Abstract
Precise and sensitive quantitation of viral RNA in specimens from human immunodeficiency virus (HIV) type 1 (HIV-1)-infected individuals has become an indispensable tool for the monitoring of the efficacy of highly active antiretroviral combination therapy. The present report describes reproducible and efficient protocols with enhanced sensitivity for quantitation of HIV-1 RNA from plasma, peripheral blood mononuclear cells, and tissues with Qiagen silica columns for RNA purification combined with the Roche Amplicor HIV-1 Monitor test for quantitative reverse transcription-PCR (RT-PCR). Extraction of RNA from 0.5 ml of plasma resulted in the detection of fewer than 20 HIV RNA copies/ml of plasma, equivalent to the centrifugation-based boosted RT-PCR assay. Silica extraction of cellular RNA resulted in the detection of fewer than 3 HIV-1 RNA copies/μg of total RNA. These techniques facilitate direct comparisons of viral loads between liquid and cellular specimens. Application of these sensitive methods may improve the assessment of the response to new antiretroviral regimens.
Highly active antiretroviral combination therapy (HAART) frequently leads to undetectable viral loads in plasma (3, 14). To evaluate new forms of therapy in the research setting and to follow therapeutic efficacy in clinic practice, increasingly sensitive diagnostic tests are needed. Reverse transcription-PCR (RT-PCR) for the detection of human immunodeficiency virus (HIV) RNA in plasma is widely used to monitor HIV type 1 (HIV-1) levels (8). In particular, the ultrasensitive or boosted Roche Amplicor HIV-1 Monitor test is among the most sensitive, reproducible, and best-documented tools for the detection of low levels of HIV-1 in plasma (7, 10). The boosted method represents a modification of the original Amplicor test, in which the virus in plasma is concentrated by centrifugation before RT-PCR, resulting in detection limits in the range of 20 to 50 HIV-1 RNA copies per ml. This test has become a new standard for the monitoring of patients receiving HAART. In addition to providing a very sensitive means of detection of HIV-1 RNA, it can be performed in most well-equipped laboratories without additional equipment, and the use of commercially available reagents facilitates standardization.
Recent studies have shown that patients on long-term HAART can reach such sustained viral suppression that their plasma virus loads drop below the limit of detection of the boosted RT-PCR (<20 HIV-1 RNA copies/ml) (4). Some investigators have turned to cellular specimens, such as peripheral blood mononuclear cells (PBMCs) and lymphoid tissue biopsy specimens, to monitor HIV transcription as a marker for viral activity (2, 6, 9, 12, 13).
This report presents novel methods for the detection of HIV-1 RNA from plasma, PBMCs, and tissue specimens. By using commercially available solid silica columns for quantitative RNA extraction and the Amplicor test for RT-PCR, highly sensitive and reproducible quantitation of residual HIV-1 RNA can be achieved from plasma and cellular specimens.
MATERIALS AND METHODS
Subjects and specimens.
Responses to combination antiretroviral therapies (zidovudine [AZT] plus lamivudine [3TC] or AZT plus 3TC plus ritonavir) were studied in subgroups of a larger study population, including 44 HIV-1-infected, antiretroviral agent-naive, asymptomatic patients attending the University of Zurich Infectious Diseases Clinic. This project was approved by the University Hospital Ethics Review Committee, and all subjects provided written informed consent. Details of the study will be reported separately.
Sequential blood samples (CPT Vacutainer tubes; Becton Dickinson, Franklin Lakes, N.J.) were collected monthly after the initiation of therapy. Plasma aliquots and PBMC “dry” pellets were stored at −75°C. Tonsil biopsy specimens were obtained at 0, 4, 24, and 48 weeks, immediately snap-frozen by immersion in liquid nitrogen, and stored in liquid nitrogen. Tissues were further processed by cryosectioning of unfixed frozen biopsy specimens (6-μm sections). Individual or multiple frozen sections were placed in 1.5-ml screw-top tubes (Sarstedt, Nurnbrecht, Germany) at −20°C, and the tubes were stored at −75°C until the samples were used for RNA extraction. Cultured HIV-uninfected H9 cells (ATCC HTB 176) or HIV-infected 8E5 cells (ATCC CRL 8993) were stored at −20°C as “dry” pellets containing 106 cells. Plasma specimens from 28 patients, PBMC specimens from 9 patients, and tonsil specimens from 7 patients were assessed to collect the data presented in this report.
