Abstract
Introduction
Most patients (70%) with limited-stage SCLC (LS-SCLC) who are treated with curative-intent therapy suffer disease relapse and cancer-related death. We evaluated circulating tumor DNA (ctDNA) as a predictor of disease relapse and death after definitive therapy in patients with LS-SCLC.
Methods
In our previous work, we developed a plasma-based ctDNA assay to sequence 14 genes (TP53, RB1, BRAF, KIT, NOTCH1-4, PIK3CA, PTEN, FGFR1, MYC, MYCL1, and MYCN) that are frequently mutated in SCLC. In this work, we evaluated 177 plasma samples from 23 patients with LS-SCLC who completed definitive chemoradiation (n = 21) or surgical resection (n = 2) and had an end-of-treatment blood collection (median 4 d, range 0–40 d from treatment completion) plus monthly surveillance blood sampling. Median overall survival (OS) and progression-free survival (PFS) were compared using a Wilcoxon test.
Results
The median OS among patients in whom we ever detected ctDNA after definitive treatment (n = 15) was 18.2 months compared with a median OS of greater than 48 months among patients in whom we never detected ctDNA after definitive treatment (n = 8; p = 0.081). The median PFS among patients in whom we ever detected ctDNA after definitive treatment was 9.1 months compared with a median PFS of greater than 48 months among patients in whom we never detected ctDNA after definitive treatment (p < 0.001).
Conclusions
Detection of ctDNA in patients with LS-SCLC after curative-intent therapy predicts disease relapse and death. Prospective trials using ctDNA as an integral biomarker for therapeutic selection should be considered in SCLC.
Keywords: Small cell lung cancer, Circulating tumor DNA, Minimal residual disease, Liquid biopsy, Next-generation sequencing
Introduction
Lung cancer is the most common cause of cancer-related death in the United States, with SCLC comprising 15% of cases and accounting for 30,000 deaths annually.1,2
Although SCLC is initially responsive to chemotherapy in most patients,3 most patients with limited-stage SCLC (LS-SCLC) (70%) have lethal disease recurrence (30% locoregional and 70% distant4) with a median overall survival (OS) of 25 to 30 months.5 Currently, after completion of concurrent chemoradiotherapy or definitive surgery and adjuvant chemotherapy, patients with LS-SCLC are monitored with conventional radiography (typically computed tomography [CT] scans) every 2 to 3 months. There is an unmet need to identify microscopic disease after definitive therapy in patients with LS-SCLC to intervene and attempt to prolong patient survival.
The detection of both circulating tumor cells (CTCs)10, 11, 12, 13, 6, 7, 8, 9 and circulating tumor DNA (ctDNA)14, 15, 16, 17, 18, 19 has been well validated in patients with SCLC. On the basis of our previous work revealing the ability of ctDNA detection to precede radiographic progression in patients with SCLC,20 we hypothesized that detection of ctDNA in patients with LS-SCLC after definitive therapy would predict disease relapse and death.
Materials and Methods
Study Design and Patients
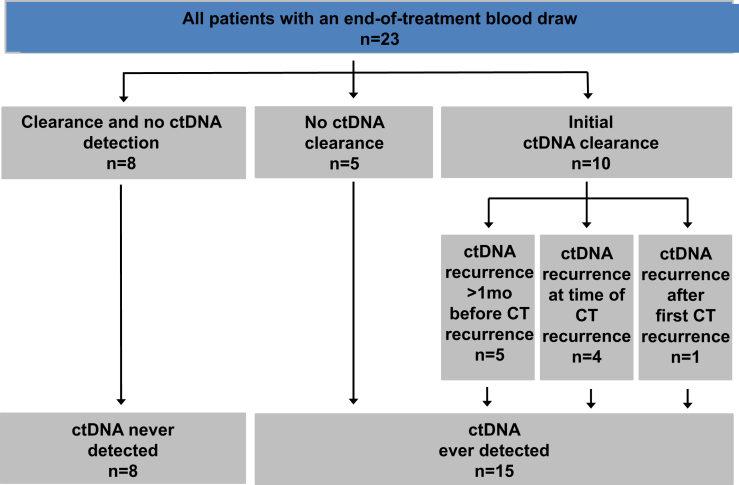
Patients with LS-SCLC treated at the Vanderbilt-Ingram Cancer Center were prospectively identified and consented using an institutional review board (IRB #030763)−approved protocol for collection of blood up to once per month plus medical record review. All samples were de-identified, and protected health information was reviewed according to the Health Insurance Portability and Accountability Act guidelines. Similar to analogous work, predominantly in patients with NSCLC,21 patients in whom blood was collected within 2 months of completion of definitive chemoradiation or surgical resection were included (Fig. 1). For eligible patients, all additional blood samples available were analyzed, resulting in the following breakup: 13 samples from treatment-naive patients, 15 samples from patients on definitive treatment, and 23 patients with analyzable end-of-treatment samples (Supplementary Table 1).
Figure 1.
