Abstract
Introduction
KRAS mutations are detected in 20% to 30% of NSCLC. However, KRAS mutation subtypes may differently influence the outcome of patients with advanced NSCLC.
Methods
In the Biomarkers France study, 4894 KRAS mutations (26.2%) were detected in 4634 patients from the 17,664 enrolled patients with NSCLC. Survival and treatment data on noncurative stage III to IV NSCLC were available for 901 patients. First- and second-line treatment effects on progression-free survival and overall survival were analyzed according to the KRAS mutations subtype.
Results
Over 95% of patients with KRAS mutation were smokers or former smokers who were white (99.5%), presenting with adenocarcinoma (82.5%). The most common KRAS mutation subtype was G12C (374 patients; 41.5%), followed by G12V (168; 18.6%), G12D (131; 14.5%), G12A (62; 6.9%), G13C (45; 5.0%), G13D (31; 3.4%), and others (10; 1%). Approximately 21% of patients had transition mutation and 68.2% had a transversion mutation. G12D and transition mutations were predominant in never-smokers. The median overall survival for patients with KRAS-mutated NSCLC was 8.1 months (95% confidence interval [CI]: 7.5–9.5), without any differences according to the different KRAS subtypes mutations. The median progression-free survival was 4.6 months (95% CI: 4.2–5.1) for first-line treatment and 4.8 months (95% CI: 4.3–6.8) for second-line treatment, without any differences according to the different KRAS subtypes mutations.
Conclusions
KRAS mutation subtypes influenced neither treatment responses nor outcomes. The KRAS G12C mutation was detected in 41.5% of patients, who are now eligible for potent and specific G12C inhibitors.
Keywords: KRAS mutation, Non–small cell lung cancer, NSCLC, Prognosis
Introduction
Over the past 10 years, the treatment of NSCLC has dramatically changed with the development of targeted therapies. Somatic EGFR, BRAF, and MET mutations, and also ALK, ROS, and RET rearrangements are oncogenic and associated with responses to targeted therapies.1
KRAS mutations are detected in 20% to 30% of NSCLC cases. These mutations are primarily located at codon 12 or 13.2 KRAS mutations lead to tumor development and growth by activating downstream signaling pathways, including the MAPK pathway involving MEK and ERK.3 KRAS mutations are associated with tobacco status and whites.3,4 In NSCLC, KRAS mutations are oncogenic but may not be addictive. In vitro tests revealed the existence of two kinds of KRAS-mutated cells, one dependent on KRAS mutations and the other independent of KRAS mutations.5
KRAS mutations were suggested to bear an adverse prognostic value. In a large meta-analysis, including 3620 patients from 28 studies, KRAS mutations were revealed to be a weak negative prognostic factor in NSCLC (hazard ratio 1.35, confidence interval [CI] 1.16–1.56).6 However, a prognostic impact of KRAS mutations was neither noted in the pooled analyses of four adjuvant NSCLC trials (Lung Adjuvant Cisplatin Evaluation),7 nor in the pooled analyses of four EGFR tyrosine kinase inhibitors (TKIs) versus placebo trials.8
Regarding metastatic stages, we previously reported in the Biomarkers France study (a prospective cohort involving 17,664 consecutive patients with NSCLC) the detection of 4634 KRAS-mutated NSCLC cases (26.2%) in 4894 patients (26.2%).9 Overall survival (OS) in patients with KRAS-mutated NSCLC was shorter than that of the entire patient cohort (11.1 mo [95% CI: 10.6–13.1] versus 13.8 mo [95% CI: 13.3–14.4], respectively).9 During first-line treatment, progression-free survival (PFS) was also shorter in patients with KRAS-mutated NSCLC than that of the entire patient cohort (7.3 mo [95% CI: 6.5–8.0] versus 8.3 mo [95% CI: 8.0–8.7], respectively).9 In addition, KRAS-mutated NSCLC is likely resistant to EGFR TKI therapy. In two meta-analyses, KRAS mutations were associated with lower response rates to EGFR TKIs.10,11
Efforts at molecular dismemberment suggest that different KRAS mutations may not all be equivalent. KRAS mutations are classified as either transition or transversion mutations. Transition mutations refer to the substitution of a purine for a purine (A to G) or a pyrimidine for a pyrimidine (C to G), whereas transversion mutations correspond to the substitution of a purine for a pyrimidine. Moreover, nonsmokers exhibit a higher frequency of transitional mutations (G13D, G12D, G12S),12,13 whereas KRAS transversion mutations (G12A, G12C, G12V, G13C) are more common among current or former smokers.12
Recently, a new oral therapy was designed to selectively and irreversibly target the KRAS G12C protein.14 Preliminary phase I clinical data concerning a small number of patients were presented to the American Society of Clinical Oncology and World Conference on Lung Cancer 2019 meetings, which revealed encouraging results for the management of patients with KRAS G12C mutation.15,16
This study primarily sought to identify differences in the outcome and response to chemotherapies according to specific KRAS mutation subtypes in patients with metastatic NSCLC from the largest prospective Biomarkers study conducted in France.
