Abstract
Previously regarded as simple fat storage particles, new evidence suggests that lipid droplets (LDs) are dynamic and functional organelles involved in key cellular processes such as membrane biosynthesis, lipid metabolism, cell signalling and inflammation. Indeed, an increased LD content is one of the most apparent features resulting from lipid metabolism reprogramming necessary to support the basic functions of cancer cells. LDs have been associated to different cellular processes involved in cancer progression and aggressiveness, such as tumorigenicity, invasion and metastasis, as well as chemoresistance. Interestingly, all of these processes are controlled by a subpopulation of highly aggressive tumoral cells named cancer stem cells (CSCs), suggesting that LDs may be fundamental elements for stemness in cancer. Considering the key role of CSCs on chemoresistance and disease relapse, main factors of therapy failure, the design of novel therapeutic approaches targeting these cells may be the only chance for long-term survival in cancer patients. In this sense, their biology and functional properties render LDs excellent candidates for target discovery and design of combined therapeutic strategies. In this review, we summarise the current knowledge identifying LDs and CSCs as main contributors to cancer aggressiveness, metastasis and chemoresistance.
Keywords: Lipids, Lipid droplets, Lipid metabolism, Stemness, Cancer stem cells, Chemoresistance
Core Tip: Increasing evidence suggests that lipid droplets (LDs) support cancer stem cells (CSCs) functionality at different levels. Indeed, an increased LD content has been linked to tumorigenicity, metastatic spread and chemoresistance in different cancer types, highlighting their value as prognostic and treatment response predictive biomarker. A deeper understanding of the molecular mechanisms by which LDs control these processes would expedite the discovery of novel potentially druggable targets and the design of more efficient therapeutic strategies aimed at eliminating highly tumorigenic CSCs.
INTRODUCTION
Cancer stem cells
The cancer stem cell concept: Consistent evidence supports that most of the heterogeneity found in both liquid and solid cancers might be originated in the context of hierarchical organisation of the tumours. Indeed, a subset of cells with self-renewal capacity and tumour-initiating properties called cancer stem cells (CSCs) undergo asymmetrical and symmetrical divisions in order to originate bulk differentiated tumour cells and identical CSCs to perpetuate its lineage. Cancer hierarchy at cellular level was first described in acute myeloid leukaemia[1], representing a huge milestone in the understanding of cancer emergence. The CSC theory has been supported since then by increasing evidence in other malignancies such as breast cancer[2], brain tumours[3] colon and colorectal cancers[4,5], as well as pancreatic cancer[6,7], among others.
The current approach to cancer therapy has been both clarified and challenged by the existence of CSCs. On the one hand, the increasing evidence of their existence and contribution to tumorigenesis and metastasis has allowed researchers and clinicians to acquire a better understanding of cancer origin and evolution. On the other hand, proof of the implication of CSCs in treatment failure due to their intrinsic chemoresistance abilities has demonstrated that specific therapeutic strategies against this tumoral subpopulation are still urgently needed.
The origin of CSCs remains unclear, since it might vary between malignancies. One hypothesis derives from the observed similarities between CSCs and their normal homologous SCs, suggesting that local SCs may suffer a malignant transformation[8]. Other theories involve the acquisition of stemness features by differentiated cells. On the one hand, it has been suggested that differentiated cancer cells undergoing epithelial-to-mesenchymal transition acquire stem-like properties under the regulation of Notch signalling[9,10]. On the other hand, microenvironmental signals from stromal cells might facilitate non-CSCs dedifferentiation. For instance, Wnt signalling conferred self-renewal and tumorigenic abilities to colorectal cancer cells[11]. Furthermore, FGF5 and collagen production induced by Hedgehog promoted triple negative breast cancer chemoresistance by acquiring self-renewal capacity[12]. In any case, a dual scenario in which both local SCs and differentiated tumour cells originate new CSCs may be present in chemoresistant pancreatic[13] and lung[14] cancer cells.
CSC metabolism
Microenvironmental selective pressure forces CSCs to adapt continuously in order to survive and progress. For instance, as the tumour grows, glucose and oxygen levels diminish, the pH becomes acidic and reactive oxygen species (ROS) and inflammatory mediators accumulate in the tumour microenvironment. Since most differentiated tumour cells are fully glycolytic in order to cope with their enhanced proliferative rates (e.g. Warburg effect), resource scarcity forces CSCs to become metabolically and functionally plastic in order to survive and detoxify their microenvironment. Theoretically, an active mitochondrial metabolism would provide CSCs with an increased plasticity since a larger array of substrates could be feeding the tricarboxylic acid cycle. However, depending on the tumour type and model systems studied, CSCs use either mitochondrial oxidative phosphorylation (OXPHOS) or glycolysis[15,16] preferentially, with varying degrees of plasticity to switch from, even within the same tumour. Indeed, although the majority of pancreatic CSCs relies on OXPHOS and is very sensitive to mitochondrial inhibition, a small portion of CSCs shows a plastic phenotype, activating glycolysis when its mitochondria are inhibited[17]. However, full metabolic plasticity comes at the expense of self-renewal capacity[17].