Study schema. All patients with limited-stage SCLC with blood collection within 2 months of completion of definitive chemoradiation or surgical resection were included. Patients were divided into the following two broad categories: (1) patients in whom we never detected tumor-associated cell-free DNA (circulating tumor DNA [ctDNA]; n = 8) after definitive therapy and (2) patients in whom we ever detected circulating tumor DNA after definitive therapy (n = 15). CT, computed tomography.
Blood Samples and Cell-Free DNA Isolation
Blood samples (7.5 mL per tube) were collected in Streck tubes (Streck Inc., Omaha, NE) at up to monthly intervals at time points before, during, and after therapy. Blood was centrifuged at 1200g for 30 minutes. Plasma was removed and recentrifuged at 500g for 10 minutes and immediately aliquoted and stored at -80°C. DNA was extracted from the patients’ plasma samples using circulating nucleic acid extraction kits following the manufacturer’s instructions except for samples that were incubated with proteinase K for 1 hour rather than 30 minutes (Qiagen, Hilden, Germany). The yield of the double-strand DNA was quantified using a Qubit fluorometer (Thermo Fisher, Waltham, MA) and the corresponding double-strand DNA quantification kit. Approximately 40 to 100 ng of ctDNA, depending on the yield of ctDNA from the sample, was used for the library construction.
Targeted Next-Generation Sequencing
A detailed description of the platform used for ctDNA sequencing has been provided in a previous publication.20 Briefly, the panel contains 1608 probes that target all coding exons of BRAF, KIT, NOTCH1-4, PIK3CA, PTEN, RB1, and TP53. The panel also contains probes for copy variation detection in the genes FGFR1, MYC, MYCL1, and MYCN, and control probes that target select regions in all 22 autosomes. Full next-generation sequencing results for all patients and time points are included in Supplementary Table 1.
Statistical Analysis
Patient demographics and clinical information were summarized with median and range for continuous variables and frequency and percentage for categorical variables. The primary study end points were progression-free survival (PFS) and OS. PFS was defined as the time from the first treatment start date to the date of radiographic relapse, previous follow-up without progression, or death. OS was defined as the date of disease diagnosis to the date of all-cause death or previous follow-up. The Kaplan-Meier method, log-rank test, and Cox proportional hazard models were used to investigate the associations between PFS and OS and ctDNA status. Estimated hazard ratios (HRs) and 95% confidence intervals (CIs) were provided to measure the effect of the association between ctDNA clearance with PFS and OS. All statistical inferences were assessed using a two-sided 5% significance level, and all summary statistics, graphics, and survival models were generated using R version 3.6 statistical software.22
Results
Patient Demographics and Treatment
We prospectively enrolled 23 participants with a median age of 70 years (Table 1) over a period of 42 months. For subsequent analyses, we divided the cohort of patients into the following two groups: patients in whom we never detected ctDNA after definitive concurrent chemoradiation or surgical resection (“never detected,” n = 8) and patients in whom we ever detected ctDNA after definitive concurrent chemoradiation (“ever detected,” n = 15) (Fig. 1). In total, 21 patients were treated with chemotherapy (platinum plus etoposide) and radiation, and two patients were treated with surgical resection. Table 1 outlines the demographics, TNM staging, year of diagnosis, and treatment strategies among all patients and within the never detected and ever detected cohorts. The primary notable difference is that there is a shift toward earlier-stage patients, including two who were treated with surgical resection, in the never detected cohort. Median follow-up was 524 days (range 70–1474 d) for the overall cohort, 748 days (range 331–1474 d) for the never detected cohort, and 510 days (range 70–860 d) for the ever detected cohort (Table 1).
Table 1.
Baseline Demographics of Patients With Limited-Stage SCLC in Our Study Cohort
| Overall (N = 23) | ctDNA Never Detected After Definitive Treatment (n = 8) | ctDNA Ever Detected After Definitive Treatment (n = 15) | |
|---|---|---|---|
| Median age, y (range) | 70 (43–82) | 66 (43–75) | 70 (53–82) |
| Median follow-up, d (range) | 524 (70–1474) | 748 (331–1474) | 510 (70–860) |
| Sex, no. (%) | |||
| Female | 16 (70) | 5 (63) | 11 (73) |
| Male | 7 (30) | 3 (37) | 4 (27) |
| Ethnicity, no. (%) | |||
| White | 20 (88) | 12 (80) | |
| Black | 2 (8) | 8 (100) | 2 (13) |
| Asian | 1 (4) | 1 (7) | |
| Year of diagnosis | |||
| 2015 | 6 | 1 | 5 |
| 2016 | 5 | 3 | 2 |
| 2017 | 8 | 2 | 6 |
| 2018 | 4 | 2 | 2 |
| 2019 | 0 | 0 | 0 |
| TNM stage at diagnosis | |||
| IA2 | 2 | 1 | 1 |
| IA3 | 1 | 1 | 0 |
| IB | 1 | 1 | 0 |
| IIA | 0 | 0 | 0 |
| IIB | 2 | 1 | 1 |
| IIIA | 11 | 2 | 9 |
| IIIB | 3 | 1 | 2 |
| IIIC | 3 | 1 | 2 |
| First-line treatment, no. (%) | |||
| Platinum/etoposide with radiation | 21 (92) | 6 (75) | 15 (100) |
| Surgical resection | 1 (4) | 1 (12.5) | |
| Surgical resection with chemotherapy | 1 (4) | 1 (12.5) | |
| Prophylactic cranial irradiation, no. (%) | 10 (43) | 4 (50) | 6 (40) |
| Clinical treatment response, no. (%) | |||
| Partial response | 21 (91) | 6 (75) | 15 (100) |
| Complete response | 2 (9) | 2 (25) | 0 (0) |
ctDNA, circulating tumor DNA.