Materials and Methods
Data Source
Molecular test results for EGFR, HER2, KRAS, BRAF, and PIK3CA mutations, and also ALK rearrangements and histologic types were provided directly to the Intergroupe Francophone de Cancérologie Thoracique by the certified molecular genetics platforms. At the same time, each patient’s treating physician was given secure access to the patient’s chart to complete the required data: sex, ethnicity, smoking history, family cancer history, Eastern Cooperative Oncology Group performance status, stage, type of treatment, the impact of molecular findings on the treatment decision, and outcomes.
All patients with KRAS mutation with NSCLC of the Biomarkers France cohort were reviewed. In the Biomarker France cohort, all consecutive patients with NSCLC who were routinely screened for molecular alterations from April 2012 to April 2013 at one of the 28 certified molecular genetics centers were eligible for this study.
Only patients with advanced NSCLC were considered (relapse, noncurative stage III and stage IV). Clinical data were collected, as previously described.9 Patients were treated according to the medical standard of 2012 to 2013. The effects of first- and second-line treatments (i.e., chemotherapy or EGFR TKI) on the objective response, disease control, PFS, and OS were analyzed. An evaluation of response and survival was completed by each clinician according to the Response Evaluation Criteria in Solid Tumors 1.1. The OS was defined as the time from diagnosis to the date of death or last follow-up (FU) censored on October 1, 2016. At the last FU, 299 patients (33.2%) were censored (FU [mo], 95% CI: 15.9 [14.9–17.4]).
A KRAS mutation analysis was performed on available tissues through standard Sanger sequencing or a more sensitive validated allele-specific technique, as previously described.9 KRAS mutation associated with another genetic alteration (EGFR or BRAF mutations, ALK translocation) were excluded from the analysis. The KRAS mutations were classified as transversion versus transition mutations.
Ethics
This study was approved by the National Committee for the Protection of Persons (Comité de protection des personnes), according to French law. All patients with NSCLC included in this program received information from their institution or referring clinician, as recommended by competent authorities, which specified that, according to French laws, they were allowed to ask for complete access or removal of their own collected data.
Statistical Analysis
Descriptive statistics were used and expressed in medians with ranges for continuous variables and percentages for categorical variables. Comparisons between categorical variables were conducted using the chi-square test. The significance level was set at p less than 0.05. The OS, first-line PFS, and second-line PFS were previously defined.9 Survival curves were estimated using the Kaplan-Meier method. The disease control rate was defined as the percentage of patients presenting stable disease, partial response, or complete response to treatment, and the overall response rate was defined as the percentage of patients with a partial or complete response. The analyses were carried out using the SAS software, Version 9.3 (SAS Institute, Cary, NC).
Results
In the Biomarkers France trial, 4894 (26.2%) KRAS mutations from 4634 patients were detected out of 18,679 molecular analyses from 17,664 enrolled patients with NSCLC. Survival and treatment data were available for 1014 patients. Data on sex, tobacco status, and TNM stage were not declared for 31 patients. Three patients were excluded because they presented multiple biopsies with distinct molecular mutations; patients with stages I to II (31 patients) and stage III with curative treatment (chemoradiation therapy or surgery, 51 patients) were also excluded. Therefore, 901 patients treated for noncurative stage III or IV NSCLC were included in this analysis (Fig. 1).
Figure 1.