Importantly, OXPHOS-dependent CSCs and therapy-resistant tumour cells from different cancer types bear higher levels of the master regulator of mitochondrial biogenesis peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α)[17-19], which supports OXPHOS metabolism and provides resistance to oxidative stress and chemotherapy[18-20]. Considering that PGC-1α is a transcriptional coactivator of the peroxisome proliferator-activated superfamily of receptors (PPARs) which controls the balance between glucose and lipid metabolism[21,22], we can hypothesise that PGC-1α enables CSCs to control a complex metabolic programme associating stemness to mitochondrial metabolism, including lipid and fatty acid (FA) oxidation (FAO). In fact, different studies have demonstrated that lipid metabolism is required to maintain the CSC pools in several tumour types[23-26].
LIPID METABOLISM AND LIPID DROPLETS
Cancer and lipid metabolism
Cancer cells have metabolic reprogramming abilities to sustain high proliferation rates as well as energy production, not only through high glycolysis (Warburg effect), but also through reprogrammed lipid metabolism[27-29]. Indeed, they enhance de novo lipid synthesis, lipogenesis and FAO, being FA synthesis one of the most important aberrations of cancer cell metabolism[30]. FAs are involved in many different aspects of tumorigenesis and tumour progression and sustain three requirements of cancer cells and CSCs: Cell membrane formation, signalling molecules and lipid-derived messengers, and energy production[31-33]. Importantly, an increased FA metabolism has been associated to poor prognosis in different types of cancer, such as pancreatic cancer or melanoma[34]. In pancreatic cancer, it is generally associated to a high expression of key regulatory enzymes like the FA synthase and sterol regulatory element-binding protein[35,36].
Cancer cells accumulate more lipids in their cytoplasm than normal cells[37]. Novikoff was the first to demonstrate the presence of cytoplasmic inclusions in the rat liver tumour cells and to identify the lipid nature of these droplets[38]. Although regarded as simple fat storage particles for long, lipid droplets (LDs) are currently considered conserved, dynamic and functional organelles involved in membrane biosynthesis, lipid metabolism, cell signalling and inflammation[39]. Indeed, they have been associated with an increased tumour aggressiveness and resistance to chemotherapy[40], considerably raising attention within the cancer biology community.
LDs
LDs, also known as lipid bodies or liposomes, are cellular organelles ranging from 20-40 nm to 100 mm, with key functions for lipid and energy homeostasis[41,42]. The quantity, size, composition and intracellular localisation differ significantly between or within cells, mainly due to their type, function and metabolic state[43]. Indeed, LDs are highly dynamic organelles which alternate periods of growth and consumption, depending on cell energy and nutritional status[39,41].
However, all LDs have a similar structure consisting of a hydrophobic core of neutral lipids, such as cholesteryl esters (CE), retinyl esters and triglycerides (TAGs)[44], separated from the aqueous cytoplasm by a monolayer of phospholipids, mainly phosphatidylcholine[45]. Additionally, LDs are coated with integral and peripheral proteins[46] derived from the cytosol or the endoplasmic reticulum (ER)[47]. These proteins can be classified into four groups: (1) Resident/structural proteins, such as members of the perilipin (PLIN)-ADRP-TIP47 family or the cell death-inducing DFF45-like effector (CIDE) family[48-50] (Figure 1); (2) Lipid metabolism enzymes, such as diacylglycerol acyltransferases 1 and 2 (DGAT1 and DGAT2), adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL); (3) Membrane trafficking proteins, including a variety of Ras related protein (Rab) GTPases, as well as soluble NSF binding protein receptor proteins; and (4) Cell signalling proteins such as mitogen-activated protein kinases and protein kinase C. Other types of proteins can be associated to the ribosome and cytoskeleton, or processes such as protein degradation[51,52].
Figure 1.
Structure and cellular functions of lipid droplets. Lipid droplets (LDs) have a hydrophobic core of neutral lipids, mainly cholesteryl esters and triglycerides surrounded by a monolayer of phospholipids. LDs are coated with peripheral and integral proteins such as cell death-inducing DFF45-like effector and perilipins proteins, and lipid metabolism enzymes implicated in lipid synthesis and lipolysis: diacylglycerol acyltransferases 1 and 2 (DGAT1 and DGAT2), acyl-CoA cholesterol acyltransferases 1 and 2 (ACAT1 and ACAT2), adipose triglyceride lipase, hormone-sensitive lipase and monoacylglycerol lipase. LDs play roles in energy supply, via fatty acid oxidation, and signalling, by producing lipid intermediates that include pro- and anti-inflammatory signalling molecules and peroxisome proliferator-activated (PPAR) ligands. Upon activation, PPARs together with the coactivator peroxisome proliferator-activated receptor gamma coactivator 1-alpha form a nuclear complex with RXR that binds to the DNA to activate the transcription of target genes. TAG: Triglycerides; CIDE: Cell death-inducing DFF45-like effector; PLIN: Perilipins; ATGL: Adipose triglyceride lipase; HSL: Hormone-sensitive lipase; MAGL: Monoacylglycerol lipase; FA: Fatty acid; CE: Cholesteryl esters; PGC-1α: Peroxisome proliferator-activated receptor gamma coactivator 1-alpha; DGAT: Diacylglycerol acyltransferases; ACAT: Acyl-CoA cholesterol acyltransferases; PPARs: Peroxisome proliferator-activated receptors.