PFS and OS by ctDNA Detection After Definitive Treatment
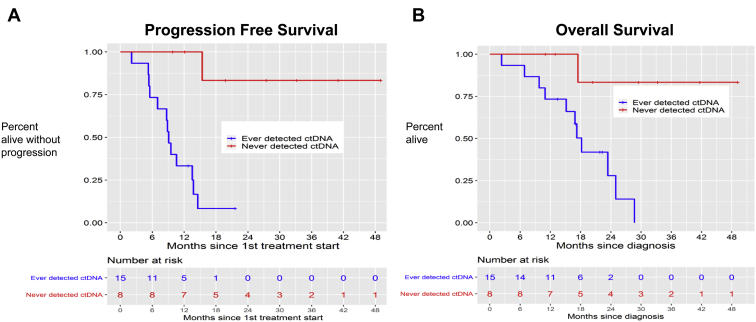
The median PFS among patients in whom we ever detected ctDNA was 9.1 months compared with a median PFS of greater than 48 months among patients in whom we never detected ctDNA (p < 0.001) (Fig. 2A). The median OS among patients in whom we ever detected ctDNA was 18.2 months compared with a current median OS of greater than 48 months among patients in whom we never detected ctDNA (p = 0.081) (Fig. 2B).
Figure 2.
Progression-free and overall survival for study cohort of patients with limited-stage SCLC. (A) Progression-free survival of the cohort of patients in whom we ever detected circulating tumor DNA (ctDNA) after definitive therapy (n = 15, blue line) versus never detected ctDNA after definitive therapy (n = 8, red line). (B) Overall survival of the cohort of patients in whom we ever detected ctDNA after definitive therapy (n = 15, blue line) versus never detected ctDNA after definitive therapy (n = 8, red line). LS-SCLC, limited-stage SCLC.
At the time of analysis, four of the 15 patients (26%) in whom ctDNA was ever detected remain alive, whereas seven of the eight patients (88%) in whom we never detected ctDNA remain alive.
Clinical Sequelae and Genomic Sequencing Results at ctDNA Detection
A summary of all patient cases with salient genomic changes is provided in Table 2. There was one lethal relapse in a patient in whom we never detected ctDNA. This individual withdrew consent for longitudinal blood collections 5 months and 18 days before disease relapse. The patient’s disease relapsed locally (in the hilar area) and was identified during hospitalization for an acute cerebrovascular accident with substantial debility. The patient enrolled in hospice care without receiving second-line systemic therapy and died 18 months after the diagnosis.
Table 2.
Overview of All Cases
| Patient ID | Age at Dx (y) | Sex | TNM Stage at Dx | Treatment-Naive Specimen | TP53 VAF (%) in Treatment-naive Specimen | Treatment Completion Specimen Peak Mutation VAF (%) | TP53 VAF (%) in First Posttreatment Specimen | Months Between ctDNA Detection and Radiographic Relapse or Death | Site(s) of Relapse | PFS (mo) | OS (mo) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 65 | Female | IIIA | No | No tx-naive sample | No detectable ctDNA | R175H 0.67% | 0 | Brain | 6 | 18 |
| 2 | 54 | Female | IIIC | Yes | Q192a 38.48% | No detectable ctDNA | Q192a 12.6% | 0 | Adrenal, thoracic LN | 6 | 23 |
| 3 | 74 | Male | IIIA | Yes | R65a 32.2% | No detectable ctDNA | R65a 1.73% | 3 | Bone marrow | 6 | 9 |
| 4 | 39 | Female | IIIA | Yes | Noneb | No detectable ctDNA | NA | No detectable ctDNA | NA | 48a | 50a |
| 5 | 74 | Male | IIIA | No | No tx-naive sample | No detectable ctDNA | Nonec | 0 | Pulmonary nodules | 9 | 23 |
| 6 | 65 | Female | IIIB | No | No tx-naive sample | BRAF K601E 0.65% | Noned | 12 | Lung, hilar LN | 12 | 29 |
| 7 | 72 | Male | IIIA | No | No tx-naive sample | TP53 K351a 0.22% | K351a 0.22% | 1 | Unknown | 1 | 3 |
| 8 | 65 | Female | IIIC | No | No tx-naive sample | No detectable ctDNA | NA | No detectable ctDNA | NA | 40a | 42a |
| 9 | 55 | Male | IIIB | Yes | E204a 26.08% | No detectable ctDNA | NA | No detectable ctDNA | NA | 32a | 40a |
| 10 | 61 | Female | IIB | Yes | E258K 2.1% | No detectable ctDNA | NA | No detectable ctDNAe | Lung | 14 | 18 |
| 11 | 79 | Female | IIIB | Yes | No detectable ctDNA | No detectable ctDNA | Nonef | 6g | Lung | 4 | 16 |
