Abstract
The present study reports on the microglial populations present in 34 regions of the human infant brain (1–11 months), and whether developmental parameters or extrinsic factors such as cigarette smoke exposure, prone sleeping and an upper respiratory tract infection (URTI) influence their expression. Further, we compare microglia populations amongst three sudden unexpected death in infancy (SUDI) sub-groups: explained SUDI (eSUDI, n = 7), sudden infant death syndrome (SIDS) I (n = 8) and SIDS II (n = 13). Ionised calcium binding adaptor molecule-1 (Iba1) was used to determine the morphology and area covered by microglia in a given brain region. Activation was explored using cluster-of-differentiation factor 68 (CD68) and human leukocyte antigen-DP,DQ,DR (HLA). We found regional heterogeneity in the area covered and activation status of microglia across the infant brain. The hippocampus, basal ganglia, white matter and dentate nucleus of the cerebellum showed larger areas of Iba1, while the brainstem had the smallest. Microglia in regions of the basal ganglia and cortex demonstrated positive correlations with infant developmental parameters, while in nuclei of the rostral medulla, negative correlations between microglia parameters were seen. URTI and cigarette smoke exposure were associated with a reduced microglial area in regions of the hippocampus and cortex (parietal and occipital), respectively. In the context of SIDS, a reduced microglial area was seen in SIDS II and fewer SIDS I infants demonstrated activated phenotypes in the hippocampus. Overall, we identify the distribution of microglia in the infant brain to be heterogenous, and influenced by intrinsic and extrinsic factors, and that the SIDS I group is a useful control group for future research into other infant CNS pathologies.
Keywords: Sudden infant death, SIDS, Growth and development, Immunohistochemistry, Iba1, HLA antigens, CD68, Infection, Cigarette, Sleep
Highlights
-
•
Distribution of microglia in the human infant brain is heterogenous & approximates 5%.
-
•
Iba1 expression is higher than CD68 and HLA.
-
•
Resting and activated levels correlate with age and growth but are region dependant.
-
•
Cigarette exposure and hypoxic-sleep had minimal impact and only in cortical regions.
-
•
The hippocampus predominated as a region of interest in the context of SIDS & URTI.
Abbreviations
- AN
arcuate nucleus
- CA
cornu ammonis
- CD68
cluster of differentiation factor 68
- Cun
cuneate nucleus
- DMNV
dorsal motor nucleus of the vagus
- eSUDI
explained sudden unexpected death in infancy
- HLA
human leukocyte antigen (DP, DQ, DR clone)
- Iba1
ionised calcium binding adaptor molecule-1
- IGL
internal granular layer (of the cerebellar cortex)
- LRt
lateral reticular formation
- ML
molecular layer (of the cerebellar cortex)
- NTS
nucleus of the solitary tract
- PCA
post-conceptional age
- PMI
post mortem interval
- SIDS
sudden infant death syndrome
- SUDI
sudden unexpected death in infancy
- URTI
upper respiratory tract infection
- Vest
vestibular nucleus
- XII
hypoglossal nucleus
1. Introduction
Microglia are a ubiquitous and highly dynamic population of housekeeping cells in the CNS (Nimmerjahn et al., 2005). Resident microglial populations are retained from early gestation through to adulthood, sustained through self-regeneration and proliferation (Ajami et al., 2007). Microglia are estimated to constitute approximately 5–20% of all cells in the adult human CNS, exhibiting regional heterogeneity in both density and phenotype (Lawson et al., 1990; Mittelbronn et al., 2001; reviewed by Tan et al., 2019). In the healthy brain, microglia are predominantly found in a surveillant state, typified by a ramified morphology with fine, regularly distributed processes extending from a flattened cell body (Streit and Graeber, 1996). These processes radiate into the surrounding parenchyma, actively monitoring synapses and the microenvironment for any disruptions to homeostasis (Davalos et al., 2005; Streit and Graeber, 1996; Wake et al., 2009). Upon disruption, microglial activation occurs, with changes in the physical and molecular phenotype of microglia and rapid proliferation to increase the local microglial population (Boche et al., 2013; Liu et al., 2001). Activation is thought to exist along a spectrum (Boche et al., 2013). Classical activation involves retraction of processes and adoption of an amoeboid configuration, however, a number of intermediate activation states characterised by unique morphologies (e.g. hypertrophied and bushy microglia) have also been reported and are thought to reflect the strength and duration of the stimulus and, the functional commitments of the cell (Boche et al., 2013; Nelson et al., 2002; Schwartz et al., 2006; Sołtys et al., 2001; Taylor et al., 2014). Molecular changes are also highly varied and include modification of surface antigen expression and/or cytokine profiles, both of which are commonly investigated with immunohistochemistry (Beynon and Walker, 2012; Boche et al., 2013; Cherry et al., 2014; Liu et al., 2001; Nelson et al., 2002).
Herein, we utilise three of the most commonly used markers of microglia: ionised calcium-binding adaptor molecule 1 (Iba1), cluster of differentiation factor 68 (CD68) and human leukocyte antigen clone-DP, DQ, DR (HLA), given their widespread use in the literature, validation for immunohistochemistry in human formalin fixed paraffin embedded tissue, and that they detect both ramified and activated microglia (Hendrickx et al., 2017; Sasaki, 2016). Specifically, Iba1 is a general marker of microglia, providing uniform staining throughout the cytoplasm and processes of all microglia, regardless of activation state (i.e. resting and activated microglia) and, thus morphology (Boche et al., 2013; Sasaki, 2016). CD68 is a transmembrane glycoprotein shown to
activated microglia (and perivascular macrophages) with a phagocytic role, through labelling of lysosomes (Guillemin and Brew, 2004; Holness et al., 1993). HLA, also known as CR3/43, has been shown to detect cells expressing the major histocompatibility complex class II, including macrophages and in the CNS, activated microglia (Graeber et al., 1994).
Microglia accumulate and undergo phenotypic changes during normal development, and as an integral component of a number of CNS pathologies (reviewed: Arcuri et al., 2017; Wolf et al., 2017). During development, microglia promote appropriate synapse and neural circuit formation through processes including; synaptic pruning, regulation and facilitation of programmed cell death (PCD) and neurogenesis (reviewed: Paolicelli and Ferretti, 2017; Tremblay et al., 2011; Wake et al., 2013). Interactions with other glial cells have also been identified, with a relationship shown between microglia and oligodendrocyte precursors in developing white matter, and a sustained role of microglia in the maintenance of myelin homeostasis into adulthood (Hagemeyer et al., 2017). Further, microglia are implicated in immune mediated processes, including; phagocytosis, cytokine release and both the up- and down-regulation of the adaptive immune response in the CNS (Arcuri et al., 2017; Boche et al., 2013; Cherry et al., 2014). Pathological perturbation of microglia function during critical development periods (i.e. the postnatal period), regardless of aetiology, might result in aberrant synaptic and neural circuit development, and thus, functional consequences in the brain regions involved (Paolicelli and Ferretti, 2017; Wake et al., 2013).
Sudden unexpected death in infancy (SUDI) is the term used to describe all deaths of a sudden, unexpected nature occurring in infants aged between one week and one year (Fleming et al., 2000), and is the commonest cause of post-natal infant death in Australia (Statistics, 2018). SUDI is a broad term encompassing deaths which may be explained (eSUDI), such as due to drowning, cardiac or infectious causes, or remain unexplained; this includes cases of sudden infant death syndrome (SIDS) (Blair et al., 2012). SIDS has been defined as “the sudden unexpected death of an infant <1 year of age, with onset of the fatal episode apparently occurring during sleep, that remains unexplained after a thorough investigation, including performance of a complete autopsy and review of the circumstances of death and the clinical history” (Krous et al., 2004). For the purposes of standardization, the San Diego Criteria may be used to sub-categorise SIDS into SIDS I and SIDS II (Krous et al., 2004). The main distinction between the SIDS categories is that SIDS I cases have the necessary investigations performed but results provide no insight into mechanisms contributing to death, while SIDS II cases are those outside of the age range of SIDS, or where factors such as unsafe sleeping conditions, resolved developmental delays or acute illness, are present but insufficient alone to result in death. The current dataset, with classification adherent to the San Diego Criteria, has been previously defined in our recent publication (Ambrose et al., 2019).
To date, the study of microglia in the CNS of the human infant (1–12months) has been limited to a few regions, including the hippocampus (Del Bigio and Becker, 1994; Kinney et al., 2015; Paine et al., 2014), caudate nucleus and medulla (Esiri et al., 1991), cerebellum (Maslinska et al., 1998), parietal cortex (Billiards et al., 2006), other regions of the neocortex (Kjær et al., 2016), and subcortical white matter and key white matter tracts (Sigaard et al., 2014). While these studies identified unique microglial populations in discrete brain regions, they did not compare microglial populations, microglial morphology or activation state in multiple brain regions in the same subject. These studies also used different microglial markers (summarized in Supplement Table 1), making direct comparisons between the studies challenging. Given the diverse roles and influence of microglia on neurons and other glia during development, simultaneous characterisation of microglial area, morphology and activation state across multiple brain regions is important to further understanding microglial phenotype and function during this period of infant brain development.
As such, in this study we first report on the microglial populations present in 34 regions of the human infant brain, including the cerebral cortex, cerebellum, brainstem, hippocampus, and basal ganglia, and determine how these populations vary with age, sex, head circumference, body weight, and brain weight. We secondly compare and contrast the microglial populations in three SUDI sub-groups: eSUDI, SIDS I and SIDS II, and investigate whether these populations are affected by known extrinsic risk factors for SIDS (reviewed by Moon and Hauck, 2018), including a history of cigarette smoke exposure, unsafe/hypercapnic hypoxic sleeping conditions (i.e. the prone position or bedsharing), and a recent upper respiratory tract infection (URTI). We hypothesised that: 1- of the three markers, expression of Iba1 will be greatest and widespread, as it detects microglia of all activation statuses, and that its expression will vary with age and brain weight as part of the developmental process; 2- expression of CD68 and HLA will be restricted to brain regions containing subsets of activated microglia and macrophages due to diagnosis of SIDS and/or presence of the risk factors, with that for CD68 accompanying regions of known heightened PCD (e.g. hippocampus and medulla; (Ambrose et al., 2019); and 3- SIDS I infants will have lower expression of activated microglia indicative of the homogeneity of this group relatively free from risk factors, including -inflammatory and immune mediated pathology.
2. Material and methods
2.1. Data and tissue collection
All infant brain tissue was collected by the NSW Forensic and Analytical Science Services (FASS; formerly, the Department of Forensic Medicine), between 2008 and 2012 as we recently detailed (Ambrose et al., 2019). Autopsy data, infant clinical characteristics and SIDS risk factor information pertaining to each case were obtained from FASS. In accordance with ethics approval, all cases were de-identified (ethical approval from the NSW Health RPAH Zone (X13-0038 and HREC/13/RPAH/54) and University of Sydney Ethic committees).
Where available, a minimum of four serial 7 μm sections were collected from 20% formalin fixed, paraffin embedded tissue blocks using rotary microtomy, and tissue was mounted on 2% 3-aminopronopyltriehoxysaline treated slides. Thirty-four microscopically defined brain regions were examined (as reported recently (Ambrose et al., 2019); listed Table 1).
Table 1.