Extraction of RNA from plasma.
Two protocols were used for extraction of RNA from plasma. The standard Amplicor protocol was performed with the reagents included in the Amplicor HIV-1 Monitor test kit (Roche Diagnostic Systems, Inc., Branchburg, N.J.), with the following modifications. Instead of the recommended 0.2 ml of plasma, 0.15 ml of plasma was used as the starting volume so that the entire procedure could be performed in 1.6-ml microtubes. The resulting precipitate was dissolved in 0.3 ml of Amplicor specimen diluent instead of the recommended 0.4 ml, to compensate for the decreased plasma input volume. In addition, the plasma was mixed with 2 μl of SeeDNA (Amersham, Little Chalfont, United Kingdom) before extraction to better visualize the precipitate. The remainder of the procedure was conducted as recommended by the manufacturer. These modifications improved the reproducibility and handling of the Amplicor HIV-1 Monitor test without affecting other aspects of the test performance (unpublished observations).
Silica column extraction of RNA from plasma was carried out with the QIAamp Viral RNA Kit (Qiagen, Hilden, Germany), except that the manufacturer’s instructions were modified as follows. Plasma (0.56 ml) was mixed with 4 volumes (2.24 ml) of QIAamp lysis buffer (AVL) supplemented with 2 μg of poly(A) carrier RNA, and the mixture was incubated for 10 min at room temperature. The Roche Amplicor Quantitation Standard (QS) from the Amplicor HIV-1 Monitor test was separately diluted 5.4-fold in AVL buffer (Qiagen). Five microliters of diluted QS (about 70 to 100 QS copies, which varied from batch to batch) and 0.8 volume (2.24 ml) of absolute ethanol were added to the plasma-lysis buffer solution. The equivalent of 0.5 ml of plasma (90% of the mixture, i.e., 4.54 ml) was passed through a QIAamp mini column at a speed of 2 ml/min by suction with a peristaltic micropump (IPS-16; Ismatec, Zurich, Switzerland). The column was washed successively with 850 μl of AW1 buffer and 700 μl of AW2 buffer (Qiagen) by applying negative pressure as described above. The column was spun dry by spinning at 2,000 × g for 5 min, loaded with 50 μl of sterile water preheated to 80°C, and incubated in a preheated oven at 80°C for 5 min. Nucleic acid was recovered by centrifugation at 6,000 × g for 1 min and, preferably, was used immediately or was stored at −70°C until it was needed for RT-PCR. Up to 12 specimens could be processed simultaneously with a 12-place manifold and multiple channels on the peristaltic pump.
Extraction of RNA from PBMCs and tissues.
Extraction of RNA from cultured cells, PBMCs, and tonsil specimens was performed with the RNeasy Kit (Qiagen) according to the manufacturer’s instructions, with minor modifications. Briefly, 106 to 107 frozen PBMCs or 0.05 to 1.0 mg of tissue (1 to 20 frozen 6-μm sections) was dissolved in 0.6 ml of Qiagen RNeasy lysis buffer. The cell-lysis buffer mixtures were further dissociated by passage through a Qiashredder column (Qiagen). Total RNA was then extracted with an RNeasy spin column (Qiagen). After elution with 100 μl of diethyl pyrocarbonate-treated water and adjustment of the eluate to 10 mM MgCl2 and 10 mM Tris HCl (pH 7.5), residual DNA was digested with 10 U of RNase-free DNase I (Boehringer, Mannheim, Germany) for 15 min at 37°C. The DNase-treated RNA was repurified with another RNeasy column and was eluted in 100 μl of diethyl pyrocarbonate-treated water. The purified RNA was stored at −75°C until it was needed.
