–121 C. albicans was reported to be one of the immunogens for ASCA.122 Standaert-Vitse et al.122 showed that the pathogenic C. albicans in human tissues may induce overexpression of ASCA major epitopes. It was also shown that patients with CD and their first-degree healthy relatives were more frequently and more heavily colonized by C. albicans than healthy controls based on traditional culture method.123 Moreover, in vivo experiments suggested that colonic inflammation facilitated C. albicans colonization in dextran sodium sulfate-induced mice colitis model.124,125
Figure 3.
Schematic summary of the gut mycobiome alterations in IBD. Compared with healthy individuals, the gut mycobiome of patients with IBD is characterized by increased fungal diversity and mycobiota dysbiosis accompanying with inflammation. Increased Basidiomycota/Ascomycota ratio and Candida albicans level, decreased proportion of Saccharomyces cerevisiae were shown in the fecal samples of patients with IBD. Similarly, increased fungus load including of C. albicans, C. tropicalis, C. glabrata, and Gibberella moniliformis were also found in the intestinal mucosa of patients with IBD. Nevertheless, pediatric patients with IBD showed a decreased fungal diversity in the gut. C. albicans, C. utilis, C. parapsilosis, S. cerevisiae, Clavispora lusitaniae, and Kluyveromyces marxianus prevailed in the feces of children, while increased abundance of Psathyrellaceae, Cortinariaceae, Psathyrella, and Gymnopilus were detected in their mucosa.
Based on denaturing gradient gel electrophoresis (DGGE) analysis, Ott et al.96 showed that fungal diversity was increased in patients with CD compared with controls. Sokol et al.126 reported a distinct fungal microbiota dysbiosis in stool of patients with IBD characterized by an increased Basidiomycota/Ascomycota ratio, decreased proportion of S. cerevisiae, and an increased C. albicans using ITS2 sequencing. Liguori et al. profiled the fungal composition in colonic mucosa of patients with CD and healthy subjects and found an increase in fungus load during CD flare. In addition, Cystofilobasidiaceae family, Dioszegia genera, and C. glabrata species were overrepresented in CD while Leptosphaeria and Trichosporon genera were reduced compared with healthy controls. Similarly, a higher richness in fungal diversity in the inflamed mucosa of treatment-naïve UC patients was detected when compared with noninflamed mucosa (Figure 3). However, after 5-ASA treatment, the richness of diversity declined.33 Furthermore, increased diversity of mucosal fungal microbiota was associated with expression of tumor necrosis factor (TNF)-α and interferon (IFN)-γ, which are key factors in the pathogenesis of IBD.4 In contrast to findings in adults, a reduced diversity in fungal microbiota was reported in the stool of pediatric patients with CD and UC.127 Besides, the represented fungal taxa detected in stool of pediatric patients with CD also differed from those of adults. Chehoud et al.127 showed that C. utilis and Candida parapsilosis were more common in stool samples of children with IBD than healthy children via ITS1 Region Gene Sequencing. El Mouzan et al.128 investigated fungal microbiota composition in treatment-naïve children with CD and showed that Psathyrellaceae, Cortinariaceae, Psathyrella, and Gymnopilus were significantly increased in the mucosa, while Cortinariaceae, Hymenochaete, and Gymnopilus were enriched in stool samples. In addition, Lewis et al. reported that S. cerevisiae, Clavispora lusitaniae, Candida utilis, C. albicans, and Kluyveromyces marxianus were present in abundance in stools from 86 pediatric subjects with CD compared to controls using shotgun metagenomic analysis (Figure 3). Pediatric and adult subjects with CD shared a small proportion of fungal taxa but there is also distinct differences in the fungal species.129 These difference may be due to different clinical phenotypes, drugs, and gut bacterial community between adults and pediatric IBD subjects. Numerous studies have highlighted the interaction between fungi and bacteria in IBD. Several correlations between bacteria and fungi were observed in a pediatric cohort, which varied between the healthy controls and patients with CD.129 Hoarau reported a significant interkingdom associations in bacteriome and mycobiome in 13 familial clusters, which included 6 bacterial-fungal genus and 13 species-level correlations.69 In a DSS-induced mice colitis model, Sovran et al.35 found that the beneficial effects of Saccharomyces boulardii and pathogenic effects of C. albicans on colitis severity could be eliminated through a broad-spectrum antibiotic cocktail treatment, including ampicillin, neomycin, metronidazole, and vancomycin. Overall, the gut fungal microbiota is altered in IBD but how fungi are involved in the occurrence and development of IBD remains unclear. Modulation of the fungal microbiota can be considered as a therapeutic approach for IBD as certain strains including S. boulardii and S. cerevisiae have shown therapeutic effects in human and murine IBD models. More studies on host–fungi interactions are necessary to enhance our understanding on how fungal microbiota interact with other constituents of the gut microbiota and the mechanisms of these relationships, in association with pathogenesis and development of IBD. Novel approaches, such as dietary interventions, probiotics, fecal microbiota transplantation (FMT), and antifungal metabolites to restore the dysbiotic states of intestinal mycobiota could be considered in the future; however, further studies are needed to assess their safety and efficacy.