LD biogenesis can be described as an evolutionary model consisting of three main steps: (1) Lipid synthesis; (2) LD formation; and (3) LD growth. In step 1, TAG and CE synthesis enzymes, such as DGAT1, DGAT2 and acyl-CoA cholesterol acyltransferases 1 and 2 (ACAT1 and ACAT2), deposit neutral lipids between the sheets of the ER bilayer[53,54]. During step 2, the lipid quantity increases and, when it reaches a certain concentration, the LD detaches from the ER[55]. Thereafter, a variety of proteins such as perilipins, are recruited to the lens structure and facilitate the growth of the nascent LD[56]. Finally, step 3 only occurs in some mammalian cells, where LDs can grow by local lipid synthesis, by transporting lipids to LDs or by fusing with other LDs[57].
LDs can be broken down for energy supply and membrane synthesis through lipolysis or lipophagy (Figure 1). The lipolysis enables the release of FAs from TAGs through the consecutive action of ATGL, HSL and monoacylglycerol lipase[58,59]. Through lipophagy, LDs are enclosed in autophagosomes, fused with lysosomes and degraded by hydrolytic enzymes[60,61].
LDs are mainly found in the cytoplasm, but also in the nucleus of some cell types[62]. Their intracellular location is determined by interacting with other organelles to promote lipid exchange, metabolic dynamics and stress adaptation[63]. LDs come into contact with the ER early in their biogenesis, as well as with the lysosome in the lipophagy process[56,61]. LDs also connect with mitochondria to enable the direct flow of FAs into the mitochondrial matrix to fulfil the cell energy requirements[64]. Their interaction with peroxisomes also allows the transport of FAs, phospholipids and TAGs[65]. Moreover, there is direct and indirect contact with nucleus and Golgi organelles[66].
Besides energy supply and membrane synthesis, LDs play additional roles to ensure proper cell functionality under stress. Prolonged nutrient deprivation upregulates autophagy, causing breakdown of proteins and membranous organelles, which release amino acids and lipids potentially toxic for the cell. In this sense, LDs store neutral lipids, inert within its structure[67]. Additionally, LDs serve as extra source of lipids for FAO under nutrient stress[31,68,69] and hypoxic stress[68]. LDs also ensure the maintenance of redox homeostasis, proper mitochondrial function and membrane and organelle homeostasis[64,70]. In addition, they protect against ER stress; that is, against imbalances in ER protein folding capacity, calcium uptake and lipid composition[41,71]. Finally, LDs produce lipid intermediates that include pro- and anti-inflammatory signalling molecules[72].
LDs IN CANCER STEMNESS AND CHEMORESISTANCE
Considering LDs regulate different cellular processes, it is not surprising that they have been strongly associated to cancer progression and aggressiveness in recent years[69,73-75]. In fact, LDs facilitate not only tumour growth, but also metastasis, chemoresistance and disease relapse in multiple types of cancers[68,74,76], all processes intimately related to CSCs.
Indeed, a direct relation between LD content and stemness has been demonstrated in different types of cancers such as pancreatic, colorectal, ovarian and breast cancer[25,77-79]: On the one hand, the isolation of cells with high LD content led to an enrichment of CSCs; on the other hand, isolated CD133+ CSCs show higher LD content than differentiated CD133- cancer cells. Interestingly, tumour-initiating pancreatic cells resistant to KRAS ablation showed an LD accumulation coupled with macrolipophagy, corresponding to the fusion of LD with autophagosomes. Correlated with a high catabolism rate of endogenous lipids and FAs, Viale et al[80] determined that these KRAS ablation-resistant cells used autophagy/macrolipophagy to maintain their energy balance. Indeed, the inhibition of either autophagy or entry of FAs in the mitochondria (using bafilomycin or etomoxir, respectively) dramatically reduced cellular oxygen consumption rate. This metabolic stress was associated with a strong decrease of survival and sphere formation capacity[80]. Functionally, Tirinato et al[81] demonstrated that sorted colorectal CSC with high or low LD content were able to form tumours after subcutaneous injection in immunocompromised mice, although cells with low LD content generated delayed small tumours less frequently. These results suggested that cells with high LD content increase tumorigenic potential, while cells with low LD content represented a more differentiated and less tumorigenic population. Thereby, LD content seems directly linked to tumorigenicity and is suggested as a marker of CSCs, in addition to molecular markers[81]. Moreover, LD-related proteins from the PLINs and CIDE families can be associated to tumorigenicity in several cancer types[82]. Nevertheless, Cao et al[82] highlighted that an increased expression of PLIN2 was associated with a better survival rate in clear cell renal cell carcinoma (ccRCC), decreased with a higher tumour grade. Indeed, PLIN2 knockdown enhanced proliferation, migration and invasion of ccRCC cells. These findings underpin that more studies are needed to clearly identify the specific roles of LD-associated proteins in tumorigenesis or tumour progression, which may be cell or context-specific.