| 12 | 67 | Female | IA2 | No | No tx-naive sample | TP53 V272L 0.4% | V272L 0.4% | 4 | Unknown (clinical POD) | 4 | 7 |
| 13 | 69 | Male | IBh | Yes | No detectable ctDNA | No detectable ctDNA | NA | No detectable ctDNA | NA | 26a | 31a |
| 14 | 74 | Female | IIIA | Yes | E298a 44.4% | No detectable ctDNA | E298a 0.96% | 2 | Paratracheal LN, renal, adrenal | 4 | 11 |
| 15 | 66 | Male | IIIC | No | No tx-naive sample | No detectable ctDNA | C176F 15.05% | 0 | Liver, bone | 3 | 17 |
| 16 | 65 | Female | IIB | Yes | Y205S 2.63% | No detectable ctDNA | Y205S 0.24% | 7 | Unknown (clinical POD) | 12 | 15 |
| 17 | 67 | Female | IIIA | No | No tx-naive sample | TP53 frameshift 0.39% | Frameshift 0.39% | 7 | Pleural, supraclav LN | 7 | 24 |
| 18 | 72 | Female | IIIA | No | No tx-naive sample | No detectable ctDNA | NA | No detectable ctDNA | NA | 18a | 22a |
| 19 | 76 | Female | IIIA | No | No tx-naive sample | NOTCH3 T272M 0.37% | Nonei | 20 (ongoing) | Nonea | 20a | 23a |
| 20 | 52 | Female | IIIA | Yes | G245C 19.89% | No detectable ctDNA | G245C 0.51% | 3 | Lung | 11 | 18a |
| 21 | 78 | Female | IIIA | Yes | Y234C 33.61% | No detectable ctDNA | R248W 0.15% | 11 (ongoing) | Nonea | 12a | 17a |
| 22 | 73 | Female | IA3 | Yes | No detectable ctDNA | No detectable ctDNA | NA | No detectable ctDNA | NA | 12a | 14a |
| 23 | 59 | Male | IA2h | Yes | No detectable ctDNA | No detectable ctDNA | NA | No detectable ctDNA | NA | 12a | 13a |
Dx, diagnosis; VAF, variant allele frequency; NA, not applicable; PFS, progression-free survival from completion of first-line therapy; OS, overall survival from date of diagnosis; POD, progression of disease; ctDNA, circulating tumor DNA; Tx, treatment; LN, lymph node.
Ongoing.
MYCL1 amplification.
NOTCH1 R2272H at 0.18%.
BRAF K601E at 0.18%.
Withdrew consent 6 months before relapse.
PIK3CA E542K at 0.24%.
First ctDNA detection 6 months after radiographic relapse.
Surgical resection with adjuvant platinum/etoposide.
NOTCH3 T272M at 0.37%.
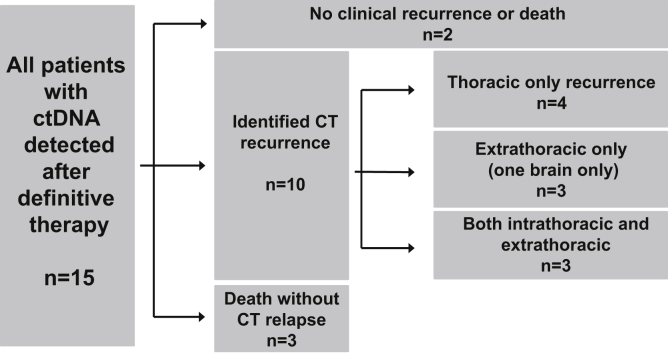
Among the 15 patients in whom we ever detected ctDNA, two have not had disease recurrence (11 and 20 mo after completion of definitive chemoradiation; patient IDs 21 and 19, respectively, in Table 2). Three patients in whom we ever detected ctDNA died without clear radiographic progression (with the most recent CT imaging 1, 2, and 7 mo before death; patient IDs 7, 12, and 16, respectively, in Table 2). In the remaining 10 patients, CT recurrence was first identified at an intrathoracic site in four patients, only at an extrathoracic site in three patients (including one patient with brain-only recurrence), and at both intra- and extrathoracic sites in three patients (Fig. 3). Among the 10 patients in whom radiographic relapse was identified, three did not receive further systemic therapy; three received nivolumab plus ipilimumab with best responses of stable disease in one patient and progression of disease at first evaluation in two patients; two patients with platinum-sensitive disease received platinum rechallenge and both initially responded; one patient enrolled on a clinical trial of a CHK1 inhibitor (LY2606368; NCT02735980)23 and responded; and one patient received paclitaxel and progressed at first disease evaluation. Only patients who clinically responded to second-line systemic therapy experienced bloodstream clearance of their ctDNA (three of seven, 43%).