Mean percentage area of Iba1 immuno-positivity in the infant brain and comparison amongst diagnoses.
| BRAIN REGION | Mean Iba1+ ± SEM |
P-values; LSD∗ |
|||||
|---|---|---|---|---|---|---|---|
| All infants (n = 28) | eSUDI (n = 7) | SIDS I (n = 8) | SIDS II (n = 13) | eSUDI vs SIDS I | eSUDI vs SIDS II | SIDS I vs SIDS II | |
| Basal Ganglia | |||||||
| Caudate nucleus (head) | 2.98 ± 0.39 | 1.81 ± 0.44 | 3.31 ± 1.06 | 3.44 ± 0.55 | 0.18 | 0.08 | 0.89 |
| Claustrum | 2.97 ± 0.26 | 2.79 ± 0.75 | 3.21 ± 0.36 | 2.92 ± 0.35 | 0.58 | 0.85 | 0.64 |
| Globus pallidus (inner + outer) | 3.24 ± 0.26 | 3.07 ± 0.50 | 3.88 ± 0.55 | 2.99 ± 0.36 | 0.28 | 0.90 | 0.16 |
| Lentiform nucleus | 3.35 ± 0.24 | 3.00 ± 0.46 | 3.98 ± 0.51 | 3.17 ± 1.1 | 0.15 | 0.78 | 0.16 |
| Putamen | 3.07 ± 0.21 | 2.31 ± 0.17 | 3.46 ± 0.50 | 3.21 ± 0.29 | 0.06 | 0.09 | 0.62 |
| Striatum | 2.79 ± 0.34 | 1.74 ± 0.40 | 2.99 ± 0.95 | 2.79 ± 0.34 | 0.19 | 0.07 | 0.75 |
| Cerebellum | |||||||
| Dentate Nucleus | 3.61 ± 0.36 | 3.45 ± 0.86 | 2.99 ± 0.68 | 4.06 ± 0.50 | 0.66 | 0.52 | 0.22 |
| Internal granular layer | 1.69 ± 0.17 | 2.19 ± 0.43 | 1.59 ± 0.27 | 1.49 ± 0.21 | 0.12 | 0.10 | 0.80 |
| Molecular layer | 1.60 ± 0.14 | 1.88 ± 0.45 | 1.76 ± 0.26 | 1.35 ± 0.11 | 0.77 | 0.15 | 0.24 |
| White Matter | 3.34 ± 0.28 | 3.33 ± 0.51 | 2.73 ± 0.35 | 3.71 ± 0.48 | 0.44 | 0.58 | 0.15 |
| Cortical regions | |||||||
| Anterior superior frontal cortex | 2.59 ± 0.22 | 2.90 ± 0.44 | 2.94 ± 0.34 | 2.26 ± 0.32 | 0.96 | 0.22 | 0.22 |
| Posterior frontal cortex | 2.38 ± 0.23 | 2.14 ± 0.29 | 2.89 ± 0.57 | 2.24 ± 0.33 | 0.25 | 0.85 | 0.26 |
| Insular cortex | 2.99 ± 0.22 | 2.76 ± 0.52 | 3.21 ± 0.46 | 2.97 ± 0.32 | 0.50 | 0.73 | 0.67 |
| Occipital cortex | 1.96 ± 0.17 | 1.76 ± 0.55 | 2.13 ± 0.23 | 1.98 ± 0.17 | 0.45 | 0.61 | 0.73 |
| Parietal cortex | 2.01 ± 0.20 | 2.52 ± 0.45 | 2.30 ± 0.34 | 1.58 ± 0.23 | 0.68 | 0.046 | 0.12 |
| Temporal cortex | 2.98 ± 0.16 | 2.66 ± 0.30 | 3.14 ± 0.24 | 3.07 ± 0.26 | 0.29 | 0.31 | 0.86 |
| Hippocampus | |||||||
| Cornu ammonis 4 | 2.80 ± 0.25 | 3.71 ± 0.32 | 3.04 ± 0.63 | 2.17 ± 0.26 | 0.29 | 0.009 | 0.12 |
| Cornu ammonis 3 | 2.35 ± 0.19 | 3.24 ± 0.22 | 2.29 ± 0.36 | 1.90 ± 0.36 | 0.05 | 0.003 | 0.34 |
| Cornu ammonis 2 | 2.33 ± 0.18 | 3.07 ± 0.17 | 2.20 ± 0.43 | 2.01 ± 0.23 | 0.07 | 0.014 | 0.64 |
| Cornu ammonis 1 | 2.54 ± 0.17 | 2.94 ± 0.24 | 2.56 ± 0.41 | 2.31 ± 0.25 | 0.44 | 0.15 | 0.55 |
| Dentate gyrus | 2.52 ± 0.19 | 2.98 ± 0.26 | 2.66 ± 0.52 | 2.19 ± 0.21 | 0.53 | 0.09 | 0.30 |
| Parahippocampal gyrus | 2.66 ± 0.14 | 3.15 ± 0.25 | 2.57 ± 0.22 | 2.44 ± 0.22 | 0.14 | 0.042 | 0.70 |
| Subiculum | 2.70 ± 0.17 | 3.29 ± 0.24 | 2.55 ± 0.35 | 2.47 ± 0.24 | 0.10 | 0.043 | 0.83 |
| Medulla (rostral) | |||||||
| Arcuate nucleus | 1.62 ± 0.13 | 1.80 ± 0.26 | 1.54 ± 0.33 | 1.57 ± 0.17 | 0.48 | 0.50 | 0.92 |
| Cuneate nucleus | 1.54 ± 0.13 | 1.97 ± 0.32 | 1.34 ± 0.12 | 1.42 ± 0.18 | 0.04 | 0.08 | 0.81 |
| Dorsal motor nucleus of the vagus | 1.45 ± 0.09 | 1.64 ± 0.13 | 1.27 ± 0.12 | 1.45 ± 0.17 | 0.15 | 0.40 | 0.41 |
| Hypoglossal nucleus | 1.54 ± 0.13 | 1.78 ± 0.28 | 1.36 ± 0.24 | 1.51 ± 0.19 | 0.25 | 0.41 | 0.62 |
| Inferior olivary nucleus | 1.96 ± 0.13 | 2.24 ± 0.27 | 1.85 ± 0.32 | 1.87 ± 0.14 | 0.28 | 0.26 | 0.94 |
| Lateral reticular formation | 1.71 ± 0.17 | 1.75 ± 0.34 | 1.63 ± 0.45 | 1.72 ± 0.18 | 0.81 | 0.95 | 0.83 |
| Nucleus of the spinal trigeminal tract | 1.84 ± 0.14 | 2.07 ± 0.30 | 1.91 ± 0.38 | 1.49 ± 0.13 | 0.68 | 0.29 | 0.52 |
| Nucleus of the solitary tract | 1.53 ± 0.09 | 1.89 ± 0.27 | 1.49 ± 0.13 | 1.36 ± 0.08 | 0.10 | 0.02 | 0.52 |
| Vestibular nucleus | 1.45 ± 0.10 | 1.75 ± 0.25 | 1.25 ± 0.11 | 1.41 ± 0.15 | 0.08 | 0.18 | 0.52 |
| Other | |||||||
| Amygdala | 2.72 ± 0.19 | 2.47 ± 0.22 | 2.82 ± 0.26 | 2.79 ± 0.25 | 0.51 | 0.50 | 0.94 |
| Lateral geniculate nucleus | 2.61 ± 0.14 | 3.05 ± 0.23 | 2.46 ± 0.24 | 2.45 ± 0.23 | 0.14 | 0.09 | 0.98 |
Bold indicates significance, which was taken at p < 0.05. Italics indicates a trend, which was taken at p = 0.05–0.1.
2.2. Immunostaining
Manual peroxidase immunohistochemistry was performed as described previously (Ambrose et al., 2018). Briefly, sections were de-paraffinized in xylene and rehydrated through a graded ethanol series prior to microwave antigen retrieval in TRIS-EDTA buffer (1 mM trisodium citrate, 1 mM EDTA, 2 mM TRIS; pH9.0). Sections were allowed to cool and endogenous peroxidase activity was quenched for 20 min in a 3% H2O2, methanol and phosphate buffered saline (PBS) solution. All sections were blocked for 1 h with 10% normal horse serum (NHS) before overnight incubation at room temperature in one of three primary antibodies: Iba1 (1:1000; 019–19741, Wako, Japan), CD68 (1:200; monoclonal, clone KP1; M-0814, Dako, USA) and, HLA-DP, -DQ, -DR (1:200, monoclonal, clone CR3/43; M-0775, Dako, USA). Sections were washed with PBS, incubated at room temperature for 1 h in an affinity purified anti-mouse/anti-rabbit secondary antibody (1:250; BA-400, Vector Laboratories) and washed in PBS. Peroxidase avidin-biotin complex (PK-4000, Vectastain ABC Kit, Vector Laboratories) was allowed to bind for 1 h, sections were then washed in PBS and chromogen 3,3′-diaminobenzidine (DAB; K3468; DakoCytomation, USA) applied. Following counterstaining with Harris’ haematoxylin, sections were dehydrated through a graded ethanol series and cleared in xylene before mounting with Di-N-Butyle Pthalate in xylene (DPX). Owing to limited tissue availability, duplicate staining, for each marker, was only performed on 20% of sections for each brain region, encompassing all SUDI sub-groups.
2.3. Quantitative analysis of IBA1 immunopositivity
All Iba1 stained sections were viewed using light microscopy (Olympus Upright BX51 Microscope, Olympus Optical Co., Ltd, Japan) and imaged using image capture software (DP Controller, Olympus Optical Co., Ltd, Japan). For each brain region, multiple, non-overlapping images were taken using either the 20x- or 40×-objective lenses; ensuring only the region of interest (ROI) covered the field of view. Where possible, captured images avoided large blood vessels, artefacts and tissue edge to maximise the valid tissue area within the captured frame.
Red Green Blue (RGB) images were exported to Fiji imaging processing software (Schindelin et al., 2012). For each image, colour de-convolution was performed using the inbuilt ‘H DAB’ plug-in to isolate three image colour layers; blue, brown and, green. The brown layer (R:0.268, G:0.570, B:0.777) corresponding to positive DAB detection and thus, Iba1 positive staining, was selected and used for analysis thereafter. Manual thresholding using the histogram was performed to ensure only positively stained Iba1 positive cell bodies and/or processes were being selected for measurement. The percentage of the total image area covered by the thresholded material was calculated using the inbuilt ‘measure’ function and these percentage values were exported and collated in an excel spreadsheet. For a ROI that extended over more than one image, the percentage of the area covered by the thresholded immune-reactive material was averaged across the multiple images of that ROI. All imaging and quantification were performed by a single observer (NA), blind to the SUDI classification.