Purified cellular RNA was quantified and checked for intactness by electrophoresis in a denaturing formaldehyde–1.5% agarose gel (1). A photograph (Polaroid instant film 667, Polaroid, St. Albans, United Kingdom) of the gel was digitized with a scanner (ScanJet IIc; Hewlett-Packard Co., Palo Alto, Calif.), and the 28S and the 18S rRNA bands were quantitated by comparison to an RNA standard by using densitometric computer software (Intelligent Quantifier, BioImage, Ann Arbor, Mich.).
RT-PCR.
RNA extracted with silica columns was analyzed by the Amplicor HIV-1 Monitor test. QIAamp-extracted plasma RNA (50 μl that already contained QS) was mixed with 50 μl of the Amplicor Master Mix. RNeasy-extracted total cellular RNA (3 to 30 μl) was mixed with 50 μl of Amplicor Master Mix, 0.83 μl of QS (70 to 90 copies of QS, depending on the batch used), and sample diluent (from the Amplicor test kit) to a final volume of 100 μl. Regardless of the extraction method, the amount of QS added to each specimen was calculated so that the numbers of copies of QS in each amplification reaction were the same as the number recommended for the unmodified Amplicor test. Thermal cycling and detection were performed according to the Amplicor test instructions. Limits of detection were determined by comparison of the QS internal controls with a constant cutoff value (0.2 optical density [OD] unit) for positive HIV results, as suggested in the manual for the Amplicor test, by the following formula: absolute detection limit (number of copies per PCR mixture) = 0.2 OD × input number of QS copies/PCR)/(total QS OD). Relative detection limits were calculated from the latter by normalizing to the sample input. Thus, relative detection limits are expressed as number of copies per milliliter for plasma and as number of copies per microgram of total RNA or as number of copies per cell equivalent for cellular specimens.
Statistics and calculations.
Statistical analyses were carried out with GraphPad Prism software, version 2.01 (AMPL Software Pty. Ltd., Turramurra, Australia). Unless stated otherwise, calculated values are followed by the standard error (SE).
Calculations of plasma HIV-1 RNA copy numbers were done according to the Amplicor instructions, except that the dilution factor was adjusted to reflect the volume of plasma represented in each reaction mixture. For cellular RNA, the number of HIV RNA copies per reaction mixture was divided by the micrograms of total RNA added to that reaction mixture, resulting in the number of HIV-1 RNA copies per microgram of total RNA (number of copies of HIV RNA per microgram of RNA).
RESULTS
Quantitation of HIV-1 RNA from plasma.
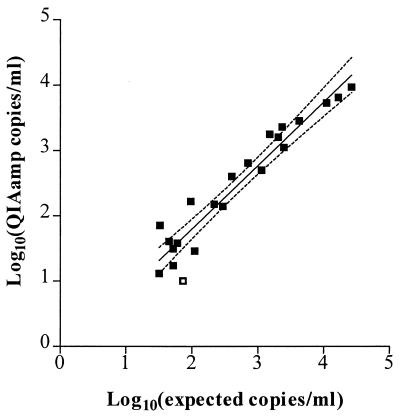
Recovery of native HIV-1 RNA from plasma was tested by spiking HIV-negative plasma with clinically obtained HIV-1-positive plasma resulting in 22 mixtures containing 30 to 27,000 HIV-1 RNA copies/ml (Fig. 1). Linear regression of log10(QIAamp viral load) versus log10(expected viral load) demonstrated a strong correlation over the range tested (r2=0.91), with a slope of 0.97 ± 0.07, which was not significantly different from 1 (P = 0.67; t test) and a y intercept of −0.14 ± 0.18. Among the nine specimens within this data set with expected viral loads of <200 copies/ml (range, 32 to 110 copies/ml), only one was negative (expected viral load, 73 copies/ml; measured load, <17 copies/ml).
FIG. 1.