Figure 3.
Sites of relapse in patients in whom circulating tumor DNA (ctDNA) was ever detected after definitive therapy. Clinical outcomes with radiographic sites of progression among the 15 patients in whom we ever detected circulating tumor DNA after definitive therapy. CT, computed tomography.
At the first time of detection of ctDNA in the ever detected cohort, the median variant allele frequency (VAF) was 0.4%, with a range of 0.15% to 15.05%. In 11 of the 15 patients in whom we ever detected ctDNA, there was a TP53 variant at greater than or equal to 0.1% allele frequency present (Table 2). In the four cases in which a TP53 variant was not present at first ctDNA detection after definitive therapy, the detected variants were in BRAF, NOTCH1, NOTCH3, and PIK3CA (patient IDs 6, 5, 19, and 11, respectively, in Table 2).
Exemplary Patient Cases
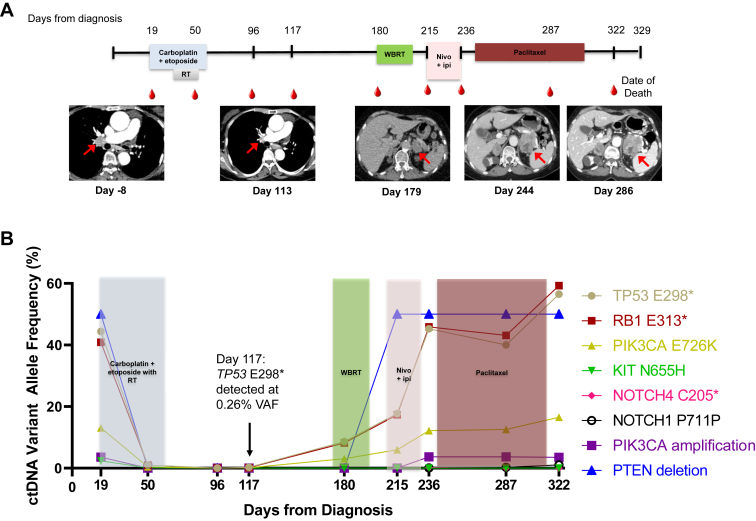
As a case demonstrative of the use of ctDNA after definitive therapy for patients with LS-SCLC, patient ID 14 (Table 2; Fig. 4A and B) was diagnosed with stage IIIA disease with a 7-mm upper lobe lung primary and biopsy-confirmed ipsilateral hilar and mediastinal lymph node involvement. She was 74 years at diagnosis, a former smoker with a 50 pack-year history, and Eastern Cooperative Oncology Group performance status score of 1. She had a ctDNA assessment at diagnosis with multiple findings: TP53 E298∗ at 44.4%, RB1 E313∗ at 40.86%, PIK3CA E726K at 13.05%, KIT N655H at 2.36%, a PIK3CA amplification, and a PTEN deletion. She was treated with concurrent chemoradiation with carboplatin plus etoposide, and she had a complicated treatment course with febrile neutropenia and pancytopenia limiting her chemotherapy to two cycles. Four days before her final first-line chemotherapy dose (day 50), she continued to have detectable ctDNA: TP53 E298∗ at 0.96%, RB1 E313∗ at 0.96%, PIK3CA E726K at 0.4%, and KIT N655H at 0.1%. She completed 68 gray thoracic radiation 6 weeks after her previous dose of chemotherapy, and at that time, she had no detectable ctDNA in her end-of-treatment blood sample (day 96). At her follow-up with medical oncology four weeks after completion of radiation (day 117), she again had detectable ctDNA (TP53 E298∗ at 0.26%, RB1 E313∗ at 0.17%, PIK3CA E726K at 0.11%), but a partial response on chest CT, with expected radiation-related changes, was observed. She proceeded to receive prophylactic cranial irradiation, and at her follow-up 3 months after therapy completion, she had multiple sites of disease recurrence (a fluorodeoxyglucose avid 1.5 cm paratracheal lymph node, an avid 3.4 cm adrenal metastasis, and an avid 1.7 cm renal metastasis) with a significant rise in her ctDNA: TP53 E298∗ at 8.63%, RB1 E313∗ at 8.17%, and PIK3CA E726K at 3.06%. She was treated with nivolumab plus ipilimumab, but she had significant disease progression at first evaluation with both enlargement of her preexisting adrenal lesion and new areas of disease (retroperitoneal, supraclavicular, and para-aortic lymph nodes, contralateral adrenal lesion) prompting a change of therapy to paclitaxel. At the time of progression on nivolumab plus ipilimumab, her ctDNA was notably increasing: TP53 E298∗ at 45.28%, RB1 E313∗ at 45.88%, PIK3CA E726K at 12.2%, NOTCH4 C205∗ at 0.24%, NOTCH1 P711P at 0.05%, a PIK3CA amplification, and a PTEN deletion. Her disease continued to progress on paclitaxel, with initial ctDNA stabilization, followed by a notable rise on a blood draw 1 week before her death (she transitioned to hospice care 1 week before death): TP53 E298∗ at 56.55%, RB1 E313∗ at 59.27%, PIK3CA E726K at 16.51%, NOTCH1 P711P at 1.05%, a PIK3CA amplification, and a PTEN deletion.