2.4. Qualitative analysis of microglia morphology
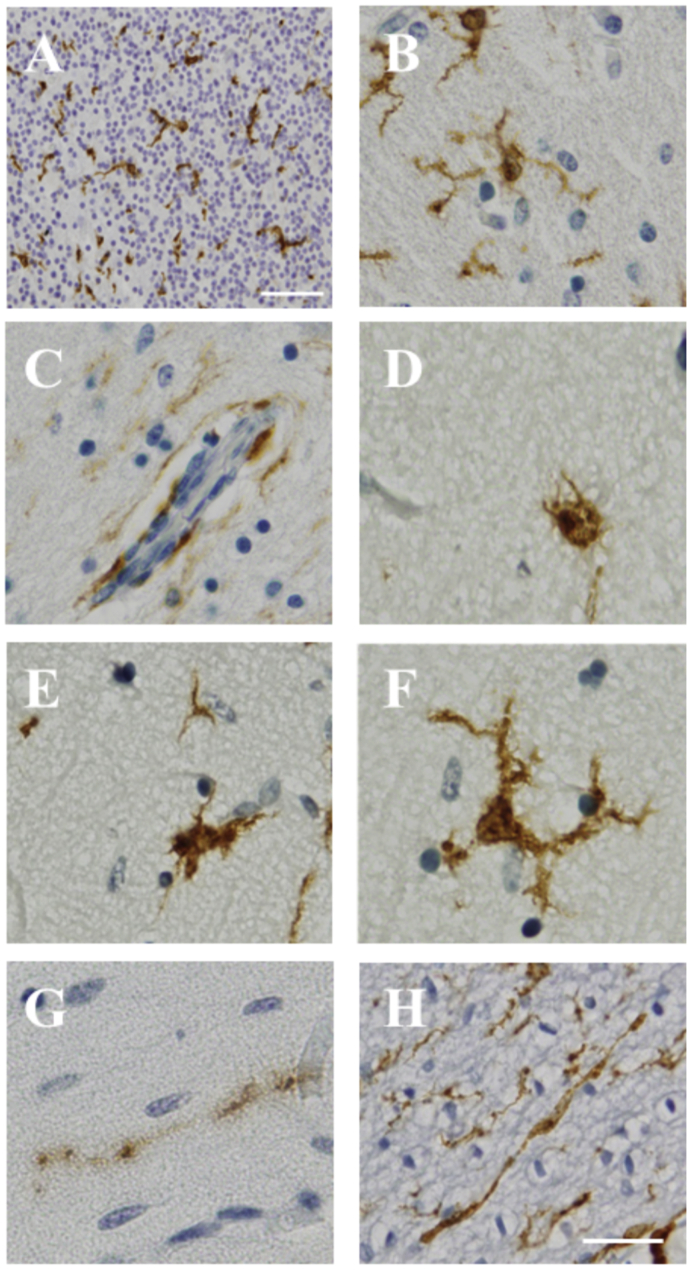
All Iba1 stained sections were manually scanned under a light microscope, using 20× and 40× objective lenses. All cases and brain regions were scored by a single observer (NA) for the presence (scored as ‘1’) or absence (scored as ‘2’) of seven morphologically defined Iba1 positive cell types, including perivascular cells, ramified microglia, hypertrophied microglia, bushy cells, amoeboid microglia, rod cells, and dystrophic microglia (Fig. 1). Scores were collated in Excel.
Fig. 1.
Iba1 positive cellular morphologies observed in the human infant brain. Iba1 positive expression, as shown in the internal granular layer of the cerebellar cortex (A), with morphologies including: ramified microglia (B), perivascular cells (C), amoeboid microglia (D), bushy microglia (E), hypertrophied microglia (F), dystrophic microglia (G) and rod cells (H). Scale bar = 50 μm (A) and scale bar = 20 μm (B–H; shown in H).
2.5. Qualitative analysis of markers of microglia activation
CD68 and HLA stained sections were scanned under a light microscope using the 20× and 40× objective lenses. The number of cells expressing each marker was scored qualitatively in each region by a single observer (NA) using a 5 tier scoring system (‘0’ = no cells, ‘1’ = only perivascular cells, ‘2’ = perivascular cells and sparse parenchymal expression, ‘3’ = perivascular cells and moderate parenchymal expression, ‘4’ = perivascular cells and frequent parenchymal expression). Independent scores for the two activated markers were determined and all scores collated in Excel.
2.6. Statistical analysis
All data was exported to SPSS for Windows (version21; SPSS (IBM) Inc., Illinois, USA) for statistical analysis. ANOVA with post-hoc least significant difference (LSD) correction was used to compare the mean area of Iba1 immunopositive expression (Iba1+ area) between SUDI subgroups. The proportion of cases demonstrating each of the Iba1 positive cell morphologies was compared amongst SUDI subgroups using Chi square testing (Fischer’s exact test), with values expressed as a percentage of cases within a SUDI subgroup. In analysis of the expression of activation markers, scores were grouped to form two bins: scores 0 and 1 were grouped (no or perivascular cells positive staining only) and those scored 2,3,4 (positive staining in the parenchyma) were grouped. The proportion of cases with positive staining in the parenchyma was compared amongst SUDI subgroups using Chi square (Fischer’s exact test).
The relationship between Iba1+ area and continuous data (infant growth characteristics) was determined using bivariate correlation, and with Student’s T-tests for nominal data (risk factors). Analysis of the impact of infant characteristics and SIDS risk factors on morphology and activated marker expression was determined using Chi-square testing (Fischer’s exact test).
Iba1+ area data is presented as the mean ± standard error of the mean (SEM), and morphology and activated marker expression (CD68 and HLA) data is presented as the proportion of cases (%). Unless otherwise noted, p < 0.05 was taken as statistically significant, and trends where 0.05< p < 0.1, are mentioned.
3. Results
3.1. Infant characteristics
The dataset, established in a previous study from our laboratory (Ambrose et al., 2019) includes 28 SUDI cases (17 males, 11 females) with the following growth related parameters: post-conceptional age (PCA) (54.2 ± 2.0wks; median = 52.0wks), birth weight (3.0 ± 0.2 kg; median = 3.0 kg), body weight (6.2 ± 0.4 kg; median = 5.8 kg), brain weight (714 ± 34 g; median = 698 g), and head circumference (40.2 ± 0.8 cm; median = 40.3 cm). Cases were classified into three SUDI sub-groups, eSUDI (n = 7), SIDS I (n = 8) and SIDS II (n = 13). The cause of death in the eSUDI group included: acute pneumonia (n = 1), thrombotic occlusion of a Blalock Taussig Shunt (n = 1), complications of congenital heart disease (n = 2), myocarditis (n = 2) and meningoencephalitis of uncertain aetiology (n = 1). Infants in the three groups were sex and age matched and did not differ in respect to clinical, developmental and autopsy related features (Ambrose et al., 2019). The prevalence of the following SIDS risk factors was similar in SIDS I and SIDS II infants: cigarette smoke exposure (38% vs 40%), an URTI in the two weeks prior to death (25% vs 46%) and being found prone sleeping (63% vs 55%). As expected on the basis of classification, bedsharing was only evident in the SIDS II group with prevalence of 69%.
3.2. Microglial marker expression
3.2.1. Iba1+ area
Homogeneous Iba1+ staining was observed in the cell bodies and processes of microglia in all 34 brain regions for all 28 cases. The Iba1+ area varied between 1.45% and 3.61% of the tissue area in each region, with the mean across all brain regions 2.39% (Table 1). Comparing brain regions, the mean area of Iba1+ was highest in components of the basal ganglia, cerebellum and hippocampus and, lowest in nuclei in the rostral medulla (Table 1).
3.2.2. Iba1 cell morphology
Seven Iba1 positive cell morphologies were present and qualitatively assessed (Fig. 1). Of the seven cell morphologies assessed, ramified microglia and perivascular cells (Fig. 1 B & C) were the most frequent, and dystrophic microglia and rod cells (Fig. 1 G & H) the least frequent in all infant brain regions.
3.2.3. Microglia activation CD68
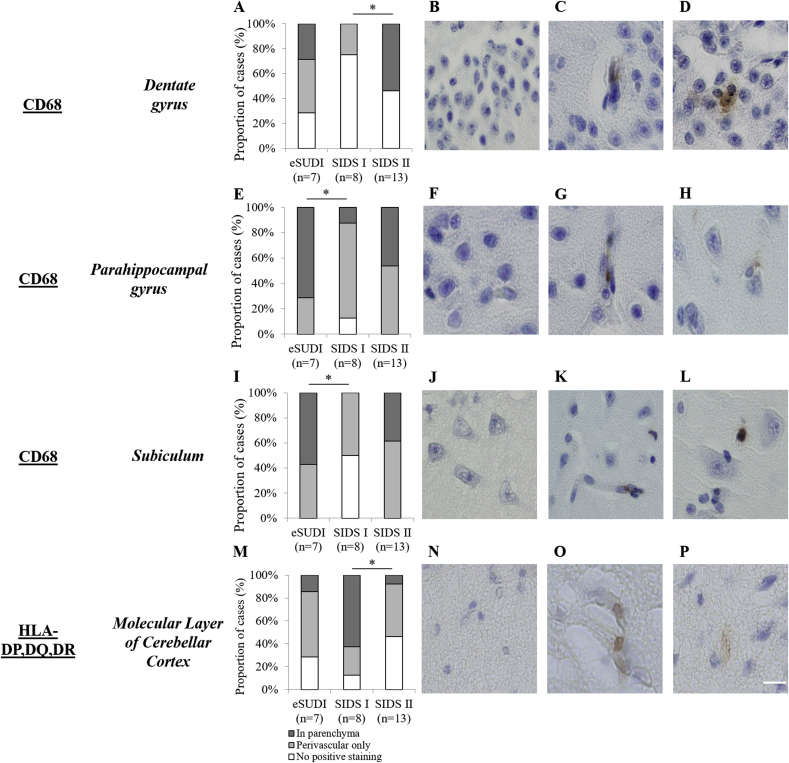
Positive CD68 staining was cytoplasmic, with a lysosomal distribution (Fig. 2). CD68 expression was frequent in perivascular cells in all brain regions, but infrequent in parenchymal microglia. Parenchymal CD68 expression was always sparse (i.e. score 2) and was not seen in all brain regions examined (Table 2). Across the infant dataset, parenchymal CD68 expression was most frequently identified in the putamen, striatum, cerebellar white matter and lentiform nucleus; seen in 93%, 88%, 71% and 65% of cases respectively (Table 2). No parenchymal CD68 expression was identified in the lateral reticular formation (LRt) and nucleus of the solitary tract (NTS) of the rostral medulla (Table 2). In a further three nuclei of the rostral medulla and in the CA1, CA2 and CA3 regions of the hippocampus, less than 10% of infants in the dataset had positive CD68 expression in the parenchyma (Table 2).
Fig. 2.
Markers of microglial activation in the human infant hippocampus and cerebellum, with comparison of expression among SUDI subgroups. The proportion of infant cases demonstrating no positive, perivascular cell, and parenchymal CD68 expression in the dentate gyrus (A), parahippocampal gyrus (E) and subiculum (I) of hippocampus, and HLA-DP,DQ,DR expression in the molecular layer of the cerebellum (M). Chi squared analysis (Fischer’s exact test) was used to compare the proportion of infants with positive staining in the parenchyma between eSUDI, SIDS I and SIDS II infants. ∗Significance taken at p < 0.05. Micrographs demonstrate no positive staining (B,F,J,N), positive perivascular cell staining only (C,G,K,O) and parenchymal positive staining (D,H,L,P). Scale bar = 20 μm for all.
Table 2.