Gut mycobiome in colorectal cancer (CRC)
Studies have shown that gut microbiota dysbiosis is associated with CRC.130 Besides bacteriome and virome, involvement of gut mycobiota in colorectal carcinogenesis has been increasingly recognized recently. Luan et al. compared mucosa-adherent fungal microbiota of paired biopsy samples of adenomas with adjacent normal colon tissue and found that gut fungal diversity in adenomas was decreased compared with adjacent normal tissues. At the genus level, the opportunistic pathogens Phoma and Candida accounted for an average of 45% of the fungal microbiota. They also found an OTU, assigned to Spizellomycetales, to be significantly enriched in adenomas compared with adjacent samples while one OTU, assigned to Paraglomerales, was significantly enriched in nonadvanced adenomas compared with adjacent tissues. More importantly, Fusarium and Trichoderma genera, respectively, were significantly enriched in adjacent biopsy samples in advanced adenoma compared with samples of nonadvanced adenoma which may be useful for early diagnosis.6 In a mycobiome study of 29 polyps, 74 CRC patients, and 28 healthy controls, fecal mycobiota composition differed among the three groups with a significant increase in the ratio of Ascomycota to Basidiomycota from control to CRC samples suggesting that changes of the gut mycobiota may be associated with progression of tumorigenesis. In addition, a higher fungal diversity was shown in late-stage CRC than in early-stage CRC.5 An increased proportion of opportunistic fungi Trichosporon and Malassezia was detected as the major contributor in the progression CRC.7 In a large multicenter study of 184 patients with CRC, 197 patients with adenoma and 204 control subjects, the ratio of Basidiomycota/Ascomycota was reported to be higher in CRC than in controls.8 In addition, six fungal features were detected to be enriched in CRC compared with controls at the genus level, including Malassezia, Moniliophthtora, Rhodotorula, Acremonium, Thielaviopsis, and Pisolithus.8 A small cohort comparing mycobiota in patients with colitis-associated cancer (CAC) and sporadic cancer showed no difference in the fungal microbiota composition.131 Overall, fungal dysbiosis has been found in adenomas, CRC, or CAC. Understanding fungal diversity and abundance in CRC is crucial to help elucidate the potential contribution of fungal species in colorectal tumorigenesis and delineation of fecal fungal dysbiosis might shed light on new opportunities for utilizing fungal species as noninvasive diagnostic biomarkers for CRC or its precursor lesions.
Gut mycobiome in metabolic diseases
Animal and human studies support a role of gut fungi in metabolic disease.17,19,132 In a mouse model, Heisel et al.133 investigated the effects of obesogenic diet on fungal composition by ITS2 sequencing and found that S. cerevisiae in the gut was significantly more abundant in lean mice than in mice fed with a high-fat diet. In an obese subject (body weight index = 48.9), a higher fecal fungal diversity was observed compared with healthy lean individuals.134 Furthermore, by using parallel aerobic culture-dependent approach on 24 subjects and ITS-based sequencing on 52 subjects, impaired fungal communities were reported in the gut of obese individuals, characterized by an increased presence of the phylum Ascomycota, the class Saccharomycetes, and the families Dipodascaceae and Saccharomycetaceae as compared with nonobese individuals.17,135 These findings suggest that obese subjects had altered gut fungi composition compared with their lean counterparts.
Mycobiome dysbiosis has also been reported in the gut of patients with diabetes mellitus.136–139 A higher diversity in fecal fungal species was found to distinguish children with type 1 diabetes mellitus (T1DM) from healthy controls.136 Moreover, a significantly increase in Candida colonization was found in the fecal samples of patients with type 1 and 2 diabetes compared with controls, which was verified by quantitative real-time PCR and medium cultures.137,138 In contrast, a separate study reported that C. albicans was significantly less prevalent in individuals with T1DM (62% of all strains identified) compared to control subjects (85% of all strains identified) based on medium cultures. Moreover, the fungal species isolated in this study were shown to be more resistant to antifungal drugs.136 Overall, the role of Candida in the pathogenesis of T1DM requires further confirmation. Using fungal ITS1 metagenomic sequencing on fecal samples from 10 healthy controls, 14 newly diagnosed T2DM, and 16 long-standing patients with T2DM, Bhute et al.139 found that opportunistic fungal pathogens such as Aspergillus and Candida were more abundant in newly diagnosed subjects compared with other groups. Jayasudha et al.19 used metagenomic sequencing to characterize fungal structure in fecal samples from 21 individuals with T2DM and 30 healthy controls. They found that patients with T2DM had increased fungal richness and evenness in the gut compared to healthy controls. An increase in the abundance of known human pathogens (genus Candida, Kodamaea, and Meyerozyma) and a decrease in the phylum Mucoromycota were noted in the gut of T2DM subjects.
Apart from altered gut mycobiome, altered fungal abundance in oral cavity was also reported in the subjects with diabetes mellitus. C. albicans and C. glabrata were both detected in the oral cavity of patients with T1DM.140,141 Nowakowska et al.141 suggested that C. glabrata resident in vagina and rectum was more than four times higher in women with diabetes than in nondiabetics. Significant associations were showed between glycemia and serum lipids with fungal abundance in patients with diabetes.137,141 Overall, these findings suggest that individuals with T2DM had gut mycobiome dysbiosis but whether this is the cause or consequence of the disease remains unknown. Most current studies are descriptive and longitudinal follow-up cohorts are important to delineate the importance of mycobiota dysbiosis in disease progression or complications in patients with diabetes mellitus.
Atherosclerosis is associated with metabolic diseases.142 Gut mycobiota dysbiosis has also been shown to be related with the development of carotid atherosclerosis.132 In a study of 33 subjects who had fecal samples collected for ITS amplicon sequencing, the subjects were divided into two groups of 12 nonobese subjects versus 21 obese subjects. The authors showed that the relative abundance of phylum Zygomycota and family Mucoraceae and Mucor racemosus in the fecal samples of participants were negatively correlated with the carotid intima-media thickness and to the risk of subclinical atherosclerosis (using the Framingham risk scores) in prospective follow-up.132 A separate study compared gut mycobiome of 48 coronary atherosclerosis patients with healthy controls, and there was no significant difference in the fungal composition of fecal samples between both groups with ITS sequencing. However, the abundance of Thermoascus and species Malassezia restricta in the patients with coronary atherosclerosis was significantly lower than in healthy individuals, and the decrease of M. restricta might have a close association with lipid metabolism disorder in atherosclerosis patients.143 Altogether these findings implied the emerging role of the gut mycobiota on atherosclerosis. As cardiovascular and metabolic risks vary in individuals and can also be influenced by external factors including diet and lifestyle, multiomics studies incorporating metabolomics, transcriptomics, and the overall gut microbiome are needed.
