Abstract
The fluorescent amplified-fragment length polymorphism (FAFLP) assay potentially amplifies a unique set of genome fragments from each bacterial clone. It uses stringently hybridizing primers which carry a fluorescent label. Precise fragment sizing is achieved by the inclusion of an internal size standard in every lane. Therefore, a unique genotype identifier(s) can be found in the form of fragments of precise size or sizes, and these can be generated reproducibly. In order to evaluate the potential of FAFLP as an epidemiological typing method with a valid phylogenetic basis, we applied it to 87 strains of Escherichia coli. These comprised the EcoR collection, which has previously been classified by multilocus enzyme electrophoresis (MLEE) and which represents the genetic diversity of the species E. coli, plus 15 strains of the clinically important serogroup O157. FAFLP with an unlabelled nonselective EcoRI primer (Eco+0) and a labelled selective MseI primer (Mse+TA) gave strain-specific profiles. Fragments of identical sizes (in base pairs) were assumed to be identical, and the genetic distances between the strains were calculated. A phylogenetic tree derived from measure of distance correlated closely with the MLEE groupings of the EcoR collection and placed the verocytotoxin-producing O157 strains on an outlier branch. Our data indicate that FAFLP is suitable for epidemiological investigation of E. coli infection, providing well-defined and reproducible identifiers of genotype for each strain. Since FAFLP objectively samples the whole genome, each strain or isolate can be assigned a place within the broad context of the whole species and can also be subjected to a high-resolution comparison with closely related strains to investigate epidemiological clonality.
To determine the relatedness between bacterial isolates, epidemiological investigations of outbreaks of infection require each isolate to be assigned a unique genotype identifier. This identifier may be a fragment or group of fragments of unique size or sizes. Strain characterization therefore places a premium on precise and reproducible ways of comparing the genomes of bacterial isolates. Many methods have been used for bacterial genotyping; they include ribotyping (9), pulsed-field gel electrophoresis (PFGE) (1), and rapid PCR-based methods such as arbitrary primed PCR or randomly amplified polymorphic DNA analysis (21, 26) or PCR-restriction fragment length polymorphism analysis. Although ribotyping and PFGE provide a firm basis for typing various bacterial pathogens (8, 10, 23), their levels of precision and discriminatory power could be improved. Randomly amplified polymorphic DNA analysis and arbitrary primed PCR are useful for preliminary intralaboratory comparisons of isolates, but they are insufficiently robust or reproducible for genotyping or interlaboratory comparisons (17). PCR-restriction fragment length polymorphism analysis can be applied only to small regions of polymorphism, such as single genes.
Amplified-fragment length polymorphism (AFLP) analysis (25) selectively amplifies by PCR a subset of restriction fragments from a digest of whole genomic DNA. In its radioactively labelled format, AFLP has been shown to generate specific profiles for small numbers of strains of Clostridium, Bacillus, Acinetobacter, Vibrio, Aeromonas, Pseudomonas, and Xanthomonas species (6, 11–15).
In the fluorescent AFLP (FAFLP) assay a 6-base restriction endonuclease (such as EcoRI) and a 4-base endonuclease (such as MseI) together digest the bacterial genomic DNA, creating fragments of a size suitable for resolution on polyacrylamide (sequencing) gels. Double-stranded linkers specific to each restriction site are ligated to the cohesive ends, generating templates for amplification when the sequences of linkers and restriction sites serve as primer-binding sites. The primer specific for the 6-base cutter enzyme site is fluorescently labelled, and only fragments cut with that enzyme will be visible to laser detection. A differentially labelled internal size standard allows precise sizing of the fragments, certain of which act as a unique identifier(s) of the genotype.
We have previously reported empirical conditions for FAFLP analysis and their use for a molecular epidemiological investigation of outbreaks caused by the gram-positive bacterium Streptococcus pyogenes (5). In the case of Escherichia coli, however, the complete genome sequence is available for strain K-12, and we have been able to use it to predict DNA fragments that would be generated by FAFLP analysis. This has enabled us to make experimental comparisons between isolates. We have designed an FAFLP method for E. coli that gives the most informative number of fragments and that gives the optimum distribution of the fragments on a laser-read sequencing gel. We now present this as a predictive FAFLP system modeled on the published sequence for strain K-12, and we evaluate its ability to type the 72 members of the EcoR reference collection, a genetically diverse group defined by multilocus enzyme electrophoresis (MLEE) (22), as well as 15 serogroup O157 isolates.