Proportion of infants demonstrating positive CD68 and HLA-DP,DQ,DR in the parenchyma of different regions of the brain comparing amongst diagnostic groups.
| BRAIN REGION | Proportion cases positive CD68 in parenchyma (%) |
P-value; Fischer’s∗ |
Proportion cases positive HLA in parenchyma (%) |
P-value; Fischer’s∗ |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All Infants (n = 28) |
eSUDI (n = 7) | SIDS I (n = 8) | SIDS II (n = 13) | eSUDI vs SIDS I | eSUDI vs SIDS II | SIDS I vs SIDS II | All Infants (n = 28) | eSUDI (n = 7) | SIDS I (n = 8) | SIDS II (n = 13) | eSUDI vs SIDS I | eSUDI vs SIDS II | SIDS I vs SIDS II | |
| Basal Ganglia | ||||||||||||||
| Caudate nucleus (head) | 14/26 (54) | 3/7 (43) | 3/6 (50) | 8/13 (62) | 1.00 | 0.64 | 1.00 | 5/26 (19) | 2/7 (29) | 1/6 (17) | 2/13 (15) | 1.00 | 0.59 | 1.00 |
| Claustrum | 4/26 (15) | 1/6 (17) | 1/7 (14) | 2/13 (15) | 1.00 | 1.00 | 1.00 | 3/26 (12) | 2/6 (33) | 1/7 (14) | 0/13 (0) | 0.56 | 0.09 | 0.35 |
| Globus pallidus (inner + outer) | 9/26 (35) | 1/6 (17) | 3/7 (43) | 5/13 (39) | 0.56 | 0.61 | 1.00 | 1/26 (4) | 1/6 (17) | 0/7 (0) | 1/13 (8) | 0.46 | 1.00 | 1.00 |
| Lentiform nucleus | 17/26 (65) | 5/6 (83) | 4/7 (57) | 8/13 (62) | 0.56 | 0.61 | 1.00 | 8/26 (31) | 2/6 (33) | 3/7 (43) | 3/13 (23) | 1.00 | 1.00 | 0.61 |
| Putamen | 25/27 (93) | 6/7 (86) | 7/7 (100) | 12/13 (92) | 1.00 | 1.00 | 1.00 | 15/27 (56) | 3/7 (43) | 6/7 (86) | 6/13 (46) | 0.27 | 1.00 | 0.16 |
| Striatum | 23/26 (88) | 5/7 (71) | 6/6 (100) | 12/13 (92) | 0.46 | 0.27 | 1.00 | 11/26 (42) | 3/7 (43) | 2/6 (33) | 6/13 (46) | 1.00 | 1.00 | 1.00 |
| Cerebellum | ||||||||||||||
| Dentate Nucleus | 10/28 (36) | 2/6 (33) | 1/8 (13) | 7/13 (54) | 0.57 | 0.37 | 0.09 | 8/28 (29) | 2/7 (29) | 2/8 (25) | 4/13 (31) | 1.00 | 1.00 | 1.00 |
| Internal granular layer | 5/28 (18) | 2/7 (29) | 2/8 (25) | 1/13 (8) | 1.00 | 0.27 | 0.53 | 2/28 (7) | 0/7 (0) | 1/8 (13) | 1/13 (8) | 1.00 | 1.00 | 1.00 |
| Molecular layer | 14/28 (50) | 3/7 (43) | 4/8 (50) | 7/13 (54) | 1.00 | 1.00 | 1.00 | 7/28 (25) | 1/7 (14) | 5/8 (63) | 1/13 (8) | 0.12 | 1.00 | 0.01 |
| White Matter | 20/28 (71) | 6/7 (86) | 3/8 (38) | 11/13 (85) | 0.12 | 1.00 | 0.06 | 18/28 (64) | 5/7 (71) | 5/8 (63) | 8/13 (62) | 1.00 | 1.00 | 1.00 |
| Cortical regions | ||||||||||||||
| Anterior superior frontal cortex | 13/26 (50) | 5/7 (71) | 1/6 (17) | 7/13 (54) | 0.10 | 0.64 | 0.18 | 8/26 (31) | 1/7 (14) | 2/6 (33) | 5/13 (38) | 0.56 | 0.35 | 1.00 |
| Posterior frontal cortex | 14/27 (52) | 5/7 (71) | 3/7 (43) | 6/13 (46) | 0.59 | 0.37 | 1.00 | 14/27 (52) | 4/7 (57) | 2/7 (29) | 8/13 (62) | 0.56 | 1.00 | 0.35 |
| Insula cortex | 3/26 (12) | 0/7 (0) | 1/7 (14) | 2/13 (15) | 1.00 | 1.00 | 1.00 | 6/26 (23) | 2/6 (33) | 2/7 (29) | 2/13 (15) | 1.00 | 0.56 | 0.59 |
| Occipital cortex | 9/27 (33) | 3/7 (43) | 2/7 (29) | 4/13 (31) | 1.00 | 0.65 | 1.00 | 13/27 (48) | 1/7 (14) | 3/7 (43) | 9/13 (69) | 0.56 | 0.06 | 0.36 |
| Parietal cortex | 15/27 (56) | 4/7 (57) | 4/7 (57) | 7/13 (54) | 1.00 | 1.00 | 1.00 | 10/27 (37) | 3/7 (43) | 2/7 (29) | 5/13 (38) | 1.00 | 1.00 | 1.00 |
| Temporal cortex | 15/27 (56) | 5/7 (71) | 3/7 (43) | 7/13 (54) | 0.59 | 0.64 | 1.00 | 3/28 (11) | 1/7 (14) | 1/8 (13) | 1/13 (8) | 1.00 | 1.00 | 1.00 |
| Hippocampus | ||||||||||||||
| Cornu ammonis 4 | 11/28 (39) | 4/7 (57) | 2/8 (11) | 5/13 (38) | 0.32 | 0.64 | 0.66 | 6/28 (21) | 1/7 (14) | 2/8 (25) | 3/13 (23) | 1.00 | 1.00 | 1.00 |
| Cornu ammonis 3 | 2/28 (7) | 1/7 (14) | 0/8 (0) | 1/13 (8) | 0.47 | 1.00 | 1.00 | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 |
| Cornu ammonis 2 | 1/28 (4) | 0/7 (0) | 0/8 (0) | 1/13 (8) | 1.00 | 1.00 | 1.00 | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 |
| Cornu ammonis 1 | 1/28 (4) | 0/7 (0) | 0/8 (0) | 1/13 (8) | 1.00 | 1.00 | 1.00 | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 |
| Dentate gyrus | 9/28 (32) | 2/7 (29) | 0/8 (0) | 7/13 (54) | 0.20 | 0.37 | 0.02 | 2/28 (7) | 0/7 (0) | 1/8 (13) | 1/13 (8) | 1.00 | 1.00 | 1.00 |
| Parahippocampal gyrus | 12/28 (43) | 5/7 (71) | 1/8 (13) | 6/13 (46) | 0.04 | 0.37 | 0.17 | 1/28 (4) | 1/7 (14) | 0/8 (0) | 1/13 (8) | 0.47 | 1.00 | 1.00 |
| Subiculum | 9/28 (32) | 4/7 (57) | 0/8 (0) | 5/13 (38) | 0.03 | 0.64 | 0.11 | 3/28 (11) | 2/7 (29) | 0/8 (0) | 1/13 (8) | 0.20 | 0.27 | 1.00 |
| Medulla (rostral) | ||||||||||||||
| Arcuate nucleus | 3/28 (11) | 0/7 (0) | 1/8 (13) | 2/13 (15) | 1.00 | 0.52 | 1.00 | 5/28 (18) | 1/7 (14) | 1/8 (13) | 3/13 (23) | 1.00 | 1.00 | 1.00 |
| Cuneate nucleus | 1/28 (4) | 0/7 (0) | 0/8 (0) | 1/13 (8) | 1.00 | 1.00 | 1.00 | 2/28 (7) | 0/7 (0) | 0/8 (0) | 2/13 (15) | 1.00 | 0.52 | 0.51 |
| Dorsal motor nucleus of the vagus | 3/28 (11) | 1/7 (14) | 0/8 (0) | 2/13 (15) | 0.47 | 1.00 | 0.51 | 3/28 (11) | 1/7 (14) | 1/8 (13) | 1/13 (8) | 1.00 | 1.00 | 1.00 |
| Hypoglossal nucleus | 2/28 (7) | 0/7 (0) | 0/8 (0) | 2/13 (15) | 1.00 | 0.52 | 0.51 | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 |
| Inferior olivary nucleus | 8/28 (29) | 1/7 (14) | 2/8 (25) | 5/13 (38) | 1.00 | 0.35 | 0.66 | 6/28 (21) | 3/7 (43) | 2/8 (25) | 1/13 (8) | 0.61 | 0.10 | 0.53 |
| Lateral reticular formation | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 |
| Nucleus of the spinal trigeminal tract | 2/28 (7) | 0/7 (0) | 2/8 (25) | 0/13 (0) | 0.47 | 1.00 | 0.13 | 1/28 (4) | 1/7 (14) | 0/8 (0) | 0/13 (0) | 0.47 | 0.35 | 1.00 |
| Nucleus of the solitary tract | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 | 2/28 (7) | 1/7 (14) | 1/8 (13) | 0/13 (0) | 1.00 | 0.35 | 0.38 |
| Vestibular nucleus | 1/28 (4) | 0/7 (0) | 0/8 (0) | 1/13 (8) | 1.00 | 1.00 | 1.00 | 4/28 (14) | 0/7 (0) | 1/8 (13) | 3/13 (23) | 1.00 | 0.52 | 1.00 |
| Other | ||||||||||||||
| Amygdala | 7/26 (27) | 2/6 (33) | 2/7 (29) | 3/13 (23) | 1.00 | 1.00 | 1.00 | 7/27 (26) | 1/7 (14) | 2/7 (29) | 4/13 (31) | 1.00 | 0.61 | 1.00 |
| Lateral geniculate nucleus | 8/26 (31) | 1/6 (17) | 1/7 (14) | 6/13 (46) | 1.00 | 0.33 | 0.33 | 0/28 (0) | 0/7 (0) | 0/8 (0) | 0/13 (0) | 1.00 | 1.00 | 1.00 |
Bold indicates significance, which was taken at p < 0.05. Italics indicates a trend, which was taken at p = 0.05–0.1.
3.2.4. Microglia activation HLA-DR-DP-DQ
Of the three markers studied, HLA was the most infrequently observed across brain regions. Cytoplasmic HLA expression (Fig. 2) in the parenchyma was identified most frequently in the cerebellar white matter, putamen, posterior frontal and occipital cortices, present in 64%, 56%, 52% and 48% of all infants respectively. No parenchymal HLA expression was identified in the lateral geniculate nucleus (LGN), the CA3, CA2 and CA1 regions of the hippocampus, the hypoglossal nucleus (XII) and LRt of the rostral medulla (Table 2). In a further three nuclei of the rostral medulla, the internal granular layer (IGL) of the cerebellum, dentate gyrus, parahippocampal gyrus and in the globus pallidus, less than 10% of infants had positive HLA expression in the parenchyma (Table 2).
3.3. Infant development and microglia marker expression
Associations between microglia characteristics and infant development parameters were explored in all cases regardless of diagnosis (Supplement Tables 2-4 and summarized in Fig. 3). Associations between microglia characteristics and sex and, post mortem interval (PMI) were also explored.
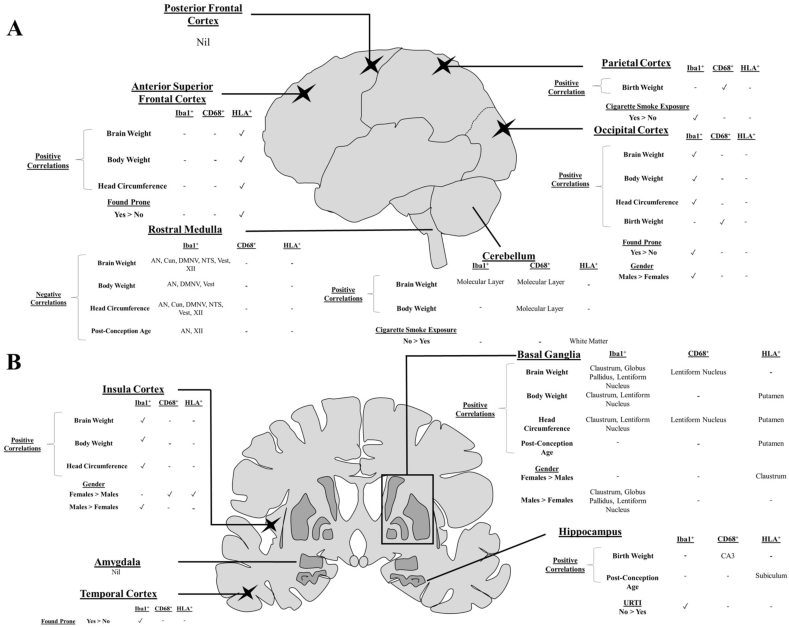
Fig. 3.