Therapeutic approaches targeting the gut mycobiome
Pathogenic fungi infections have been a major challenge to global health and leads to significant morbidity and mortality. Recently, study suggested that host adaptive immune system could suppress harmful fungal effectors of pathogenic fungi in the gut to improve their commensal fitness, so as to maintain intestinal homeostasis in healthy state.144 However, pathogens such as C. albicans, C. neoformans, and Aspergillus fumigatus could present a threat on immunocompromised individuals.145 In subjects with fungal infections, antifungal drugs are generally used as the first choice to clear the pathogens. Current antifungal drugs in clinical use consist of azoles (disruption of fungal ergosterol synthesis), polyenes (breakdown of membranes), echinocandins (inhibition of cell wall synthesis), and pyrimidines (inhibition of DNA synthesis and miscoding of RNA).146 Invasive candidiasis is a frequent health-care-associated fungal infection caused by C. albicans, C. glabrata, and C. tropicalis. Mortality of this fungi infection is up to 40% annually.147,148 Amphotericin B deoxycholate and micafungin treatments are effective to control the infections.149,150 Recently alterations in the gut mycobiome have been reported in fecal samples of patients admitted to hospital with coronavirus disease (COVID-19) with increased proportions of opportunistic fungal pathogens, C. albicans, C. auris, and A. flavus compared with controls. Importantly, two respiratory-associated fungal pathogens, A. flavus and Aspergillus niger, were detected in fecal samples from a subset of patients with COVID-19, even after clearance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from nasopharyngeal samples and resolution of respiratory symptoms.63 In sick individuals, Aspergillus sp. (A. fumigatus, A. penicillioides, A. niger and A. flavus) were detected in the endotracheal aspirate or bronchoalveolar lavage fluid imposing a high risk developing complicating invasive pulmonary aspergillosis. Antifungal therapies, such asvoriconazole and isavuconazole, were commonly used for preventing pulmonary aspergillosis and reducing mortality.151–153 However, toxicities and drug–drug interactions limit the utility of current antifungal drugs. Ubiquity use of antifungal drugs has contributed to our reliance on fungicides and the emergence of multidrug-resistant pathogenic fungi.154
Diet could be one of the major driving force influencing gut fungal mycobiota structure.90 A high-fat diet changed the gut fungal communities in murine models. It was reported that the abundances of the Alternaria, Saccharomyces, Septoriella, and Tilletiopsis genera were higher in mice administrated with normal chow compared with those fed with high-fat diet.133 Moreover, diet rich in plant-derived carbohydrate can support Candida in the gut of Indian subjects, while saturated fatty acid and coconut oil–rich diet negatively correlated with the load of Candida in the gut.155–157 In addition, dietary short-chain fatty acids correlated with decreased colonization of Aspergillus spp.90
Probiotics and prebiotics have been designed to provide health benefit. Multiple studies have identified several bacteria species with antifungal effects in vitro experiments, like Lactobacillus spp. and Bifidobacterium spp.158–160 In mice models, oral heat-killed Lactobacillus acidophilus (HKLA) and heat killed Lactobacillus casei could decrease the colonization of viable C. albicans in the tract.161 Colitis would induce overgrowth of opportunistic yeast pathogen C. glabrata in murine gut. Mice with colitis treated with β-glucans presented a decreased level of C. glabrata in the gut compared the colitis mice.162 Mycobiota regulation with probiotics and prebiotics in human study is rare, increasing evidences provide the possibility that the antagonistic relationships between bacterial and fungal species may decrease the perturbations and enhance the cross-talks in the gut to establish a balanced microbial community.163
Other than bacteria, fungal probiotics could also confer beneficial effects to support human health. S. cerevisiae and S. boulardii are the most common yeast. S. boulardii has shown the potential abilities to alleviate gastrointestinal disorder caused by Helicobacter pylori, Salmonella, and Clostridium difficile.164–166 In the gut of patients with IBD, the environment may favor fungi over bacteria, leading to both fungal and bacterial microbiome dysbiosis.129,167 Guslandi has demonstrated that S. boulardii could be used to treat IBD effectively, modulating the microbial composition in the gut.168,169 However, whether S. boulardii was involved in the restoration of mycobiome composition in the gut remains unknown. Understanding how probiotics interplay with fungal community to maintain or restore a stable ecosystem and improve human health remains a challenge for future work.
Fecal microbiome transplantation (FMT) is the transfer of stool from a healthy individual to the gastrointestinal tract of another individual to re-establish the balance of microbiome.170 Up to date, the effective application of FMT in treating patients with recurrent Clostridium difficile-associated diarrhea (CDI),171 IBD,172 irritable bowel syndrome (IBS)173 have been successively demonstrated. The efficacy of FMT is also associated with mycobiota. CDI is accompanied by outgrowth of C. albicans and dysbiosis in fungal diversity. High levels of Saccharomyces and Aspergillus in donor stool were reported in FMT responders, while nonresponders were related to a high abundance of C. albicans. The results implied that FMI is a promising therapy to improve gut mycobiome dysbiosis in CDI patients, while the fungi also have a tight association with FMT treatment outcomes.
Future perspective
Here we review the existing literature on the human gut mycobiome in order to provide a comprehensive insight into both the methodologies available to research the gut mycobiota and also to highlight the latest research findings. We also draw on research into what is known about the human mycobiome composition at diseases and early life in order to provide both comparative insight and productive direction for future studies in this burgeoning research area. Studies performed so far have shown potential links of mycobiome in health and diseases, it is important to demonstrate the causation rather than association. An increasing body of evidence suggests the alteration of interkingdom microbial community alterations contributing to the detrimental consequences to the host. To expand our knowledge and obtain deeper insight into the role of the microbiome in health and disease, future studies should characterize the different microorganisms (bacteria, fungi, and viruses) in the same sample types and inter-kingdom microbial community. Future challenge will be to understand these interactions on both the molecular level and in their complexity. To meet this challenge, improved approaches and collaborations among bacteriologists, mycologists, immunologists, and clinicians are required to develop the foundation for personalized microbiome medicine. A better understanding of the fungi–bacteria–host interactions can allow identification of patients who are at risk and improvement of patient care by tailored manipulation of the microbiota.
Footnotes
Author contributions: Lin Zhang and Hui Zhan were joint first authors.