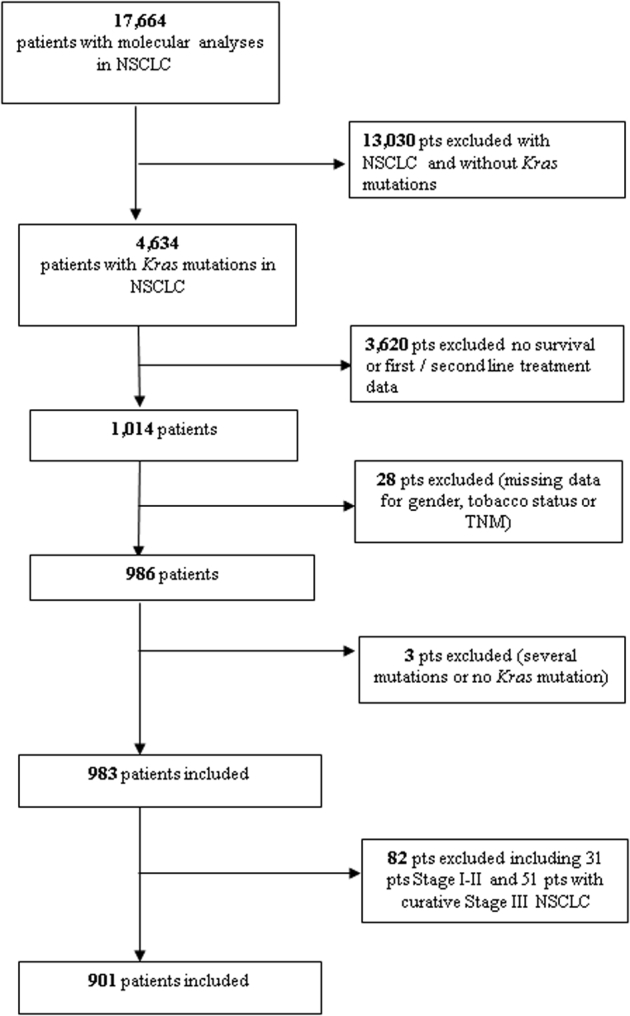
Flow chart exhibiting patient selection.
Clinical Characteristics
Patient demographics are summarized in Table 1. The median age was 61.6 years [30.0–87.7], and 66.6% were men. Almost all patients were smokers or former smokers (95.2%) and white (99.5%). The predominant histology was adenocarcinoma (82.5%).
Table 1.
Patients Characteristics
| Patients Characteristics | N = 901 | |
|---|---|---|
| Sex | ||
| Male | N (%) | 600 (66.6) |
| Female | N (%) | 301 (33.4) |
| Age (y) | ||
| Median | 61.65 | |
| Range | 30.0–87.7 | |
| Asian origin | ||
| Yes | N (%) | 4 (0.5) |
| No | N (%) | 797 (99.5) |
| Smoking | ||
| Smoker/former smoker | N (%) | 858 (95.2) |
| Nonsmoker | N (%) | 43 (4.8) |
| PSa | ||
| 0–1 | N (%) | 604 (70.4) |
| 2 | N (%) | 198 (23.1) |
| 3–4 | N (%) | 50 (6.5) |
| Histology | ||
| Squamous | N (%) | 10 (1.1) |
| Adenocarcinoma | N (%) | 743 (82.5) |
| Large cell | N (%) | 27 (3.0) |
| Other | N (%) | 121 (13.4) |
| OS | ||
| Median (95% CI) | 8.1 (7.5–9.5) | |
| First-line PFS | ||
| Chemotherapy | N Median (95% CI) |
831 4.6 (4.2–5.1) |
| Clinical trial | N Median (95% CI) |
41 4.2 (3.2–7.8) |
| EGFR TKI/other | N Median (95% CI) |
23 2.5 (1.1–4.3) |
| No treatment | N Median (95% CI) |
5 0.8 [0.5-NR] |
| Second-line PFS | ||
| Chemotherapy | N Median (95% CI) |
316 4.8 (4.3–6.8) |
| Clinical trial | N Median (95% CI) |
33 3.9 (2.3–6.3) |
| EGFR TKI/other | N Median (95% CI) |
106 3.8 (2.8–4.7) |
| No treatment | N Median (95% CI) |
342 0.8 (0.6–1.1) |
CI, confidence interval; OS, overall survival; PFS, progression-free survival; PS, performance status; TKI, tyrosine kinase inhibitor.
