Abstract
Introduction
A subset of lung adenocarcinomas (ADs) has been found to have somatic activating mutations in the tyrosine kinase domain of the EGFR gene, associated with response to EGFR tyrosine kinase inhibitor therapy. Rare germline mutations within this domain, including EGFR T790M, have been associated with genetic susceptibility to lung ADs. Using high-throughput sequencing, we elucidate the genomic evolution in tissues from a patient with lung AD carrying a germline EGFR T790M mutation.
Methods
We performed microdissection, targeted panel, and whole-exome sequencing to molecularly characterize multiple foci of atypical adenomatous hyperplasia (AAH), in situ and invasive components of AD, normal lung tissue, and whole blood from the patient. Normal lung tissue was analyzed for potential acquired somatic genome alterations (“field effect”).
Results
All lesions harbored a secondary somatic EGFR mutation, either L858R or L861Q, in addition to the germline T790M mutation. Clear overlap was observed between the somatic profiles of in situ and invasive AD components, confirming clonal relatedness. AAH lesions shared few to no somatic alterations with the AD, suggesting clonal independence. No robust evidence of field effect was identified in the normal lung tissue.
Conclusions
Somatic EGFR mutations are early events in the pathogenesis of lung ADs arising in the context of germline EGFR T790M. Synchronous AAH lesions seem to be independent. Stepwise genomic evolution is observed in association with invasiveness of the neoplastic cell population.
Keywords: NSCLC, Lung adenocarcinoma, EGFR germline mutation, High-throughput sequencing, Genome evolution
Introduction
A subset of lung adenocarcinomas (ADs) is characterized by somatic activating mutations in the tyrosine kinase domain of the EGFR gene.1 These mutations typically predict response to treatment with EGFR tyrosine kinase inhibitors. Responsive tumors are most common in women, nonsmokers, and East Asians. Lung ADs arising in the context of EGFR germline mutations are less well characterized. Germline mutations in the tyrosine kinase domain of EGFR have been reported, primarily consisting of T790M mutations.2, 3, 4 Other observed germline alterations in EGFR include R776G/H, V834L, V843I, and P848L. Studies of families carrying the EGFR germline mutations have revealed that many affected individuals develop a secondary somatic EGFR mutation.5
EGFR germline variants are rare; germline T790M mutations have an estimated prevalence of 0.1% among patients with lung cancer and 0.003% in the general population.6,7 As a result, the clinicopathologic features that distinguish these patients are not well defined. However, germline EGFR mutations seem to correlate with development of multiple lung ADs and precursor lesions in nonsmokers.4 To better understand the pathogenesis of lung ADs arising from EGFR germline mutations, we used high-throughput sequencing to characterize precursor and invasive lesions, as well as normal tissue, from the lungs of a patient with AD with a germline EGFR T790M mutation.
Materials and Methods
Patient Samples
The patient is a 65-year-old female nonsmoker with a suspicious lung nodule detected on computed tomography imaging. She had an extensive family history of lung cancer and underwent germline testing on the Investigating Hereditary Risk From T790M study (NCT01754025) after her two nonsmoking sisters had died of lung cancer before the age of 50 years, one of whom had been tested and found to carry a germline EGFR T790M mutation (Fig. 1A).8 Computed tomography scans revealed persistent, bilateral ground-glass and solid nodules, including a dominant, left lower lobe solid nodule that grew from 0.9 cm to 1.5 cm in 4 years (Fig. 1B). The patient, therefore, underwent a left lower lobe pulmonary wedge resection and was diagnosed with having a 2.3-cm acinar-predominant lung AD with minor (20%) lepidic (in situ) component. In addition, five discrete foci of atypical adenomatous hyperplasia (AAH) were detected in the background lung, measuring less than 0.1 cm to 0.4 cm. Sampled lymph nodes were negative for metastatic tumor. Formalin-fixed, paraffin-embedded (FFPE) tissue samples were obtained from two synchronous AAH foci of adequate size and cellularity to allow for microdissection and from in situ and invasive components of AD (Fig. 1B–F). AAH and both components of AD were microdissected or macrodissected to maximize tumor content. Genomic DNA was extracted from the neoplastic tissues and morphologically normal lung tissue (Fig. 1G) using the QIAamp DNA FFPE Tissue Kit (FFPE; Qiagen, Hilden, Germany) and from whole blood using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. This study was approved by the Partners Human Research Committee, and the patient provided written informed consent.
Figure 1.