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Siew C. Ng  https://orcid.org/0000-0002-6850-4454
https://orcid.org/0000-0002-6850-4454
Contributor Information
Lin Zhang, Center for Gut Microbiota Research, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Li Ka Shing Institute of Health Science, The Chinese University of Hong Kong, Shatin, Hong Kong, China; State Key Laboratory for Digestive disease, Institute of Digestive Disease, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Department of Medicine and Therapeutics, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Department of Anaesthesia and Intensive Care and Peter Hung Pain Research Institute, The Chinese University of Hong Kong, Shatin, Hong Kong, China.
Hui Zhan, Center for Gut Microbiota Research, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Li Ka Shing Institute of Health Science, The Chinese University of Hong Kong, Shatin, Hong Kong, China; State Key Laboratory for Digestive disease, Institute of Digestive Disease, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Department of Medicine and Therapeutics, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China.
Wenye Xu, Center for Gut Microbiota Research, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Li Ka Shing Institute of Health Science, The Chinese University of Hong Kong, Shatin, Hong Kong, China; State Key Laboratory for Digestive disease, Institute of Digestive Disease, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Department of Medicine and Therapeutics, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China.
Shuai Yan, Center for Gut Microbiota Research, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Li Ka Shing Institute of Health Science, The Chinese University of Hong Kong, Shatin, Hong Kong, China; State Key Laboratory for Digestive disease, Institute of Digestive Disease, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Department of Anaesthesia and Intensive Care and Peter Hung Pain Research Institute, The Chinese University of Hong Kong, Shatin, Hong Kong, China.
Siew C. Ng, Center for Gut Microbiota Research, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Li Ka Shing Institute of Health Science, The Chinese University of Hong Kong, Shatin, Hong Kong, China; State Key Laboratory for Digestive disease, Institute of Digestive Disease, The Chinese University of Hong Kong, Shatin, Hong Kong, China; Department of Medicine and Therapeutics, Faculty of Medicine, The Chinese University of Hong Kong, 9/F, Lui Che Woo Clinical Sciences Building, Prince of Wales Hospital, Shatin, Hong Kong, China; Microbiota I-Center (MagIC) Limited, The Chinese University of Hong Kong, Shatin, Hong Kong, China.
References
- 1.Iliev ID, Leonardi I.Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol 2017; 17: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soverini M, Turroni S, Biagi E, et al. HumanMycobiomeScan: a new bioinformatics tool for the characterization of the fungal fraction in metagenomic samples. BMC Genomics 2019; 20: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liguori G, Lamas B, Richard ML, et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J Crohns Colitis 2016; 10: 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, Wang C, Tang C, et al. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clin Gastroenterol 2014; 48: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Zuo T, Yeoh YK, et al. Longitudinal dynamics of gut bacteriome, mycobiome and virome after fecal microbiota transplantation in graft-versus-host disease. Nat Commun 2021; 12: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luan C, Xie L, Yang X, et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep 2015; 5: 7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao R, Kong C, Li H, et al. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur J Clin Microbiol Infect Dis 2017; 36: 2457–2468. [DOI] [PubMed] [Google Scholar]
- 8.Coker OO, Nakatsu G, Dai RZ, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019; 68: 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botschuijver S, Roeselers G, Levin E, et al. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology 2017; 153: 1026–1039. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Dai B, Tang Y, et al. Altered bacterial-fungal interkingdom networks in the guts of ankylosing spondylitis patients. Msystems 2019; 4: e00176-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang HY, Pan LY, Zhang X, et al. Altered gut bacterial–fungal interkingdom networks in patients with current depressive episode. Brain Behav 2020; 10: e01677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Pan L-y, Zhang Z, et al. Analysis of gut mycobiota in first-episode, drug-naïve Chinese patients with schizophrenia: a pilot study. Behav Brain Res 2020; 379: 112374. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Leonardi I, Semon A, et al. Response to fungal dysbiosis by gut-resident CX3CR1+ mononuclear phagocytes aggravates allergic airway disease. Cell Host Microbe 2018; 24: 847–856.e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonardi I, Li X, Semon A, et al. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 2018; 359: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limon JJ, Tang J, Li D, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 2019; 25: 377–388.e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012; 336: 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez MM, Pérez D, Chaves FJ, et al. Obesity changes the human gut mycobiome. Sci Rep 2015; 5: 14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinert RE, Rehman A, Souto Lima EJ, et al. Roux-en-Y gastric bypass surgery changes fungal and bacterial microbiota in morbidly obese patients – a pilot study. PLoS One 2020; 15: e0236936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayasudha R, Das T, Kalyana Chakravarthy S, et al. Gut mycobiomes are altered in people with type 2 diabetes mellitus and diabetic retinopathy. PLoS One 2020; 15: e0243077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al Bataineh MT, Dash NR, Lassen PB, et al. Revealing links between gut microbiome and its fungal community in type 2 diabetes mellitus among Emirati subjects: a pilot study. Sci Rep 2020; 10: 9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Merwe M, Sharma S, Caldwell JL, et al. Time of feeding alters obesity-associated parameters and gut bacterial communities, but not fungal populations, in C57bl/6 male mice. Curr Dev Nutr 2020; 4: nzz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arrieta M-C, Arévalo A, Stiemsma L, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol 2018; 142: 424–434.e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honkanen J, Vuorela A, Muthas D, et al. Fungal dysbiosis and intestinal inflammation in children with beta-cell autoimmunity. Front Immunol 2020; 11: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boix-Amorós A, Puente-Sánchez F, Du Toit E, et al. Mycobiome profiles in breast milk from healthy women depend on mode of