Missing data N = 44.
KRAS Mutation Subtypes
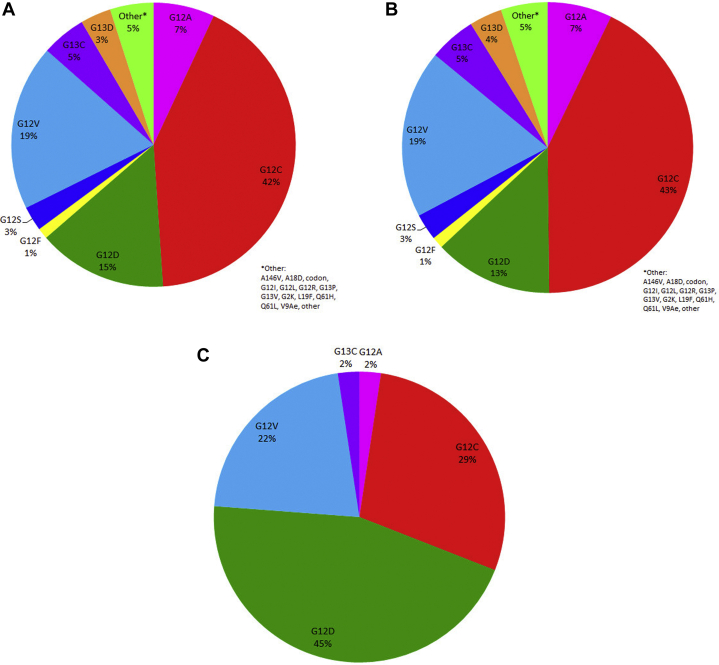
The most frequent KRAS mutation was a codon 12 guanine to cytosine (G > C) mutation in 374 patients (41.5%) (Fig. 2, Table 2). Other nucleotide changes in tumor samples were found on codon 12, which includes the following: G12V in 168 patients (18.6%), G12D in 131 patients (14.5%), G12A in 62 patients (6.9%), G12S in 25 patients (2.8%), G12F in 11 patients (1.2%), and others in 44 patients (4.9%). Overall, mutations were found at codon 12 in 771 patients (85%). Mutations at codon 13 were found in 81 patients (9.0%) including G13C in 45 (5.0%) and G13D in 31 (3.4%).
Figure 2.
Distribution of KRAS mutation subtypes (A) in all patients, (B) in smokers, and (C) in never smokers.
Table 2.
Patients Characteristics According to Different KRAS Point Mutations
| Patients Characteristics | G12C (N = 374) | G12V (N = 168) | G12D (N = 131) | G12A (N = 62) | G12S (N = 25) | G12F (N = 11) | G13C (N = 45) | G13D (N = 31) | Other (N = 44) | Total (N = 901) |
|---|---|---|---|---|---|---|---|---|---|---|
| Sex | ||||||||||
| Male | ||||||||||
| N (%) | 253 (67.6) | 107 (63.7) | 92 (70.2) | 38 (61.3) | 13 (52.0) | 5 (45.5) | 35 (77.8) | 21 (67.7) | 29 (65.9) | 593 (66.6) |
| Female | ||||||||||
| N (%) | 121 (32.4) | 61 (36.3) | 39 (29.8) | 24 (38.7) | 12 (48.0) | 6 (54.5) | 10 (22.2) | 10 (32.3) | 15 (34.1) | 298 (33.4) |
| Age (years) | ||||||||||
| Median | 61.01 | 61.06 | 64.28 | 62.62 | 61.36 | 61.34 | 59.90 | 60.16 | 59.40 | 61.61 |
| Range | 38.9–87.7 | 30.0–87.2 | 32.6–85.2 | 41.3–82.4 | 45.3–84.4 | 46.4–80.5 | 39.7–83.3 | 41.8–80.0] | 39.3–82.5 | 30.0–87.7 |
| Asian origin | ||||||||||
| Yes | ||||||||||
| N (%) | 4 (1.2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 (0.4) |
| No | ||||||||||
| N (%) | 329 (98.8) | 152 (100.0) | 115 (100.0) | 57 (100.0) | 21 (100.0) | 9 (100.0) | 38 (100.0) | 26 (100.0) | 40 (100.0) | 787 (88.3) |
| Missing | ||||||||||
| N | 41 | 16 | 16 | 5 | 4 | 2 | 7 | 5 | 4 | 100 |
| Smoking | ||||||||||
| Nonsmoker | ||||||||||
| N (%) | 12 (3.2) | 9 (5.4) | 19 (14.5) | 1 (1.6) | 0 | 0 | 1 (2.2) | 0 | 0 | 42 (4.7) |