Comparison of plasma viral load determination by using QIAamp extraction versus the standard Amplicor protocol. Aliquots of clinically obtained HIV-1-positive plasma specimens were thawed and measured by the standard Amplicor test. Other aliquots of these specimens were thawed, diluted 10-fold in HIV-negative control plasma, and extracted by the QIAamp procedure, and the purified RNA was measured by the Amplicor test. Expected viral loads were calculated as 10% of the value measured for the undiluted aliquot by the standard Amplicor test. Closed squares, data points for specimens with a detectable measurement in the QIAamp test. One diluted specimen (open square) was negative for HIV RNA (expected readout, 73 copies/ml; measured value, <17 copies/ml). Linear regression (r2 = 0.91; n = 22), as shown by the solid line, and the 95% confidence intervals (dotted lines) result in an equation of y = (0.97 ± 0.07)x − (0.14 ± 0.18) (the slope and the y-intercept values are indicated as the mean ± SE).
To obtain an estimate of the sensitivity of the measurement of the viral load in plasma specimens, relative detection limits (expressed as number of HIV copies per milliliter of plasma) were determined with clinical specimens from which RNA was extracted by the standard Amplicor or by the QIAamp procedures (Table 1). The mean relative detection limit of the QIAamp procedure (14.9 ± 0.6 copies/ml) was 11 times lower than that of the standard Amplicor protocol (163.6 ± 5.9 copies/ml).
TABLE 1.
Absolute and relative detection limits of RT-PCR
| Specimen type | Extraction method | Specimen input/PCR mixture | Absolute detection limit (no. of copies/PCR mixture)a | Relative detection limitab | Sample no. |
|---|---|---|---|---|---|
| Plasma | Amplicor | 25 μl | 4.1 ± 0.1 | 163.6 ± 5.9 | 278 |
| QIAamp | 500 μl | 7.4 ± 0.3 | 14.9 ± 1.6 | 252 | |
| Cellsc | RNeasy | 20–2,600 ng of RNA | 3.1 ± 0.2 | NDd | 97 |
| RNeasy | 20–500 ng of RNA | 3.5 ± 0.5 | ND | 19 | |
| RNeasy | 900–1,100 ng of RNA | 2.3 ± 0.2 | 2.4 ± 0.2 | ||
| RNeasy | 1,500–2,600 ng of RNA | 3.7 ± 0.3 | ND | 20 |
Values are averages ± standard errors of the means.
For plasma, the detection limit is number of copies per milliliter; for cellular specimens, the detection limit is number of copies per microgram of RNA.
PBMCs and tonsil biopsy specimens.
ND, not determined.
Quantitation of HIV-1 RNA from cells.
The RNeasy RNA extraction kit (Qiagen) was used to extract total RNA from cultured cells, PBMCs, and tonsil biopsy specimens. With 106 H9 cells per extraction, the RNeasy kit produced an average RNA yield of 12.1 ± 0.4 μg/106 H9 cells (11 specimens were tested). In experiments with 106 to 107 cells per extraction with PBMCs from two HIV-negative donors, the average yield was 1.1 ± 0.1 μg of total RNA/106 PBMCs (six specimens were tested). The average yield of RNA from tonsil tissue was 2.8 ± 0.4 μg/mg of tissue (27 specimens tested).
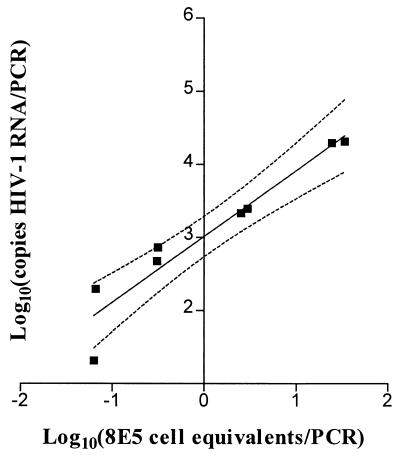
To assess the linearity of HIV-1 RNA quantitation, fresh HIV-infected 8E5 cells were mixed with uninfected H9 cells to create controls containing 2, 10, 100, or 1,000 8E5 cells per 106 H9 cells. Aliquots of silica-extracted RNA from each mixture were amplified with the Amplicor test kit (Fig. 2). The correlation between input the number of 8E5 cell equivalents and the number of HIV-1 RNA copies was linear over the range tested (0.06 to 34 infected cell equivalents per PCR; 20 to 19,000 HIV-1 RNA copies per PCR; r2 = 0.91), and the slope (0.90) was not significantly different from 1 (P = 0.40; t test).