Histopathologic analysis of the patient samples. (A) Genetic pedigree of the patient. Two individuals in the pedigree have been tested and found to carry a germline EGFR T790M mutation. The index patient is marked with a black arrowhead. (B) CT scan result revealing the dominant, subpleural left lower lobe lung mass (AD; 1.5 cm × 0.9 cm) 3 months before the surgery. A 0.6-cm solid nodule is also visible in the right upper lobe. Representative H&E staining images of (C) AAH1, (D) AAH2, (E) in situ component of AD, (F) invasive component of AD, and (G) the normal lung. Scale bar in H&E stainings: (C–F) 100 μm and (G) 500 μm. AAH, atypical adenomatous hyperplasia; AD, adenocarcinoma; COPD, chronic obstructive pulmonary disease; CT, computed tomography; H&E, hematoxylin and eosin.
High-Throughput Sequencing
Targeted cancer gene panel sequencing (Oncopanel version 3.19) was performed for all samples (n = 6) and whole-exome sequencing (WES) for the AD components, normal lung, and whole blood samples (n = 4) at the Center for Cancer Genomics at Dana-Farber Cancer Institute. Genomic libraries were constructed using the KAPA Hyper Prep Kit (Roche, Pleasanton, CA) and target regions enriched with the SureSelect Human All Exome v5 Kit (Agilent Technologies, Santa Clara, CA). Paired-end sequencing was performed on HiSeq 2500 (Illumina, San Diego, CA), and sequenced reads were aligned to GRCh37 (hg19) reference assembly. Mean target coverage for cancer gene panel data was 421× (136–564×) and for WES 133× (116–140×) (Supplementary Table 1). The details of our somatic variant calling pipeline are described in the Supplementary Methods.
Results
EGFR Mutations
On the basis of both the cancer gene panel and WES data, EGFR T790M mutation was present in all patient samples (mean allele fraction [AF] = 0.47 [0.39–0.53]), consistent with its germline origin (Supplementary Table 2). EGFR L858R mutation was observed in the AAH1 (AF = 0.02), in situ (mean AF = 0.14 [0.12–0.16]), and invasive (AF = 0.22) AD samples. Another EGFR alteration, L861Q, was identified in AAH2 (AF = 0.07). No copy-number variants (CNVs) affecting EGFR were observed. Thus, both AAH specimens, including the in situ and invasive components of AD, harbored a secondary somatic EGFR mutation.
Landscape of Somatic Genomic Alterations
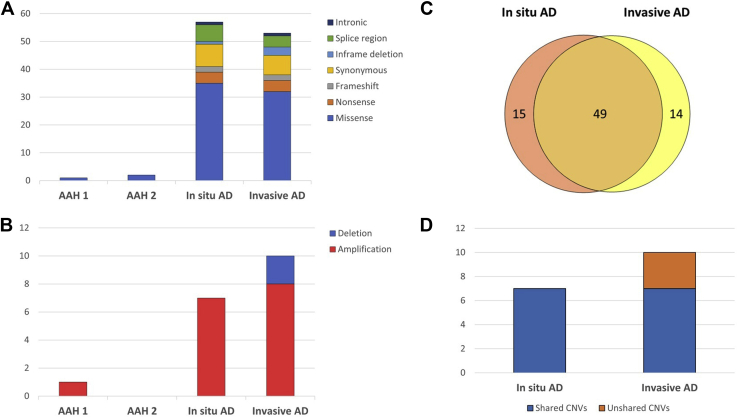
The number of observed somatic single-nucleotide variants (SNVs) and indels ranged from 1 to 57 within the patient lesions (Supplementary Table 3). The mutational spectrum is illustrated in Fig. 2A. In addition to EGFR, two other genes, SMARCA4 and FLT3, previously linked to lung AD displayed somatic alterations. A nonsynonymous S937N alteration in SMARCA4 was identified in the invasive component of AD, and an intronic variant 7 base pair downstream of FLT3 exon 4 was observed in both AD components (Supplementary Table 4). Both alterations were present at low AFs (SMARCA4: 0.08, FLT3: 0.03 [0.01–0.05]). Furthermore, three other known cancer genes, CPEB3 (AF = 0.07), FAS (mean AF = 0.04 [0.02–0.06]), and SLC34A2 (AF = 0.07), harbored somatic alterations with low AFs.
Figure 2.
Mutational spectrum of the identified SNVs, indels, and CNVs. (A) SNVs and indels. For AAH lesions, the observed SNVs and indels came from the targeted cancer gene panel data, whereas for in situ and invasive components of AD, they were based on both targeted cancer gene panel and WES data. (B) Identified CNVs included both amplifications and deletions. For AAH lesions, targeted cancer gene panel data were analyzed for CNVs, whereas for in situ and invasive AD samples, WES data were analyzed. (C) The Venn diagram reveals that in situ and invasive components of AD shared a considerable fraction of somatic variants. (D) All the CNVs identified in the in situ component of AD were present also in the invasive component. AAH, atypical adenomatous hyperplasia; AD, adenocarcinoma; CNV, copy-number variant; SNV, single-nucleotide variant; WES, whole-exome sequencing.