Figure 4.
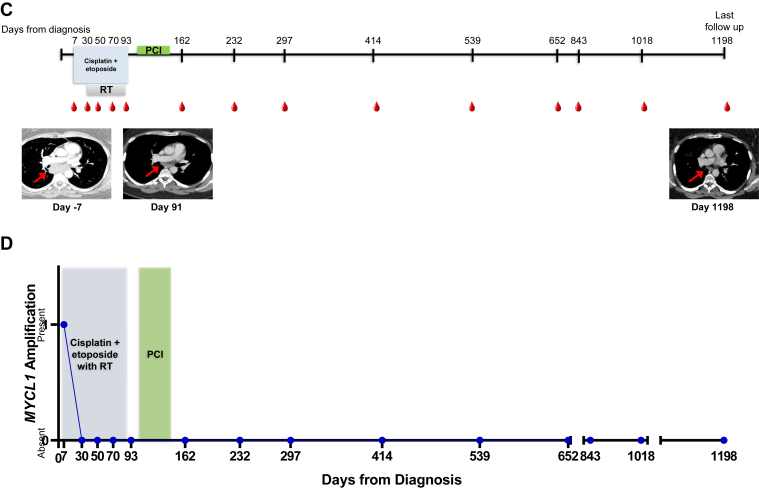
Exemplary patient cases. (A) A timeline for patient ID 14’s clinical treatments and outcomes from the day of diagnosis until death with radiographic images is revealed. The shaded bars represent the treatment time frames, radiation therapy (RT), whole-brain radiation therapy, and nivolumab plus ipilimumab therapy (nivo + ipi). Radiographic images are presented from day 8 before diagnosis and at days 113, 179, 244, and 286. The red arrows indicate the patient’s right hilar disease in the left two panels and the enlarging left adrenal mass in the right three panels. (B) The patient’s circulating tumor DNA (ctDNA) variants and variant allele frequencies (VAFs) are plotted in accord with their treatment course. The black arrow denotes the detection of TP53 E298∗ mutation, VAF. (C) A timeline for patient ID 4’s clinical treatments and outcomes from the day of diagnosis until previous follow-up with radiographic images is revealed. The shaded bars represent the treatment time frames, RT, and prophylactic cranial irradiation (PCI). Radiographic images are presented from day 7 before diagnosis and at days 91 and 1198. The red arrows indicate the patient’s mediastinal disease that has notably improved and not increased with prolonged surveillance. (D) The patient’s circulating tumor DNA variants and VAFs are plotted in accord with their treatment course.
In contrast to the aforementioned case, patient ID 4, who we described in our previous publication20 and have had subsequent follow-up, (Table 2; Fig. 4C and D) was diagnosed with stage IIIA LS-SCLC with a 3.2-cm right lower lobe primary and bulky mediastinal lymph node involvement. She was 39 years at diagnosis and had a 10 pack-year history of smoking and Eastern Cooperative Oncology Group performance status score of 0. She had a MYCL1 amplification detected in a treatment-naive blood sample but cleared this finding after her first cycle of cisplatin plus etoposide. She completed four cycles of chemotherapy plus thoracic radiation and prophylactic cranial irradiation, and has had 11 peripheral blood assessments since her end-of-treatment draw (up to 36 mo after treatment completion) with no ctDNA findings and no evidence of disease recurrence.
Prognostic Value of Treatment-Naive VAF and ctDNA Clearance on First-Line Treatment
Of the 23 patients included in the full analysis, 13 had treatment-naive samples available for analysis. Among these 13 patients, we did not observe any prognostic significance for progression or death on the basis of maximum diagnostic ctDNA VAF (HR for progression: 1.01, CI: 0.97–1.05; HR for death: 1, CI: 0.95–1.04) or mean diagnostic ctDNA VAF (HR for progression: 1.01, CI: 0.94–1.08; HR for death: 0.99, CI: 0.91–1.07). Nevertheless, of the patients with clearance of ctDNA during first-line therapy (n = 9), delayed time to ctDNA clearance was a significant predictor of progression (HR for progression 1.1, CI: 1.01–1.19) and death (HR for death 1.07, CI: 1.01–1.15), with a median time to clearance of 63 days (range 29–92 d) among all patients. Notably, three patients had cleared ctDNA at their first on-treatment draw at approximately 30 days (29 d, 29 d, and 32 d, respectively). These three patients have had no evidence of relapse and all remain alive at greater than 1 year (median 965 d) since the start of their first-line treatment. Of the patients that had disease recurrence, the median time to clearance on first-line therapy was 65 days and median time to progression was 249 days, with all but one patient having died of their disease (median time to death 437 d).