MATERIALS AND METHODS
EcoR collection.
The EcoR collection is a set of 72 strains from humans and 16 other mammalian species selected to be broadly representative of the enzyme genotypic diversity in E. coli as a whole (19). Members of the collection were defined by electrophoretic analysis of 35 enzymes (22).
O157 strains.
Fifteen strains of E. coli serogroup O157 included 11 isolates that produced both VT1 and VT2 or VT2 alone. They possessed either the flagellar antigen H7 (serotype O157:H7) or were nonmotile (O157:H−). One further strain of O157:H7 (NCTC12900) was a naturally occurring verocytotoxin (VT)-negative derivative of VT-producing E. coli (VTEC) O157. The 15 O157 strains described above belonged to eight phage types and were obtained from epidemiologically unlinked outbreaks or incidents between 1995 and 1997; some of the strains have been described previously (24, 28). The remaining three strains were biochemically distinct from the O157 VTEC group and were of serotypes O157:H8, O157:H19, and O157:H42 (27). They were from humans with infections, cattle, and beef, respectively. All of these strains were VT negative.
Computer methods.
The complete genome sequence of E. coli MG1655 (accession nos. ECAE000111 to ECAE000510) was analyzed with Lasergene (DNAStar, Madison, Wis.) and MacVector (Oxford Molecular, Oxford, United Kingdom). Data concerning the size and number of fragments predicted following an MseI-EcoRI digest of the genome were imported into a spreadsheet. The fragment size data were then adjusted to allow for the addition of primers during PCR, and those fragments predicted to be amplified with each of the chosen selective primers were identified.
FAFLP.
The DNAs of the O157 strains were prepared by the method of Ausubel et al. (2). DNAs from strains in the EcoR collection were extracted from colonies on plate cultures (3), and 500 ng was digested in a total volume of 22 μl consisting of 5 U of MseI (New England Biolabs), 2 μl of 10× MseI buffer, 0.2 μl of 10× bovine serum albumin, and 1.0 μl of DNase-free RNase A (10 μg/μl) for 1 h at 37°C. To this digest was added 5 U (1.0 μl) of EcoRI (Life Technologies), 1.68 μl of 0.5 M Tris-HCl (pH 7.6), and 2.1 μl of 0.5 M NaCl (total volume, 26 μl), and the reaction mixtures were incubated for a further hour at 37°C. Endonucleases were inactivated at 65°C for 10 min prior to ligation.
To the double-digested DNA was added 25 μl of a solution containing 40 U of T4 DNA ligase (New England Biolabs), 5 pmol of EcoRI adaptor, 50 pmol of MseI adaptor, and 5 μl of 10× T4 ligase buffer. The reaction mixture was incubated at 12°C for 17 h, heated at 65°C for 10 min to inactivate the ligase, and stored at −20°C.
The nonselective forward primer for the EcoRI adaptor site was labelled with the blue fluorescent dye 5-carboxyfluorescein (Genosys Biotechnologies). The reverse primer for the MseI adaptor site, which contained the selective bases T and A, was obtained from an AFLP kit (PE Biosystems, Foster City, Calif.). PCRs were performed in 25-μl volumes containing 2.5 μl of ligated DNA, 16.6 pmol of labelled EcoRI primer, 100 pmol of MseI primer, 2.5 μl of 10× Taq polymerase buffer, each of the four deoxynucleoside triphosphates at a concentration of 10 mM, 1.0 μl of 100× bovine serum albumin (New England Biolabs), 1.5 mM MgCl2, and 0.625 U of Taq DNA polymerase. To minimize PCR artifacts, “touchdown” PCR was performed as follows: a 2-min denaturation step at 94°C (one cycle), followed by 30 cycles of denaturation at 94°C for 20 s, a 30-s annealing step (see below), and a 2-min extension step at 72°C. The annealing temperature for the first cycle was 66°C; for the next nine cycles, the temperature was decreased by 1°C at each cycle. The annealing temperature for the remaining 20 cycles was 56°C. This was followed by a final extension at 60°C for 30 min. PCR was performed in a PE-9600 thermocycler (Perkin-Elmer Corp., Norwalk, Conn.). The amplification products were stored at −20°C.