| Smoker/former smoker | ||||||||||
| N (%) | 362 (96.8) | 159 (94.6) | 112 (85.5) | 61 (98.4) | 25 (100.0) | 11 (100.0) | 44 (97.8) | 31 (100.0) | 44 (100.0) | 849 (95.3) |
| PS | ||||||||||
| 0–1 | ||||||||||
| N (%) | 243 (68.3) | 119 (73.0) | 91 (71.7) | 42 (71.2) | 14 (63.6) | 9 (81.8) | 35 (83.3) | 15 (55.6) | 27 (65.9) | 595 (66.8) |
| 2 | ||||||||||
| N (%) | 84 (23.6) | 36 (22.1) | 27 (21.3) | 13 (22.0) | 5 (22.7) | 1 (9.1) | 7 (16.7) | 12 (44.4) | 12 (29.3) | 197 (22.1) |
| 3–4 | ||||||||||
| N (%) | 29 (8.1) | 8 (4.9) | 9 (7.1) | 4 (6.8) | 3 (13.6) | 1 (9.1) | 0 | 0 | 2 (4.9) | 56 (6.3) |
| Missing | ||||||||||
| N | 18 | 5 | 4 | 3 | 3 | 0 | 3 | 4 | 3 | 43 |
| Histology | ||||||||||
| Squamous | ||||||||||
| N (%) | 5 (1.3) | 2 (1.2) | 3 (2.3) | 0 | 0 | 0 | 0 | 0 | 0 | 10 (1.1) |
| Adenocarcinoma | ||||||||||
| N (%) | 310 (82.9) | 135 (80.4) | 104 (79.4) | 54 (87.1) | 20 (80.0) | 8 (72.7) | 40 (88.9) | 26 (83.9) | 37 (84.1) | 734 (82.4) |
| Large cell | ||||||||||
| N (%) | 8 (2.1) | 8 (4.8) | 2 (1.5) | 1 (1.6) | 0 | 0 | 2 (4.4) | 2 (6.5) | 3 (6.8) | 26 (2.9) |
| Other | ||||||||||
| N (%) | 51 (13.6) | 23 (13.7) | 22 (16.8) | 7 (11.3) | 5 (20.0) | 3 (27.3) | 3 (6.7) | 3 (9.7) | 4 (9.1) | 121 (13.6) |
PS, performance status.
Overall, 21% of patients (187 of 901) displayed transition mutations (G12D, G12S, G13D, G13S), and 68.2% (615 of 901) transversion mutations (G12A, G12C, G12F, G12V) (Supplementary Table 1).
Specific point mutations differed between never-smokers and former/current smokers (Fig. 3, Table 2). Among never-smokers, the most common KRAS mutation was G12D (19 of 42; 45%), whereas G12C was the most frequent mutation among former/current smokers (362 of 849; 42.6%). Patients with transition mutations were more likely to be nonsmokers than patients exhibiting transversion mutations (10.2% versus 3.6%; p < 0.001). Other clinical characteristics were similar between patients with KRAS transition versus transversion mutations.
Figure 3.
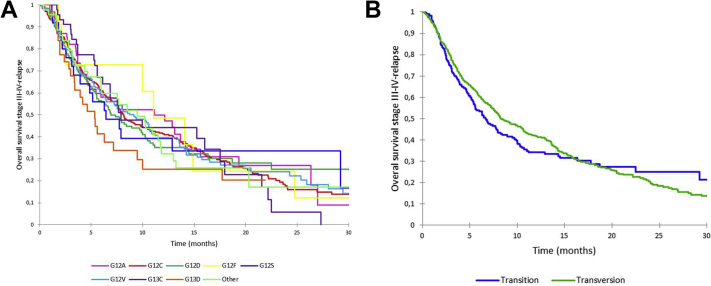
Overall survival (A) according to KRAS mutations and (B) according to KRAS transition versus transversion.
