Abstract
Angiotensin-converting enzyme 2 (ACE2), also known as peptidyl-dipeptidase A, belongs to the dipeptidyl carboxydipeptidases family has emerged as a potential antiviral drug target against SARS-CoV-2. Most of the ACE2 inhibitors discovered till now are chemical synthesis; suffer from many limitations related to stability and adverse side effects. However, natural, and selective ACE2 inhibitors that possess strong stability and low side effects can be replaced instead of those chemicals' inhibitors. To envisage structurally diverse natural entities as an ACE2 inhibitor with better efficacy, a 3D structure-based-pharmacophore model (SBPM) has been developed and validated by 20 known selective inhibitors with their correspondence 1166 decoy compounds. The validated SBPM has excellent goodness of hit score and good predictive ability, which has been appointed as a query model for further screening of 11,295 natural compounds. The resultant 23 hits compounds with pharmacophore fit score 75.31 to 78.81 were optimized using in-silico ADMET and molecular docking analysis. Four potential natural inhibitory molecules namely D-DOPA (Amb17613565), L-Saccharopine (Amb6600091), D-Phenylalanine (Amb3940754), and L-Mimosine (Amb21855906) have been selected based on their binding affinity (−7.5, −7.1, −7.1, and −7.0 kcal/mol), respectively. Moreover, 250 ns molecular dynamics (MD) simulations confirmed the structural stability of the ligands within the protein. Additionally, MM/GBSA approach also used to support the stability of molecules to the binding site of the protein that also confirm the stability of the selected four natural compounds. The virtual screening strategy used in this study demonstrated four natural compounds that can be utilized for designing a future class of potential natural ACE2 inhibitor that will block the spike (S) protein dependent entry of SARS-CoV-2 into the host cell.
Keywords: ACE2, COVID-2019, Molecular dynamics simulation, Molecular docking, MM/GBSA, Structure-based pharmacophore model, Virtual screening
Abbreviations: +ssRNA, positive single strand RNA; ICTV, International Committee on Taxonomy of Viruses; 2D, two dimensional; MD, molecular dynamics; 3D, three dimensional; MERS, Middle East Respiratory Syndrome; AA, amino acids; PBVS, pharmacophore based virtual screening; ACE2, angiotensin-converting enzyme 2; PDB, Protein Data Bank; ADMET, absorption, distribution, metabolism, excretion, and toxicity; RMSD, root mean square deviation; ADT, Auto Dock Tools; RMSF, root mean square fluctuation; COVID-19, coronavirus disease 2019; RO5, rule of five; DUDE, Database of Useful Decoys; SARS-CoV2, sever acute respiratory syndrome coronavirus 2; HTS, High Throughput Screening; SB, structure-based; SBPM, structure-based pharmacophore model; TMPRSS2, transmembrane protease serine protease 2; T.E.S.T., Toxicity Estimation Software Tool; WHO, World Health Organization
1. Introduction
The ongoing novel coronavirus 2019 (nCoV-2019) outbreak has been recently taken place and hit almost all over the world. The disease has been named COVID-19 (coronavirus disease 2019) by the World Health Organization (WHO) recently after the outbreak started in Wuhan, Hubei province of China on 31 December 2019 [1]. The International Committee on Taxonomy of Viruses (ICTV) renamed the outbreak-causing virus as sever acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2], which is responsible for lower respiratory tract disease in humans [3]. COVID-19 pandemic is a great threat to both tropical and polar regions of the world [4], and WHO decreed the disease as a sixth public health emergency. There are rapidly growing numbers of cases globally, and most of the countries have already reported nationwide community transmission [5]. The transmissible and pathogenic virus infects an estimated >223,475,490 people and caused 4,611,347 confirmed deaths (September 09, 2021, 09:41 GMT) across 221 countries and territories around the world [6]. The emergence of new COVID-19 has led to increased demand for new antiviral strategies [7]. But, to date, no specific proven drugs, and therapeutics have been developed that, can prevent or treat infections resulting from these pathogens [8], [9].
SARS-CoV-2 is morphologically oval, round, or often polymorphic shape in nature with a diameter being 60-140 nm [10]. From a genomic perspective, the virus consists of a positive sense single-strand RNA [+ssRNA] and belongs to the lineage of β (beta)-coronavirus [11], [12]. The +ssRNA genome of the virus carries a length of around 29.8 kilobases (kb) formed by 29.86% adenosine, 18.39% cytosine, 19.63% guanine, and 32.12% thymine's [13]. Phylogenetically the virus belonging to the β-genus of coronavirus and their virion consists of the major surface spike (S) protein, integral membrane (M) and envelope (E) protein, and complexes genomic RNA forming nucleocapsid (N) proteins [14]. Early genomic sequence analyses of the SARS-CoV-2 indicated that the virus encodes similar structural proteins (such as spike glycoprotein) and enzymes (helicase, and RNA-dependent RNA polymerase) as of SARS and Middle East Respiratory Syndrome (MERS) virus [7], [15]. Furthermore, the SARS-CoV-2 genome shares 79.6% and 96% sequences similarity to SARS-CoV and bat coronavirus, respectively [16], which previously utilizes the ACE2 protein as a receptor to get entrance into the host [17], [18], [19].
It has been reported that ACE2 a member of the dipeptidyl carboxy-dipeptidases family has a great impact on supporting the SARS-CoV-2 viral entry into the human host cell [20], [21], [22]. The S proteins of the virus are the main target for the neutralization antibody, attach to the host cellular ACE2 receptor, and allow it to enter into the targeted host [12], [20], [23]. SARS-CoV-2 engages their S protein into two molecular subunits as S1 and S2 [24], where S1 is responsible for attachment to the entry receptor and S2 to viral particle infusions [25]. S1 subunit of the virus is composed of receptor-binding domain and receptor binding motif [26], and this organizational structure help to contacts the receptor ACE2 of the host [20]. The S proteins of the virus also known as a type of class I viral fusion proteins require protease cleavage for their activation. The activation of the S protein is a two-way process known as priming cleavage, efficiently cleaved at the boundary between the S1 and S2 subunits [24], and activating cleavage that activated at the boundary to the S2′ priming site [25]. Once S protein comes in contact with the ACE2, a serine protease called transmembrane protease serine protease-2 (TMPRSS2) help to cleaves the ACE2 for initiating the S protein priming [20], and the resulting activation of the cleavage site mostly leading to the viral fusion and infectivity to the pathogens [20], [24], [27]. Being an attentional host cellular target, different quantitative kinetics studied through surface plasmon resonance revealed that ACE2 has had 10-20 folds higher affinity to the S protein of the SARS-CoV-2 than other coronaviruses [28]. Additionally, the human ACE2 known as I integral membrane protein [29], holds 17 amino acids (AA) residue to the N-terminal domain, 22 AA in the C-terminal domain, and 43 AA residue in the cytoplasmic domain [4], which contains potential phosphorylation sites have major impacts on SARS-CoV-2 viral infection [30]. For example, first helix and lysine 353 and proximal residues of the N terminus of ACE2 extracellular portion, interacts with viral spike glycoprotein has a major role in the virus infectivity. Since host cell entry of the SARS-CoV-2 depends on the receptor ACE2 [20], and compounds responses against this enzyme could at least partially protect against the virus. Although ACE2 was suggested to be a novel SARS-CoV-2 target [18], [32], [33], only a few and effective inhibitory compounds were identified hitherto.
Natural products, often defined as compounds or substances produced by a living organism are derived from nature and historically use as active components of many traditional medicines [34]. The compounds derived from natural sources have great therapeutic value and represent more than half of FDA-approved drugs [35], which also received attention for their extensive pharmacological and biological activities [36]. However, the chemically synthesized compound has poor biological activities or obvious side effects [37], [38], hence developing novel ACE2 inhibitors from natural resources for the treatment of COVID-19 is an urgent matter.
The conventional process of developing new drugs normally involves lengthy, expensive, and requires intense effort [39]. For example, high-throughput screening (HTS) is a process that aids drug discovery by assaying a large number of potential drug-like compounds, whereby the technology combines multiple-well microplate and robotic processing [40]. Nevertheless, HTS requires huge resources, as the cost to process a single HTS program is relatively high and necessitates advance robotic systems [37]. Computer-aided drug design also known as in silico drug design on the other hand is a relatively new method compared to a high-throughput approach, to screen large databases of compounds [41], [42], [43]. The in silico virtual screening process help to generate hits to lead compounds in the way to the discovery of new drugs at a faster time and at a lower cost [44]. Advance in silico drug design option tends to decrease the amount of time to develop, design, and optimize a new drug. Over the past few decades, the virtual screening process was engaged to identify the best lead compounds with different structural features for combination with a specific biological target [45], [46], [47]. Moreover, the computer-aided drug design by using pharmacophore-based virtual screening (PBVS), molecular docking and dynamics simulation approaches has been identified diverse promising drug targets and hits [14]. Therefore, this study aimed to screen potential natural anti-COVID-19 compounds by combining SBPM, virtual screening, molecular docking, ADMET (absorption, distribution, metabolism, excretion, and toxicity), and MD simulation approaches.