LDs seem to be necessary for CSCs functionality[40], not only to sustain energy demands and biomass production but also to regulate several important oncogenic signalling pathways such as Wnt/β-catenin and Hippo/Yes-associated protein 1 pathways[79] (Figure 2). In this sense, the PPARs superfamily directly associates signalling with LDs, since most lipid-derived second messengers produced in LDs act mainly through these nuclear receptors. Recently, Kuramoto et al[77] demonstrated that PPARα was activated in CSCs that accumulated LDs from pancreatic and colorectal cancer. At the same time, PPARα induced the expression of lipolytic factors like ATGL, leading to the release of FAs that supported stemness characteristics in a positive feedback loop. Indeed, a decreased PPARα activity, by using inhibitors or siRNAs, reduced sphere formation as well as pluripotency-related genes expression (SOX2, OCT4 and NANOG) in pancreatic cells in vitro[77]. These results suggest that pharmacological agents modulating PPARs activity could represent interesting compounds in order to target CSCs.
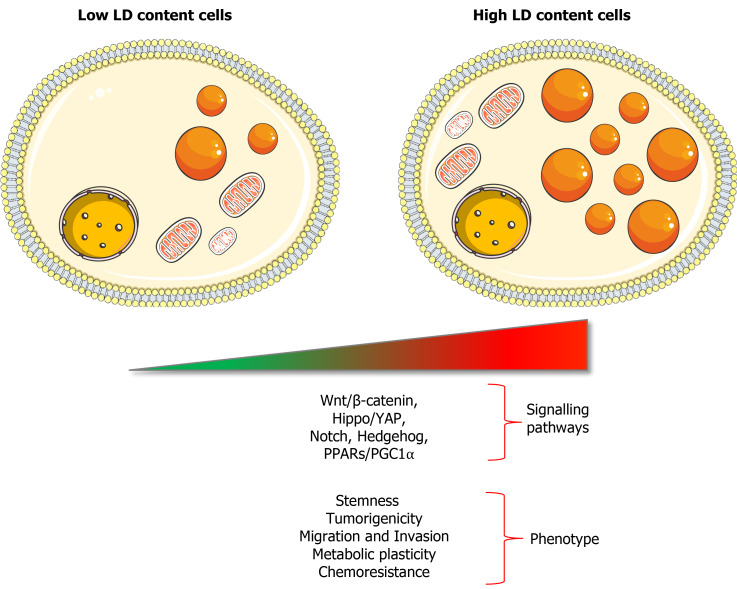
Figure 2.
Features of cells with high content of lipid droplets. Cells with high lipid droplets (LD) content show activation of different signalling pathways such as Wnt/β-catenin, Hippo/Yes-associated protein, Notch, Hedgehog and PPARs/PGC-1α. Increased LD content has been linked to aggressive phenotypes in tumour cells, such as stemness, tumorigenicity, migration and invasion, metabolic plasticity and chemoresistance. LD: Lipid droplets; PGC-1α: Peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PPARs: Peroxisome proliferator-activated receptors; YAP: Yes-associated protein.
Several studies have demonstrated the importance of LDs and the associated lipase HSL in invasion and metastasis regulation, with special relevance in pancreatic cancer[83]. For instance, oncogenic KRAS down-regulates HSL to control lipid storage and utilisation, leading to LD accumulation and tumour invasion[84,85]. Disruption of the KRAS-HSL axis or overexpression of HSL reduces lipid storage and suppresses invasive migration in vitro and metastasis in vivo[83,84]. Interestingly, Mitra et al[86] demonstrated by Raman spectrometry that circulating tumour cells isolated from the peripheral blood of patients with metastatic prostate cancer, accumulated LDs[86], further strengthening the relation between metastasis and LD accumulation.
Increasing evidence links lipid metabolism with chemoresistance in different cancer types[74]. For instance, FAO-derived adenosine triphosphate has been shown to drive chemoresistance in breast cancer and leukemic stem cells[87,88]. In addition, Incio et al[89] showed that 5-Fluorouracil (5-FU) uptake and efficacy in pancreatic cancer cells decreased significantly in an obese context, indicating that large obesity-caused accumulation of LDs resulting from obesity can reduce drug delivery and chemotherapy efficiency.
The contribution of LDs to chemoresistance is twofold: On the one hand, intrinsic presence of LDs has been widely reported to be a characteristic of chemoresistant cancer cell lines[68,69,74,76]. For instance, prostate cancer cells survive androgen deprivation therapy by metabolising lipids present in LDs[90]. On the other hand, chemotherapy treatments may induce de novo LD biogenesis. For example, doxorubicin and 5-FU induced TAG biosynthesis, accumulated in LDs in human colon carcinoma cells[74,91]. Moreover, direct or indirect pharmacological inhibition of FAO or OXPHOS is sufficient to drive LD formation in cancer cells[74]. Indeed, treatment with the c-MYC/Max inhibitor 10058-F4 induced LD accumulation resulting from mitochondrial dysfunction[92]. Interestingly, a combination of both LD presence and accumulation has been described in colorectal cancer cells. For instance, high LD content identified cancer cell lines with increased chemoresistance to 5-FU and oxaliplatin. These cells further accumulated LDs in response to chemotherapy in a process facilitated by lysophosphatidyl-choline acyltransferase 2 (LPCAT2), an LD-associated enzyme essential for phosphatidylcholine synthesis[93]. An elevated expression of LPCAT2 prevented chemotherapy-induced ER stress, further highlighting the protective role of LDs against cellular stresses[74,93]. Importantly, it has been recently reported that LDs can also act as a sink to sequester hydrophobic compounds impairing drug-induced apoptosis, resulting in chemoresistance of cancer cells[68,69].