Schematic overview of the significant regional contributions of intrinsic and extrinsic factors on microglia marker expressions. Lateral (A) and coronal (B) views highlighting the key areas examined in the present study and summary of the influences on expression of Iba1, CD68 and HLA-DP,DQ,DR. Schematics not shown to scale.
3.3.1. Iba1+ area
A number of moderately-strong negative correlations (r-value ranging from −0.41 to −0.64, p value from <0.001 to 0.04) were observed between Iba1+area and infant growth parameters in the rostral medulla, including: post conceptional age (PCA) in the AN and XII, body weight in the AN, DMNV and Vest, head circumference in the AN, Cun, DMNV, NTS, Vest and XII, and brain weight in the AN, Cun, DMNV, NTS, Vest and XII (Supplement Table 2).
Moderately-strong positive correlations (r-value ranging from −0.40 to −0.54, p value from <0.01 to 0.04) were observed between body weight and Iba1+ area in the occipital and insula cortices, molecular layer (ML) of the cerebellar cortex, lentiform nucleus and claustrum, between brain weight and Iba1+ area in the occipital and insula cortices, globus pallidus, lentiform nucleus and claustrum, and between Iba1+ area and head circumference in the occipital and insular cortices, lentiform nucleus and claustrum (Supplement Table 2).
Iba1+ area was greater in males compared to females in the occipital (2.24 ± 0.21% vs 1.50 ± 0.23%; p = 0.033) and insular (3.47 ± 0.29% vs 2.21 ± 0.22%; p = 0.005) cortices; globus pallidus (3.65 ± 0.36% vs 2.60 ± 0.29%; p = 0.048), lentiform nucleus (3.80 ± 0.29% vs 2.62 ± 0.28%; p = 0.012) and claustrum (3.50 ± 0.32% vs 2.12 ± 0.25%; p = 0.006). Iba1+ area did not correlate with PMI or birth weight.
3.3.2. Microglia activation CD68
As compared to infants with no or only perivascular cell CD68 expression, infants with positive CD68 in the parenchyma had a greater mean brain weight (ML of the cerebellum and lentiform nucleus), a greater mean head circumference (lentiform nucleus), a greater mean birth weight (parietal cortex and CA3 region of the hippocampus) and, a greater mean body weight at death (ML of the cerebellum) (Supplement Table 3). Compared to males, more females demonstrated positive CD68 expression in the insular cortex (30% vs 0%; p = 0.046). No differences were observed according to PMI or PCA.
3.3.3. Microglia activation HLA-DP,DQ,DR
The mean PCA, body weight and head circumference were greater in infants with HLA expression in the parenchyma of the putamen, compared to infants with no or only perivascular cell HLA expression (Supplement Table 4). As compared to infants with no or only perivascular cell HLA expression, infants with parenchymal HLA expression in the anterior superior frontal cortex had a greater mean body weight, brain weight and head circumference (Supplement Table 4). In the subiculum, the mean PCA was greater in infants with no or perivascular cell expression of HLA compared to infants with parenchymal expression (Supplement Table 4). Compared to males, more females demonstrated positive HLA expression in the insular cortex (50% vs 6%; p = 0.046) and claustrum (30% vs 0%; p = 0.046). HLA expression in the parenchyma was not associated with differences in PMI or birth weight.
3.4. Comparison of SUDI subgroups
Although age and growth parameters were associated with differences in expression (as detailed above), given these were not consistent for all brain regions, we did not include them as covariates in our analysis. Further, covariate analysis was not warranted given the three SUDI subgroups did not differ statistically for these parameters (Ambrose et al. (2019).
3.4.1. Iba1+ area
Compared to eSUDI, Iba1+ area in SIDS II infants was reduced in the parietal cortex (p = 0.046), NTS (p = 0.020), parahippocampal gyrus (p = 0.042), subiculum (p = 0.043), CA2 (p = 0.014), CA3 (p = 0.003) and CA4 (p = 0.009) of the hippocampus (Table 1). There were no differences in Iba1+ area when comparing SIDS I to either SIDS II or eSUDI (Table 1).
3.4.2. Iba1 cell morphologies
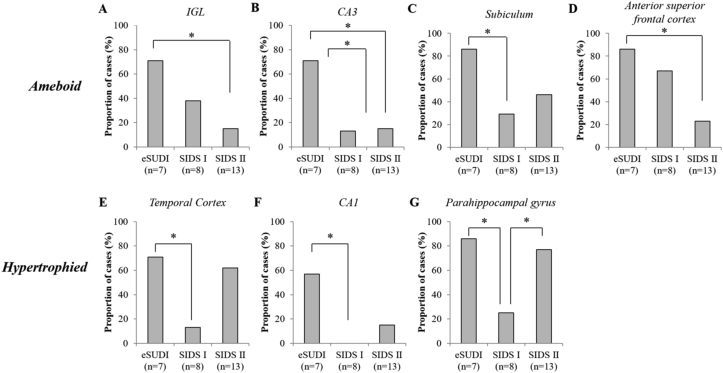
Fewer SIDS I infants demonstrated amoeboid microglia compared to eSUDI infants (Fig. 4B and C) in the CA3 (13% vs 71%; p = 0.041) and subiculum (29% vs 80%; p = 0.041) of the hippocampus. Compared to eSUDI, fewer SIDS II infants demonstrated amoeboid microglia in the IGL of the cerebellum (71% vs 15%; p = 0.022), CA3 (71% vs 15%; p = 0.022) and anterior superior frontal cortex (86% vs 23%; p = 0.017) (Fig. 4A, B, D).
Fig. 4.
Proportion of infants with activated microglial morphologies in regions of the infant brain showing differences between SUDI subgroups. Identified are regions of the infant brain where the proportion of cases with amoeboid (A–D) and hypertrophied (E–G) microglia differs between SUDI subgroups. ∗ Indicates significance, taken at p < 0.05 following Chi-square testing; Fischer’s exact test. Abbreviations: CA = cornu ammonis; IGL = internal granular layer (of the cerebellar cortex); eSUDI = explained sudden unexpected death in infancy; SIDS = sudden infant death syndrome.
Compared to eSUDI, fewer SIDS I infants demonstrated hypertrophic microglia (Fig. 4E–G) in the temporal cortex (71% vs 13%; p = 0.041), CA1 region (57% vs 0%; p = 0.026) and the parahippocampal gyrus (86% vs 25%; p = 0.041). More SIDS II infants demonstrated hypertrophied microglia compared to SIDS I infants in the parahippocampal gyrus (77% vs 25%; p = 0.032) (Fig. 4G).
The proportion of infants with ramified microglia, perivascular cells, bushy microglia, rod cells and dystrophic microglia were not different between SUDI subgroups (data not shown).
3.4.3. Microglia activation CD68
Compared to eSUDI infants, fewer SIDS I infants demonstrated positive parenchymal expression of CD68 in the subiculum (57% vs 0%; p = 0.026) and parahippocampal gyrus (71% vs 13%; p = 0.041), and fewer SIDS II infants demonstrated positive parenchymal CD68 expression in the dentate gyrus of the hippocampus (54% vs 0%; p = 0.018) (Fig. 2).
3.4.4. Microglia activation HLA-DR-DP-DQ
Compared to SIDS II infants, more SIDS I infants demonstrated positive HLA expression in the parenchyma of the ML of the cerebellum (63% vs 8%; p = 0.014) (Fig. 2M–P).
3.5. Extrinsic SIDS risk factors and microglia
To explore whether known SIDS risk factors are associated with changes in microglia, analysis was performed for the risk factors of an URTI and cigarette smoke exposure across the entire dataset, while sleep related parameters (being found prone and bedsharing/co-sleeping at the time of death), were only studied amongst the SIDS cohort (SIDS I and II). Sleep data was not relevant to the eSUDI infants given their mode of death was not sleep related.
3.5.1. URTI
3.5.1.1. Iba1+ area
Compared to infants without an URTI (n = 19), infants with an URTI (n = 9) had a reduced Iba1+ area in the CA4 (3.15 ± 0.9% vs 1.95 ± 0.32%; p = 0.024) and subiculum (2.93 ± 0.19% vs 2.16 ± 0.24%; p = 0.031) of the hippocampus, and parahippocampal gyrus (2.88 ± 0.17% vs 2.13 ± 0.18%; p = 0.015) with trends to reduced Iba1+ area (p = 0.05–0.07) in the remaining hippocampal sub-regions (Supplement Table 5, Fig. 3).
3.5.1.2. Iba1 cell morphologies
Compared to those without an URTI, infants with an URTI more commonly demonstrated amoeboid microglia in the nucleus of the spinal trigeminal tract (33% vs 0%; p = 0.026), and less commonly demonstrated amoeboid microglia in the LGN (68% vs 11%; p = 0.013). More infants with an URTI had hypertrophied microglia in the amygdala (6% vs 50%; p = 0.020), compared to those without an URTI. The proportion of infants with each of the other five morphologies was similar in infants with or without an URTI (data not shown).
3.5.1.3. Microglia activation (CD68 and HLA-DP,DQ,DR)
The proportion of infants with positive expression of CD68 and/or HLA in the parenchyma of all brain regions examined was not different between infants with and without an URTI (Supplement Table 5).
3.5.2. Cigarette smoke exposure
3.5.2.1. Iba1+ area
Compared to non-smoke exposed infants (n = 15), Iba1+ area was lower in cigarette smoke exposed (n = 8) infants in the parietal (2.52 ± 0.25% vs 1.46 ± 0.32%; p = 0.020) and occipital (2.39 ± 0.19% vs 1.61 ± 0.29%; p = 0.030) cortices (Supplement Table 6, Fig. 3).
3.5.2.2. Iba1 cell morphologies
Compared to non-smoke (n = 15) exposed infants, no infant exposed to cigarette smoke (n = 8) demonstrated amoeboid microglia in the IGL of the cerebellum (53% vs 0%; p = 0.019) and the parietal cortex (67% vs 0%; p = 0.005). The proportion of infants with each of the other six morphologies was similar in infants with, and without cigarette smoke exposure (data not shown).
3.5.2.3. Microglia activation (CD68 and HLA-DP,DQ,DR)
Parenchymal CD68 expression did not differ according to cigarette smoke exposure status in any region, yet for HLA, fewer cigarette smoke exposed infants had positive expression in the white matter of the cerebellum (87% vs 38%; p = 0.026, Supplement Table 6, Fig. 3).
3.5.3. Prone sleeping
3.5.3.1. Iba1+ area
Iba1+ area was greater in the temporal cortex of SIDS infants found prone (n = 11), compared to those found in other positions (n = 8) (3.29 ± 0.20% vs 2.55 ± 0.19%; p = 0.018, Supplement Table 7).
3.5.3.2. Iba1 cell morphology
The presence of each of the seven morphologies investigated, were not associated with found sleep position.
3.5.3.3. Microglia activation (CD68 and HLA-DP,DQ,DR)
Parenchymal CD68 expression did not differ according to the sleep position of SIDS infants (Supplement Table 7), but HLA expression in the anterior superior frontal cortex was more frequent in SIDS infants found prone (0% vs 60%; p = 0.035, Supplement Table 7, Fig. 3).
3.5.4. BED-SHARE/CO-SLEEPING
3.5.4.1. Iba1+ area
Among SIDS infants, there was no differences in Iba1+ area between those who were (n = 9) and were not (n = 12) bed-sharers/co-sleepers (Supplement Table 8).
