Abstract
A rapid procedure for the diagnosis of malaria infections directly from dried blood spots by PCR amplification was evaluated with samples from 52 patients. Plasmodium infections were identified with a genus-specific primer set, and species differentiation between Plasmodium falciparum and Plasmodium vivax was analyzed by multiplex PCR. The PCR test with any of the three primer sets was able to detect as few as four parasites per microliter by gel electrophoresis or by nonisotopic paper hybridization chromatography. The diagnoses obtained by PCR correlated closely with those obtained by Giemsa staining except for two samples observed to have mixed P. falciparum-P. vivax infections. These were initially missed by microscopic analysis. In comparison with antigen-capture assays for P. falciparum, the PCR assays were able to detect three infections that were missed by the ParaSight-F test. The PCR test was negative for nine ParaSight-F-positive samples and one ICT Malaria Pf-positive sample, and these were confirmed to be false-positive results. The PCR thus gave no false-negative or false-positive results. Patients undergoing antimalarial therapy were also monitored by the PCR assay. Four of seven patients who were PCR positive for P. vivax at the time of discharge were later readmitted to the hospital with a recurrence of P. vivax infection. We would like to propose that PCR is a sensitive and easy method that can serve as a useful addition to microscopy for the diagnosis and the clinical monitoring of treatment of malaria.
Microscopy has historically been the mainstay of the diagnosis of malaria. A clinical diagnosis of malaria currently depends on the visualization of parasites by light microscopy of Giemsa-stained thick and thin blood smears. This procedure is cheap and simple, but it is labor intensive and requires personnel who are well trained in the morphological differentiation of the Plasmodium species (11) for successful diagnosis, which leads to proper treatment.
In recent years, alternative methods for the identification of malaria infections have been developed, and these have had various specificities and sensitivities. Several malaria diagnostic kits based on antigen detection of Plasmodium falciparum have been developed, such as ParaSight-F (Becton Dickinson, Cockeysville, Md.) and ICT Malaria Pf (ICT Diagnostics, Sydney, Australia). At the same time, several PCR assays have been developed for the diagnosis of malaria. The 18S rRNA gene has been used as a DNA target for the differentiation of plasmodial species by nested PCR (14, 15) and reverse transcription-PCR (1). Other DNA targets such as the circumsporozoite protein gene (5, 13, 15) have also been investigated for species-specific regions. Tan et al. (16) demonstrated that the large-subunit rRNA gene is extensively conserved within Plasmodium species and is suitable as a genus-specific DNA target region. In this paper, we describe a sensitive and reliable two-step PCR-based amplification assay for the diagnosis of malaria. Plasmodium infections were diagnosed by use of a genus-specific primer set. In addition, two distinct primer sets were designed to specifically detect either P. falciparum or Plasmodium vivax, the two major Plasmodium species which infect and cause malaria in humans. We also present the results of a clinical study of this PCR-based assay in which it was compared with the ParaSight-F and ICT Malaria Pf diagnostic kits performed with samples from patients in Singapore. One advantage of studying malaria in Singapore is the virtual absence of local transmission of malaria, thereby excluding the compounding factor of reinfection as a source of relapse.
MATERIALS AND METHODS
Microscopic diagnosis and sample collection.
All patients who had a history of fever and persistent headaches and who had recently traveled outside of Singapore were examined for malaria at the National University Hospital of Singapore. Thick and thin blood films were prepared from venous blood collected in tubes containing EDTA. The use of heparin was avoided because of its known high-level inhibitory effect on Taq polymerase (5). The slides were stained with Giemsa and analyzed for the presence of parasites and parasite species. Quantitative Buffy Coat (QBC; Becton Dickinson) analysis for malaria, which is a fluorescent microscopic examination of capillary-centrifuged blood, was performed in tandem with the thick-film Giemsa stain analysis. Parasites were quantified by counting the number of parasites per 200 leukocytes (3, 7). The parasite density (number of parasites per microliter) was then calculated from the automated leukocyte count obtained with a Bayer Technicon H*3 automated cell counter (Bayer Tarrytown, N.Y.) Five microliters of blood from each patient was spotted onto grade 1 Whatman filter paper (Whatman International Ltd., Maidstone, United Kingdom) and air dried at room temperature for PCR analysis. Blood was obtained daily from the patients for monitoring the progress of the antimalarial therapy. Four methods were used to examine these samples: microscopy, tests with two commercially available test kits (ParaSight-F and ICT Malaria Pf), and PCR amplification.