CONCLUSION
Even if our knowledge about the mechanisms by which LDs support cancer stemness is still very limited, it seems clear now that high levels of LDs are strongly associated with cancer aggressiveness and chemotherapy resistance in different tumour types. Considering this, measurement of LD accumulation could be potentially used as a prognostic biomarker, also with predictive value in terms of treatment response to conventional therapies. A deeper understanding of the molecular mechanisms dictating their implication in essential processes of the CSC biology, such as tumorigenicity, metastatic spread and chemoresistance, should pave the way to discover novel LD-related targets and therapeutic approaches for more effective cancer treatment.
ACKNOWLEDGEMENTS
We want to thank Laura Sancho for proofreading the manuscript.
Footnotes
Conflict-of-interest statement: There is no conflict of interest associated with any of the senior author or other co-authors who contributed their efforts in this manuscript.
Manuscript source: Invited manuscript
Peer-review started: March 12, 2021
First decision: May 5, 2021
Article in press: August 18, 2021
Specialty type: Cell and tissue engineering
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lara Riegos JC S-Editor: Fan JR L-Editor: A P-Editor: Xing YX
Contributor Information
Alba Royo-García, Hospital Universitario Miguel Servet, IIS Aragón, Zaragoza 50009, Spain.
Sarah Courtois, Hospital Universitario Miguel Servet, IIS Aragón, Zaragoza 50009, Spain.
Beatriz Parejo-Alonso, Hospital Universitario Miguel Servet, IIS Aragón, Zaragoza 50009, Spain.
Pilar Espiau-Romera, Hospital Universitario Miguel Servet, IIS Aragón, Zaragoza 50009, Spain.
Patricia Sancho, Hospital Universitario Miguel Servet, IIS Aragón, Zaragoza 50009, Spain. psancho@iisaragon.es.
References
- 1.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 8.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabbah M, Emami S, Redeuilh G, Julien S, Prévost G, Zimber A, Ouelaa R, Bracke M, De Wever O, Gespach C. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11:123–151. doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Takam Kamga P, Bassi G, Cassaro A, Midolo M, Di Trapani M, Gatti A, Carusone R, Resci F, Perbellini O, Gottardi M, Bonifacio M, Nwabo Kamdje AH, Ambrosetti A, Krampera M. Notch signalling drives bone marrow stromal cell-mediated chemoresistance in acute myeloid leukemia. Oncotarget. 2016;7:21713–21727. doi: 10.18632/oncotarget.7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, Sprick MR, Kemper K, Richel DJ, Stassi G, Medema JP. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 12.Cazet AS, Hui MN, Elsworth BL, Wu SZ, Roden D, Chan CL, Skhinas JN, Collot R, Yang J, Harvey K, Johan MZ, Cooper C, Nair R, Herrmann D, McFarland A, Deng N, Ruiz-Borrego M, Rojo F, Trigo JM, Bezares S, Caballero R, Lim E, Timpson P, O'Toole S, Watkins DN, Cox TR, Samuel MS, Martín M, Swarbrick A. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat Commun. 2018;9:2897. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, Ali S, Abbruzzese JL, Gallick GE, Sarkar FH. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie M, Zhang L, He CS, Xu F, Liu JL, Hu ZH, Zhao LP, Tian Y. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem. 2012;113:1501–1513. doi: 10.1002/jcb.24019. [DOI] [PubMed] [Google Scholar]
- 15.Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. 2016;114:1305–1312. doi: 10.1038/bjc.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peiris-Pagès M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18:55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Graña O, Viera CR, Yuneva M, Sainz B Jr, Heeschen C. MYC/PGC-1α Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, Puigserver P. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz-Bermúdez A, Laza-Briviesca R, Vicente-Blanco RJ, García-Grande A, Coronado MJ, Laine-Menéndez S, Palacios-Zambrano S, Moreno-Villa MR, Ruiz-Valdepeñas AM, Lendinez C, Romero A, Franco F, Calvo V, Alfaro C, Acosta PM, Salas C, Garcia JM, Provencio M. Cisplatin resistance involves a metabolic reprogramming through ROS and PGC-1α in NSCLC which can be overcome by OXPHOS inhibition. Free Radic Biol Med. 2019;135:167–181. doi: 10.1016/j.freeradbiomed.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, Wargo JA, Song JS, Fisher DE, Arany Z, Widlund HR. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong F, Pan S, Guo Y, Xu P, Zhai Y. PPARs as Nuclear Receptors for Nutrient and Energy Metabolism. Molecules. 2019;24 doi: 10.3390/molecules24142545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takada I, Makishima M. Peroxisome proliferator-activated receptor agonists and antagonists: a patent review (2014-present) Expert Opin Ther Pat. 2020;30:1–13. doi: 10.1080/13543776.2020.1703952. [DOI] [PubMed] [Google Scholar]