Discussion
Using a custom 14-gene SCLC next-generation sequencing panel, we have reported that detection of ctDNA at any time point after curative-intent therapy is a poor prognostic finding. We have reported that residual ctDNA can be detected before radiographic relapse, and it presages relapse at both isolated intrathoracic and extrathoracic sites. These findings are consistent with those of reports in patients with NSCLC21 and stage III colorectal cancer.24 We have also reported that in a cohort of patients with LS-SCLC, time to ctDNA clearance during first-line therapy is a significant predictor of PFS and OS. This finding is consistent with those of previous studies revealing the negative prognostic significance of delayed CTC clearance in patients with SCLC.12,13 Our finding that there was no association between peak or median diagnostic VAF and clinical outcomes (PFS, OS) among 13 of our patients is not consistent with that of a previous publication demonstrating that patients with SCLC with a higher-than-median VAF at diagnosis (0.18%) have inferior PFS and OS. Importantly, our analysis cohort was smaller (n = 14) and not powered specifically for a diagnostic ctDNA VAF analysis, and the comparator study analyzed a larger cohort of patients (n = 22) with approximately half LS-SCLC and half extensive-stage SCLC.16 It has also been reported that in only patients with LS-SCLC, detection of 15 or more CTCs per 7.5 mL at diagnosis is a poor prognostic finding independent of therapy.25 The prognostic significance of ctDNA VAF requires further study to draw definitive conclusions.
The limitations of the current study include its single-center accrual and moderate number of patients. A larger cohort of patients exclusively managed with concurrent chemoradiation is the ideal cohort in which to further validate these findings.
Although the field of SCLC CTC6, 7, 8 and ctDNA analysis14, 15, 16, 17, 18, 19, 20, 21 has progressed rapidly from assay validation and characterization of the disease’s dynamic genomic evolution throughout a patient’s treatment course to identification of potential targetable mutations, there remains a significant unmet need in evaluating the clinical utility of the integration of ctDNA into routine patient care, such as for residual disease monitoring.3 On the basis of our analysis, a randomized, prospective clinical trial that evaluates the initiation of second-line systemic therapy at the time of detection of ctDNA rather than measurable lesions on conventional imaging in patients with LS-SCLC should be considered.
Acknowledgments
Dr. Lovly was supported in part by a Vanderbilt-Ingram Cancer Center Young Ambassadors Award, a Lung Cancer Foundation of America/ International Association for the Study of Lung Cancer Lori Monroe Scholarship, P30-CA086485, UG1CA233259, U54CA217450-01, and U01CA224276-01. Dr. Iams was supported by the National Institutes of Health (NIH) and National Cancer Institute (NCI) Vanderbilt Clinical Oncology Research Career Development Award 2K12CA090625-17 and an American Society of Clinical Oncology/ Conquer Cancer Foundation Young Investigator Award. Mr. Zhao was supported in part by CCSG NCI/ NIH 2P30CA068485-19. All data storage for this research project utilized Vanderbilt’s Redcap data storage infrastructure, funded by the National Center for Advancing Translational Sciences/ NIH grant UL1TR000445. The authors would like to thank the patients and their families. The authors would also like to thank all the members of the Lovly Laboratory (authors Kopparapu, Yan, Brandon Williams, Yunkai Zhang, Huan Qiao, Henry Henderson, and Portia Thomas) and the Cancer Systems Biology Consortium U54 Research Team at Vanderbilt for valuable project discussions. Dr. Iams, Dr. Kopparapu, Ms. Yan, Dr. Lim, and Dr. Lovly designed the experiments. Dr. Kopparapu and Ms. Yan performed the experiments. Dr. Iams, Mr. Zhao, Dr. Chen, Dr. Cann, Ms. Bertucci, Mr. Shaffer, Dr. Horn, Dr. Garg, Dr. Hosseini, Dr. Lim, and Dr. Lovly generated and analyzed the data. Dr. Horn, Dr. York, Dr. Ancell, and Dr. Wyman provided direct patient care. Dr. Iams and Dr. Lovly wrote the manuscript. Zhao and Dr. Chen performed the statistical analysis. All the authors reviewed the data and final manuscript.
Footnotes
Disclosure: Dr. Lovly has served as a consultant for Pfizer, Novartis, AstraZeneca, Genoptix, Sequenom, ARIAD, Takeda, Foundation Medicine, Blueprints Medicine, Achilles, and Cepheid; has been an invited speaker for Abbott and Qiagen; and has received research funds (to her university) from Novartis, AstraZeneca, and Xcovery. Dr. Horn is a consultant for AstraZeneca, EMD Serono, Genentech-Roche, Tesaro, Pfizer, Incyte AbbVie, Bristol-Myers Squibb, Merck & Co., and Xcovery and has received research support from Xcovery, Bristol-Myers Squibb, and Boehringer Ingelheim. Dr. Iams reports consulting for Genentech, Outcomes Insights, and Defined Health and clinical trial funding from EMD Serono. Ms. Bertucci, Mr. Shaffer, Ms. Hodsdon, Dr. Garg, Dr. Hosseini, and Dr. Lim are employees and shareholders of Resolution Bioscience. The remaining authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2020.100024.