delivery, geographic location, and interaction with bacteria. Appl Environ Microbiol 2019; 85: e02994-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heisel T, Nyaribo L, Sadowsky MJ, et al. Breastmilk and NICU surfaces are potential sources of fungi for infant mycobiomes. Fungal Genet Biol 2019; 128: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willis KA, Purvis JH, Myers ED, et al. Fungi form interkingdom microbial communities in the primordial human gut that develop with gestational age. FASEB J 2019; 33: 12825–12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James SA, Phillips S, Telatin A, et al. Preterm infants harbour a rapidly changing mycobiota that includes Candida pathobionts. J Fungi 2020; 6: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doron I, Leonardi I, Iliev ID.Profound mycobiome differences between segregated mouse colonies do not influence Th17 responses to a newly introduced gut fungal commensal. Fungal Genet Biol 2019; 127: 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller KD, Zhang H, Serrano CR, et al. Gastrointestinal microbiota alteration induced by Mucor circinelloides in a murine model. J Microbiol 2019; 57: 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panpetch W, Somboonna N, Palasuk M, et al. Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLoS ONE 2019; 14: e0210798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skalski JH, Limon JJ, Sharma P, et al. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog 2018; 14: e1007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arunasri K, Mahesh M, Sai Prashanthi G, et al. Mycobiome changes in the vitreous of post fever retinitis patients. PLoS ONE 2020; 15: e0242138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jun X, Ning C, Yang S, et al. Alteration of fungal microbiota after 5-ASA treatment in UC patients. Inflamm Bowel Dis 2020; 26: 380–390. [DOI] [PubMed] [Google Scholar]
- 34.Di Paola M, Rizzetto L, Stefanini I, et al. Comparative immunophenotyping of Saccharomyces cerevisiae and Candida spp. strains from Crohn’s disease patients and their interactions with the gut microbiome. J Transl Autoimmun 2020; 3: 100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sovran B, Planchais J, Jegou S, et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 2018; 6: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong G, Li Y, Yang M, et al. Gut fungal dysbiosis and altered bacterial-fungal interaction in patients with diarrhea-predominant irritable bowel syndrome: an explorative study. Neurogastroenterol Motil 2020; 32: e13891. [DOI] [PubMed] [Google Scholar]
- 37.Stewart DB, Wright JR, Fowler M, et al. Integrated meta-omics reveals a fungus-associated bacteriome and distinct functional pathways in Clostridioides difficile infection. mSphere 2019; 4: e00454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee PK, Sendid B, Hoarau G, et al. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 2015; 12: 77–87. [DOI] [PubMed] [Google Scholar]
- 39.Fiedorová K, Radvanský M, Bosák J, et al. Bacterial but not fungal gut microbiota alterations are associated with common variable immunodeficiency (CVID) phenotype. Front Immunol 2019; 10: 1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Annavajhala MK, Khan SD, Sullivan SB, et al. Oral and gut microbial diversity and immune regulation in patients with HIV on antiretroviral therapy. mSphere 2020; 5: e00798-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu H, Duan Y, Lang S, et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J Hepatol 2020; 72: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcelino VR, Holmes EC, Sorrell TC.The use of taxon-specific reference databases compromises metagenomic classification. BMC Genomics 2020; 21: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suhr MJ, Banjara N, Hallen-Adams HE.Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol 2016; 62: 209–215. [DOI] [PubMed] [Google Scholar]
- 44.Gouba N, Raoult D, Drancourt M. Eukaryote culturomics of the gut reveals new species. PLoS One 2014; 9: e106994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frau A, Kenny JG, Lenzi L, et al. DNA extraction and amplicon production strategies deeply influence the outcome of gut mycobiome studies. Sci Rep 2019; 9: 9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamad I, Ranque S, Azhar EI, et al. Culturomics and amplicon-based metagenomic approaches for the study of fungal population in human gut microbiota. Sci Rep 2017; 7: 16788. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Fiedorová K, Radvanský M, Němcová E, et al. The impact of DNA extraction methods on stool bacterial and fungal microbiota community recovery. Front Microbiol 2019; 10: 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arfken AM, Frey JF, Ramsay TG, et al. Yeasts of burden: exploring the mycobiome–bacteriome of the piglet GI tract. Front Microbiol 2019; 10: 2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huseyin CE, Rubio RC, O’Sullivan O, et al. The fungal frontier: a comparative analysis of methods used in the study of the human gut mycobiome. Front Microbiol 2017; 8: 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aykut B, Pushalkar S, Chen R, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019; 574: 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagpal R, Neth BJ, Wang S, et al. Gut mycobiome and its interaction with diet, gut bacteria and Alzheimer’s disease markers in subjects with mild cognitive impairment: a pilot study. EBioMedicine 2020; 59: 102950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seelbinder B, Chen J, Brunke S, et al. Antibiotics create a shift from mutualism to competition in human gut communities with a longer-lasting impact on fungi than bacteria. Microbiome 2020; 8: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol 2016; 1: 16113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Jurdi N, Filali-Mouhim A, Salem I, et al. Gastrointestinal microbiome and mycobiome changes during autologous transplantation for multiple myeloma: results of a prospective pilot study. Biol Blood Marrow Transplant 2019; 25: 1511–1519. [DOI] [PubMed] [Google Scholar]
- 55.Hagan T, Cortese M, Rouphael N, et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 2019; 178: 1313–1328.e1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rush TA, Puech-Pagès V, Bascaules A, et al. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat Commun 2020; 11: 3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calusinska M, Marynowska M, Bertucci M, et al. Integrative omics analysis of the termite gut system adaptation to Miscanthus diet identifies lignocellulose degradation enzymes. Commun Biol 2020; 3: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motooka D, Fujimoto K, Tanaka R, et al. Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front Microbiol 2017; 8: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bajaj JS, Liu EJ, Kheradman R, et al. Fungal dysbiosis in cirrhosis. Gut 2018; 67: 1146–1154. [DOI] [PubMed] [Google Scholar]