2. Materials and methods
2.1. Pharmacophore modeling
The X-ray-derived crystal 3D structures of human ACE2 protein (PDB ID: 1R4L) in complex with XX5 (PDB ID) were obtained from the Protein Data Bank (PDB) [29], and an SBPM was generated automatically using the LigandScout 4.3 advance software [48]. The advanced version of the software was used to determine and visualize the main pharmacophore features of the protein-ligand interaction like hydrogen bond donor and acceptor, negative and positive ionizable area, and hydrophobic interactions of the compound. Multiple chemical features like the number of aromatic rings, binding location of zinc, magnesium, and manganese, halogen bond donor were also detected and mapped for the characterization of all or specific pharmacophore patterns of ligands [49]. In addition, LigandScout's excluded volume feature has been added to the 3D structure-based models to retain the sterical circumference of the macromolecule. These excluded volume features ensure and match the sterical requirements of the active site and help to identify desire compounds [48] and increase the selectivity of the virtual screen compounds.
2.2. Pharmacophore model validation
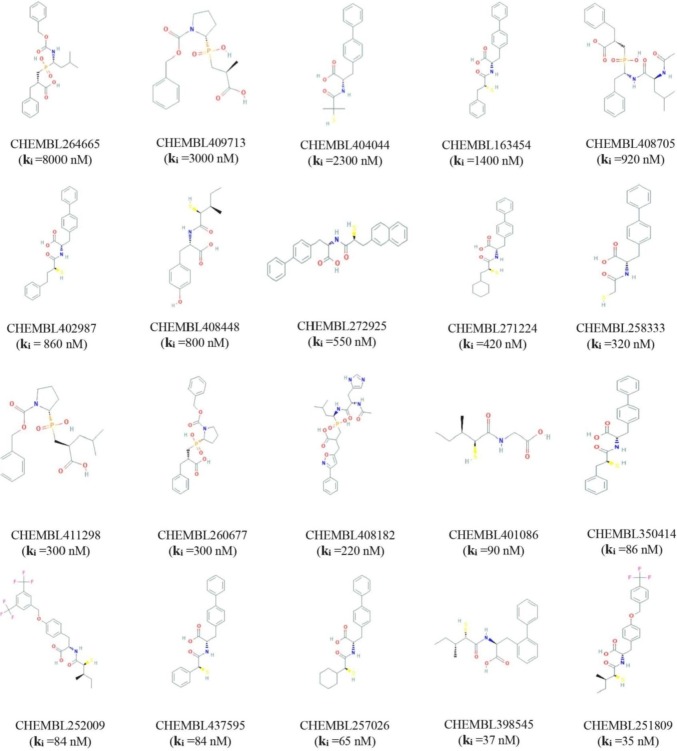
The pharmacophore model generated from complex protein-ligand interaction was validated using a set of known active inhibitors by evaluating their ability to distinguish between known potential active and inactive compounds [19]. A set of known active ACE2 inhibitors were extracted by combining ChEMBL (www.ebi.ac.uk/chembl/) database and an extensive literature search shown in Fig. 1 . Compounds with large molecular weight and peptides like molecules have been removed as well as non-ACE2 inhibitors with false activity labels [45]. The remaining set of known active ACE2 inhibitors and their correspondence decoy compounds were retrieved from the enhanced Database of Useful Decoys (DUDE) (http://dude.docking.org), which have been used to validate the 3D-interaction feature model. The decoy set of compounds obtained from the DUDE database was assigned into a multi-conformational virtual screening library by utilizing the command line executable tools “idbgen” in LigandScout 4.3, advance. The main purpose of using the executable tools “idbgen” is to create compound databases for virtual screening, and the idbgen derived LDB file format also speeds up processes for compute clusters of the compounds and annotates each conformation with generated 3D SB-pharmacophore features. To evaluate the preferential efficiency of the pharmacophore, model a 3D molecular structural database screening process was performed [48]. Valuable parameters like active hits (AH), decoy compounds (DC), early enrichment factor (EF), the total compounds in the database (D), the total number of hits retrieved (TH), and goodness of hit score (GH) has been considered to evaluate the performance of the model. The EF of a pharmacophore model is a widely used metric generated during randomly screening of compounds library, which describes the number of active hits found by utilizing an appointed PM model as inverse to the number hypothetically active compounds found [50].
Fig. 1.
Known inhibitors of ACE2 with their correspondence ChEMBL identifier and Ki value of the compounds.
The GH statistical hypothesis test used to compare how well do the observed hits correspond to fit with the assumed pharmacophore model. The GH value ranging from 0 to 1, where 0 indicates acceptance of the null hypothesis model with low or no fit with the model, and 1 indicates most significant with the highest fit to the pharmacophore model [51]. The EF and GH score were calculated from Eqs. (1), (2) accordingly [50], [51], to validate the model performance in our rational drug design approaches. The EF score Eq. (1) and GH score Eq. (2) is given below:
| (1) |
| (2) |
where, AH is the active hits in the database, TH is the total number of hits retrieved, D is the total compounds in the database, A is the total number of actives in the database, (TH − AH) = FC is the number of false positives compounds, and (D − A) = DC is the decoy compounds of the database.
2.3. Dataset generation
Ambinter (www.Ambinter.com) is a brand and worldwide supplier of advanced chemicals that supporting the scientific community by providing active compounds for drug discovery. Ambinter is a public access database that contains 36 million purchasable compounds that are ready to dock. Initially, the library was designed and developed for easy access to molecules and drug-like compounds for virtual screening, now it is widely used for VS, PBVS, and force field development [14]. The Ambinter database supply compounds from different vendors by utilizing a compounds code known as SMILES key. For new compounds discovery, the database contains a targeted library of SARS-CoV-2 that has been retrieved for the further screening process. In the present study, chemical features that were generated from the SBPM were used as a query for searching the chemical library. The molecules that fit with the query pharmacophore features were retained and retrieved for further validation.
2.4. Virtual screening
The dataset generated from the Ambinter database was virtually screened based on the validated 3D SB-pharmacophores features. The load screening database features of the LigandScout 4.3 advanced use to convert the compounds into a (*.ldb) database file format, have been uploaded to the molecule database list for quick pharmacophore features based virtual screening [48]. The screening has been done with the relative pharmacophore-fit as a scoring function with a maximum of four omitted pharmacophore features. The hit compounds fitted with the geometry and features of the 3D model were ranked according to the pharmacophore fit score and retrieved for further validation.
2.5. Protein and ligand preparation
Pharmacophore based virtual screening process can generate many ‘hits’ compounds with disproportionate quality [52]. To optimize hits and sorting the better interaction features in comparison with the XX5 inhibitor, and the selected virtually screened compounds have been docked to the binding site of the ACE2 protein. The crystal X-ray structure of the ACE2 protein (PDB ID: 1R4L) in complex with the ligand XX5 was chosen as the positive control system for the study [29]. The ligand in a complex with the protein ACE2 was separated using the Biovia Discovery Studio visualizer version (16.1.0). The PDB structure of proteins was prepared by the following steps (i) bond order has been assigned, (ii), metal ions, cofactors, and water beyond 5 Å from het groups has been removed, (iii) het state has been generated by using Epik at pH 7 ± 2 [53], (ii) non-polar H has been merged and polar H-atoms has been assigned by using PROPKA at pH 7, (iv) the protein structure integrity has adjusted, and the missing side-chain atoms of the protein residues has been predicted by Prime, and (v) the default charges and solvation parameters have been allocated by using the Protein Preparation Wizard in the Schrödinger suite [54].
The ligands that were generated by pharmacophore-based virtual screening have been prepared and optimized by the LigPrep module of Schrodinger Suit [55]. Initially, the OPLS_2005 force field has been chosen to minimize the energy. The ionization state was set at a pH range of 7 ± 2, by using the Epik ionization tool. All possible deprotonated states and ionization states, tautomers, stereochemistry, and ring conformations have been identified. Stereoisomers were generated by retaining specified chiralities with various other chiral centers, considering a maximum of 32 stereoisomers per ligand. Only the lowest energy conformation of each ligand has been kept for further validation.