KRAS Mutation and Survival
The median OS for all patients with KRAS-mutated locally advanced or metastatic lung cancer was 8.1 months (95% CI: 7.5–9.5 mo). No difference was noted when comparing survival according to the different KRAS point mutations (Fig. 3A, Supplementary Table 1). There were no differences in outcome for patients with KRAS transition (6.6 mo [95% CI: 5.4–8.2] versus transversion mutations (8.4 mo [95% CI: 7.6–10.3], p = 0.46, Fig. 3B, Supplementary Table 2). The OS in patients with KRAS mutations did not differ according to tobacco status (smoker/former smoker versus nonsmoker) (data not shown).
KRAS Mutation and First-Line Chemotherapy
First-line treatment for patients with advanced NSCLC consisted of chemotherapy for 831 patients (92.3%), a clinical trial for 41 patients (4.5%), other for 23 patients (6.5%), and supportive care alone for five patients (0.5%).
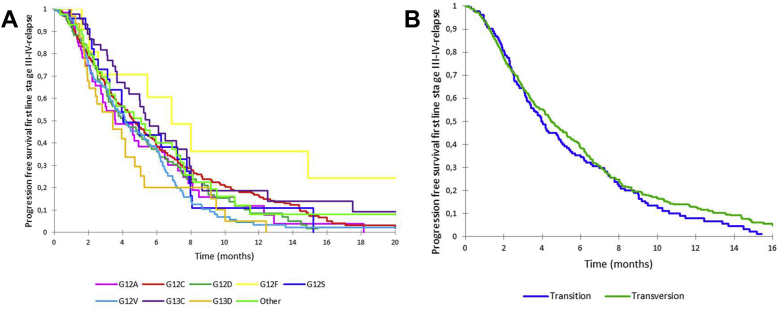
The median PFS on chemotherapy was 4.6 months (95% CI: 4.2–5.1), without any differences according to the KRAS point mutation subtype, whether transition or transversion (Fig. 4, Supplementary Tables 2 and 3).
Figure 4.
Progression-free survival on first-line chemotherapy (A) according to KRAS mutations and (B) according to KRAS transition versus transversion mutation.
Second-line treatment for patients with advanced NSCLC consisted of chemotherapy for 316 patients (35.1%), supportive care alone for 388 patients (38.0%), and other treatment for 95 patients (10.5%). The median PFS for chemotherapy was 4.8 months (95% CI: 4.3–6.8) without any differences according to the KRAS point mutation type as to whether transition or transversion.
Discussion
Of the 17,664 Biomarkers France patients, 4894 (26.2%) KRAS mutations were detected in 4634 patients.9 To our knowledge, this cohort represents the largest stages III to IV KRAS-mutated NSCLC cohort ever analyzed.
Over 95% of the patients with KRAS mutations were smokers or former smokers and white (99.5%), with adenocarcinoma as their predominant histology, which is in line with previous publications.3 The most common point mutation was a codon 12 guanine to cytosine (G > C) mutation in 374 patients (41.5%). In nonsmoker patients, G12D mutations and transition mutations were predominant, consistent with previous reports.13
This project was primarily aimed to identify differences in response and outcome according to the different KRAS mutations in metastatic NSCLC cases in this real-life cohort. The median OS in this cohort was 8.1 months. No survival impact was noted regarding the different KRAS point mutations.
We hypothesized that KRAS mutations subtypes may not all be equivalent to outcome or treatment response. Today, it remains speculative whether different KRAS mutation subtypes are prognostic or predictive. The main reason is that most studies were not powered to detect differences in outcome or treatment response according to KRAS mutation subtypes.