- 23.Chen CL, Uthaya Kumar DB, Punj V, Xu J, Sher L, Tahara SM, Hess S, Machida K. NANOG Metabolically Reprograms Tumor-Initiating Stem-like Cells through Tumorigenic Changes in Oxidative Phosphorylation and Fatty Acid Metabolism. Cell Metab. 2016;23:206–219. doi: 10.1016/j.cmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandi J, Dando I, Pozza ED, Biondani G, Jenkins R, Elliott V, Park K, Fanelli G, Zolla L, Costello E, Scarpa A, Cecconi D, Palmieri M. Proteomic analysis of pancreatic cancer stem cells: Functional role of fatty acid synthesis and mevalonate pathways. J Proteomics. 2017;150:310–322. doi: 10.1016/j.jprot.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Hershey BJ, Vazzana R, Joppi DL, Havas KM. Lipid Droplets Define a Sub-Population of Breast Cancer Stem Cells. J Clin Med. 2019;9 doi: 10.3390/jcm9010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tirinato L, Pagliari F, Di Franco S, Sogne E, Marafioti MG, Jansen J, Falqui A, Todaro M, Candeloro P, Liberale C, Seco J, Stassi G, Di Fabrizio E. ROS and Lipid Droplet accumulation induced by high glucose exposure in healthy colon and Colorectal Cancer Stem Cells. Genes Dis. 2020;7:620–635. doi: 10.1016/j.gendis.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espiau-Romera P, Courtois S, Parejo-Alonso B, Sancho P. Molecular and Metabolic Subtypes Correspondence for Pancreatic Ductal Adenocarcinoma Classification. J Clin Med. 2020;9 doi: 10.3390/jcm9124128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122:4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73:377–392. doi: 10.1007/s00018-015-2070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz ALS, Barreto EA, Fazolini NPB, Viola JPB, Bozza PT. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020;11:105. doi: 10.1038/s41419-020-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagust P, de Luxán-Delgado B, Parejo-Alonso B, Sancho P. Metabolism-Based Therapeutic Strategies Targeting Cancer Stem Cells. Front Pharmacol. 2019;10:203. doi: 10.3389/fphar.2019.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimoto M, Matsuzaki I, Nishitsuji K, Yamamoto Y, Murakami D, Yoshikawa T, Fukui A, Mori Y, Nishino M, Takahashi Y, Iwahashi Y, Warigaya K, Kojima F, Jinnin M, Murata SI. Adipophilin expression in cutaneous malignant melanoma is associated with high proliferation and poor clinical prognosis. Lab Invest. 2020;100:727–737. doi: 10.1038/s41374-019-0358-y. [DOI] [PubMed] [Google Scholar]
- 35.Alo PL, Amini M, Piro F, Pizzuti L, Sebastiani V, Botti C, Murari R, Zotti G, Di Tondo U. Immunohistochemical expression and prognostic significance of fatty acid synthase in pancreatic carcinoma. Anticancer Res. 2007;27:2523–2527. [PubMed] [Google Scholar]
- 36.Sun Y, He W, Luo M, Zhou Y, Chang G, Ren W, Wu K, Li X, Shen J, Zhao X, Hu Y. SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumour Biol. 2015;36:4133–4141. doi: 10.1007/s13277-015-3047-5. [DOI] [PubMed] [Google Scholar]
- 37.Apffel CA, Baker JR. Lipid droplets in the cytoplasm of malignant cells. Cancer. 1964;17:176–184. doi: 10.1002/1097-0142(196402)17:2<176::aid-cncr2820170207>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 38.Novikoff AB. A transplantable rat liver tumor induced by 4-dimethylaminoazobenzene. Cancer Res. 1957;17:1010–1027. [PubMed] [Google Scholar]
- 39.Farese RV Jr, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tirinato L, Pagliari F, Limongi T, Marini M, Falqui A, Seco J, Candeloro P, Liberale C, Di Fabrizio E. An Overview of Lipid Droplets in Cancer and Cancer Stem Cells. Stem Cells Int. 2017;2017:1656053. doi: 10.1155/2017/1656053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiam AR, Beller M. The why, when and how of lipid droplet diversity. J Cell Sci. 2017;130:315–324. doi: 10.1242/jcs.192021. [DOI] [PubMed] [Google Scholar]
- 44.Cheng J, Fujita A, Ohsaki Y, Suzuki M, Shinohara Y, Fujimoto T. Quantitative electron microscopy shows uniform incorporation of triglycerides into existing lipid droplets. Histochem Cell Biol. 2009;132:281–291. doi: 10.1007/s00418-009-0615-z. [DOI] [PubMed] [Google Scholar]
- 45.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 46.Czabany T, Wagner A, Zweytick D, Lohner K, Leitner E, Ingolic E, Daum G. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J Biol Chem. 2008;283:17065–17074. doi: 10.1074/jbc.M800401200. [DOI] [PubMed] [Google Scholar]