2.6. Binding site identification and grid generation
The binding site of the protein was visualized and analyzed to predict the binding affinity and selectivity between the complex crystal protein-ligand interaction [38]. The binding position of the complex interaction was characterized based on hydrogen bond donor-acceptor features, hydrophobic interaction features, negative-positive ionizable area, and zinc chelation in addition to unspecific halogen bond donor-acceptor features of the compounds. Additionally, the PrankWeb (http://prankweb.cz/) is an online resource and server-based tool used to complex protein-ligand binding site prediction and an extensive literature search also confirmed the binding site position of the protein [56]. After identification, the binding sites of the protein a receptor grid was generated using the predicted binding site position of the complex protein-ligand structure.
2.7. Molecular docking (MD) simulation
The docking positions between the selected compounds and ACE2 proteins were predicted by PyRx software [57]. PyRx is open-source software that utilizes Auto Dock 4 (AD4) and Auto Dock Vina (ADV) tools for molecular docking simulation [58]. In this study, the PyRx tools Autodock vina (version 1.1.2) a commonly docking program for molecular docking simulation, has been used to predict the protein-ligand interaction [57]. The crystal structure of ACE2 was downloaded and prepared before molecular docking simulation, and docking positions were visualized by BIOVA Discovery Studio Visualizer Tool (16.1.0). The top 20% of compounds with the highest binding energy (negative sign value), were considered for further investigation.
2.8. Absorption, distribution, metabolism, and excretion (ADME) test
The drug design and development process involves the assessment of ADME to identify molecules with the highest chance to become an effective drug for a specific disease [39]. ADME of a compound provides information about their physicochemical, pharmacokinetics, metabolism, and excretion properties of molecules into urine and feces [59]. Due to numerous compounds with limited access to the physical samples, physicochemical and pharmacokinetic properties of compounds are critical for their initial selection [60]. Early-stage evaluation of ADME in the drug design and discovery process mitigates the fraction of pharmacokinetics-related failure during clinical trials [61]. Nowadays, computer-generated models have complied as a potent substitute for experimental methods for early-stage prediction of ADME. For the studies, a freely accessible Swiss-ADME server (http://www.swissadme.ch/) was used to predict the various pharmacokinetic and pharmacodynamics properties [62]. The results of ADME for all the ligand molecules are represented in Table 3.
Table 3.
ADME properties like physicochemical properties, lipophilicity, water solubility, pharmacokinetics, drug likeness, and medicinal chemistry of selected 4 compounds.
| Properties | Amb6600091 | Amb17613565 | Amb3940754 | Amb21855906 | |
|---|---|---|---|---|---|
| Physico-chemical properties | MW (g/mol) | 276.29 | 197.19 | 165.19 | 198.18 |
| Heavy atoms | 19 | 14 | 12 | 14 | |
| Arom. heavy atoms | 0 | 6 | 6 | 6 | |
| Rotatable bonds | 11 | 3 | 3 | 3 | |
| H-bond acceptors | 8 | 5 | 3 | 5 | |
| H-bond donors | 5 | 4 | 2 | 3 | |
| Lipophilicity | Log Po/w | 0.85 | 0.72 | 1.08 | 0.69 |
| Water solubility | Log S (ESOL) | High | High | High | High |
| Pharmacokinetics | GI absorption | Low | High | High | High |
| Drug likeness | Lipinski, violation | No | No | No | No |
| Medi. chemistry | Synth. accessibility | Very easy | Very easy | Very easy | Very easy |
2.9. Toxicity test
An early assessment of compound's toxicity is very much important in the field of drug discovery and development [63]. In-silico evaluation of toxicity is exceptionally arising as an integral stage for the determination toxicity of a chemical compound that could be potentially harmful to humans and animals [64]. The in-silico toxicity of the selected compounds was evaluated using the Toxicity Estimation Software Tool (T.E.S.T.) version 4.2.1 [65]. The software predicts the toxicity of selected compounds by using Quantitative Structure-Activity Relationships (QSARs) methodologies. This model can predict toxicity by comparing the physical characteristics features of a chemical compound entered by the user. The toxicity of a chemical compound can be access in terms of toxicity endpoints like mutagenicity, carcinogenicity, and other features, and this endpoint can be measured both quantitatively and qualitatively [64]. In our studies, the quantitative toxicity endpoints, and drug relevant properties like 96-hour fathead minnow LC50, 48-hour Daphnia magna LC50, 48-hour Tetrahymena pyriformis IGC50, oral rat LD50, bioaccumulation factor, have been evaluated using the T.E.S.T. software. The organ toxicity like hepatotoxicity and toxicity endpoints like carcinogenicity, immunotoxicity, mutagenicity, and cytotoxicity of the compounds were evaluate qualitatively using the ProTox-II (http://tox.charite.de/) web server. The ProTox-II is freely accessible virtual lab for the prediction of toxicities of small compounds and user can be accessed the server without registration. Various toxicity endpoints such as acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, immunotoxicity of the compounds can be retrieved from the server by knowing the two-dimensional structure (2D) of the input compounds. ProTox-II incorporates molecular similarity, fragment propensities and machine-learning features that helps to predict various toxicity endpoints of the compounds, indicating their possible applicability for other compound classes.
2.10. Molecular dynamics (MD) simulation
Molecular dynamics (MD) simulations were used to validate the structural stability and conformational flexibility of a protein-ligand complex [44], [66]. MD simulations provide powerful tools for the prediction of each atomic movement of protein, ligands, and physics governing interatomic interactions over a specific time [67]. To validate the structural stability and conformational flexibility protein ligands complex generated from the molecular docking study were subjected to 250 (ns) nanoseconds of MD simulations [68]. The Desmond module of Schrödinger (Release 2020-3) software package with OPLS-2005 force field for generating system topology has been selected to simulate the complex protein-ligand structure [68], [69]. A protein to box minimum distance of 0.8 nm was generated to solvate the systems and the protein was centered in the box. The solvent box was filled with the simple point charge water (SPC) type of solvent with a 3-point solvent model. Na+ and Cl− ions were used to neutralize the system to reach a 0.15 M molar concentration. Energy minimization was done by gradient optimization to reduce the net force on each movable atom with a minimization step size of 0.1 (ns) and a maximum force of 500 kJ/mol·nm2 to achieve the static state of the system, and minimization graph was generated from the system declared that the optimized construction was solvent saturated and geometrically stable. Equilibration of protein water system ensemble for 10 ns was done for both NVT (constant number of particles, volume, and temperature) and NPT (constant number of particles, pressure, and temperature) ensemble process. The NPT of the system was set at a constant temperature of 300 K and NVT of the system pressure (1.01325 bar) to sustain the stability of the system. Well, an equilibrated dynamics system was performed in the NPT (constant pressure and temperature) ensemble involved for 250 ns production run with integrator time 2 fs and resulted in production were saved every picosecond for further analysis. Analysis of molecular dynamic simulation generated the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and protein-ligand contact map was used to examine the interactions and stability between the ACE2 proteins and natural compounds.
2.11. Calculation of MM/GBSA
For calculating the binding free energy of ligands to the macromolecules “molecular mechanics generalized Born surface area” (MM/GBSA) methods have become popular methods [70]. The MM/GBSA has been used to estimate the binding free energy of the compounds by using the Maestro package that incorporated in Schrödinger (Release 2020-3) by using default parameters [71].
3. Results
3.1. Structure-based pharmacophore modeling
The first step of our rational drug design approach was the building of a 3D structure-based pharmacophore model. For this purpose, we utilize both theoretical bits of knowledge and experimentally validated 3D structural model storage in the different protein databases. To identify new inhibitor compounds, the crystal X-ray structure of ACE2 protein (PDB ID: 1R4L) in complex with ligand XX5 was retrieved from the PDB, and a 3D-SBPM to the enzymatic cavity was generated. The binding activity of the XX5 inhibitor with the ACE2 protein has confirmed experimentally IC50: 0.44 nM (100), and validated by X-ray diffraction, resolution: 3.00 Å, R-value free: 0.337 R-value work: 0.253 [29], highlighted that this series of inhibitors could bind the active site of the enzyme, followed by inhibition of their biological activity. To determine the active series of natural inhibitors with similar or better chemical features than XX5, a 3D-SBPM was generated by automatically derives key chemical features of LigandScout 4.3 advanced tools. The automatic key chemical features option of the advance tool determines and displayed a total of 16, including 12 main and four dedicated pharmacophore features (Fig. 2 ), where two hydrogen bond donors (HBD), five hydrogen bond acceptor (HBA), two negative ionizable (NI), one positive ionizable (PI) area, two hydrophobic (H) interactions features have defined as main, and two aromatic rings (AR), one zinc-binding location (ZNB), and one halogen bond donor (XBD) features have defined as dedicated pharmacophore features of the protein-ligand complex interaction. Fifteen excluded volumes feature, also known as the program, increases selectivity generated features, derived from the sterical circumference of the protein has not displayed in both Fig. 2(a) & (b), the excluded volumes feature has shown in Fig. S1.