- 60.Wu L, Zeng T, Deligios M, et al. Age-related variation of bacterial and fungal communities in different body habitats across the young, elderly, and centenarians in Sardinia. mSphere 2020; 5: e00558-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahnic A, Rupnik M.Different host factors are associated with patterns in bacterial and fungal gut microbiota in Slovenian healthy cohort. PLoS One 2018; 13: e0209209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagen LH, Brooke CG, Shaw CA, et al. Proteome specialization of anaerobic fungi during ruminal degradation of recalcitrant plant fiber. ISME J 2021; 15: 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuo T, Zhan H, Zhang F, et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology 2020; 159: 1302–1310.e1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arfken AM, Frey JF, Summers KL.Temporal dynamics of the gut bacteriome and mycobiome in the weanling pig. Microorganisms 2020; 8: 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 2010; 6: e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee PK, Chandra J, Retuerto M, et al. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 2014; 10: e1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y, Chen Z, Guo R, et al. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 2011; 70: 492–498. [DOI] [PubMed] [Google Scholar]
- 68.Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA 2012; 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoarau G, Mukherjee P, Gower-Rousseau C, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 2016; 7: e01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valinsky L, Della Vedova G, Jiang T, et al. Oligonucleotide fingerprinting of rRNA genes for analysis of fungal community composition. Appl Environ Microbiol 2002; 68: 5999–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Auchtung TA, Fofanova TY, Stewart CJ, et al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 2018; 3: e00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fiers WD, Gao IH, Iliev ID.Gut mycobiota under scrutiny: fungal symbionts or environmental transients? Curr Opin Microbiol 2019; 50: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreno-Sabater A, Autaa G, Sterlin D, et al. Systemic anti-commensal response to fungi analyzed by flow cytometry is related to gut mycobiome ecology. Microbiome 2020; 8: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris VC, Haak BW, Handley SA, et al. Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: a human, randomized-control proof-of-concept trial. Cell Host Microbe 2018; 24: 197–207.e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosshart SP, Herz J, Vassallo BG, et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 2019; 365: eaaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morrison GA, Fu J, Lee GC, et al. Nanopore sequencing of the fungal intergenic spacer sequence as a potential rapid diagnostic assay. J Clin Microbiol 2020; 58: e01972-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loit K, Adamson K, Bahram M, et al. Relative performance of MinION (Oxford Nanopore Technologies) versus Sequel (Pacific Biosciences) third-generation sequencing instruments in identification of agricultural and forest fungal pathogens. Appl Environ Microbiol 2019; 85: e01368-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leidenfrost RM, Pöther D-C, Jäckel U, et al. Benchmarking the MinION: evaluating long reads for microbial profiling. Sci Rep 2020; 10: 5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nilsson RH, Anslan S, Bahram M, et al. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol 2019; 17: 95–109. [DOI] [PubMed] [Google Scholar]
- 80.D’Andreano S, Cuscó A, Francino O.Rapid and real-time identification of fungi up to species level with long amplicon nanopore sequencing from clinical samples. Biol Methods Protoc 2020; 6: bpaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cui L, Morris A, Ghedin E.The human mycobiome in health and disease. Genome Med 2013; 5: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li S, Deng Y, Wang Z, et al. Exploring the accuracy of amplicon-based internal transcribed spacer markers for a fungal community. Mol Ecol Resour 2020; 20: 170–184. [DOI] [PubMed] [Google Scholar]
- 83.Bellemain E, Carlsen T, Brochmann C, et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 2010; 10: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smirnov E, Chmúrčiaková N, Liška F, et al. Variability of human rDNA. Cells 2021; 10: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wagner K, Springer B, Pires V, et al. Molecular detection of fungal pathogens in clinical specimens by 18S rDNA high-throughput screening in comparison to ITS PCR and culture. Sci Rep 2018; 8: 6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao B, Chi L, Zhu Y, et al. An introduction to next generation sequencing bioinformatic analysis in gut microbiome studies. Biomolecules 2021; 11: 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nash AK, Auchtung TA, Wong MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017; 5: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Strati F, Di Paola M, Stefanini I, et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol 2016; 7: 1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perfect J, Casadevall A.Molecular principles of fungal pathogenesis. Washington, DC: ASM Press, 2006. [Google Scholar]
- 90.Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 2013; 8: e66019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hallen-Adams HE, Kachman SD, Kim J, et al. Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecol 2015; 15: 9–17. [Google Scholar]
- 92.Raimondi S, Amaretti A, Gozzoli C, et al. Longitudinal survey of fungi in the human gut: its profiling, phenotyping, and colonization. Front Microbiol 2019; 10: 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khatib R, Riederer KM, Ramanathan J, et al. Faecal fungal flora in healthy volunteers and inpatients. Mycoses 2001; 44: 151–156. [DOI] [PubMed] [Google Scholar]
- 94.Sonnenburg JL, Angenent LT, Gordon JI.Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol 2004; 5: 569–573. [DOI] [PubMed] [Google Scholar]
- 95.Sam QH, Chang MW, Chai LYA. The fungal mycobiome and its interaction with gut bacteria in the host. Int J Mol Sci 2017; 18: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol 2008; 43: 831–841. [DOI] [PubMed] [Google Scholar]
- 97.Merchant HA, McConnell EL, Liu F, et al. Assessment of gastrointestinal pH, fluid and lymphoid tissue in the guinea pig, rabbit and pig, and implications for their use in drug development. Eur J Pharm Sci 2011; 42: 3–10. [DOI] [PubMed] [Google Scholar]