Few data on the prognostic impact of KRAS mutation subtypes are available. Similar to our findings, no prognostic impact of different KRAS mutation subtypes was noted in two cohorts of NSCLC, a metastatic one, and an early-stage one.7,12 Only one study noted a favorable prognosis for patients with KRAS G12C or G12V mutations compared with other codon 12 mutations in the control arm of a pooled analysis of four clinical trials.8
Few data on the predictive impact of KRAS mutation subtypes on therapy are available. In vitro analysis from the BATTLE (Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination) trial suggests a greater sensitivity to MEK inhibitor in NSCLC cell lines harboring KRAS G12C or G12V mutations.17 This in vitro result about KRAS G12C or KRAS G12V mutations was not confirmed in the phase III trial comparing docetaxel plus selumetinib versus docetaxel alone.18 In the pooled analysis of four EGFR TKIs versus placebo trials, Zer et al.8 noted that EGFR TKI was harmful in patients with G12V mutations, whereas EGFR TKIs may be effective in transition mutations. Our observation regarding EGFR TKIs was not conclusive as only 114 patients (12.7%) received EGFR TKIs.
This study displays some limitations. It was a prospective nonrandomized cohort study with no information collected regarding the chemotherapy used in first- or second-line treatment. As data collection for this cohort occurred before the availability of immunotherapy, the impact of KRAS mutations on immune checkpoint inhibitors (ICIs) could not be assessed. Identifying other concurrent mutations is also critical as it may influence outcome and treatment response. Comutations of TP53, STK11, or CDKN2A/B seems promising as their inactivation permits defining different KRAS-mutated subgroups.19 KRAS mutations and STK11 inactivation led to a more aggressive tumor phenotype and ICI resistance, whereas KRAS/TP53 or KRAS/CDKN2a/B were ICI-sensitive.20 KRAS alone neither seems to be a sufficient prognostic nor predictive biomarker, particularly for ICI.21
Until recently, KRAS mutations were considered “undruggable.”22 Different clinical trials with selumetinib as MEK inhibitor did not reveal any efficacy in phase 2 or 3 trials.23 Even with a greater objective response rate in G12C and G12V mutations in the selumetinib group compared with the placebo group, no relevant PFS benefit was noted.23
Oncogenic KRAS mutations result in activation by impairing guanosine triphosphate (GTP) hydrolysis, with an advantage on GTP over guanosine diphosphate.14 Crystallographic studies reveal a new pocket in the KRAS structure that disrupts the nucleotide preference guanosine diphosphate to GTP and disable binding to Raf. This allosteric regulatory site on KRAS is targetable in a mutant-specific manner.14 Small molecules irreversibly inhibiting the mutant cysteine residue G12C have revealed promising in vitro and in vivo results.14 AMG510 (Amgen) is the first KRAS G12C inhibitor to reach clinical development. AMG510 revealed interesting antitumor activity in a phase I trial in heavily pretreated patients with KRAS G12C–mutated solid tumor.15,16 In this trial, 34 patients with NSCLC were enrolled, and 13 of 23 evaluable patients with NSCLC treated with AMG510 at a 960 mg dose (dose selected for the ongoing phase 2 trial). Seven patients experienced a partial response (7 of 13; 54%), and another six had stable disease (6 of 13; 46%), for a disease control rate of 100% (23 of 23). Treatment-related undesirable effects at 960 mg daily were limited: grade 1 to 2 diarrhea (12%), nausea (6%), and aspartate aminotransferase/alanine aminotransferase increases (6%). At least two other molecules targeting the G12C mutant variant of KRAS are in development (MRTX849 Mirati Therapeutics, ARS-3248 Bioscience). Two other molecules that may be effective in all patients with KRAS mutation are in development, namely: KRAS-SOS1 inhibitor (BI 1701963, Boehringer Ingelheim) or the cancer vaccine for G12C, G12D, G13D, and G12V (mRNA-5671, Moderna Therapeutics).
In the Biomarkers France study, a large real-life cohort, KRAS mutations were determined to be a negative prognostic factor in metastatic NSCLC. However, no differences in response and outcome were identified according to the different KRAS mutation subtypes. Recently, a promising oral therapy was assessed to selectively inhibit the KRAS G12C protein, that is, the most common KRAS mutation, which accounted for 41.5% of KRAS mutations in this cohort and approximately 13% of NSCLC cases in the Biomarkers France study.