Fig. 2.
(a) The 3D structure-based pharmacophore model of ACE2 protein in complex with XX5 ligand derived from the X-ray derived crystal structure of the protein retrieved from the PDB (PDB code: 1R4L). (b) Merge pharmacophore features of the complex interaction, where two hydrophobic feature has indicated by yellow spherical shape  , one positive ionizable by blue star shape
, one positive ionizable by blue star shape  , two negative ionizable by red star shape
, two negative ionizable by red star shape  , two hydrogen bond donor by green spherical or green arrow shape
, two hydrogen bond donor by green spherical or green arrow shape  , five hydrogen bond acceptor by red spherical or red arrow shape
, five hydrogen bond acceptor by red spherical or red arrow shape , one zinc-binding location by royal blue star or cone shape
, one zinc-binding location by royal blue star or cone shape  , and one halogen bond donor by violet spherical or arrow shape
, and one halogen bond donor by violet spherical or arrow shape have represented within the protein-ligand complex interaction. Fifteen excluded volume areas generated by the pharmacophore model have not displayed in this figure. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
have represented within the protein-ligand complex interaction. Fifteen excluded volume areas generated by the pharmacophore model have not displayed in this figure. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The pharmacophore feature derived from the crystal complex structure of the ACE2 protein stated that the ligand formed hydrogen bond predominantly with the amino acid residue of the protein (Fig. S2). Five HBAs are determined, and they are possessed mainly by oxygen atoms. The acceptor features generated by the interaction of ARG273 and TYR515 residues occupied the trigonal planar molecular geometry oxygen atoms, and the THR371 occupied the nitrogen atoms of the benzene ring in the ligand.
HBD features have been found to form, where tetrahedral nitrogen atoms were interacting with the HIS345 and PRO346 residue. The tetrahedral carbon atom interacts with hydrophobic PHE347, PHE504, and TYR510 residues, and the chlorines on the pyridine ring interact with hydrophobic residues THR371 to the ACE2 binding pocket. A zinc finger is a small protein structural motif that has been found to form in the ZNB residue ZN803. It covered several functional groups produce from other metalloprotease inhibitors. Chlorines on the pyridine ring produce halogen donor features by interacting with the residues ASP368. Negative and positive ionizable pharmacophore features have also been found to be formed in ARG273, ZN803, and GLU375 accordingly to the complex protein-ligand structure.
3.2. Pharmacophore model validation
Pharmacophore models generated with distinguished chemical features were subject to validation to evaluate the quality of the model. Validation is an important attribute of a reliable pharmacophore model, which portends the capability to exactly predict internal and particularly external data sets of molecules [19]. The structure-based pharmacophore models were validated using the testing set that included 20 actives known ACE2 inhibitors with correspondence 1166 decoy molecules retrieved from the enhanced database DUDE.
The active test set with inhibitor constant Ki values (0.13 nM to 10,000 nM) were merged with the decoy compounds, to observe how well the pharmacophore model can distinguish the active compounds from inactive compounds. A 3D molecular structural database screening process was performed and subsequently, the EF value and GH value of the compounds were calculated (Table 1 ). The GH score of 0.75 (Out of 1) and the EF score of 1.1 were found from Eqs. (1), (2) accordingly, AUC (area under the ROC curve) value found 0.75 (Fig. S3), indicating that the model was very good and is rational for virtual screening.
Table 1.
Validation of pharmacophore model using GH scoring method includes enrichment factor, goodness of hit score of the model.
| Parameter | Symbol | Calculation | Model |
|---|---|---|---|
| Total compounds in database | D | – | 12,561 |
| Total actives in database | A | – | 11,295 |
| Decoy compounds | DC | D-A | 1166 |
| Total hits retrieved | TH | – | 23 |
| Active hits retrieved | AH | – | 23 |
| % yield of active hits | – | (AH/TH)×100 | 100% |
| % ratio of active hits | – | (AH/A)×100 | 0.20% |
| Enrichment factor | EF | Eq. (1) | 1.1 |
| False negatives | FN | A − AH | 11,272 |
| False positives | FP | TH − AH | 0 |
| Goodness of hit score | GH | Eq. (2) | 0.75 |
3.3. Dataset preparation
The Ambinter database contains a mix of natural products with synthetic and/or semisynthetic compounds along with their chemical structures and physicochemical properties. A total of 11,295 natural compounds small molecules were obtained from the Ambinter natural compounds database library (Ambinter and Greenpharma). Lipinski's rule of five (RO5) also known as Pfizer's rule of five is used to evaluate drug-likeness properties of compounds [44], which were applied for the subsequent phase of screening.
3.4. Pharmacophore-based virtual screening
Compounds with no violation of RO5 were used for pharmacophore-based virtual screening. The final key pharmacophore interaction features generated from the protein-ligand complex were applied to the 11,295 natural compounds, and a total of 23 derivatives was generated with a pharmacophore fit score ranging between 78.81 and 75.31 (Table S1). During pharmacophore-based virtual screening not all the features of the model could be matched, therefore four features of the model have been omitted, resulting in lower pharmacophore fit scores of some derivatives [48]. A higher fit score generated by measuring the geometric fit of the features of a molecule indicates a better fit for the 3D-SBPM. As the higher fit score indicates a better fit for the model, therefore molecules that fit the pharmacophore model should also show activity to the ACE2 protein.
3.5. Binding site identification and receptor grid generation
Identifying binding sites within a protein is an important part of molecular docking, and the virtual screening process [72]. The pocket on a target protein is a combination of the different binding sites that present compatible size, shape, and suitable chemical complementarity with ligands. To determine the binding site of the protein the co-crystal structure of ACE2 bound to XX5 was analyzed through the BIOVA Discovery Studio Visualizer Tool (16.1.0). The Discovery studio visualizer tool revealed that the XX5 of ACE2 formed 4 conserved hydrogen bonds with ARG273, THR371, PRO346, and TYR515 shown in Fig. 3 . Another 4 residues namely CYS344, MET360, LYS363, and PHE504 formed an alkyl bond by interacting with the ligand XX5. Attractive charge and salt bridge bonding have also been observed in ARG 514, ARG518, ZN803, and ARG273 positions accordingly.
Fig. 3.
Describing the protein binding sites generated from the protein-ligand complex structure (PDB ID: 1R4L).
TYR510 formed both Pi-Sigma and Pi-Alkyl bonds with the protein, where only one halogen bond with Cl has formed in the protein ASP368 position. Unfavorable acceptor-acceptor and Pi-Pi-T-shape bonds both have formed in the protein HIS345 residual position. The PrankWeb binding site finder and an extensive literature search were also used to confirm the binding site residue and position of the protein. Protein-ligand complex analysis from the server also revealed another 2-binding site position of the protein. Atomic coordinates of the protein and ligand were obtained from the discovery studio and PrankWeb analysis tools used for receptor grid generation. A grid box comprised of X = 57.30, Y = 51.20 and Z = 25 points spaced dimension by 0.350 Å was centered on the ACE2 binding site with a position of X = 40.03, Y = 0.29, and z = 22.45.
3.6. Molecular docking simulation
Molecular docking is an increasingly important key tool in structure-based virtual screening and computer-aided drug design approaches [73]. The molecular docking approach can be used to predict the predominant binding mode(s) of a ligand with a protein at the atomic level [4]. The goal of ligand-protein docking is to perform virtual screening, rank the results according to their binding energy, understanding the protein-ligand mechanism of action and propose a structural hypothesis of how the ligands inhibit the target [58]. To understand the binding interaction of the targeted protein with the selected 23 compounds, the PyRx tools Autodock vina (version 1.1.2) molecular docking program has been run with an exhaustiveness value 8 [38], [57]. The top four compounds have been chosen based on their binding energies ranging between −7.0 kcal/mol and −7.5 kcal/mol (Table 2 ). Molecular docking scores of selected 4 compounds Amb17613565, Amb6600091, Amb3940754, and Amb21855906 were found −7.5 kcal/mol, −7.1 kcal/mol, −7.1 kcal/mol, and −7.0 kcal/mol respectively, which have better binding energy than the control compounds XX5 (-6.4 kcal/mol).
Table 2.
Molecular docking score of ACE2 protein and natural compounds, retrieved using the PyRx tools Autodock vina.
| Ambinter ID | Formula | Molecular weight | Docking score (kcal/mol) | Pharm.-Fit score |
|---|---|---|---|---|
| Amb17613565 | C9H11NO4 | 197.188 | -7.5 | 78.81 |
| Amb6600091 | C11H20N2O6 | 276.286 | -7.1 | 78.75 |
| Amb3940754 | C9H11NO2 | 165.189 | -7.1 | 77.71 |
| Amb21855906 | C8H10N2O4 | 198.176 | -7.0 | 77.12 |
| XX5 (PDB ID) | C19H23Cl2 N3O4 | 428.31 | -6.4 | N/A |
A higher docking score indicates better stability of the ligand with the target protein. The pharmacophore fit score of each compound is provided for comparison.