Dipstick antigen-capture assays.
The blood samples were also assayed with the ParaSight-F (Becton Dickinson) and ICT Malaria Pf (ICT Diagnostics) test kits. Both test kits are based on immunological detection of the P. falciparum histidine-rich protein 2. The assays were performed in parallel according to the manufacturer’s instructions. The dipsticks were independently examined, and the results of each assay were recorded as positive or negative on the basis of the observation of the precipitated band.
PCR amplification.
Detection of PCR was performed as a two-step procedure. The Plasmodium genus-specific primers were run separately from the species-specific primers, which in turn were run as a multiplex PCR system. Both types of amplifications used dried blood spot samples as the DNA template and were performed independently of each other. Amplification was carried out as described previously (16), with minor modifications. Dried blood samples on filter paper (1 by 2 mm) were placed in a PCR mixture containing 70 mM Tris (pH 8.8), 20 mM (NH2)4SO4, 1 mM dithiothreitol, 0.1% gelatin, 2.5 mM MgCl2, 0.4 μg of each primer, 2.5 U of Taq DNA polymerase (Promega, Madison, Wis.), and each deoxynucleoside triphosphate at a concentration of 0.2 mM. All amplifications were initiated by the hot-start technique (5 min at 95°C followed by 5 min at 80°C); both the Taq DNA polymerase and deoxynucleoside triphosphates were added during the 80°C incubation period. The amplification involved 40 cycles of 1 min of denaturation at 90°C, 2 min of annealing at 56°C, and 1 min of primer extension at 72°C. A further extension at 72°C for 5 min was included following the final cycle.
Primers.
Three sets of oligonucleotide primers that have been developed were used in this study. Primer set L1 (biotin-5′-GAC CTG CAT GAA AGA TG-3′) and L2 (5′-GTA TCG CTT TAA TAG GCG-3′) is genus specific (16) and was used to detect the presence of the Plasmodium parasite. The species-specific primers were designed to amplify regions from the mitochondrial coxI gene. The P. falciparum species-specific primers were Pf1 (biotin-5′-GGA ATG TTA TTG CTA ACA C-3′) and Pf2 (5′-AAT GAA GAG CTG TGT ATC-3′), and the species-specific primers for P. vivax were Pv1 (biotin-5′-CAC CAT TAA GTA CAT CAC-3′) and Pv2 (5′-TGT TAA TAC AAC TCC AAT-3′). These species-specific primers for P. falciparum and P. vivax were used together in a single multiplex PCR.
Detection of PCR products.
Amplified PCR products were detected by running 10 μl of the PCR mixture on a standard 1.0% agarose gel, which was subsequently stained with a 0.5-μg/ml ethidium bromide solution and visualized under UV light. Amplified products were also detected by DNA hybridization by paper chromatography as described by Reinhartz et al. (12). Hybridization probes (25 ng) were spotted onto 5-mm-wide plastic-backed nitrocellulose strips (8 μm; Micron Separations, Westboro, Mass.). Hybridization with the L1 and L2 (genus-specific) probes (5′-GCG ATA AGC GCA CAT CGA GGT GCC-3′) was performed on a strip that was independent of the system with the species-specific probes. The PR-Pf (P. falciparum-specific) probe (5′-CGA CTA CCA TTT TAA TAT CAA TAC CTA CCG GTA-3′) and the PR-Pv (P. vivax-specific) probe (5′-GAT GTT ATC ATT GTT GGT CTT TTA GTA TCT GGT ATT-3′) were spotted onto the top right corner and bottom left corner, respectively, of the proximal end of the nitrocellulose strip. The probes were immobilized by UV cross-linking at 312 nm for 5 min. The probe strips were attached at the distal end with an absorbent filter paper. These probe strips could be stored for 6 months at 4°C. Each of the PCR-amplified products (10 μl of a 1:5 dilution with sterile H2O) was denatured with 2 μl of a 0.5 M NaCl–0.15 M NaOH solution for 5 min at room temperature. Fifty microliters of the hybridization buffer (0.6 M NaCl in 20 mM sodium phosphate [pH 7.5], 0.02% Ficoll-400–0.02% gelatin) was added, and the product mix was transferred to a microtiter plate well. Capillary hybridization with the probe strip was performed at 37°C for 15 min. The strips were then transferred to the conjugate buffer (streptavidin-alkaline phosphatase conjugate [1:2,000 dilution] in 1% gelatin–0.3% Tween 20 in 58 mM NaHPO4–17 mM NaH2PO4–68 mM NaCl [phosphate-buffered saline]; Boehringer Mannheim GmbH, Mannheim, Germany) and were allowed to incubate for 5 min at room temperature. The strips were then incubated in chromogenic detection solution (nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate toluidinium in 0.3% Tween 20–phosphate-buffered saline; Boehringer Mannheim GmbH) for 10 min at room temperature. The reaction was stopped with 10 mM Tris-HCl–1 mM EDTA (pH 8.0).