Supplementary Data
References
- 1.Torre L.A., Siegel R.L., Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt E.B., Jalal S.I. Small cell lung cancer. Cancer Treat Res. 2016;170:301–322. doi: 10.1007/978-3-319-40389-2_14. [DOI] [PubMed] [Google Scholar]
- 3.Blackhall F., Frese K.K., Simpson K., Kilgour E., Brady G., Dive C. Will liquid biopsies improve outcomes for patients with small-cell lung cancer? Lancet Oncol. 2018;19:e470–e481. doi: 10.1016/S1470-2045(18)30455-8. [DOI] [PubMed] [Google Scholar]
- 4.Salem A., Mistry H., Hatton M. Association of chemoradiotherapy with outcomes among patients with stage I to II vs stage III small cell lung cancer: secondary analysis of a randomized clinical trial. JAMA Oncol. 2019;5 doi: 10.1001/jamaoncol.2018.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faivre-Finn C., Snee M., Ashcroft L. Concurrent once-daily versus twice-daily chemoradiotherapy in patients with limited-stage small-cell lung cancer (CONVERT): an open-label, phase 3, randomised, superiority trial. Lancet Oncol. 2017;18:1116–1125. doi: 10.1016/S1470-2045(17)30318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter L., Rothwell D.G., Mesquita B. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med. 2017;23:114–119. doi: 10.1038/nm.4239. [DOI] [PubMed] [Google Scholar]
- 7.Hou J.M., Krebs M.G., Lancashire L. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 8.Mohan S., Foy V., Ayub M. Profiling of circulating free DNA using targeted and genome-wide sequencing in patients with SCLC. J Thorac Oncol. 2020;15:216–230. doi: 10.1016/j.jtho.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Wang H.T., Li B.G. Prognostic significance of circulating tumor cells in small-cell lung cancer patients: a meta-analysis. Asian Pac J Cancer Prev. 2014;15:8429–8433. doi: 10.7314/apjcp.2014.15.19.8429. [DOI] [PubMed] [Google Scholar]
- 10.Naito T., Tanaka F., Ono A. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J Thorac Oncol. 2012;7:512–519. doi: 10.1097/JTO.0b013e31823f125d. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y., Liu X.Q., Fan Y. Circulating tumor cell counts/change for outcome prediction in patients with extensive-stage small-cell lung cancer. Future Oncol. 2016;12:789–799. doi: 10.2217/fon.15.346. [DOI] [PubMed] [Google Scholar]
- 12.Hiltermann T.J., Pore M.M., van den Berg A. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol. 2012;23:2937–2942. doi: 10.1093/annonc/mds138. [DOI] [PubMed] [Google Scholar]
- 13.Normanno N., Rossi A., Morabito A. Prognostic value of circulating tumor cells’ reduction in patients with extensive small-cell lung cancer. Lung Cancer. 2014;85:314–319. doi: 10.1016/j.lungcan.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Cuesta L., Perdomo S., Avogbe P.H. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine. 2016;10:117–123. doi: 10.1016/j.ebiom.2016.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du M., Thompson J., Fisher H., Zhang P., Huang C.C., Wang L. Genomic alterations of plasma cell-free DNAs in small cell lung cancer and their clinical relevance. Lung Cancer. 2018;120:113–121. doi: 10.1016/j.lungcan.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Nong J., Gong Y., Guan Y. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9:3114. doi: 10.1038/s41467-018-05327-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Board R.E., Williams V.S., Knight L. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci. 2008;1137:98–107. doi: 10.1196/annals.1448.020. [DOI] [PubMed] [Google Scholar]
- 18.Devarakonda S., Sankararaman S., Herzog B.H. Circulating tumor DNA profiling in small-cell lung cancer identifies potentially targetable alterations. Clin Cancer Res. 2019;25:6119–6126. doi: 10.1158/1078-0432.CCR-19-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgensztern D., Devarakonda S.H., Masood A. Circulating cell-free tumor DNA (cfDNA) testing in small cell lung cancer. J Clin Oncol. 2016;34(suppl 15) [Google Scholar]
- 20.Almodovar K., Iams W.T., Meador C.B. Longitudinal cell-free DNA analysis in patients with small cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J Thorac Oncol. 2018;13:112–123. doi: 10.1016/j.jtho.2017.09.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri A.A., Chabon J.J., Lovejoy A.F. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2010. R: A Language and Environment for Statistical Computing: Reference Index. [Google Scholar]
- 23.Byers L.A., Golden L., Zhang W., Lin A.B., Forster M. P2.06-028 A phase 2 study of prexasertib in patients with extensive stage small cell lung cancer: topic mesothelioma and SCLC. J Thorac Oncol. 2017;12(suppl) S1088-1089. [Google Scholar]
- 24.Tie J., Cohen J.D., Wang Y. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5:1710–1717. doi: 10.1001/jamaoncol.2019.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tay R.Y., Fernandez-Gutierrez F., Foy V. Prognostic value of circulating tumour cells in limited-stage small-cell lung cancer: analysis of the concurrent once-daily versus twice-daily radiotherapy (CONVERT) randomised controlled trial. Ann Oncol. 2019;30:1114–1120. doi: 10.1093/annonc/mdz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.