Chromosome arm-level amplifications on 1q, 5p, and 16p were identified in both in situ and invasive components of AD (Supplementary Fig. 1). Furthermore, we observed various shorter CNVs across the genome (Fig. 2B). Several CNVs contained lung AD-associated genes, such as AKT3, DDR2, MYC, NTRK1, SDHA, SDHC, and RICTOR, which as in our data, have previously been reported to be amplified in this cancer type (Supplementary Table 5). Especially, amplifications of MYC (8q24.21) and RICTOR (5p13.1), present in both components of AD, have been frequently identified in lung ADs.10,11 Each lung AD-associated gene, however, was part of a larger CNV affecting multiple genes simultaneously. No structural variants were identified in the patient’s lesions. Moreover, no definitive somatic alterations were detected in the morphologically normal lung tissue (Supplementary Table 6).
Genomic Tumor Evolution
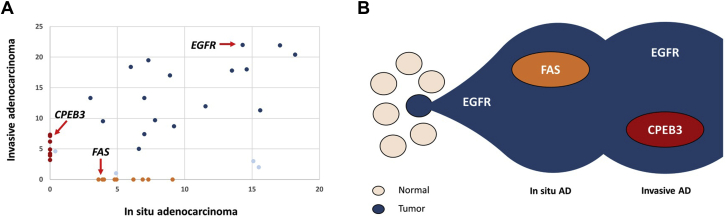
The somatic genomic profiles of in situ and invasive components of AD clearly overlapped, indicating a common clonal origin (Fig. 2C and Supplementary Tables 4–5). Most of the shared somatic alterations were nonsynonymous mutations. Although the number of SNVs was slightly higher in the in situ AD, increase in the number of indels and CNVs in invasive AD indicates a stepwise genome evolution (Fig. 2D and Supplementary Tables 4–5). The variant allele frequency distribution of somatic SNVs and indels reveals a common clone between the AD components, including EGFR L858R mutation as one of the earliest genomic events, and two component-specific clones with distinct sets of alterations (Fig. 3A and B). The only shared somatic alteration between AAH1 and AD components was the EGFR L858R mutation. Furthermore, no shared somatic alterations between AAH2 and other lesions were identified. Given the high frequency of EGFR L858R mutations in general, and the disparate geographic location of the AAH lesions, they are both almost certainly independent in origin.
Figure 3.
Clonal evolution of the in situ and invasive components of AD. (A) Diagonal plot reveals the distribution of allele frequencies (VAF) of variants predicted to have a damaging effect on protein function listed in Supplementary Table 4 in both in situ and invasive AD. The founding clones in the in situ and invasive AD are represented by blue, whereas burgundy and orange represent component-specific clones. The EGFR driver mutation and two component-specific variants present in known cancer genes are indicated by red arrows. Outliers are marked with light blue. (B) The “founder” clone in the in situ AD contained somatic mutation in the EGFR gene. Both in situ and invasive AD developed specific clones. AD, adenocarcinoma; VAF, variant allele frequency.
Clinical Follow-Up
The pathologic stage after resection was pT1bN0 (American Joint Committee on Cancer eighth edition) on the basis of an overall size of 2.3 cm with an estimated 1.8 cm of invasion. No additional systemic therapy was offered after the left lower lobe resection. Since the time of surgery, the patient has been managed with interval scans with the plan to offer additional surgery if one or more lesions grow or develop an increased solid component. The patient has developed multiple additional ground-glass lesions; however, most of the existing nodules are stable or minimally increased in size.
Discussion
Lung ADs originating from EGFR germline mutations are rare, and there is limited understanding of the molecular genetic characteristics underlying their pathogenesis. Here, we used high-throughput sequencing to perform a systematic genomic analysis across various lesions and normal tissue of a patient with lung AD with a germline EGFR T790M mutation. The sequencing data revealed that each neoplastic lesion harbored a secondary somatic EGFR mutation, either L858R or L861Q. Somatic L858R mutations have frequently been reported in patients with germline EGFR mutations, whereas L861Q has been observed once before together with a germline V843I mutation.5,12 A clear majority of patients with germline EGFR T790M have been reported to develop a secondary somatic EGFR mutation, suggesting T790M is not sufficient and a secondary EGFR mutation is required for tumor development.
AAH represents the initial step in the progression to a fully invasive AD. Our data support the widely accepted concept that somatic EGFR mutations are early events in the pathogenesis of lung AD.13 That said, we did not observe somatic EGFR mutations in the morphologically normal lung tissue, as previously described in the case of sporadic lung AD.14 The distinct EGFR mutations found in the different AAHs indicate that these lesions can arise independently and develop into molecularly distinct tumors.