3.7. Interpretation of protein-ligand binding interaction
Amb17613565, Amb6600091, Amb3940754, and Amb21855906 displayed better pharmacophore fit scores compared to the other compounds and molecular docking simulation determines the binding energy −7.5 kcal/mol, −7.1 kcal/mol, −7.1 kcal/mol, and −7.0 kcal/mol, respectively. Amb17613565 formed 3 conventional hydrogen bonds with ARG273 (2.73 Å distance) and 2 with ASP269 (2.09 Å and 2.03 Å distance) positions, one pi-alkyl bond with a distance 5.21 Å has also been observed in ALA153 residual position shown in Fig. 4 .
Fig. 4.
Showing the 3D interaction of Amb17613565 within the binding site of ACE2 protein.
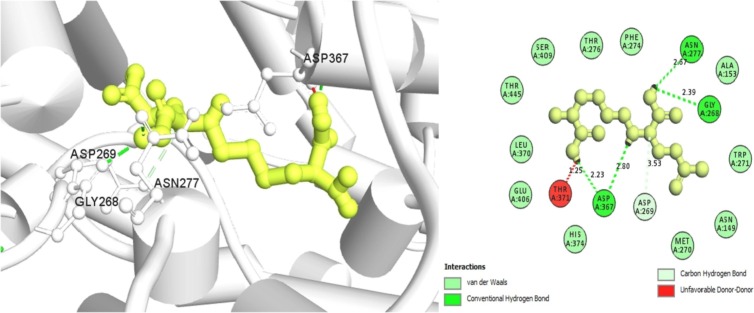
Amb6600091 has formed four conventional hydrogen bonds with GLY268 (2.39 Å), ASN277 (2.67 Å), and two with both ASP367 (2.23 Å and 2.80 Å) residues. A carbon-hydrogen bond (C—H) and an unfavorable donor-donor bond have also been established in ASP269 (3.53 Å) and THR371 (1.25 Å) residues, accordingly, depicted in Fig. 5 .
Fig. 5.
Showing the 3D interaction of Amb6600091 within the binding site of ACE2 protein.
Amb3940754 also formed four conventional hydrogen bonds with ACE2 protein, 2 with ASP350 (2.34 Å and 3.03 Å), and another two with ARG393 (2.49 Å and 2.82 Å) residue. One van-der-Waals, one pi-pi stacked and one pi-pi T-shaped bond also have been found to formed with GLY352 (3.61 Å), PHE390 (4.06 Å), and PHE40 (5.08 Å) respectively Fig. 6 .
Fig. 6.
Showing the 3D interaction of Amb3940754 within the binding site of ACE2 protein.
Another natural compound Amb21855906 have interacted with the target protein residues by forming four conventional hydrogen bonds with LYS363 (2.11 Å), THR365 (2.67 Å), ASN277 (2.46 Å), and ASN149 (2.72 Å) residues, where one attractive charge bond with ASP368 (5.39 Å) and one salt bridge bond with ASP367 (2.66 Å) have also noticed to formed Fig. 7 . Conventional hydrogen bonds also are known as classical bonds are formed when hydrogen covalently bound to elements or a molecular fragment X–H (X = N, O, or F; X is more electronegative than H) forms a second bond to another electronegative atom. All the natural compounds Amb17613565, Amb6600091, Amb3940754, and Amb21855906 formed hydrogen bonds with the desired protein in agreement with the previous binding interaction analysis of our protein-ligand complex studies.
Fig. 7.
Showing the 3D interaction of Amb21855906 within the binding site of ACE2 protein.
Ion-ion interactions are an attractive force that occurs between two oppositely charged ions that hold together ionic molecules that have been found to form in Amb21855906. In pi-alkyl interactions π-hold a bond over an aromatic group and electron group of an alkyl group that stays around the ring, leading to holding strong stability of the compounds have found to form in the compounds Amb17613565 and Amb21855906.
3.8. Pharmacophore features analysis
Pharmacophore features of a compound play a significant role in identifying the specific active site of the protein. The pharmacophore of a compound can be described based on the H, AR, HBA or HBD, PI, NI features that helps design a new drug candidate against a specific disease. These features retain the necessary geometric arrangement of atoms requires to producing a specific biological response. Therefore, the pharmacophore features of the selected four compounds include Amb17613565, Amb6600091, Amb3940754, and Amb21855906 compounds have been analyzed and compared with the query pharmacophore features shown in Fig. 8 . Each of the compounds has similar or better pharmacophore properties than the query pharmacophore features. Therefore, the selected compounds should be effective for our target protein.
Fig. 8.
Showing the hydrophobic (yellow color), positive ionizable (blue color), and negative ionizable (red color) features of the (a). Amb17613565, (b). Amb6600091, (c). Amb3940754 and (d). Amb21855906 compounds, thus resulting in a higher fit score during structure-based virtual screening and molecular docking approaches. Compound XX5 denoted by (E) is the ligand to the binding site of ACE2 protein was used to generate the main pharmacophore features, which is included in this figure for comparison purposes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.9. ADME prediction
The collaboration between drugs and the human body is a bidirectional procedure, drugs influence the human body, bringing about receptor inhibition or activation and the human body disposes of drugs by absorption, distribution, metabolism, and excretion (ADME) [39], [50]. Influence and disposes of drugs are interactional that occur simultaneously in the human body after administration and can lead to desired pharmacological function or may lead to undesirable side effects [62]. Drug design research and development (R&D) is an expensive, slow, and risky process that is generally faced with some unrehearsed even catastrophic failures in various stages of drug discovery [60]. The efficacy and safety deficiencies during drug design are thought to be the main cause of R&D-related failures, which depend mainly on compounds' ADME properties [50]. Therefore, the ADME of the compounds needs to evaluate to minimize the pharmacological failures in the drug discovery process. In this study, the Swiss-ADME an in silico ADME predictions server was used to evaluate the pharmacokinetics and drug-likeness properties of the selected 4 compounds [62]. The ADME profiles like lipophilicity known as partition coefficient between n-octanol and water (Log Po/w), water solubility an important property influencing absorption, drug-likeness that determines the chance of a molecule to become an oral drug and medicinal chemistry that evaluates the synthetic accessibility of the compounds were evaluated, and all of the 4 compounds were found to be suitable as listed in Table 3 .
3.10. Toxicity test
During drug development toxicity evaluation is an important part and should be assessed in preclinical and clinical trial phases. Toxicity and adverse effects of a drug can be evaluated using in vitro and in vivo tests, which are laborious, costly, and time-consuming and even involve animal welfare issues [74]. In the comparison of in vitro and in vivo experimental approaches computational methods developed for drug toxicity prediction have shown great advantages due to their accuracy, rapidity, accessibility and most importantly it can be done before a compound being synthesized. To access the toxicity and adverse effects of selected 4 compounds the computational methods have been used in this study. The T.E.S.T. software and the ProTox-II server were used to predict the various toxicity endpoints of the compounds. The ProTox-II server determines the acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, immunotoxicity of the selected 4 compounds, and classified the compounds in different classes based on the server predicted LD50 (Table 4 ). Compound Amb6600091, Amb17613565, Amb3940754, Amb21855906 were classified in classes 6, 4, 5, and 4, respectively based on their LD50 shown in Table 4.
Table 4.
Toxicity properties like organ toxicity, toxicity endpoints, 96-hour fathead minnow LC50, 48-hour D. magna LC50, 48-hour T. pyriformis IGC50, oral rat LD50, and bioaccumulation factor of selected 4 compounds.
| Endpoint | Target | Amb6600091 | Amb17613565 | Amb3940754 | Amb21855906 |
|---|---|---|---|---|---|
| Organ toxicity | Hepatotoxicity | Inactive | Inactive | Inactive | Inactive |
| Toxicity Endpoints | Carcinogenicity | Inactive | Inactive | Inactive | Inactive |
| Immunotoxicity | Inactive | Inactive | Inactive | Inactive | |
| Mutagenicity | Inactive | active | Inactive | Inactive | |
| Cytotoxicity | Inactive | Inactive | Inactive | Inactive | |
| LD50 (mg/kg) | 5500 | 1460 | 2400 | 2000 | |
| Toxicity class | 6 | 4 | 5 | 4 | |
| 96-hour fathead minnow LC50 | mg/L | 103.64 | 57.05 | 141.40 | 758.01 |
| 48-hour D. magna LC50 | mg/L | 464.57 | 6.26 | 31.02 | 19.6 |
| 48-hour T. pyriformis IGC50 | mg/L | 389.25 | 410.07 | 572.19 | 212.98 |
| Oral rat LD50 | mg/kg | 1768.26 | 3205.31 | 1631.99 | 1747.49 |
| Bioaccumulation factor | Log10 | -1.38 | -0.48 | 0.09 | -1.49 |
For the compound Amb6600091, the LD50 was found 5500 mg/kg, which is classified as non-toxic (LD50 > 5000) in the ProTox-II server. Amb17613565 (LD50; 1460 mg/kg) and Amb21855906 (LD50; 2000 mg/kg) were both in class 4 indicated as harmful if swallowed (300 < LD50 ≤ 2000), where compound Amb3940754 (LD50; 2400 mg/kg) found in class 5 may be harmful if swallowed (2000 < LD50 ≤ 5000). The acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, immunotoxicity of all compounds found inactive except the compound Amb17613565, which mutagenicity found to be active.