FIG. 2.
Cell mixing experiment to assess the linearity of HIV-1 RNA measurement in cellular specimens. RNA was extracted from frozen cell pellets representing mixtures of HIV-infected cells (8E5) and uninfected cells (H9). Specimens had been mixed to contain 2, 10, 100, or 1,000 8E5 cells per 106 H9 cells and 106 cells per pellet (n = 8; four duplicate measurements). Aliquots containing 297 to 409 ng of total RNA were measured by PCR. Linear regression analysis (r2 = 0.91; n = 8) as shown by the solid line and the 95% confidence intervals (dotted lines) results in an equation of y = (0.90 ± 0.11)x + (3.01 ± 0.11) (the slope and the y-intercept values are indicated as mean ± SE).
To determine whether the amount of RNA input into the test mixture had any inhibitory effects on the PCR, the absolute detection limits (expressed as number of copies per PCR mixture) of these reactions were analyzed. In contrast to measurements with plasma, with which detection limits give an indication of the efficacies of both the extraction and the PCR, absolute detection limits for measurements with cellular specimens by our protocol give an estimate of the efficiency of the PCR and its detection because the QS RNA is added to the RNA only after extraction. The readout for the internal standard therefore provides a good measure of PCR inhibition. Experiments with RNA inputs ranging from 20 to 2,600 ng produced a mean absolute detection limit of 3.1 ± 0.2 copies/PCR mixture (Table 1). Subsets of these data obtained from experiments with low (20 to 500 ng), intermediate (800 to 1,100 ng), and high (1,500 to 2,600 ng) RNA inputs (Table 1) showed similar absolute detection limits of 3.5 ± 0.5, 2.3 ± 0.2, and 3.7 ± 0.3 copies/PCR mixture, respectively, suggesting that no significant PCR inhibition was present even when large amounts of total RNA were used. In good agreement with these numbers, the relative detection limit for the subset of data for intermediate RNA inputs, which can be used as an estimate of the sensitivity of the procedure, was 2.4 ± 0.2 copies/μg of RNA (Table 1).
Elimination of DNA from cellular RNA.
Considering that very small amounts of cellular HIV-1 RNA are to be measured to monitor HIV-infected patients receiving HAART, we addressed the question of whether low-positive measurements obtained from cellular RNA reflect viral RNA or proviral DNA contaminating the RNA preparation. To avoid false-positive results due to the presence of HIV DNA, a DNase digestion step was included in the protocol for the preparation of cellular RNA. The absence of contaminating genomic DNA was routinely monitored by gel electrophoresis of PBMC and tissue RNA preparations (data not shown).
The efficacy of the DNase digestion was documented by spiking HIV-negative RNA preparations with known numbers of copies of cloned HIV-1 DNA and then digesting the specimen with DNase by the routine procedure and measuring the residual DNA by the Amplicor test. DNase digestion completely removed 200 copies of amplifiable HIV-1 DNA (Table 2). With 20,000 HIV-1 DNA input copies, the spiked HIV-1 DNA level was reduced by more than 7,000-fold (Table 2). Furthermore, in patient specimens which had been treated with DNase twice, the second treatment had no measurable effect on the result of the RT-PCR (Table 2). The finding that the plasmid DNA copy numbers measured by the Amplicor test were, in some instances, higher than the expected value (Table 2) may reflect the higher degree of chemical stability of DNA compared to that of the QS RNA.
TABLE 2.