Acknowledgments
This work was supported by Intergroupe Francophone de Cancérologie Thoracique through unrestricted grants from the Institut National Du Cancer, France (grant number 2011-038) and by Novartis, France. The authors thank Quân Tran, Antoine Deroy, Sandy Dos Santos, and Fabienne Hirchaud (French Cooperative Thoracic Intergroup, Paris, France) for their participation in data collection, monitoring, and computing.
Footnotes
Disclosure: Dr. Beau-Faller reports receiving nonfinancial support from Bristol-Myers Squibb and AstraZeneca; and grant and personal fees from AstraZeneca. Dr. Rouquette served as an expert consultant for AstraZeneca, Merck Sharp & Dohme, Bristol-Myers Squibb, Takeda, and Boehringer Ingelheim. Dr. Mazières reports receiving nonfinancial support from Pfizer and Roche Holdings AG. Dr. Ricordel has received grants from Roche Holdings AG. Dr. Barlesi reports receiving personal fees and nonfinancial support from AstraZeneca, Bayer, Bristol-Myers Squibb, Boehringer Ingelheim, Eli Lilly Oncology, F. Hoffmann–La Roche Ltd., Novartis, Merck, Merck Sharp & Dohme, Pierre Fabre, Pfizer, and Takeda; and nonfinancial support from AbbVie, Acea Pharmaceutical, Amgen, Eisai, Genentech, Ipsen, Ignyta, Innate Pharma, Loxo Oncology, Medimmune, and Sanofi-Aventis. Dr. Wislez reports receiving personal fees from Boehringer Ingelheim, Roche Holdings AG, Merck Sharp & Dohme, Bristol-Myers Squibb, AstraZeneca, Amgen; and nonfinancial support from Roche, Merck Sharp & Dohme. The remaining authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2020.100052.
Supplementary Data
References
- 1.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 2.Adderley H., Blackhall F.H., Lindsay C.R. KRAS-mutant non-small cell lung cancer: converging small molecules and immune checkpoint inhibition. EBiomedicine. 2019;41:711–716. doi: 10.1016/j.ebiom.2019.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood K., Hensing T., Malik R., Salgia R. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol. 2016;2:805–812. doi: 10.1001/jamaoncol.2016.0405. [DOI] [PubMed] [Google Scholar]
- 4.Dogan S., Shen R., Ang D.C. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh A., Greninger P., Rhodes D. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mascaux C., Iannino N., Martin B. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepherd F.A., Domerg C., Hainaut P. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zer A., Ding K., Lee S.M. Pooled analysis of the prognostic and predictive value of KRAS mutation status and mutation subtype in patients with non-small cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. J Thorac Oncol. 2016;11:312–323. doi: 10.1016/j.jtho.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Barlesi F., Mazieres J., Merlio J.P. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic InterGroup (IFCT) Lancet Lond Engl. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 10.Linardou H., Dahabreh I.J., Bafaloukos D., Kosmidis P., Murray S. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 2009;6:352–366. doi: 10.1038/nrclinonc.2009.62. [DOI] [PubMed] [Google Scholar]
- 11.Mao C., Qiu L.X., Liao R.Y. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer. 2010;69:272–278. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Yu H.A., Sima C.S., Shen R. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol. 2015;10:431–437. doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riely G.J., Kris M.G., Rosenbaum D. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govindan R, Faikih M, Price T. Phase 1 study of safety, tolerability, PK and efficacy of AMG 510, a novel KRASG12C inhibitor, evaluated in NSCLC. Paper presented at: 20th World Conference on Lung Cancer. September 8, 2019; Barcelona, Spain.
- 16.Fakih M., O Neil B., Price T. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol. 2019;37(suppl 15) 3003–3003. [Google Scholar]
- 17.Ihle N.T., Byers L.A., Kim E.S. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jänne P.A., van den Heuvel M.M., Barlesi F. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: the SELECT-1 randomized clinical trial. JAMA. 2017;317:1844–1853. doi: 10.1001/jama.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skoulidis F., Byers L.A., Diao L. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skoulidis F., Goldberg M.E., Greenawalt D.M. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C.K., Man J., Lord S. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non-small cell lung carcinoma: a systematic review and meta-analysis. JAMA Oncol. 2018;4:210–216. doi: 10.1001/jamaoncol.2017.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.