The 96-hour fathead minnow LC50 value, 48-hour D. magna LC50, 48-hour T. pyriformis IGC50, oral rat LD50, indicate the concentration of the chemical compounds in water (mg/L) responsible for 50% of fathead minnow, D. magna and T. pyriformis to die after 96, 48 and 48 h, respectively, where oral rat LD50 indicate the number of chemical compounds (mg/kg body weight) that causes 50% of rats to die after oral ingestion were predicted using the T.E.S.T. software listed in Table 4. To estimate the value and average of the predicted toxicities from all the QSAR methods mentioned in the T.E.S.T. tools called consensus method was applied in this study.
3.11. Molecular dynamics simulation
Molecular dynamics (MD) simulation was performed to understand the dynamic behavior of our compounds within the protein. The MD simulation also determined the effect of explicit solvent molecules on the ACE2 protein and their fluctuations and conformational changes to obtain time-averaged features of the complex system in different timescales [44]. In this study, the results of MD simulation were analyzed based on three major physical properties comprising root mean square deviation (RMSD), root mean square fluctuation (RMSF), and protein-ligands contact mapping of the compounds in a specific time.
3.12. RMSD analysis
The protein-ligand complex structure variation generated from molecular docking was predicted by RMSD values obtained from 0 to 250 ns simulation run. The success in molecular dynamics simulation can be evaluated based on RMSD value when the RMSD value within an arbitrary threshold rang 0.3 nm or 3 Å the true pose is considered as a hit [50]. The RMSD of the complex structure indicated the stability and deviation in the average distance of Cα-atoms as a function of simulation time [75]. The average RMSD values of Amb3940754, Amb6600091, Amb17613565, and Amb21855906 were found 2.35 Å, 2.75 Å, 2.5 Å, and 2.45 Å, respectively compared to control compound XX5 in complex with ACE2 protein (Fig. 9 ). The compound Amb6600091 showed maximum fluctuations from 200 ns to 235 ns and smoothly turned into a stable phase after the 240 ns run. The RMSD value obtained from the MD simulation showed minimal fluctuation throughout the 250 ns run and remaining stable until the end of the simulation, where higher fluctuations have observed only at the starting points during the simulation run of the complex structure.
Fig. 9.
Depicted the RMSD values extracted from the Cα atoms of the selected four compounds in complex with the ACE2 protein. Herein, showing the RMSD of reference ACE2-XX5 (blue) protein complex with the compounds (A) Amb3940754 (orange), (B) Amb6600091 (gray), and (C) Amb17613565 (yellow), where (D) Amb21855906 (red) of the compounds. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.13. RMSF analysis
The RMSF is a measure of the deviation between the position of residues contributing to protein structure and the binding site residues of the complex structure. The AA residues with low or no RMSF values are considered more stable because of their limited movement capabilities during the MD simulations [76]. The change of RMSF (ΔRMSF) value within an arbitrary threshold range of >3 Å is considered tangible and an important change in AA residue-specific flexibility [50]. The RMSF graph was calculated for ACE2 in complex with XX5 compound from 0 to 570 AA residue of Cα-atoms along with four natural compounds as potential drug candidates. The overall complex ACE2 structure seen roughly in RMSF plots exhibited an advanced fluctuation level over the 250 ns timeframe. The RMSF graph demonstrated averaged low and significant values of the ACE2-Amb3940754 complex (2-2.3 Å), ACE2-Amb6600091 complex (2-2.1 Å), ACE2-Amb17613565 complex (2- 2.2 Å), and ACE2-Amb21855906 complex (1.9-2.1 Å) compare to the reference compound XX5, commencing the natural compounds were closely bound to ACE2 protein concerning their average positions shown in Fig. 10 .
Fig. 10.
Depicted the RMSF values extracted from the Cα atoms of the selected four complex structures. Herein, showing the RMSF of ACE2-XX5 protein (blue) in complex with the compounds (A) Amb3940754 (orange), (B) Amb6600091 (gray), (C) Amb17613565 (yellow), and (D) Amb21855906 (red) of the compounds. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.14. Protein-ligand contact mapping
Protein interactions with the selected four compounds Amb3940754, Amb6600091, Amb17613565, and Amb21855906 have been monitored throughout the simulation interaction diagram (SID) of the Schrödinger (Release 2020-3). The hydrogen bonds, hydrophobic, ionic, and water bridge interactions found during the interaction analysis have been shown in the stacked bar charts (Fig. 11 ). Different types of bonding play an important role in stable binding to the targeted protein, where hydrogen bonding helps to determine influence drug specificity, metabolization, and adsorption. The hydrogen bonding interaction found for all the four compounds was observable until the last AA residue of the protein. Hydrogen bonds and their relative strength in aqueous ionic solutions at ambient conditions are necessary to initiate the protein-ligand binding interaction [51], [77]. A hydrogen bond is a weak type of dipole-dipole traction between molecules forms when a strongly electronegative atom in H-bond acceptor exists in the vicinity of another electronegative atom with a lone pair of electrons known H-bond donor [78]. In addition, other bonding interactions like hydrophobic, ionic, and water bridges bond at the same residue position of the protein. In this study, intermolecular hydrogen bonding interactions and other bonding interactions like hydrophobic, ionic, and water bridges bonds of the protein-ligands complex were determined and depicted in Fig. 11. During the MD simulation, the ACE2-Amb17613565 complex, ACE2-Amb6600091 complex, ACE2-Amb3940754 complex, and ACE2-Amb21855906 complex provided the highest number of hydrogen-bonding interaction for all the four protein-ligand complexes until the last residue of the simulation run. The analysis of the number of H-bonds formed in the ACE2-Ligands complexes indicating the improved stability of ligands to the binding site of the protein.
Fig. 11.
The stacked bar charts showing the protein-ligands interactions found during the 250 ns simulation run. Herein, showing the selected four compounds (A) Amb3940754, (B) Amb6600091, (C) Amb17613565, and (D) Amb21855906 contact mapping with ACE2 protein during the 250 ns simulation time.
3.15. MM/GBSA analysis
MM/GBSA methods have been used in this study to estimate the ligand-binding free energy to the desired protein. The MM/GBSA of the protein-ligand complex structure has been calculated from the few snapshots (∼200) of the MD simulations trajectory. The analysis of the complex structure found higher net negative binding free energy values -38.47 kcal/mol, -33.75 kcal/mol, -32.54 kcal/mol, and -35.18 kcal/mol for the selected four compounds Amb3940754, Amb6600091, Amb17613565, and Amb21855906, respectively with the targeted protein (Fig. 12 ). Additionally, physico-chemical components analysis of the four compounds identified a significant contribution in the GBind Coulomb (Coulomb energy) and GBind vdW (Van der Waals interaction energy) shown in Fig. 12. Therefore, it can be considered that the selected compounds will be able to maintain a long-term interaction with the desired ACE2 protein and result in inhibition of the desired protein.
Fig. 12.
Representing different energy components and net MM/GBSA binding free energy (kcal/mol) and standard deviation values generated from extracted snapshots of ACE2 protein in complex with selected compounds, i.e., (A) Amb3940754, (B) Amb6600091, (C) Amb17613565, and (D) Amb21855906.
4. Discussion
SARS-CoV-2 is a recently originated pandemic virus, against which vaccines and new drugs are constantly being tested [75], [76]. For thousands of years, natural products and their derivatives isolated from various sources have been demonstrated to be effective therapeutic agents, and thus play an important role in treating diverse infectious diseases [34], [79]. The chemical structure and extensive biological activities of these compounds vary comprehensively, that's why natural compounds incessantly offer inspiration to innovations in drug discovery and medical sciences [35]. Therefore, we aim to identify potential natural ACE2 inhibitors through computational approaches such as pharmacophore modeling, virtual screening, molecular docking, ADMET, and MD simulation to overcome the present demonic situation originated through the SARS-CoV-2.