- 47.Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 49.Slayton M, Gupta A, Balakrishnan B, Puri V. CIDE Proteins in Human Health and Disease. Cells. 2019;8 doi: 10.3390/cells8030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puri V, Konda S, Ranjit S, Aouadi M, Chawla A, Chouinard M, Chakladar A, Czech MP. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282:34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- 51.Xu S, Zhang X, Liu P. Lipid droplet proteins and metabolic diseases. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1968–1983. doi: 10.1016/j.bbadis.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 52.Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, Olzmann JA. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev Cell. 2017;42:9–21.e5. doi: 10.1016/j.devcel.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kassan A, Herms A, Fernández-Vidal A, Bosch M, Schieber NL, Reddy BJ, Fajardo A, Gelabert-Baldrich M, Tebar F, Enrich C, Gross SP, Parton RG, Pol A. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol. 2013;203:985–1001. doi: 10.1083/jcb.201305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walther TC, Farese RV Jr. The life of lipid droplets. Biochim Biophys Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choudhary V, Ojha N, Golden A, Prinz WA. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol. 2015;211:261–271. doi: 10.1083/jcb.201505067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol. 2011;195:953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sztalryd C, Xu G, Dorward H, Tansey JT, Contreras JA, Kimmel AR, Londos C. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol. 2003;161:1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyers A, Weiskittel TM, Dalhaimer P. Lipid Droplets: Formation to Breakdown. Lipids. 2017;52:465–475. doi: 10.1007/s11745-017-4263-0. [DOI] [PubMed] [Google Scholar]
- 60.Khawar MB, Gao H, Li W. Autophagy and Lipid Metabolism. Adv Exp Med Biol. 2019;1206:359–374. doi: 10.1007/978-981-15-0602-4_17. [DOI] [PubMed] [Google Scholar]
- 61.Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015;34:2117–2131. doi: 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohsaki Y, Kawai T, Yoshikawa Y, Cheng J, Jokitalo E, Fujimoto T. PML isoform II plays a critical role in nuclear lipid droplet formation. J Cell Biol. 2016;212:29–38. doi: 10.1083/jcb.201507122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pu J, Ha CW, Zhang S, Jung JP, Huh WK, Liu P. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2011;2:487–496. doi: 10.1007/s13238-011-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ueno M, Shen WJ, Patel S, Greenberg AS, Azhar S, Kraemer FB. Fat-specific protein 27 modulates nuclear factor of activated T cells 5 and the cellular response to stress. J Lipid Res. 2013;54:734–743. doi: 10.1194/jlr.M033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV Jr, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A, Loda M, Kinlaw WB, Swinnen JV. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv Drug Deliv Rev. 2020;159:245–293. doi: 10.1016/j.addr.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, Liu H, Luo X. Lipid droplet and its implication in cancer progression. Am J Cancer Res. 2020;10:4112–4122. [PMC free article] [PubMed] [Google Scholar]
- 70.Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li JL, Zhang Q, Wakelam MJO, Karpe F, Schulze A, Harris AL. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 71.Velázquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol. 2016;212:621–631. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS, Kaufmann C, Morgado-Diaz JA, Bozza PT, Viola JP. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res. 2008;68:1732–1740. doi: 10.1158/0008-5472.CAN-07-1999. [DOI] [PubMed] [Google Scholar]
- 73.Sunami Y, Rebelo A, Kleeff J. Lipid Metabolism and Lipid Droplets in Pancreatic Cancer and Stellate Cells. Cancers (Basel) 2017;10 doi: 10.3390/cancers10010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shyu P Jr, Wong XFA, Crasta K, Thibault G. Dropping in on lipid droplets: insights into cellular stress and cancer. Biosci Rep. 2018;38 doi: 10.1042/BSR20180764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O Connor D, Byrne A, Berselli GB, Long C, Keyes TE. Mega-stokes pyrene ceramide conjugates for STED imaging of lipid droplets in live cells. Analyst. 2019;144:1608–1621. doi: 10.1039/c8an02260g. [DOI] [PubMed] [Google Scholar]
- 76.Rak S, De Zan T, Stefulj J, Kosović M, Gamulin O, Osmak M. FTIR spectroscopy reveals lipid droplets in drug resistant laryngeal carcinoma cells through detection of increased ester vibrational bands intensity. Analyst. 2014;139:3407–3415. doi: 10.1039/c4an00412d. [DOI] [PubMed] [Google Scholar]