3.5.4.2. Iba1 cell morphology
The presence of each of the seven morphologies investigated were not associated with bed-share/co-sleeping (Supplement Table 8).
3.5.4.3. Microglia activation (CD68 and HLA-DP,DQ,DR)
The proportion of infants with positive expression of CD68 and/or HLA in the parenchyma of all brain regions examined was not different between infants who were and were not bed-sharers/co-sleepers (Supplement Table 8).
4. Discussion
Using the area of tissue occupied by microglia and their activation state, as assessed by morphology and immunohistochemistry (CD68 and HLA-DP,-DQ,-DR expression) we document microglial regional heterogeneity in the infant brain and the association of this heterogeneity with various developmental parameters (summarized in Fig. 3).
In the context of SIDS, the hippocampus predominated as a region of interest. Compared to infants with a known cause of death, fewer SIDS I infants demonstrated activated microglia, and a reduced area was covered by microglia in SIDS II infants. The presence of an URTI may be the contributing factor for the reduced area of Iba1+ in the SIDS II hippocampus.
4.1. Area covered by microglial populations (Iba1+)
To our knowledge, this is the first study to report on the distribution of microglia in as many regions of the human infant brain (1–11months). We report regional heterogeneity, with sub-regions of the cerebellum and hippocampus demonstrating the highest areas of Iba1+, and nuclei of the rostral medulla the lowest. The area covered by microglia in the dentate nucleus of the cerebellum was near double that of the DMNV. Across the infant brain, the area covered by microglia was less than 5% of the total area of any region examined, and is consistent with the findings of both Kjær et al. and Sigaard et al. who show that microglia (determined morphologically) contribute less than 2% of the total cellular distribution in the infant neocortex and white matter (Kjær et al., 2016; Sigaard et al., 2014). Further, as is consistent with the literature, intra-region variability was also noted, within the medulla, hippocampus and, between the dentate nucleus and layers of the cerebellar cortex (as reviewed Tan et al., 2019).
In several regions of the SIDS II hippocampus, the Iba1+ area was less than in eSUDI cases, despite similarities in microglial morphology. This implies that morphological changes may not underpin the reduced microglia territory observed in the SIDS II hippocampus, but rather could be due to a change in the overall number of microglia. Fewer microglia could result from either impaired migration of microglia progenitors, loss of microglia, or reduced self-renewal capacity – be it inherent or secondary to noxious insults (discussed in the context of ageing by Streit, 2006). Regardless of aetiology, a potentially reduced territory of surveillance, due to fewer microglia in the hippocampus may result in a diminished ability to detect and respond to irregularities in synapses and perturbations to homeostasis (Kettenmann et al., 2011; Nimmerjahn et al., 2005).
Porcine models suggest peripheral viral infection during development may alter the sensitivity of microglia in the hippocampus to future stimuli (Ji et al., 2016). Interestingly, we found the presence of an URTI to be independently associated with a smaller area covered by microglia in the same implicated hippocampus regions. Indeed, the hippocampus was the only structure associated with changes in the face of an URTI (summarized in Fig. 3). Conclusions on the relationship between URTI and microglia would require exploration of cytokine profiles, both peripherally and in the CNS, to determine why this was only seen in the hippocampus, and why only for Iba1 expression.
Less striking effects attributed to the two leading modifiable SIDS risk factors, cigarette smoke exposure and unsafe sleeping conditions, were observed. The microglial area in the temporal cortex was greater in SIDS infants found prone than in infants found in other positions. Conversely, in the parietal and occipital cortices, cigarette smoke exposure was associated with a reduced area of microglia, and no expression of CD68. Combined, these data suggest cortical microglia may be particularly vulnerable to extrinsic factors during development. While prenatal exposure to environmental toxins such as diesel exhaust have been shown to have long-lasting effects on microglial development in a number of brain regions (Bolton et al., 2017), similar evidence on the effects of cigarette smoke exposure or hypoxia on cortical microglial function is lacking in this age group. However, recent findings from our laboratory in a mouse model of pre-into-postnatal cigarette smoke exposure, found that Iba1+ area was not altered in the medullary nuclei (Machaalani et al., 2019) supporting the medulla findings of our cohort herein, with no associations observed between cigarette smoke exposure and the examined microglial parameters.
4.2. Morphology of microglial populations
In the surveillant state, microglia contribute to developmental processes and monitor the CNS for any perturbations to homeostasis (Paolicelli and Ferretti, 2017; Tremblay et al., 2011; Wake et al., 2013). Based off this, as expected, the most common morphology observed in all SUDI subgroups and all regions, was ramified (i.e. “resting”) microglia.
The presence of rod or dystrophic microglia was rare in all brain regions and SUDI groups (Streit et al., 2014). Rod microglia are a relatively novel morphology, characterised by a length-to-width ratio ≥1.5, retraction of planar processes and the potential to align themselves to form “trains”. Their function is yet to be elucidated but they are postulated to insulate damaged neurons following acute neurological injury (Taylor et al., 2014). Dystrophic microglia demonstrate a fragmented appearance with loss, shortening and beading of processes, and are reported in the normal ageing brain (Streit et al., 2004). Thus, given our dataset is composed of infants, none of whom have a known acute neurological injury, the scarcity of these morphologies is not surprising.
Hypertrophic or “primed” microglia reflect an intermediate state between ramified, or “resting” microglia and amoeboid, or “activated” microglia. Microglia may transition between phenotypes, however ramified microglia do not necessarily require this intermediate state to become activated (Ransohoff and Perry, 2009). Rather, in the hypertrophic phenotype, microglia might best be thought of as being in a hypervigilant state – prepared to transition into a fully activated, amoeboid microglia. Thus, the lower proportion of SIDS I infants with hypertrophic microglia in the parahippocampal gyrus compared to eSUDI and SIDS II, may suggest a less reactive/vigilant microglia population. Aberrant surveillance, may not only render an infant unable to respond appropriately to noxious stimuli, but may also result in consequences on neurodevelopment due to erroneous synapse interaction and an altered microenvironment (Tremblay et al., 2011; Wake et al., 2009). Alternatively, increased or preferential activation along the hypertrophied-intermediate pathway in eSUDI and SIDS II, as compared to SIDS I, might reflect a difference in the microglia activating stimulus (or cumulative stimuli) between the SUDI subgroups, with the eSUDI group comprised of predominantly cardiogenic and infectious causes of death.
4.3. Activation status of microglial populations (CD68+, HLA+, morphology)
Morphology and CD68 and HLA immunohistochemistry were used to investigate microglial activation status. Compared to Iba1 staining which provided recognition of microglia irrespective of activation status, in an extensive mosaic-like pattern of expression in all brain regions, parenchymal CD68 and HLA expression was limited to only a few areas. Across brain regions and SUDI sub-groups, congruency between CD68, HLA and the amoeboid morphology was poor (Fig. 5); these observations re-enforce previous warning by Hendrickx et al., that Iba1, CD68, and HLA are not interchangeable markers in the human brain and are best utilised in different neuropathological contexts (Hendrickx et al., 2017). Throughout the eSUDI brain, varying morphologies and parenchymal CD68 and HLA expression were observed, as might be expected as part of the maintenance of the infant CNS.
Fig. 5.
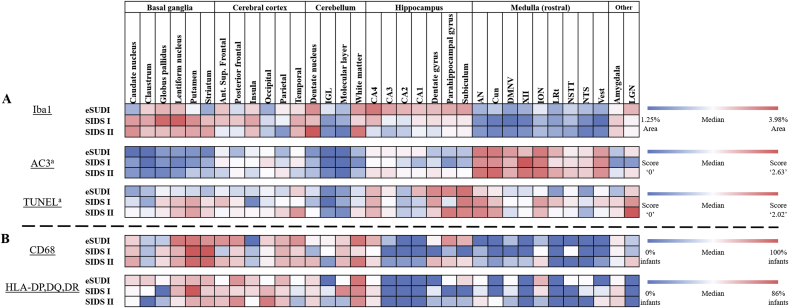
Distribution of microglial and cell death marker expression across regions of the infant brain. Visual comparison of immunohistochemistry findings, with conditional formatting applied to the scoring schema for each marker. Dark blue represents the lowest value, regardless of diagnosis and across all brain regions. Dark red represents the highest value observed. White indicates values at or approximating the median. For (A) the data shown represents the quantitative values for Iba1, AC3 and TUNEL. Iba1 values are the percentage area of the given region covered by Iba1 positive material. Values for AC3 and TUNEL were adapted from our previous findings and follow a semi-quantitative five tier scoring schema. CD68 and HLA-DP,DQ,DR values reflect the proportion of cases with positive marker expression in the parenchyma of the given region, they are shown in (B) to allow comparison. Abbreviations: AC3 = active caspase-3, Ant. = anterior, AN = arcuate nucleus, CA = cornu ammonis, CD68 = cluster of differentiation factor 68, Cun = cuneate nucleus, DMNV = dorsal motor nucleus of the vagus, eSUDI = explained sudden unexpected death in infancy, HLA-DP,DQ,DR = human leukocyte antigen – isotopes DP,DQ,DR, IGL = internal granular layer, ION = inferior olivary nucleus, LRt = lateral reticular formation, LGN = lateral geniculate nucleus, NSTT = nucleus of the spinal trigeminal tract, NTS = nucleus of the solitary tract, SIDS = sudden infant death syndrome, Sup. = superior, TUNEL = terminal deoxynucleotidyl transferase dUTP nick end labeling, Vest = vestibular nucleus, XII = hypoglossal nucleus.a Adapted from our previous findings (Ambrose et al., 2019). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The SIDS hippocampus again yielded itself as a region of interest in the context of activated microglia. Although there were no differences between eSUDI and SIDS I infants with regards to the hippocampal area covered by microglia, from the perspectives of both morphology and activated marker expression, eSUDI infants more frequently demonstrated microglial activation (CD68+) in the hippocampus compared to SIDS I infants. To date, only 3 studies have looked at microglia in the infant hippocampus; one in the context of SIDS. While Del Bigio and Becker (1994) reported an increase in the number of microglia in the dentate gyrus of 26 SIDS infants based on morphological appearance of rod cells as microglia using H+E/Luxol fast blue and immunohistochemistry for ricinus communis agglutinin-1 lectin (RCA-1), leukocyte common antigen and HAM56, Paine et al., question whether these are microglia, suggesting they are neural progenitor cells (Paine et al., 2014). A recent study by Kinney et al., utilised CD68 to study cell populations in the dentate gyrus of three SIDS infants but did not undertake further comparison between diagnostic groups (Kinney et al., 2015). In our study, the only differential we saw for CD68 between the groups was a significantly greater proportion of SIDS II infants demonstrating CD68+ expression in the dentate gyrus compared to SIDS I infants. Thus, it can be concluded that the dentate gyrus is a region of interest for further study related to ‘activated’ microglia in the context of SIDS. Given the activation markers included in this study have the propensity to also detect monocyte derived macrophages, which may infiltrate regions of the CNS in response to injury (Bennett et al., 2016; Mildner et al., 2017), future studies should consider co-localisation with highly specific markers of microglia such as TMEM119 or P2RY12.