Efficacy of DNase treatment of cellular RNA extracted from cellular specimens
| Step | No. of HIV-1 RNA copies/PCR mixture for the following specimens with the indicated no. of DNA copiesa:
|
||||||
|---|---|---|---|---|---|---|---|
| Amplicor RNA diluent
|
Control, 20,000 DNA copiesbc | Patient Ad
|
Patient B, no DNAd | Patient C, no DNAd | |||
| 200 DNA copiese | 20,000 DNA copiese | 200 DNA copiesc | No DNA | ||||
| 1. No processing | 206 | 42,730 | NDf | ND | ND | ND | ND |
| 2. RNeasy protocol without DNase I | ND | ND | 70,503 | ND | ND | ND | ND |
| 3. Standard protocol with 1× DNase I | ND | ND | 9 | 49 | 65 | 13 | 5 |
| 4. Extended protocol with 2× DNase I | ND | ND | <2 | 51 | 66 | 9 | 6 |
| No. of samplesg | 1 | 1 | 2 | 2 | 2 | 1 | 1 |
Copies of restriction enzyme (PvuII)-digested diluted plasmid pBH10 (5); calculation included the consideration that one molecule of double-stranded DNA represents two templates for the PCR.
PBMCs from an HIV-negative donor.
DNA was added after step 2.
PBMCs from HIV-positive donors receiving HAART.
DNA was added after step 1.
ND, not determined.
Quantitation of HIV-1 RNA in clinical specimens.
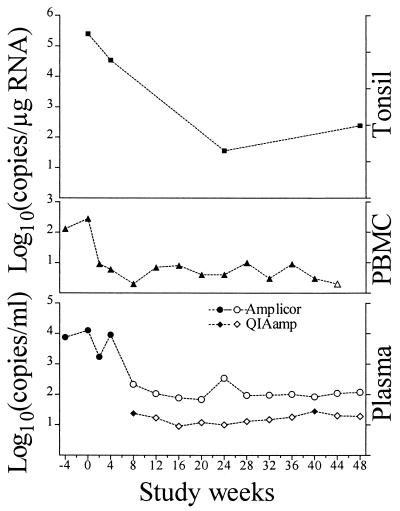
To assess the applicability of these methods to the monitoring of antiretroviral drug therapy, plasma virus loads, the number of HIV-1 RNA copies in PBMCs, and the number of HIV-1 RNA copies in tonsil biopsy specimens were measured over time in specimens from a representative HIV-infected patient taking AZT, 3TC, and ritonavir (Fig. 3). When measuring the plasma virus load by the standard Amplicor test, the last time point with detectable viremia was week 4. All specimens collected beyond that time point were assessed by both the standard Amplicor test and the silica-based (QIAamp) Amplicor test. The silica-based Amplicor procedure detected plasma HIV-1 RNA on weeks 8 and 40 (24 and 28 copies/ml, respectively). In agreement with the analysis whose results are presented in Table 1, relative detection limits (presented here only when no viral RNA could be measured) were about 1 log lower for the QIAamp procedure than for the standard Amplicor test. The levels of the cellular HIV-1 RNA in both the PBMC and the tonsil compartments showed clear decreases after the onset of therapy. In the initial phase before and shortly after the initiation of therapy, when virus was detectable in plasma, the kinetics of the PBMC and plasma HIV-1 RNA levels were similar, albeit at an overall lower (1 to 2 logs) level, and the dynamic range for PBMCs was smaller. At later time points, when the amount of virus in plasma was usually below the limit of detection, HIV-1 RNA persisted in PBMCs at 3 to 10 copies/μg of RNA.
FIG. 3.
Kinetics of HIV-1 RNA levels in plasma and PBMCs during antiretroviral treatment. An antiretroviral agent-naive, asymptomatic, HIV-infected individual started AZT-3TC-ritonavir therapy at time zero. Plasma virus loads (number of copies per milliliter) are shown for the standard Amplicor test (circles) and the silica-based (QIAamp) Amplicor test (diamonds). Levels of cellular HIV-1 RNA (number of copies per microgram of RNA) are shown for PBMCs (triangles) and tonsil tissue (squares). Filled symbols signify detectable viral RNA, whereas open symbols indicate lower detection limits for specimens devoid of measurable HIV-1 RNA.