- 98.Zhong M, Xiong Y, Zhao J, et al. Candida albicans disorder is associated with gastric carcinogenesis. Theranostics 2021; 11: 4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wampach L, Heintz-Buschart A, Hogan A, et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol 2017; 8: 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schei K, Avershina E, Øien T, et al. Early gut mycobiota and mother-offspring transfer. Microbiome 2017; 5: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ward TL, Dominguez-Bello MG, Heisel T, et al. Development of the human mycobiome over the first month of life and across body sites. mSystems 2018; 3: e00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yassour M, Jason E, Hogstrom LJ, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 2018; 24: 146–154.e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ferretti P, Pasolli E, Tett A, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018; 24: 133–145.e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mueller NT, Bakacs E, Combellick J, et al. The infant microbiome development: mom matters. Trends Mol Med 2015; 21: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010; 107: 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grice EA, Segre JA.The skin microbiome. Nat Rev Microbiol 2011; 9: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Azevedo MJ, de Lurdes Pereira M, Araujo R, et al. Influence of delivery and feeding mode in oral fungi colonization – a systematic review. Microb Cell 2020; 7: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ward TL, Knights D, Gale CA.Infant fungal communities: current knowledge and research opportunities. BMC Med 2017; 15: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhu T, Duan Y-Y, Kong F-Q, et al. Dynamics of skin mycobiome in infants. Front Microbiol 2020; 11: 1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Walker A.Breast milk as the gold standard for protective nutrients. J Pediatr 2010; 156(Suppl. 2): S3–S7. [DOI] [PubMed] [Google Scholar]
- 111.Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016; 8: 343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shao Y, Forster SC, Tsaliki E, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019; 574: 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boix-Amorós A, Martinez-Costa C, Querol A, et al. Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci Rep 2017; 7: 13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barrera-Vázquez OS, Gomez-Verjan JC.The unexplored world of human virome, mycobiome, and archaeome in aging. J Gerontol A Biol Sci Med Sci 2020; 75: 1834–1837. [DOI] [PubMed] [Google Scholar]
- 115.Ahmad HF, Mejia JLC, Krych L, et al. Gut mycobiome dysbiosis is linked to hypertriglyceridemia among home dwelling elderly Danes. bioRxiv. Epub ahead of print 17 April 2020. DOI: 10.1101/2020.04.16.044693. [DOI] [Google Scholar]
- 116.Alonso R, Pisa D, Fernández-Fernández AM, et al. Infection of fungi and bacteria in brain tissue from elderly persons and patients with Alzheimer’s disease. Front Aging Neurosci 2018; 10: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ye Y, Pang Z, Chen W, et al. The epidemiology and risk factors of inflammatory bowel disease. Int J Clin Exp Med 2015; 8: 22529–22542. [PMC free article] [PubMed] [Google Scholar]
- 118.McKenzie H, Main J, Pennington C, et al. Antibody to selected strains of Saccharomyces cerevisiae (baker’s and brewer’s yeast) and Candida albicans in Crohn’s disease. Gut 1990; 31: 536–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Annese V, Andreoli A, Andriulli A, et al. Familial expression of anti-Saccharomyces cerevisiae mannan antibodies in Crohn’s disease and ulcerative colitis: a GISC study. Am J Gastroenterol 2001; 96: 2407–2412. [DOI] [PubMed] [Google Scholar]
- 120.Ruemmele FM, Targan SR, Levy G, et al. Diagnostic accuracy of serological assays in pediatric inflammatory bowel disease. Gastroenterology 1998; 115: 822–829. [DOI] [PubMed] [Google Scholar]
- 121.Sendid B, Quinton J-F, Charrier G, et al. Anti-Saccharomyces cerevisiae mannan antibodies in familial Crohn’s disease. Am J Gastroenterol 1998; 93: 1306–1310. [DOI] [PubMed] [Google Scholar]
- 122.Standaert-Vitse A, Jouault T, Vandewalle P, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology 2006; 130: 1764–1775. [DOI] [PubMed] [Google Scholar]
- 123.Standaert-Vitse A, Sendid B, Joossens M, et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am J Gastroenterol 2009; 104: 1745–1753. [DOI] [PubMed] [Google Scholar]
- 124.Panpetch W, Hiengrach P, Nilgate S, et al. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes 2020; 11: 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jawhara S, Thuru X, Standaert-Vitse A, et al. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J Infect Dis 2008; 197: 972–980. [DOI] [PubMed] [Google Scholar]
- 126.Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2017; 66: 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chehoud C, Albenberg LG, Judge C, et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis 2015; 21: 1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.El Mouzan M, Wang F, Al Mofarreh M, et al. Fungal microbiota profile in newly diagnosed treatment-naive children with Crohn’s disease. J Crohns Colitis 2017; 11: 586–592. [DOI] [PubMed] [Google Scholar]
- 129.Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015; 18: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Louis P, Hold GL, Flint HJ.The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014; 12: 661–672. [DOI] [PubMed] [Google Scholar]
- 131.Richard ML, Liguori G, Lamas B, et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018; 9: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chacon M, Lozano-Bartolome J, Portero-Otin M, et al. The gut mycobiome composition is linked to carotid atherosclerosis. Benef Microbes 2018; 9: 185–198. [DOI] [PubMed] [Google Scholar]
- 133.Heisel T, Montassier E, Johnson A, et al. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere 2017; 2: e00351-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gouba N, Raoult D, Drancourt M. Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PLoS ONE 2013; 8: e59474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Borges FM, de Paula TO, Sarmiento MRA, et al. Fungal diversity of human gut microbiota among eutrophic, overweight, and obese individuals based on aerobic culture-dependent approach. Curr Microbiol 2018; 75: 726–735. [DOI] [PubMed] [Google Scholar]
- 136.Kowalewska B, Zorena K, Szmigiero-Kawko M, et al. Higher diversity in fungal species discriminates children with type 1 diabetes mellitus from healthy control. Patient Prefer Adherence 2016; 10: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gosiewski T, Salamon D, Szopa M, et al. Quantitative evaluation of fungi of the genus Candida in the feces of adult patients with type 1 and 2 diabetes-a pilot study. Gut Pathog 2014; 6: 43–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Soyucen E, Gulcan A, Aktuglu-Zeybek AC, et al. Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatr Int 2014; 56: 336–343. [DOI] [PubMed] [Google Scholar]