RESULTS
Fifty-two patients were identified as being infected with malaria by blood film analysis and QBC analysis. By light microscopy, 34 were identified as having P. vivax infections, 16 were identified as having P. falciparum infections, and 2 were identified as having mixed P. falciparum-P. vivax infections (Table 1). All blood samples were also tested by two commercially available malaria antigen-capture diagnostic tests, the ParaSight-F and the ICT Malaria Pf tests, and were then subjected to PCR detection.
TABLE 1.
Comparison of PCR assays with Giemsa staining and ParaSight-F and ICT Malaria Pf (P. falciparum antigen-capture) tests for detection of Plasmodium infection in patients
| Cause of infection | No. of samples in which an organism was detected by the following methoda:
|
|||||
|---|---|---|---|---|---|---|
| Microscopy | PCR detection
|
P. falciparum antigen-capture assays
|
||||
| Genus specific | Pf specific | Pv specific | ParaSight-F | ICT Malaria Pf | ||
| P. falciparum | 16 | 16 | 16 | 0 | 13 | 16 |
| P. vivax | 34 | 34 | 0 | 34 | 9 | 1 |
| Mixed (P. falciparum and P. vivax) | 2 | 2 | 2 | 2 | 2 | 2 |
| Total Plasmodium spp. detected | 52 | 52 | 18 | 36 | 24 | 19 |
Pf, P. falciparum; Pv, P. vivax.
By the ParaSight-F assay, 24 of 52 samples were observed to be P. falciparum positive. When the results were compared with those of film diagnosis, nine samples had false-positive results and three samples had false-negative results. By the ICT Malaria Pf assay 19 samples were P. falciparum positive, and only 1 of these was observed to have a false-positive result, while the results for the remaining 18 positives samples correlated with those of blood film diagnosis.
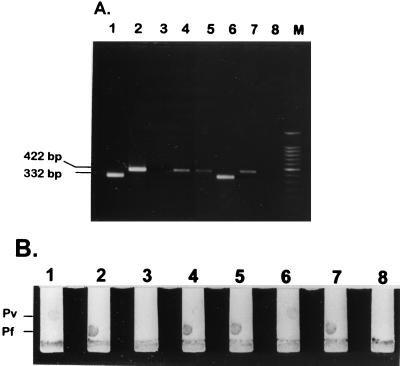
PCR analysis for detection of the Plasmodium genus and species determination were run as independent assays. With the genus-specific primers L1 and L2, a 100% detection rate for the presence of Plasmodium infection was achieved. With the L1 and L2 primers, a 595-bp PCR product was obtained. Amplification with the species-specific primers gave rise in 16 samples to a 422-bp PCR product which correlates to the P. falciparum primer set Pf1-Pf2. In the other 34 samples, a 332-bp PCR product was observed and this product correlates to the P. vivax primer set Pv1-Pv2. In the course of these diagnostic tests, two samples were observed to have both the 422-bp and the 332-bp fragments. These two blood samples were from the two patients with mixed infections, which were initially misdiagnosed as only P. falciparum infections by light microscopy. Paper chromatography hybridization detection of the multiplex PCR products obtained with the species-specific primers was performed with probes designed to detect the two plasmodial species fixed on nitrocellulose strips (Fig. 1). Thus, colorimetric detection was able to differentiate the two species in the same PCR.
FIG. 1.
Detection of multiplex PCR products with species-specific primers. Random samples of patients’ blood were analyzed. Lanes 1 and 6, blood infected with P. vivax (parasitemias, 1.3 and 0.3%, respectively); lanes 2, 4, 5, and 7, blood infected with P. falciparum (parasitemias, 1.1, 0.8, 0.075, and 0.15%, respectively); lanes 3 and 8, blood from non-malaria-infected patients; lane M, 100-bp marker (Promega). (A) PCR-amplified products were run on a 1% agarose gel in 1× TAE electrophoresis buffer. (B) Paper chromatography hybridization. Oligonucleotide probes specific for P. falciparum (Pf) and P. vivax (Pv) were spotted onto 5-mm-wide nitrocellulose strips. The P. vivax-specific probe was spotted on the top right corner. The P. falciparum-specific probe was spotted on the bottom left corner.