Initially, a validated SBPM was applied to the virtual screening of 11,295 natural compounds that retrieved 23 similar scaffolds as hits with a maximum fit value of 78.81. The filtered compounds, which contain all the chemical features attendant in the SBPM, were retrieved for molecular docking simulation to avoid false-positive hits generated from the structure-based pharmacophore screening. Molecular docking simulation was performed to observe the complex structure of the small natural compounds with ACE2, calculate the binding energy of the complex interaction, and finding the best geometrical arrangements. The best four compounds with binding affinity range between −7.5 and −7.0 kcal/mol have been chosen for further evaluation.
Hereafter these four compounds have been submitted for in silico ADME, where ADME properties like lipophilicity, water-solubility, drug-like effectiveness, pharmacokinetic and physicochemical properties were evaluated and found optimum. After that, the in-silico toxicity properties like acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, immunotoxicity along with LD50 score were also evaluated. Based on evaluation we found that all four compounds were nontoxic to the host. The MD simulation was performed on the selected four compounds and demonstrated good stability and affinity to the protein binding site.
Then we performed a molecular dynamics simulations study for investigating the stability of these four compounds with ACE2 protein. Because if the ligands do not dorm stable interaction with protein, then the inhibition of protein may hinder. By using the Desmond module of Schrödinger, we run the MD simulation for 250 ns for the selected four natural compounds. Here we observed the RMSD, RMSF, and protein-ligand contact of the complex system. The RMSD, RMSF, and protein-ligand contact study found for all the selected compounds showed enhance stability and optimized fluctuations with the ACE2 proteins.
5. Conclusions
Structure-based drug design is becoming an essential, efficient, and exterior approach to identify inhibitory compounds against a specific target protein. In this study, we describe the quick and successful identification of novel natural ACE2 inhibitors by a computer-aided drug design approach. The CADD approaches include pharmacophore modeling, virtual screening, molecular docking, ADMET, and MD simulation, which identified four natural compounds Amb17613565, Amb6600091, Amb3940754, and Amb21855906 can potentially inhibit the activity of ACE2 and resulting blocking the entry of SARS-CoV-2 into the human host cell.
Funding
This research received no external funding.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
CRediT authorship contribution statement
FA, AS, MSR designed the project; TAB, AS, FA, MMIT, and ZN performed the experiments; FA, SP, TAB, and AS evaluated and interpreted the data; SP, TAB, FA, SMN, MA-Al-M, RA, RRI, MEK, MSH, MMIT, IQ, TMK, and MSR prepared the draft manuscript; FA, AS, ZN, and MSH finalized the manuscript. All authors approved the final version of the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We thank the Deanship of Scientific Research (DSR) at King Abdulaziz University and Biological Solution Centre (BioSol Centre) for their technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2021.09.146.
Appendix A. Supplementary data
Supplementary material
References
- 1.Mackenzie J.S., Smith D.W. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol. Aust. 2020;41:45. doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-NCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahanta S., Chowdhury P., Gogoi N., Goswami N., Borah D., Kumar R., Chetia D., Borah P., Buragohain A.K., Gogoi B. Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: an in silico based approach. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1768902. [DOI] [PubMed] [Google Scholar]
- 5.Yen M.Y., Schwartz J., Chen S.Y., King C.C., Yang G.Y., Hsueh P.R. Interrupting COVID-19 transmission by implementing enhanced traffic control bundling: implications for global prevention and control efforts. J. Microbiol. Immunol. Infect. 2020;53:377–380. doi: 10.1016/j.jmii.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worldometers.info Coronavirus Update (Live) https://www.worldometers.info/coronavirus/
- 7.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robson B. COVID-19 coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed Achilles' heel conserved region to minimize probability of escape mutations and drug resistance. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahammad F., Abd Rashid T.R.T.T.R.T., Mohamed M., Tanbin S., Fuad F.A.A.F.A.A. Contemporary strategies and current trends in designing antiviral drugs against dengue fever via targeting host-based approaches. Microorganisms. 2019;7:296. doi: 10.3390/microorganisms7090296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng S.-Q., Peng H.-J. Characteristics of and public health responses to the coronavirus disease 2019 outbreak in China. J. Clin. Med. 2020;9:575. doi: 10.3390/jcm9020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L., Huang T., Wang Y., Wang Z., Liang Y., Huang T., Zhang H., Sun W., Wang Y. 2019 novel coronavirus patients’ clinical characteristics, discharge rate and fatality rate of meta-analysis. J. Med. Virol. 2020;92:577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sah R., Rodriguez-Morales A.J., Jha R., Chu D.K.W., Gu H., Peiris M., Bastola A., Lal B.K., Ojha H.C., Rabaan A.A., Zambrano L.I., Costello A., Morita K., Pandey B.D., Poon L.L.M. 2020. Complete Genome Sequence of a 2019 Novel Coronavirus (SARS-CoV-2) Strain Isolated in Nepal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-NCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X.-G., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., Moore M.J., Vasllieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greeneugh T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rella M., Rushworth C.A., Guy J.L., Turner A.J., Langer T., Jackson R.M. Proceedings of the Journal of Chemical Information and Modeling. vol. 46. 2006. Structure-based pharmacophore design and virtual screening for novel angiotensin converting enzyme 2 inhibitors; pp. 708–716. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Li W., Farzan M., Harrison S.C. Structural biology: structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 22.Liu X., Li Z., Liu S., Sun J., Chen Z., Jiang M., Zhang Q., Wei Y., Wang X., Huang Y.-Y., Shi Y., Xu Y., Xian H., Bai F., Ou C., Xiong B., Lew A.M., Cui J., Fang R., Huang H., Zhao J., Hong X., Zhang Y., Zhou F., Luo H.-B. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm. Sin. B. 2020;10:1205–1215. doi: 10.1016/j.apsb.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong N., Yang X., Ye L., Chen K., Chan E.W.-C., Yang M., Chen S. Genomic and protein structure modelling analysis depicts the origin and pathogenicity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. F1000Research. 2020;9:121. [Google Scholar]
- 24.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian Z., Ou X., Góes L.G.B., Osborne C., Castano A., Holmes K.V., Dominguez S.R. Identification of the receptor-binding domain of the spike glycoprotein of human betacoronavirus HKU1. J. Virol. 2015;89:8816–8827. doi: 10.1128/JVI.03737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-NCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner A.J., Hooper N.M. Handbook of Proteolytic Enzymes. Second edition. vol. 1. Elsevier Inc.; 2004. Angiotensin-converting enzyme 2; pp. 349–352. [Google Scholar]
- 32.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman S.M.M., Atikullah M., Islam M.N.S., Mohaimenul M., Ahammad F., Islam M.N.S., Saha B., Rahman M.H. Anti-inflammatory, antinociceptive and antidiarrhoeal activities of methanol and ethyl acetate extract of hemigraphis alternata leaves in mice. Clin. Phytosci. 2019;5:16. [Google Scholar]
- 35.B.B. M., V.K. T. Natural products: an evolving role in future drug discovery. Eur. J. Med. Chem. 2011;46:4769–4807. doi: 10.1016/j.ejmech.2011.07.057. [DOI] [PubMed] [Google Scholar]
- 36.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 37.Szymański P., Markowicz M., Mikiciuk-Olasik E. Adaptation of high-throughput screening in drug discovery-toxicological screening tests. Int. J. Mol. Sci. 2012;13:427–452. doi: 10.3390/ijms13010427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahammad F., Fuad F.A.A. Proceedings of the Proceedings of 5th International Electronic Conference on Medicinal Chemistry. MDPI; Basel, Switzerland: 2020. The in silico identification of potent natural bioactive anti-dengue agents by targeting the human hexokinase 2 enzyme; p. 6342. [Google Scholar]