- 139.Bhute SS, Suryavanshi MV, Joshi SM, et al. Gut microbial diversity assessment of Indian type-2-diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Front Microbiol 2017; 8: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aly F, Blackwell C, MacKenzie D, et al. Chronic atrophic oral candidiasis among patients with diabetes mellitus–role of secretor status. Epidemiol Infect 1991; 106: 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nowakowska D, Kurnatowska A, Stray-Pedersen B, et al. Species distribution and influence of glycemic control on fungal infections in pregnant women with diabetes. J Infect 2004; 48: 339–346. [DOI] [PubMed] [Google Scholar]
- 142.Marín de Evsikova C, Raplee ID, Lockhart J, et al. The transcriptomic toolbox: resources for interpreting large gene expression data within a precision medicine context for metabolic disease atherosclerosis. J Pers Med 2019; 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu J, Zhang Y, Wang X, et al. Changes and roles of intestinal fungal microbiota in coronary heart disease complicated with nonalcoholic fatty liver disease. Am J Transl Res 2020; 12: 3445–3460. [PMC free article] [PubMed] [Google Scholar]
- 144.Ost KS, O’Meara TR, Stephens WZ, et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature 2021; 596: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med 2012; 4: 165rv113. [DOI] [PubMed] [Google Scholar]
- 146.Robbins N, Wright GD, Cowen LE.Antifungal drugs: the current armamentarium and development of new agents. In: Heitman J, Stukenbrock EH, Gow NAR, et al. (eds) The fungal kingdom. New York: Wiley, 2017, pp. 903–922. [Google Scholar]
- 147.Kullberg BJ, Arendrup MC.Invasive candidiasis. N Engl J Med 2015; 373: 1445–1456. [DOI] [PubMed] [Google Scholar]
- 148.Peçanha-Pietrobom PM, Colombo AL.Mind the gaps: challenges in the clinical management of invasive candidiasis in critically ill patients. Curr Opin Infect Dis 2020; 33: 441–448. [DOI] [PubMed] [Google Scholar]
- 149.Pappas PG, Kauffman CA, Andes D, et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 2009; 48: 503–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pappas PG, Rotstein CM, Betts RF, et al. Micafungin versus caspofungin for treatment of candidemia and other forms of invasive candidiasis. Clin Infect Dis 2007; 45: 883–893. [DOI] [PubMed] [Google Scholar]
- 151.Prattes J, Valentin T, Hoenigl M, et al. Invasive pulmonary aspergillosis complicating COVID-19 in the ICU—a case report. Med Mycol Case Rep 2021; 31: 2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bartoletti M, Pascale R, Cricca M, et al. Epidemiology of invasive pulmonary aspergillosis among COVID-19 intubated patients: a prospective study. Clin Infect Dis. Epub ahead of print 28 July 2020. DOI: 10.1093/cid/ciaa1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sharma A, Hofmeyr A, Bansal A, et al. COVID-19 associated pulmonary aspergillosis (CAPA): an Australian case report. Med Mycol Case Rep 2021; 31: 6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fisher MC, Hawkins NJ, Sanglard D, et al. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018; 360: 739–742. [DOI] [PubMed] [Google Scholar]
- 155.Vargas SL, Patrick C, Ayers G, et al. Modulating effect of dietary carbohydrate supplementation on Candida albicans colonization and invasion in a neutropenic mouse model. Infect Immun 1993; 61: 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gunsalus KT, Tornberg-Belanger SN, Matthan NR, et al. Manipulation of host diet to reduce gastrointestinal colonization by the opportunistic pathogen Candida albicans. mSphere 2016; 1: e00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pareek S, Kurakawa T, Das B, et al. Comparison of Japanese and Indian intestinal microbiota shows diet-dependent interaction between bacteria and fungi. NPJ Biofilms Microbiomes 2019; 5: 37–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Payne S, Gibson G, Wynne A, et al. In vitro studies on colonization resistance of the human gut microbiota to Candida albicans and the effects of tetracycline and Lactobacillus plantarum LPK. Curr Issues Intest Microbiol 2003; 4: 1–8. [PubMed] [Google Scholar]
- 159.de Barros PP, Scorzoni L, Ribeiro FC, et al. Lactobacillus paracasei 28.4 reduces in vitro hyphae formation of Candida albicans and prevents the filamentation in an experimental model of Caenorhabditis elegans. Microb Pathog 2018; 117: 80–87. [DOI] [PubMed] [Google Scholar]
- 160.Ghazvini RD, Kouhsari E, Zibafar E, et al. Antifungal activity and aflatoxin degradation of Bifidobacterium bifidum and Lactobacillus fermentum against toxigenic Aspergillus parasiticus. Open Microbiol J 2016; 10: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wagner R, Pierson C, Warner T, et al. Probiotic effects of feeding heat-killed Lactobacillus acidophilus and Lactobacillus casei to Candida albicans-colonized immunodeficient mice. J Food Prot 2000; 63: 638–644. [DOI] [PubMed] [Google Scholar]
- 162.Charlet R, Bortolus C, Barbet M, et al. A decrease in anaerobic bacteria promotes Candida glabrata overgrowth while β-glucan treatment restores the gut microbiota and attenuates colitis. Gut Pathogens 2018; 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.García-Bayona L, Comstock LE.Bacterial antagonism in host-associated microbial communities. Science 2018; 361: eaat2456. [DOI] [PubMed] [Google Scholar]
- 164.Corthier G, Dubos F, Ducluzeau R.Prevention of Clostridium difficile induced mortality in gnotobiotic mice by Saccharomyces boulardii. Can J Microbiol 1986; 32: 894–896. [DOI] [PubMed] [Google Scholar]
- 165.Duman DG, Bor S, Ozütemiz O, et al. Efficacy and safety of Saccharomyces boulardii in prevention of antibiotic-associated diarrhoea due to Helicobacterpylori eradication. Eur J Gastroenterol Hepatol 2005; 17: 1357–1361. [DOI] [PubMed] [Google Scholar]
- 166.Martins FS, Vieira AT, Elian SD, et al. Inhibition of tissue inflammation and bacterial translocation as one of the protective mechanisms of Saccharomyces boulardii against Salmonella infection in mice. Microbes Infect 2013; 15: 270–279. [DOI] [PubMed] [Google Scholar]
- 167.Ni J, Wu GD, Albenberg L, et al. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol 2017; 14: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Guslandi M, Mezzi G, Sorghi M, et al. Saccharomyces boulardii in maintenance treatment of Crohn’s disease. Dig Dis Sci 2000; 45: 1462–1464. [DOI] [PubMed] [Google Scholar]
- 169.Guslandi M, Giollo P, Testoni PA.A pilot trial of Saccharomyces boulardii in ulcerative colitis. Eur J Gastroenterol Hepatol 2003; 15: 697–698. [DOI] [PubMed] [Google Scholar]
- 170.Zhang F, Luo W, Shi Y, et al. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol 2012; 107: 1755; author reply p. 1755–1756. [DOI] [PubMed] [Google Scholar]
- 171.Bakken JS, Borody T, Brandt LJ, et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 2011; 9: 1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Costello SP, Hughes PA, Waters O, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA 2019; 321: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu D, Chen VL, Steiner CA, et al. Efficacy of fecal microbiota transplantation in irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol 2019; 114: 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]