When analyzing the sensitivity of the PCR assay with serially diluted infected blood, it was found that the PCR assay could detect as few as three to four parasites per microliter of blood with either the genus- or the species-specific primers (Fig. 2). This corresponds to parasitemias of 0.0005 to 0.0015% obtained with fresh infected venous blood diluted with uninfected human blood. This level of sensitivity was observed both by agarose gel electrophoresis and by the paper chromatography hybridization method. PCR amplification with the three primer sets was also performed with 115 random hospital blood samples that were not infected with malaria parasites. No amplicons were observed. Human β-actin primers were used as a positive control as described previously (16).
FIG. 2.
Assay of sensitivities of genus- and species-specific primer sets for the detection of malaria by PCR. Infected blood was serially diluted with noninfected blood. (A) Genus-specific L1 and L2 primers (initial parasitemia, 0.016%). (B) P. vivax-specific Pv1 and Pv2 primers (initial parasitemia, 0.05%). (C) P. falciparum-specific Pf1 and Pf2 primers (initial parasitemia, 0.075%). The numbers above each lane indicate the number of parasites present per microliter of blood; lane M, 100-bp ladder marker (Promega).
Blood samples from 21 patients receiving treatment were examined by PCR with the genus- and the species-specific primers. The patients were monitored for an average of 4 days following initial presentation. Response to treatment could readily be monitored by agarose gel electrophoresis and ethidium bromide staining (Fig. 3). No PCR amplification products were observed from the blood of 14 patients upon discharge from the hospital. However, for the remaining 7 patients, the PCR product obtained with the L1-L2 primer set was still detectable when they were discharged from the hospital. For all of these patients, the corresponding Giemsa-stained films and QBC test were negative at the time of discharge. Four of these patients were later readmitted to the hospital with a recurrence of P. vivax infection. One of these patients had initially been diagnosed with a P. falciparum infection by thick and thin blood film analysis. However, the PCR assay originally detected a mixed P. falciparum and P. vivax infection. The patient was initially treated only for P. falciparum infection and was then discharged from the hospital. When the patient was readmitted to the hospital 3 weeks later, P. vivax infection with a parasite load of 0.1% was diagnosed. Microscopy detected a predominance of gametocytes. Analysis by the ParaSight-F, ICT Malaria Pf, and PCR assays showed no P. falciparum infection.
FIG. 3.
PCR monitoring of blood samples from patients undergoing treatment for malaria by using the genus-specific primers L1 and L2. Five microliters of blood spotted on filter paper was assayed in each reaction. Blood samples were obtained daily. For patient A (A; initial parasitemia, 0.35%) and patient B (B; initial parasitemia, 0.5%), PCR products could still be detected after day 5. No PCR product was observed after 4 days for patient C (C; initial parasitemia, 0.3%). Lane M, 100-bp ladder (Promega) used as a molecular size marker.
DISCUSSION
Here we have reported on the development and application of a PCR-based test for the diagnosis of malaria and the differentiation between P. falciparum and P. vivax infections in a clinical environment. This method permits the detection of four parasites per microliter, which is equivalent to a 0.0015% parasitemia. One of the major advantages of the technique is the minimal need for sample preparation. Infected blood directly spotted on filter paper was used immediately for PCR amplification. Within our assay system, no inhibition of the PCR by any of the blood components was observed. It was possible to amplify old blood spot samples that had been stored at room temperature up to 3 years by using the L1 and L2 primers as well as the multiplex Pf1-Pf2 and Pv1-Pv2 primers (17).
Our strategy for the PCR amplification was to use two regions from the extrachromosomal DNA of Plasmodium. Detection of a malarial infection was done with genus-specific primers made from the conserved large-subunit rRNA gene, and detection of the two main human Plasmodium species, P. falciparum and P. vivax, was done with primers made from the coxI gene. These primers were then used in a multiplex PCR system. The ability to perform multiplex PCR to differentiate the species decreases the number of PCR assays required to be performed with each blood sample. Most nonisotopic PCR methods require liquid hybridization and capture on microtiter plates for detection (7, 10). The microtiter plate system has limited application in that each well is assigned to detect a single amplicon. This is not suitable for the detection of the multiplex PCR products. Detection of our multiplex PCR products was by paper hybridization chromatography. This method is based on capillary movement of the PCR product along a membrane strip and across the immobilized oligonucleotide probes (12). Hybridization of the biotinylated PCR product with the probe can easily be detected with the strepavidin-alkaline phosphatase conjugates.