Positive CD68 staining in the CNS identifies lysosomes in phagocytic microglia and macrophages (Guillemin and Brew, 2004; Holness et al., 1993). The role of microglia in phagocytosis following cell death and the reciprocal regulatory effects of cell death on microglia have been well characterised (Burguillos et al., 2011; Kettenmann et al., 2011). Since the postnatal brain is known to undergo programmed cell death (PCD) in a number of regions, it was hypothesised that CD68 expression might mirror the distribution of PCD. However, this was not the case. When compared to the distribution of active caspase-3 (AC3) and terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) expression which we recently explored in the same dataset (Ambrose et al., 2019), the distribution of activated microglia was not mirrored; for example, an inverted distribution of AC3 and Iba1+ was observed where AC3 was greatest in nuclei of the rostral medulla and lowest in sub-regions of the basal ganglia, while Iba1+ was greatest in the basal ganglia and lowest in the rostral medulla (summarized in Fig. 5). Such incongruence between microglial activation and cell death distribution has previously been reported in the cerebellum and hippocampus during development, with microglia localisation and PCD shown to be independent processes (Ashwell, 1990; Eyo et al., 2016). Thus, the dynamic between neuronal cell death and microglia in the infant brain is far more complex than we had anticipated, and likely reflects a number of factors including regional microenvironments, cytokine profiles, temporal parameters and strength of relevant stimuli, with chemokines such as fractalkine suggested to be crucial in microglial dynamics and neuronal-microglia interactions (reviewed: Ransohoff and El Khoury, 2015).
Although the pattern of distribution was similar, compared to CD68, expression of HLA was sparser and limited to 28 of 34 regions examined. HLA recognises microglia with an antigen-presenting capacity and so is useful in identifying immune activation and inflammation in the CNS (Collawn and Benveniste, 1999; Hendrickx et al., 2017). In the present study, the white matter of the cerebellum was the brain region with the greatest proportion of infants with HLA+ expression, suggesting this area to be the most likely in an immune or inflammation primed state. This is consistent with findings in the normal adult human brain, with the white matter of the cerebellum reported as one of the brain regions with the greatest number of MHC II expressing microglia (Mittelbronn et al., 2001). For the remaining brain regions, HLA expression was otherwise low or not present, as might be expected in the immune-privileged CNS (Collawn and Benveniste, 1999). Compared to eSUDI cases, no significant differences were observed in the SIDS groups, but a difference was noted between SIDS I and SIDS II in the ML of the cerebellum, and this was independent of the risk factors of URTI, unsafe sleeping conditions, and cigarette smoke exposure. The cerebellar ML shapes the rate and activity of Purkinje cells, which are the key neurons of cerebellar efferent pathways (Brown et al., 2019). Through projections to the deep cerebellar nuclei and brainstem, Purkinje cells are implicated in autonomic and respiratory regulation (Harvey and Napper, 1991; Napper and Harvey, 1988). Particularly relevant in the context of SIDS, alterations (namely loss of neurons) in the Purkinje layer are hypothesised to impair responses to hypercapnic environments; potentially contributing to sudden death in the face of cardiorespiratory challenge (Calton et al., 2014, 2016). It is surprising that SIDS I infants had higher expression of microglial activation markers compared to SIDS II, as it might be expected that the presence of risk factors leading to respiratory challenge (e.g. prone sleeping/unsafe sleeping conditions) might result in neuropathological changes. Thus, detection of activated microglia in the cerebellar cortex of SIDS I cases suggests an innate intrinsic vulnerability possibly linked to the pathophysiological mechanism of SIDS, highlighting this region as one that should be explored further.
4.4. Associations between microglia and infant characteristics
Infant growth and development parameters of brain/body weight and head circumference, were associated with differences in the area of microglia populations in certain brain regions, with positive correlations in the basal ganglia, insula and occipital cortices, and the ML of the cerebellum, and negative correlations in the brainstem. The period of infancy characterised by growth is accompanied by local expansion of the self-renewing microglial population; as reviewed by Lenz and Nelson (2018) in rodent studies microglial proliferation is reported to continue until postnatal day (PND) 14 (equivalent to 1 postnatal year in human infants (Sengupta, 2013)), with a stable microglia population achieved by PND28 (equivalent to 2 postnatal years in human infants (Sengupta, 2013)). Thus, the demonstrated impact of infant growth on microglia populations within these structures is unsurprising. However, it is curious that PCA was only a contributor for the brainstem, with younger infants and infants with smaller measurements demonstrating a greater area of Iba1+, activated morphologies and activated marker expression in medullary nuclei. This suggests the brainstem microenvironment and subsequent microglia population may be unique from a developmental perspective, when compared to the rest of the brain, and that it may be more prone to inflammatory responses in the face of parameters affecting normal growth and development such as nutrition and socio-economic conditions. Although we could not address the role of these conditions in our cohort since these were not always reported on in the autopsy investigation, animal studies do show that maternal diet, feeding and nutrition affect microglia phenotypes in the developing (Bilbo and Tsang, 2010) and adult (Baufeld et al., 2016) brain.
Sexual dimorphism was also observed, with males having a greater area of Iba1+ in regions of the basal ganglia. Rodent studies have previously reported sex-specific changes in multiple brain regions during development, with differences in microglia activation between the sexes postulated to be related to chemokines, steroid hormones, and cell-surface receptor expression (reviewed: Lenz and McCarthy, 2015; Pierre et al., 2017).
4.5. Limitations
Heterogeneity of the control (eSUDI) population is a common issue in SUDI research and this applied to our study where death was attributed to various cardiogenic and infectious causes. Thus, our eSUDI group is not necessarily indicative of the “normal” infant brain. Infant deaths of a sudden nature, such as motor vehicle accidents (with no neurological damage or head injury) might best represent the “normal” postnatal infant brain, however no such cases were available for inclusion. Given the greatest homogeneity, characteristically and from a microglial expression level, was observed amongst SIDS I cases, this population may be useful as a control group for future studies investigating microglia in the context of infant CNS pathologies, as it may be the closest available representation of the “normal infant brain” for human post-mortem studies.
Due to limited availability of tissue, the present study was restricted to the use of a dichotomous scoring method based on the presence or absence of varying morphologies in a given region rather than a stereotactic approach which would have enabled quantification and characterisation of microglia processes and their branching pattern. Moreover, we could not study for dynamic parameters such as microglia cell death and proliferation. As it yields itself as a region of interest, future studies should look to better characterise the microglial populations present in the SIDS hippocampus through thorough stereological quantification techniques and use of additional markers such as proliferation markers (e.g. Ki67), putative pro- and anti-inflammatory markers (e.g. IL-1β and CD206), novel specific markers of microglia (e.g. Tmem119 and P2RY12) and, markers of myeloid cell origin (e.g. PU.1). Further, co-localisation studies using Iba1 and such markers might provide insight into the kinetics and exposures of these microglial populations.
5. Conclusion
In this study, we showed regional heterogeneity in both the area covered by and the activation status of microglia in 34 regions of the human infant brain. Developmental parameters (brain weight, body weight and head circumference) correlated with both: the Iba1+ area in the basal ganglia, frontal cortex, cerebellum and medulla, and microglial activation in the basal ganglia, frontal cortex and cerebellum. Between SUDI sub-groups, key differences were observed in the hippocampus, but may be confounded by a history of an URTI. Known extrinsic SIDS risk factors, including cigarette smoke exposure and the prone sleeping position, had minimal effect on microglia activation. Further, we identify SIDS I to be a relatively homogenous infant population with respect to microglia, suggesting their utility as a control for future research into other infant CNS pathologies. Overall, we show the distribution and functional state of microglia in the human infant brain to be heterogeneous, and influenced by intrinsic and extrinsic factors.
Funding
This study was funded by philanthropy (The Miranda Belshaw Foundation, Australia).
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethical approval
Ethical approval from the NSW Health RPAH Zone (X13-0038 and HREC/13/RPAH/54) and University of Sydney Ethic committees.
Declaration of competing interest
All authors have seen and approved the manuscript. There is no conflict of interest to declare.
Acknowledgements
The tissue used in this study was obtained from NSW Forensic and Analytical Science Services.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2020.100117.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ajami B., Bennett J.L., Krieger C., Tetzlaff W., Rossi F.M. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007;10:1538. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- Ambrose N., Rodriguez M., Waters K.A., Machaalani R. Cell death in the human infant central nervous system and in sudden infant death syndrome (SIDS) Apoptosis. 2019;24:46–61. doi: 10.1007/s10495-018-1509-0. [DOI] [PubMed] [Google Scholar]
- Ambrose N., Waters K.A., Rodriguez M.L., Bailey K., Machaalani R. Neuronal apoptosis in the brainstem medulla of sudden unexpected death in infancy (SUDI), and the importance of standardized SUDI classification. Forensic Sci. Med. Pathol. 2018;14:42–56. doi: 10.1007/s12024-018-9954-1. [DOI] [PubMed] [Google Scholar]
- Arcuri C., Mecca C., Bianchi R., Giambanco I., Donato R. The pathophysiological role of microglia in dynamic surveillance, phagocytosis and structural remodeling of the developing CNS. Front. Mol. Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell K. Microglia and cell death in the developing mouse cerebellum. Dev. Brain Res. 1990;55:219–230. doi: 10.1016/0165-3806(90)90203-b. [DOI] [PubMed] [Google Scholar]
- Baufeld C., Osterloh A., Prokop S., Miller K.R., Heppner F.L. High-fat diet-induced brain region-specific phenotypic spectrum of CNS resident microglia. Acta Neuropathol. 2016;132:361–375. doi: 10.1007/s00401-016-1595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon S., Walker F. Microglial activation in the injured and healthy brain: what are we really talking about? Practical and theoretical issues associated with the measurement of changes in microglial morphology. Neuroscience. 2012;225:162–171. doi: 10.1016/j.neuroscience.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Bilbo S.D., Tsang V. Enduring consequences of maternal obesity for brain inflammation and behavior of offspring. Faseb. J. 2010;24:2104–2115. doi: 10.1096/fj.09-144014. [DOI] [PubMed] [Google Scholar]
- Billiards S.S., Haynes R.L., Folkerth R.D., Trachtenberg F.L., Liu L.G., Volpe J.J., Kinney H.C. Development of microglia in the cerebral white matter of the human fetus and infant. J. Comp. Neurol. 2006;497:199–208. doi: 10.1002/cne.20991. [DOI] [PubMed] [Google Scholar]
- Blair P.S., Byard R.W., Fleming P.J. Sudden unexpected death in infancy (SUDI): suggested classification and applications to facilitate research activity. Forensic Sci. Med. Pathol. 2012;8:312–315. doi: 10.1007/s12024-011-9294-x. [DOI] [PubMed] [Google Scholar]