- 77.Kuramoto K, Yamamoto M, Suzuki S, Togashi K, Sanomachi T, Kitanaka C, Okada M. Inhibition of the Lipid Droplet-Peroxisome Proliferator-Activated Receptor α Axis Suppresses Cancer Stem Cell Properties. Genes (Basel) 2021;12 doi: 10.3390/genes12010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J, Condello S, Thomes-Pepin J, Ma X, Xia Y, Hurley TD, Matei D, Cheng JX. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell. 2017;20:303–314.e5. doi: 10.1016/j.stem.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yi M, Li J, Chen S, Cai J, Ban Y, Peng Q, Zhou Y, Zeng Z, Peng S, Li X, Xiong W, Li G, Xiang B. Emerging role of lipid metabolism alterations in Cancer stem cells. J Exp Clin Cancer Res. 2018;37:118. doi: 10.1186/s13046-018-0784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tirinato L, Liberale C, Di Franco S, Candeloro P, Benfante A, La Rocca R, Potze L, Marotta R, Ruffilli R, Rajamanickam VP, Malerba M, De Angelis F, Falqui A, Carbone E, Todaro M, Medema JP, Stassi G, Di Fabrizio E. Lipid droplets: a new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells. 2015;33:35–44. doi: 10.1002/stem.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cao Q, Ruan H, Wang K, Song Z, Bao L, Xu T, Xiao H, Wang C, Cheng G, Tong J, Meng X, Liu D, Yang H, Chen K, Zhang X. Overexpression of PLIN2 is a prognostic marker and attenuates tumor progression in clear cell renal cell carcinoma. Int J Oncol. 2018;53:137–147. doi: 10.3892/ijo.2018.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Man J, Pajic M, Joshua AM. Fats and Mets, KRAS-Driven Lipid Dysregulation Affects Metastatic Potential in Pancreatic Cancer. Cancer Res. 2020;80:4886–4887. doi: 10.1158/0008-5472.CAN-20-3082. [DOI] [PubMed] [Google Scholar]
- 84.Rozeveld CN, Johnson KM, Zhang L, Razidlo GL. KRAS Controls Pancreatic Cancer Cell Lipid Metabolism and Invasive Potential through the Lipase HSL. Cancer Res. 2020;80:4932–4945. doi: 10.1158/0008-5472.CAN-20-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu M, Chang HH, Jung X, Moro A, Chou CEN, King J, Hines OJ, Sinnett-Smith J, Rozengurt E, Eibl G. Deficiency in hormone-sensitive lipase accelerates the development of pancreatic cancer in conditional KrasG12D mice. BMC Cancer. 2018;18:797. doi: 10.1186/s12885-018-4713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mitra R, Chao O, Urasaki Y, Goodman OB, Le TT. Detection of lipid-rich prostate circulating tumour cells with coherent anti-Stokes Raman scattering microscopy. BMC Cancer. 2012;12:540. doi: 10.1186/1471-2407-12-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang T, Fahrmann JF, Lee H, Li Y-J, Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, Somlo G, Jandial R, Ann D, Hanash S, Jove R, Yu H. JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 2018;27:136–150.e5. doi: 10.1016/j.cmet.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, Bosc C, Sugita M, Stuani L, Fraisse M, Scotland S, Larrue C, Boutzen H, Féliu V, Nicolau-Travers ML, Cassant-Sourdy S, Broin N, David M, Serhan N, Sarry A, Tavitian S, Kaoma T, Vallar L, Iacovoni J, Linares LK, Montersino C, Castellano R, Griessinger E, Collette Y, Duchamp O, Barreira Y, Hirsch P, Palama T, Gales L, Delhommeau F, Garmy-Susini BH, Portais JC, Vergez F, Selak M, Danet-Desnoyers G, Carroll M, Récher C, Sarry JE. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017;7:716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Incio J, Liu H, Suboj P, Chin SM, Chen IX, Pinter M, Ng MR, Nia HT, Grahovac J, Kao S, Babykutty S, Huang Y, Jung K, Rahbari NN, Han X, Chauhan VP, Martin JD, Kahn J, Huang P, Desphande V, Michaelson J, Michelakos TP, Ferrone CR, Soares R, Boucher Y, Fukumura D, Jain RK. Obesity-Induced Inflammation and Desmoplasia Promote Pancreatic Cancer Progression and Resistance to Chemotherapy. Cancer Discov. 2016;6:852–869. doi: 10.1158/2159-8290.CD-15-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaini RR, Sillerud LO, Zhaorigetu S, Hu CA. Autophagy regulates lipolysis and cell survival through lipid droplet degradation in androgen-sensitive prostate cancer cells. Prostate. 2012;72:1412–1422. doi: 10.1002/pros.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mehdizadeh A, Bonyadi M, Darabi M, Rahbarghazi R, Montazersaheb S, Velaei K, Shaaker M, Somi MH. Common chemotherapeutic agents modulate fatty acid distribution in human hepatocellular carcinoma and colorectal cancer cells. Bioimpacts. 2017;7:31–39. doi: 10.15171/bi.2017.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zirath H, Frenzel A, Oliynyk G, Segerström L, Westermark UK, Larsson K, Munksgaard Persson M, Hultenby K, Lehtiö J, Einvik C, Påhlman S, Kogner P, Jakobsson PJ, Henriksson MA. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc Natl Acad Sci U S A. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cotte AK, Aires V, Fredon M, Limagne E, Derangère V, Thibaudin M, Humblin E, Scagliarini A, de Barros JP, Hillon P, Ghiringhelli F, Delmas D. Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat Commun. 2018;9:322. doi: 10.1038/s41467-017-02732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]