Our comparative study of microscopy, the two antigen-capture kits, and the PCR test showed that the results obtained by PCR were equivalent or superior to those obtained by microscopy, in that all microscopy-positive samples were positive by PCR. In addition, the PCR test was able to detect mixed infections that were missed by microscopy. This could probably be due to the tendency for one species to be dominant over another species (7), as well as the fact that the antigen-capture kits could detect only P. falciparum. The ICT Malaria Pf kit produced only one false-positive result and no false-negative results. The ParaSight-F kit failed to detect three P. falciparum infections and had false-positive results for nine samples. In contrast, similar false-negative results with the ParaSight-F kit had previously been reported by Kodisinghe et al. (8) when the test was routinely used for the diagnosis of malaria.
The ability to detect the presence of the parasite during the course of treatment could be demonstrated by PCR amplification. It had been observed that PCR-positive results were obtained only when the DNA was extracted from samples containing live parasites. Dead parasites or parasites cleared by drug treatment or immune system pressure did not register as positive by PCR amplification. In our hands, no PCR product could be detected after successful treatment. Similar results have been reported when PCR was used to assess the response to antibiotic treatment for Borrelia burgdorferi infection (9) and acyclovir treatment of herpes simplex encephalitis (2). In both cases, PCR results were negative following the course of successful drug therapy (2, 9). This was also demonstrated when P. vivax-infected patients undergoing chloroquine treatment were monitored by PCR amplification of the circumsporozoite protein gene, whereby no amplicons were observed after the 4th day of treatment (6).
The amplification of the targeted regions could be applied to detect the persistence of very low grade parasitemia. PCR assays have been used to study patients whose P. falciparum parasite densities were below the microscopic threshold. Sethabutr et al. (13) demonstrated by the PCR technique that P. falciparum DNA in the blood of infected patients could be detected transiently at a time when the parasite could no longer be detected microscopically. The recurrence of P. vivax infections cannot be theoretically predicted by PCR assays because PCR cannot detect relapses of the P. vivax hypnozoite liver stage (7). On the other hand, monitoring of treatment by PCR can be used to detect the efficacy of drug treatment. In this study, more than half of the patients who were still PCR positive but blood film negative were readmitted to the hospital with malaria infection. This recurrence of malaria infection is not due to a new Plasmodium infection because Singapore does not have locally transmitted malaria.
In summary, we have described a direct PCR test system for the diagnosis of malaria. The test detects plasmodial infection and can differentiate P. falciparum from P. vivax via a multiplex reaction. No preparation or treatment of the blood samples is required prior to the amplification. The blood spots on filter paper were found to be very stable and can be stored without any noticeable effects on the sensitivity of the assay. The test can be completed within 3 h with a high degree of sensitivity. Although our PCR test for the detection and differentiation of malaria parasites requires more time than the antigen-capture dipstick assays (approximately 10 min), it has been shown to be a more sensitive assay with the ability to differentiate plasmodial species and detect mixed infections. The production cost of our PCR assay for the diagnosis of malaria is comparable to that of the commercially available antigen-capture assays. Further development and evaluation of the dipstick detection system are under way. This PCR diagnostic assay can easily be developed for mass screenings through automation and could thus be an effective diagnostic tool that is sensitive, specific, and less labor intensive than currently used methods. We would like to present this system as a simple and reliable test for the diagnosis of malaria.
ACKNOWLEDGMENTS
We acknowledge the technical expertise provided by Esther Wong and Alice Tay.
This work was supported by a grant from the National Science and Technology Board, Singapore, and research grant RP940361 from the National University of Singapore to U.A.K.K.