Apparent overlap between the somatic genomic profiles of in situ and invasive components of AD is consistent with a shared clonal origin. An increased number of detected unique alterations in invasive AD compared with in situ AD are likely to reflect genomic progression but may also be influenced by greater tumor percentage in AD. Interestingly, most CNVs containing lung AD-associated genes were observed in both in situ and invasive ADs.
Although we performed a systematic characterization of somatic genome alterations across various lesions and normal tissue of the patient, we were only able to perform targeted cancer gene panel sequencing for the AAHs owing to their small size and low amount of DNA. Thus, we had limited power to study the full spectra of somatic genome alterations in these lesions. Small amounts of tissue material continue to impede our ability to study the pathogenesis of lung AD precursors. The use of alternative methodologies, such as single cell-based approaches, may allow more comprehensive genomic analysis of these lesions in the future. Moreover, further studies with a larger number of patients with germline EGFR T790M mutations harboring synchronous preneoplastic lesions are important to identify those mutational features which characterize the lesions that progress to lung ADs.
In conclusion, our study sheds light on the somatic genomic alterations that initiate and drive the progression of a lung AD arising from a germline EGFR T790M mutation. Management of patients with germline EGFR mutations remains a challenge. Comprehensive genomic analysis of early molecular events in lung cancer pathogenesis is important to better predict the fate of these early lesions and tailor therapy to prevent progression.
Acknowledgments
This study was supported by the National Institutes of Health/National Cancer Institute (2R01CA116020) and Stand Up To Cancer-LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Cancer Research Grant (grant no. SU2C-AACR-DT23-17 to S.M. Dubinett and A.E. Spira). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. Dr. Mäkinen acknowledges support from the Academy of Finland (295693). The authors thank Marina Milan and Safiya Aijazi for their help in obtaining the whole blood sample.
Footnotes
Disclosure: Dr. Meyerson declares receiving research support from Bayer, Ono, NoVo, and Janssen; having patent licensing royalties from LabCorp; and serving as scientific advisory board chair and consultant for OrigiMed. Dr. Sholl reports receiving consulting fees or honoraria from Loxo Oncology, AstraZeneca, Foghorn Therapeutics, and EMD Serono and research funding to her institution from Roche/Genentech. Dr. Oxnard is an employee of Foundation Medicine, Inc., and holds equity in Roche. The remaining authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2021.100146.
Supplementary Data
References
- 1.Paez J.G., Jänne P.A., Lee J.C. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 2.Bell D.W., Gore I., Okimoto R.A. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 3.Oxnard G.R., Miller V.A., Robson M.E. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol. 2012;7:1049–1052. doi: 10.1097/JTO.0b013e318250ed9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gazdar A., Robinson L., Oliver D. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol. 2014;9:456–463. doi: 10.1097/JTO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto H., Yatabe Y., Toyooka S. Inherited lung cancer syndromes targeting never smokers. Transl Lung Cancer Res. 2018;7:498–504. doi: 10.21037/tlcr.2018.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y., Alden R.S., Odegaard J.I. Discrimination of germline EGFR T790M mutations in plasma cell-free DNA allows study of prevalence across 31,414 cancer patients. Clin Cancer Res. 2017;23:7351–7359. doi: 10.1158/1078-0432.CCR-17-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karczewski K.J., Francioli L.C., Tiao G. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oxnard G., Heng J., Chen R. OA 06.02 final report of the INHERIT EGFR study—33 unrelated kindreds carrying germline EGFR mutations. J Thorac Oncol. 2017;12(suppl 2):S1758. [Google Scholar]
- 9.Sholl L.M., Do K., Shivdasani P. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1 doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beroukhim R., Mermel C.H., Porter D. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H., Zou Y., Ross J.S. RICTOR amplification defines a novel subset of patients with lung cancer who may benefit from treatment with mTORC1/2 inhibitors. Cancer Discov. 2015;5:1262–1270. doi: 10.1158/2159-8290.CD-14-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda K., Nomori H., Mori T., Sasaki J., Kobayashi T. Novel germline mutation: EGFR V843I in patient with multiple lung adenocarcinomas and family members with lung cancer. Ann Thorac Surg. 2008;85:1430–1432. doi: 10.1016/j.athoracsur.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Izumchenko E., Chang X., Brait M. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun. 2015;6:8258. doi: 10.1038/ncomms9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X., Shigematsu H., Bekele B.N. EGFR tyrosine kinase domain mutations are detected in histologically normal respiratory epithelium in lung cancer patients. Cancer Res. 2005;65:7568–7572. doi: 10.1158/0008-5472.CAN-05-1705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.