- Boche D., Perry V., Nicoll J. Activation patterns of microglia and their identification in the human brain. Neuropathol. Appl. Neurobiol. 2013;39:3–18. doi: 10.1111/nan.12011. [DOI] [PubMed] [Google Scholar]
- Bolton J.L., Marinero S., Hassanzadeh T., Natesan D., Le D., Belliveau C., Mason S.N., Auten R.L., Bilbo S.D. Gestational exposure to air pollution alters cortical volume, microglial morphology, and microglia-neuron interactions in a sex-specific manner. Front. Synaptic Neurosci. 2017;9 doi: 10.3389/fnsyn.2017.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.M., Arancillo M., Lin T., Catt D.R., Zhou J., Lackey E.P., Stay T.L., Zuo Z., White J.J., Sillitoe R.V. Molecular layer interneurons shape the spike activity of cerebellar Purkinje cells. Sci. Rep. 2019;9:1742. doi: 10.1038/s41598-018-38264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguillos M.A., Deierborg T., Kavanagh E., Persson A., Hajji N., Garcia-Quintanilla A., Cano J., Brundin P., Englund E., Venero J.L. Caspase signalling controls microglia activation and neurotoxicity. Nature. 2011;472:319. doi: 10.1038/nature09788. [DOI] [PubMed] [Google Scholar]
- Calton M., Dickson P., Harper R.M., Goldowitz D., Mittleman G. Impaired hypercarbic and hypoxic responses from developmental loss of cerebellar Purkinje neurons: implications for sudden infant death syndrome. Cerebellum. 2014;13:739–750. doi: 10.1007/s12311-014-0592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calton M.A., Howard J.R., Harper R.M., Goldowitz D., Mittleman G. The cerebellum and SIDS: disordered breathing in a mouse model of developmental cerebellar Purkinje cell loss during recovery from hypercarbia. Front. Neurol. 2016;7:78. doi: 10.3389/fneur.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J.D., Olschowka J.A., O’Banion M.K. Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J. Neuroinflammation. 2014;11:98. doi: 10.1186/1742-2094-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collawn J.F., Benveniste E.N. Regulation of MHC class II expression in the central nervous system. Microb. Infect. 1999;1:893–902. doi: 10.1016/s1286-4579(99)00228-2. [DOI] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., Gan W.-B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005;8:752. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Del Bigio M., Becker L. Microglial aggregation in the dentate gyrus: a marker of mild hypoxic-ischaemic brain insult in human infants. Neuropathol. Appl. Neurobiol. 1994;20:144–151. doi: 10.1111/j.1365-2990.1994.tb01173.x. [DOI] [PubMed] [Google Scholar]
- Esiri M.M., al Izzi M.S., Reading M.C. Macrophages, microglial cells, and HLA-DR antigens in fetal and infant brain. J. Clin. Pathol. 1991;44:102–106. doi: 10.1136/jcp.44.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo U.B., Miner S.A., Weiner J.A., Dailey M.E. Developmental changes in microglial mobilization are independent of apoptosis in the neonatal mouse hippocampus. Brain Behav. Immun. 2016;55:49–59. doi: 10.1016/j.bbi.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming P., Blair P., Bacon C., Berry J. The Stationery Office; London: 2000. Sudden Unexpected Deaths in Infancy: the CESDI SUDI Studies 1993–1996; pp. 1–5. [Google Scholar]
- Graeber M.B., Bise K., Mehraein P. CR3/43, a marker for activated human microglia: application to diagnostic neuropathology. Neuropathol. Appl. Neurobiol. 1994;20:406–408. doi: 10.1111/j.1365-2990.1994.tb00987.x. [DOI] [PubMed] [Google Scholar]
- Guillemin G.J., Brew B.J. Microglia, macrophages, perivascular macrophages, and pericytes: a review of function and identification. J. Leukoc. Biol. 2004;75:388–397. doi: 10.1189/jlb.0303114. [DOI] [PubMed] [Google Scholar]
- Hagemeyer N., Hanft K.-M., Akriditou M.-A., Unger N., Park E.S., Stanley E.R., Staszewski O., Dimou L., Prinz M. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 2017;134:441–458. doi: 10.1007/s00401-017-1747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R., Napper R. Quantitatives studies on the mammalian cerebellum. Prog. Neurobiol. 1991;36:437–463. doi: 10.1016/0301-0082(91)90012-p. [DOI] [PubMed] [Google Scholar]
- Hendrickx D.A., van Eden C.G., Schuurman K.G., Hamann J., Huitinga I. Staining of HLA-DR, Iba1 and CD68 in human microglia reveals partially overlapping expression depending on cellular morphology and pathology. J. Neuroimmunol. 2017;309:12–22. doi: 10.1016/j.jneuroim.2017.04.007. [DOI] [PubMed] [Google Scholar]
- Holness C.L., da Silva R.P., Fawcett J., Gordon S., Simmons D.L. Macrosialin, a mouse macrophage-restricted glycoprotein, is a member of the lamp/lgp family. J. Biol. Chem. 1993;268:9661–9666. [PubMed] [Google Scholar]
- Ji P., Schachtschneider K.M., Schook L.B., Walker F.R., Johnson R.W. Peripheral viral infection induced microglial sensome genes and enhanced microglial cell activity in the hippocampus of neonatal piglets. Brain Behav. Immun. 2016;54:243–251. doi: 10.1016/j.bbi.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H., Hanisch U.-K., Noda M., Verkhratsky A. Physiology of microglia. Physiol. Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- Kinney H.C., Cryan J.B., Haynes R.L., Paterson D.S., Haas E.A., Mena O.J., Minter M., Journey K.W., Trachtenberg F.L., Goldstein R.D., Armstrong D.D. Dentate gyrus abnormalities in sudden unexplained death in infants: morphological marker of underlying brain vulnerability. Acta Neuropathol. 2015;129:65–80. doi: 10.1007/s00401-014-1357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjær M., Fabricius K., Sigaard R.K., Pakkenberg B. Neocortical development in brain of young children—a stereological study. Cerebr. Cortex. 2016;27:5477–5484. doi: 10.1093/cercor/bhw314. [DOI] [PubMed] [Google Scholar]
- Krous H.F., Beckwith J.B., Byard R.W., Rognum T.O., Bajanowski T., Corey T., Cutz E., Hanzlick R., Keens T.G., Mitchell E.A. Sudden infant death syndrome and unclassified sudden infant deaths: a definitional and diagnostic approach. Pediatrics. 2004;114:234–238. doi: 10.1542/peds.114.1.234. [DOI] [PubMed] [Google Scholar]
- Lawson L., Perry V., Dri P., Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- Lenz K.M., McCarthy M.M. A starring role for microglia in brain sex differences. Neuroscientist. 2015;21:306–321. doi: 10.1177/1073858414536468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz K.M., Nelson L.H. Microglia and beyond: innate immune cells as regulators of brain development and behavioral function. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00698. 698-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Wang K., Gao H.M., Mandavilli B., Wang J.Y., Hong J.S. Molecular consequences of activated microglia in the brain: overactivation induces apoptosis. J. Neurochem. 2001;77:182–189. doi: 10.1046/j.1471-4159.2001.t01-1-00216.x. [DOI] [PubMed] [Google Scholar]
- Machaalani R., Thawley M., Huang J., Chen H. Effects of prenatal cigarette smoke exposure on BDNF, PACAP, microglia and gliosis expression in the young male mouse brainstem. Neurotoxicology. 2019;74:40–46. doi: 10.1016/j.neuro.2019.05.009. [DOI] [PubMed] [Google Scholar]
- Maslinska D., Laure-Kamionowska M., Kaliszek A. Morphological forms and localization of microglial cells in the developing human cerebellum. Folia Neuropathol. 1998;36:145–151. [PubMed] [Google Scholar]
- Mildner A., Huang H., Radke J., Stenzel W., Priller J. P2Y12 receptor is expressed on human microglia under physiological conditions throughout development and is sensitive to neuroinflammatory diseases. Glia. 2017;65:375–387. doi: 10.1002/glia.23097. [DOI] [PubMed] [Google Scholar]
- Mittelbronn M., Dietz K., Schluesener H., Meyermann R. Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol. 2001;101:249–255. doi: 10.1007/s004010000284. [DOI] [PubMed] [Google Scholar]
- Moon R.Y., Hauck F.R. University of Adelaide Press; 2018. Risk Factors and Theories. SIDS Sudden Infant and Early Childhood Death: the Past, the Present and the Future. [PubMed] [Google Scholar]
- Napper R., Harvey R. Number of parallel fiber synapses on an individual Purkinje cell in the cerebellum of the rat. J. Comp. Neurol. 1988;274:168–177. doi: 10.1002/cne.902740204. [DOI] [PubMed] [Google Scholar]
- Nelson P.T., Soma L.A., Lavi E. Microglia in diseases of the central nervous system. Ann. Med. 2002;34:491–500. doi: 10.1080/078538902321117698. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Paine S.M.L., Willsher A.R., Nicholson S.L., Sebire N.J., Jacques T.S. Characterization of a population of neural progenitor cells in the infant hippocampus. Neuropathol. Appl. Neurobiol. 2014;40:544–550. doi: 10.1111/nan.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R.C., Ferretti M.T. Function and dysfunction of microglia during brain development: consequences for synapses and neural circuits. Front. Synaptic Neurosci. 2017;9 doi: 10.3389/fnsyn.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre W.C., Smith P.L., Londono I., Chemtob S., Mallard C., Lodygensky G.A. Neonatal microglia: the cornerstone of brain fate. Brain Behav. Immun. 2017;59:333–345. doi: 10.1016/j.bbi.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Ransohoff R.M., El Khoury J. Microglia in Health and disease. Cold Spring Harbor Perspect. Biol. 2015;8 doi: 10.1101/cshperspect.a020560. a020560-a020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff R.M., Perry V.H. Microglial physiology: unique stimuli, specialized responses. Annu. Rev. Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Sasaki A. Neuropathology; 2016. Microglia and Brain Macrophages: an Update. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Butovsky O., Brück W., Hanisch U.-K. Microglial phenotype: is the commitment reversible? Trends Neurosci. 2006;29:68–74. doi: 10.1016/j.tins.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Sengupta P. The laboratory rat: relating its age with human’s. Int. J. Prev. Med. 2013;4:624–630. [PMC free article] [PubMed] [Google Scholar]
- Sigaard R.K., Kjær M., Pakkenberg B. Development of the cell population in the brain white matter of young children. Cerebr. Cortex. 2014;26:89–95. doi: 10.1093/cercor/bhu178. [DOI] [PubMed] [Google Scholar]
- Sołtys Z., Ziaja M., Pawliński R., Setkowicz Z., Janeczko K. Morphology of reactive microglia in the injured cerebral cortex. Fractal analysis and complementary quantitative methods. J. Neurosci. Res. 2001;63:90–97. doi: 10.1002/1097-4547(20010101)63:1<90::AID-JNR11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Statistics A.B.o. vol. 2017. 2018. (3303.0: Causes of Death). Australia. [Google Scholar]
- Streit W.J. Microglial senescence: does the brain’s immune system have an expiration date? Trends Neurosci. 2006;29:506–510. doi: 10.1016/j.tins.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Streit W.J., Graeber M.B. Microglia A pictorial. Prog. Histochem. Cytochem. 1996;31:XII–89. doi: 10.1016/s0079-6336(96)80004-0. [DOI] [PubMed] [Google Scholar]
- Streit W.J., Sammons N.W., Kuhns A.J., Sparks D.L. Dystrophic microglia in the aging human brain. Glia. 2004;45:208–212. doi: 10.1002/glia.10319. [DOI] [PubMed] [Google Scholar]
- Streit W.J., Xue Q.-S., Tischer J., Bechmann I. Microglial pathology. Acta Neuropathol. Commun. 2014;2 doi: 10.1186/s40478-014-0142-6. 142-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-L., Yuan Y., Tian L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatr. 2019:1–17. doi: 10.1038/s41380-019-0609-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.E., Morganti-Kossmann C., Lifshitz J., Ziebell J.M. Rod microglia: a morphological definition. PloS One. 2014;9 doi: 10.1371/journal.pone.0097096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M.-È., Stevens B., Sierra A., Wake H., Bessis A., Nimmerjahn A. The role of microglia in the healthy brain. J. Neurosci. 2011;31:16064–16069. doi: 10.1523/JNEUROSCI.4158-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H., Moorhouse A.J., Jinno S., Kohsaka S., Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci. 2009;29:3974–3980. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H., Moorhouse A.J., Miyamoto A., Nabekura J. Microglia: actively surveying and shaping neuronal circuit structure and function. Trends Neurosci. 2013;36:209–217. doi: 10.1016/j.tins.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Wolf S.A., Boddeke H.W.G.M., Kettenmann H. Microglia in physiology and disease. Annu. Rev. Physiol. 2017;79:619–643. doi: 10.1146/annurev-physiol-022516-034406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.