REFERENCES
- 1.Abdullah N R, Furuta T, Taib R, Kita K, Kojima S, Wah M J. Short report: development of a new diagnostic method for Plasmodium falciparum infection using a reverse transcriptase-polymerase chain reaction. Am J Trop Med Hyg. 1996;54:162–163. doi: 10.4269/ajtmh.1996.54.162. [DOI] [PubMed] [Google Scholar]
- 2.Aurelius E, Johansson B, Skoldenberg B, Staland A, Forsgren M. Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet. 1991;337:189–192. doi: 10.1016/0140-6736(91)92155-u. [DOI] [PubMed] [Google Scholar]
- 3.Craig M H, Sharp B L. Comparative evaluation of four techniques for the diagnosis of Plasmodium falciparum infections. Trans R Soc Trop Med Hyg. 1997;91:279–282. doi: 10.1016/s0035-9203(97)90074-2. [DOI] [PubMed] [Google Scholar]
- 4.Holodniy M, Kim S, Katzenstein D, Konrad M, Groves E, Merigan T C. Inhibition of human immunodeficiency virus gene amplification by heparin. J Clin Microbiol. 1991;29:676–679. doi: 10.1128/jcm.29.4.676-679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kain K C, Brown A E, Mirabelli L, Webster H K. Detection of Plasmodium vivax by polymerase chain reaction in a field study. J Infect Dis. 1993;168:1323–1326. doi: 10.1093/infdis/168.5.1323. [DOI] [PubMed] [Google Scholar]
- 6.Kain K C, Brown A E, Lanar D E, Ballou W R, Webster H K. Response of Plasmodium vivax variants to chloroquine as determined by microscopy and quantitative polymerase chain reaction. Am J Trop Med Hyg. 1993;49:478–484. doi: 10.4269/ajtmh.1993.49.478. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M, Miyake H, Kim H-S, Tanabe M, Arai M, Kawai S, Yamane A, Wataya Y. Species-specific PCR detection of malaria parasites by microtiter plate hybridization: clinical study with malaria patients. J Clin Microbiol. 1995;33:2342–2346. doi: 10.1128/jcm.33.9.2342-2346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodisinghe H M, Perera K L, Premawansa S, Naotunne T, Wickramasinghe A R, Mendis K N. p6e ParaSight-F dipstick test as a routine diagnostic tool for malaria in Sri Lanka. Trans R Soc Trop Med Hyg. 1997;91:398–402. doi: 10.1016/s0035-9203(97)90255-8. [DOI] [PubMed] [Google Scholar]
- 9.Nocton J J, Dressler F, Rutledge B J, Rys P N, Persing D H, Steere A C. Detection of Borrelia burgdorferi DNA by polymerase chain reaction in synovial fluid from patients with Lyme arthritis. N Engl J Med. 1994;330:229–234. doi: 10.1056/NEJM199401273300401. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira D A, Holloway B P, Durigon E L, Collins W E, Lal A A. Polymerase chain reaction and a liquid-phase, non-isotopic hybridization for species-specific and sensitive detection of malaria infection. Am J Trop Med Hyg. 1995;52:139–144. doi: 10.4269/ajtmh.1995.52.139. [DOI] [PubMed] [Google Scholar]
- 11.Payne D. Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Bull W H O. 1988;66:621–626. [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhartz A, Alajem S, Samson A, Herzberg M. A novel rapid hybridization technique: paper chromatography hybridization assay (PACHA) Gene. 1993;136:221–226. doi: 10.1016/0378-1119(93)90468-i. [DOI] [PubMed] [Google Scholar]
- 13.Sethabutr O, Brown A E, Panyim S, Kain K C, Webster H K, Echeverria P. Detection of Plasmodium falciparum by polymerase chain reaction in a field study. J Infect Dis. 1992;166:145–148. doi: 10.1093/infdis/166.1.145. [DOI] [PubMed] [Google Scholar]
- 14.Snounou G. Detection and identification of the four malaria parasite species infecting humans by PCR amplification. Methods Mol Biol. 1996;50:263–291. doi: 10.1385/0-89603-323-6:263. [DOI] [PubMed] [Google Scholar]
- 15.Tahar R, Ringwald P, Basco L K. Diagnosis of Plasmodium malariae infection by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1997;91:410–411. doi: 10.1016/s0035-9203(97)90259-5. [DOI] [PubMed] [Google Scholar]
- 16.Tan T M C, Nelson J S, Ng H C, Ting R C Y, Kara U A K. Direct PCR amplification and sequence analysis of extrachromosomal Plasmodium DNA from dried blood spots. Acta Trop. 1997;68:105–114. doi: 10.1016/s0001-706x(97)00080-6. [DOI] [PubMed] [Google Scholar]
- 17.Tham, J. M. 1999. Unpublished data.