DISCUSSION
Use of the methods presented here resulted in a highly sensitive means of detection of HIV-1 RNA in plasma, with an average lower detection limit of 14.9 copies per ml, and in cellular specimens, with an average lower detection limit of 2.4 copies per μg of total cellular RNA. This was achieved by silica column extraction of total RNA from plasma, PBMC, or tissue specimens followed by the addition of the purified RNA to the Amplicor HIV-1 Monitor test.
Some previously reported boosted methods for measurement of the plasma virus load rely on virus concentration by high-speed centrifugation followed by Amplicor RT-PCR (7, 10). However, this approach does not control for the possibility of incomplete recovery of virus by the centrifugation step because the QS RNA is added after centrifugation, when the lysis buffer added at this stage protects the QS RNA from nucleases. In the silica-based Amplicor method, RNA is extracted directly from plasma with the QIAamp Viral RNA Kit (Qiagen). In this protocol the QS RNA is added before extraction, which, like the standard Amplicor test, controls for the efficiency of extraction as well as that of amplification.
The linearity of the silica-based method was similar to that for the unmodified Amplicor test, which is claimed by the manufacturer to extend from approximately 8 to 18,000 HIV-1 RNA copies per amplification. When using 0.025 ml of plasma per PCR mixture (standard Amplicor test), this represents approximately 320 to 720,000 HIV-1 RNA copies per ml of plasma. When using 0.5 ml of plasma, a range from approximately 16 to 32,000 HIV-1 RNA copies per ml can be expected. In good accordance with those expected ranges, our data for specimens from which RNA was extracted with silica show linearity in a range from 30 to 27,000 HIV-1 copies/ml and an average lower relative detection limit of 16 copies/ml. If needed, plasma virus load measurements might be boosted further. For instance, Shafer and colleagues (11) boosted their nested HIV RT-PCR by combining centrifugation-based concentration with silica extraction. Also, the silica extraction method presented here may be developed further to accommodate larger plasma volumes. In preliminary experiments, we successfully performed silica extraction with up to 3 ml of plasma (unpublished observations).
Total cellular RNA purified with RNeasy silica columns was added to the Amplicor test to quantitate HIV-1 RNA from PBMCs and tonsil biopsy specimens. In contrast to the extraction of plasma, the extraction procedure itself is not controlled by coextraction of the QS RNA. Appropriate normalization of the input is, however, achieved by the measurement of the concentration of total RNA. In our hands, up to 2.6 μg of extracted total RNA could be added to an amplification reaction without significant inhibition, resulting in an average lower detection limit of 2.4 HIV-1 RNA copies per μg of total RNA. Addition of purified cellular RNA to the Amplicor test was previously reported by Tamalet et al. (12), who monitored HAART by measuring HIV transcription in PBMCs and lymphoid cells. Their method differs from our protocol in the following two main ways: (i) QS RNA was added to lysed cells before extraction (as is done with the standard Amplicor extraction and the boosted method with QIAamp silica for plasma RNA), and (ii) about 5- to 10-fold less RNA (250,000 cell equivalents, or approximately 0.28 μg of total RNA [our calculations based on their data]) extracted by a guanidine-phenol-based method was added to the Amplicor test mixture.
Those investigators reported detectable HIV-1 RNA levels of less than 50 copies per 106 cells in patients receiving HAART for 8 weeks. Thus, their method appears to give results similar to those achieved by the protocol for cellular specimens reported here. However, our method offers the advantage of increasing the amount of input RNA, resulting in improved sensitivity.
The expected sensitivity and precision obtained by combining silica-based extraction with the Amplicor test were realized in clinical plasma, PBMC, and tonsil biopsy tissue specimens obtained from our ongoing study of antiretroviral therapy (EARTH study). Data for a representative patient are reported here. Detectable levels of HIV-1 RNA from PBMCs in the face of undetectable HIV-1 RNA in plasma (<20 copies/ml) attest to the sensitivity of the Amplicor RT-PCR with silica-extracted cellular specimens. The usefulness of these methods was substantiated by the following observations. In a subset of 16 patients receiving double- or triple-drug therapy, the numbers of copies of HIV-1 RNA in plasma, PBMCs, and, where available (13 patients), tonsil tissue were assessed by the methods described here during 1 year (unpublished data). That analysis revealed that the levels of HIV-1 transcripts were significantly correlated (P > 0.001) between PBMCs and plasma, PBMCs and tonsil tissue, and plasma and tonsil tissue (Spearman r values = 0.71, 0.61, and 0.62, respectively).