- 39.Zhou W., Wang Y., Lu A., Zhang G. Systems pharmacology in small molecular drug discovery. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes J.P., Rees S.S., Kalindjian S.B., Philpott K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang P.H., Cheng W.C., Lee Y.L., Yu H.P., Wu Y.T., Lin Y.L., Wong C.H. Novel five-membered iminocyclitol derivatives as selective and potent glycosidase inhibitors: new structures for antivirals and osteoarthritis. ChemBioChem. 2006;7:165–173. doi: 10.1002/cbic.200500321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samad A., Haque F., Nain Z., Alam R., Al Noman M.A., Rahman Molla M.H., Hossen M.S., Islam M.R., Khan M.I., Ahammad F. Computational assessment of MCM2 transcriptional expression and identification of the prognostic biomarker for human breast cancer. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabassum A., Samdani M.N., Dhali T.C., Alam R., Ahammad F., Samad A., Karpiński T.M. Transporter associated with antigen processing 1 (TAP1) expression and prognostic analysis in breast, lung, liver, and ovarian cancer. J. Mol. Med. 2021;99:1293–1309. doi: 10.1007/s00109-021-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wichapong K., Nueangaudom A., Pianwanit S., Sippl W., Kokpol S. Identification of potential hit compounds for dengue virus NS2B/NS3 protease inhibitors by combining virtual screening and binding free energy calculations. Trop. Biomed. 2013;30:388–408. [PubMed] [Google Scholar]
- 45.Wu J., Hu B., Sun X., Wang H., Huang Y., Zhang Y., Liu M., Liu Y., Zhao Y., Wang J., Yu Z. In silico study reveals existing drugs as α-glucosidase inhibitors: structure-based virtual screening validated by experimental investigation. J. Mol. Struct. 2020;1218 [Google Scholar]
- 46.Rafi J.H., Jafar T., Pathan M.T., Reza R., Islam S., Sourna I.J., Alam R., Samad A., Ahammad F. High expression of bone morphogenetic protein 1 (BMP1) is associated with a poor survival rate in human gastric cancer, a dataset approaches. Genomics. 2021;113:1141–1154. doi: 10.1016/j.ygeno.2020.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Rahman F., Mahmood T.Bin, Amin A., Alam R., Jharna J.F., Samad A., Ahammad F. A multi-omics approach to reveal the key evidence of GDF10 as a novel therapeutic biomarker for breast cancer. Informatics Med. Unlocked. 2020;21 [Google Scholar]
- 48.Wolber G., Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 49.Seidel T., Ibis G., Bendix F., Wolber G. Strategies for 3D pharmacophore-based virtual screening. Drug Discov. Today Technol. 2010;7:e221–e228. doi: 10.1016/j.ddtec.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Jana S., Ganeshpurkar A., Singh S.K. Multiple 3D-QSAR modeling, e-pharmacophore, molecular docking, and in vitro study to explore novel AChE inhibitors. RSC Adv. 2018;8:39477–39495. doi: 10.1039/c8ra08198k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu S.H., Wu J.W., Liu H.L., Zhao J.H., Liu K.T., Chuang C.K., Lin H.Y., Tsai W.B., Ho Y. The discovery of potential acetylcholinesterase inhibitors: a combination of pharmacophore modeling, virtual screening, and molecular docking studies. J. Biomed. Sci. 2011;18:1–13. doi: 10.1186/1423-0127-18-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.T. Seidel S. Bryant G. Ibis … G.P.-T. 2017, undefined. 3D Pharmacophore Modeling Techniques in Computer-Aided Molecular Design Using LigandScout. books.google.com.
- 53.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 54.Ahammad F., Alam R., Mahmud R., Akhter S., Talukder E.K., Tonmoy A.M., Fahim S., Al-Ghamdi K., Samad A., Qadri I. Pharmacoinformatics and molecular dynamics simulation-based phytochemical screening of neem plant (Azadiractha Indica) against human cancer by targeting MCM7 protein. Brief. Bioinform. 2021;2021:1–15. doi: 10.1093/bib/bbab098. [DOI] [PubMed] [Google Scholar]
- 55.Schrödinger, Release, S. (2015). 2 (2015) LigPrep, Version 3.4, 2015.
- 56.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 58.Shaker B., Yu M.S., Lee J., Lee Y., Jung C., Na D. User guide for the discovery of potential drugs via protein structure prediction and ligand docking simulation. J. Microbiol. 2020;58:235–244. doi: 10.1007/s12275-020-9563-z. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Meng Q., Yang M., Liu D., Hou X., Tang L., Wang X., Lyu Y., Chen X., Liu K., Yu A.M., Zuo Z., Bi H. Current trends in drug metabolism and pharmacokinetics. Acta Pharm. Sin. B. 2019;9:1113–1144. doi: 10.1016/j.apsb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eddershaw P.J., Beresford A.P., Bayliss M.K. ADME/PK as part of a rational approach to drug discovery. Drug Discov. Today. 2000;5:409–414. doi: 10.1016/s1359-6446(00)01540-3. [DOI] [PubMed] [Google Scholar]
- 61.Zhuang X., Lu C. PBPK modeling and simulation in drug research and development. Acta Pharm. Sin. B. 2016;6:430–440. doi: 10.1016/j.apsb.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li C., Wang J., Wang Y., Gao H., Wei G., Huang Y., Yu H., Gan Y., Wang Y., Mei L., Chen H., Hu H., Zhang Z., Jin Y. Recent progress in drug delivery. Acta Pharm. Sin. B. 2019;9:1145–1162. doi: 10.1016/j.apsb.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Segall M.D., Barber C. Addressing toxicity risk when designing and selecting compounds in early drug discovery. Drug Discov. Today. 2014;19:688–693. doi: 10.1016/j.drudis.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 65.Martin T. 2016. User's Guide for T.E.S.T. (Version 4.2) (Toxicity Estimation Software Tool): A Program to Estimate Toxicity from Molecular Structure. [Google Scholar]
- 66.Aljahdali M.O., Molla M.H.R., Ahammad F. Compounds identified from marine mangrove plant (Avicennia Alba) as potential antiviral drug candidates against WDSV, an in-silico approach. Mar. Drugs. 2021;19:253. doi: 10.3390/md19050253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coelho C.M., Dos Santos T., Freitas P.G., Nunes J.B., Marques M.J., Padovani C.G.D., Júdice W.A.S., Camps I., Da Silveira N.J.F., Carvalho D.T., Veloso M.P. Design, synthesis, biological evaluation and molecular modeling studies of novel eugenol esters as leishmanicidal agents. J. Braz. Chem. Soc. 2018;29:715–728. [Google Scholar]
- 68.Opo F.A.D.M., Rahman M.M., Ahammad F., Ahmed I., Bhuiyan M.A., Asiri A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci. Rep. 2021;11:4049. doi: 10.1038/s41598-021-83626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bharadwaj S., Dubey A., Yadava U., Mishra S.K., Kang S.G., Dwivedi V.D. Exploration of natural compounds with anti-SARS-CoV-2 activity via inhibition of SARS-CoV-2 mpro. Brief. Bioinform. 2021;2021:1–17. doi: 10.1093/bib/bbaa382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yadav D.K., Khan F., Negi A.S. Pharmacophore modeling, molecular docking, QSAR, and in silico ADMET studies of gallic acid derivatives for immunomodulatory activity. J. Mol. Model. 2012;18:2513–2525. doi: 10.1007/s00894-011-1265-3. [DOI] [PubMed] [Google Scholar]
- 74.Toropov A.A., Toropova A.P., Raska I., Leszczynska D., Leszczynski J. Comprehension of drug toxicity: software and databases. Comput. Biol. Med. 2014;45:20–25. doi: 10.1016/j.compbiomed.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 75.Cruz J.N., Costa J.F.S., Khayat A.S., Kuca K., Barros C.A.L., Neto A.M.J.C. Molecular dynamics simulation and binding free energy studies of novel leads belonging to the benzofuran class inhibitors of mycobacterium tuberculosis polyketide synthase 13. J. Biomol. Struct. Dyn. 2019;37:1616–1627. doi: 10.1080/07391102.2018.1462734. [DOI] [PubMed] [Google Scholar]
- 76.Fang J., Wu P., Yang R., Gao L., Li C., Wang D., Wu S., Liu A.L., Du G.H. Inhibition of acetylcholinesterase by two genistein derivatives: kinetic analysis, molecular docking and molecular dynamics simulation. Acta Pharm. Sin. B. 2014;4:430–437. doi: 10.1016/j.apsb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anuar N.F.S.K., Wahab R.A., Huyop F., Amran S.I., Hamid A.A.A., Halim K.B.A., Hood M.H.M. Molecular docking and molecular dynamics simulations of a mutant acinetobacter haemolyticus alkaline-stable lipase against tributyrin. J. Biomol. Struct. Dyn. 2020;39:2079–2091. doi: 10.1080/07391102.2020.1743364. [DOI] [PubMed] [Google Scholar]
- 78.Sindhu T., Srinivasan P. Exploring the binding properties of agonists interacting with human TGR5 using structural modeling, molecular docking and dynamics simulations. RSC Adv. 2015;5:14202–14213. [Google Scholar]
- 79.Bhuiyan M.A., Quayum S.T., Ahammad F., Alam R., Samad A., Nain Z. Discovery of potential immune epitopes and peptide vaccine design - a prophylactic strategy against Rift Valley fever virus. F1000Res. 2020;9:999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material