The relative ease of performance of these procedures with clinical specimens makes the tests well-suited for large-scale screening in the context of clinical studies, and our laboratory uses these methods routinely. Another potential advantage of measuring RNA in specimens of both cellular and cell-free origin by the same well-characterized RT-PCR method is that the levels of viral RNA in different compartments can be directly compared.
In summary, the methods described here provide highly sensitive, reproducible, and simplified tests for the measurement of HIV-1 RNA in plasma, PBMC, and frozen tissue biopsy specimens. Application to clinical research specimens may enhance the throughput and quality of the data, especially for patients in whom residual viral RNA levels are very low due to HAART.
ACKNOWLEDGMENTS
This work was supported by the Kanton of Zurich and by Swiss National Science Foundation grants 32-46016 and 3239-043654.
We thank the following members of the Infectious Diseases Division, University Hospital Zurich: Friederike Burgener for laboratory assistance, Christina Grube for clinical assistance, and Beda Joos for helpful discussions. We also thank Ruth Martin (Roche, Switzerland) and Kurt Zoller (Qiagen, Switzerland) for helpful cooperation.
REFERENCES
- 1.Ausubel S M, Brent R M, Kingston R E, Moore D D, Seldmann J G, Smith J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Cavert W, Notermans D W, Staskus K, Wietgrefe S W, Zupancic M, Gebhard K, Henr K, Zhang Z Q, Mills R, McDade H, Goudsmit J, Danner S A, Haase A T. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 4.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 5.Hahn B H, Shaw G M, Arya S K, Popovic M, Gallo R C, Wong Staal F. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature. 1984;312:166–169. doi: 10.1038/312166a0. [DOI] [PubMed] [Google Scholar]
- 6.Michael N L, Mo T, Merzouki A, O’Shaughnessy M, Oster C, Burke D S, Redfield R R, Birx D L, Cassol S A. Human immunodeficiency virus type 1 cellular RNA load and splicing patterns predict disease progression in a longitudinally studied cohort. J Virol. 1995;69:1868–1877. doi: 10.1128/jvi.69.3.1868-1877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder J, Resnick R, Saget B, Scheibel S, Herman S, Payne H, Harrigan R, Kwok S. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma with enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Revets H, Marissens D, de Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLICOR-HIV monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saksela K, Stevens C E, Rubinstein P, Taylor P E, Baltimore D. HIV-1 messenger RNA in peripheral blood mononuclear cells as an early marker of risk for progression to AIDS Ann. Intern Med. 1995;123:641–648. doi: 10.7326/0003-4819-123-9-199511010-00001. [DOI] [PubMed] [Google Scholar]
- 10.Schockmel G A, Yerly S, Perrin L. Detection of low HIV-1 RNA levels in plasma. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;14:179–183. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 11.Shafer R W, Levee D J, Winters M A, Richmond K L, Huang D, Merigan T C. Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1 RNA from plasma. J Clin Microbiol. 1997;35:520–525. doi: 10.1128/jcm.35.2.520-522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamalet C, Lafeuillade A, Fantini J, Poggi C, Yahi N. Quantification of HIV-1 viral load in lymphoid and blood cells; assessment during four-drug combination therapy. AIDS. 1997;11:895–901. doi: 10.1097/00002030-199707000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Wong J K, Gunthard H F, Havlir D V, Zhang T Q, Haase A T, Ignacio C C, Kwok S, Emini E, Richman D D. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc Natl Acad Sci USA. 1997;94:12574–12579. doi: 10.1073/pnas.94.23.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong J K, Hezareh M, Gunthard H F, Havlir DV D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]