Gel analysis.
The amplification products were separated on a 5% denaturing (sequencing) polyacrylamide gel on an ABI Prism 377 DNA automated sequencer (Perkin-Elmer Corp.). The gel was prepared by using 5% acrylamide (Amresco and FMC LongRanger) and 6.0 M urea in 1× TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA). To 50 ml of the gel solution was added 250 μl of 10% ammonium persulfate and 35 μl of N,N,N′,N′-tetramethylethylenediamine (TEMED; Amresco). Spacers and sharks-tooth combs were 0.2 mm in thickness. Gels were poured with a PE Biosystems 377 casting frame and gel pourer and were allowed to polymerize at room temperature for at least 2 h. The sample (1.5 μl) was added to 1.5 μl of loading dye, which was a mixture containing 1.25 μl of formamide and 0.25 μl of blue dextran–50 mM EDTA loading solution, and to 0.5 μl of the internal lane standard, Genescan 2500, labelled with the red fluorophore ROX (PE Biosystems). The sample mixture was heated at 95°C for 2 min, cooled on ice, and immediately loaded onto the gel. Electrophoresis conditions were 2.5 kV, 51°C, and 7 h, with 1× TBE used as the buffer.
Data capture and analysis.
Genescan collection software (PE Biosystems) was used to automatically size and quantify individual fragments by using the internal lane standards. Results were viewed in the form of a gel image, an electropherogram, tabular data, or a combination of all three. Genotyper software (PE Biosystems) automatically interpreted the Genescan data after the analysis parameters were set to medium smoothing and the baseline fluorescence was set to 150 units. The presence or absence of precisely sized fragments was ascertained, and these digital data were transferred to spreadsheets for further analysis. Pairwise comparisons were made between all strains with the coefficient of Nei and Li (18) since this method does not infer direction or weight of DNA change, i.e., the acquisition or loss of a restriction site which would change an FAFLP profile. The distance matrix thus generated was used as input for the Fitch tree-building program in PHYLIP (7).
RESULTS
Predictive modeling of FAFLP and experimental evaluation.
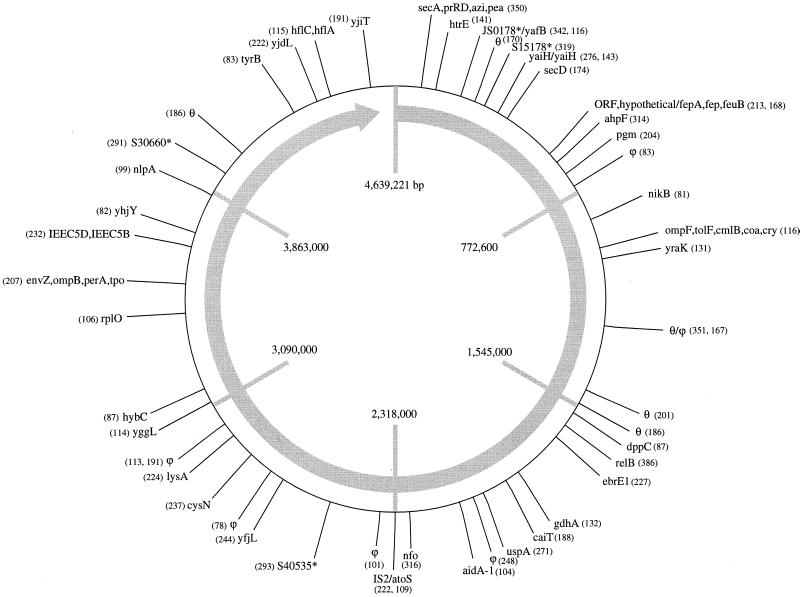
The sizes of the predicted fragments and their locations in the E. coli K-12 MG1655 genome are shown in Fig. 1. To determine the accuracy and reproducibility of the FAFLP assay, reactions with the primer pair MseI+TA–EcoRI+0 were performed three times with the same DNA extract, and the experimental data were compared with the predicted values. All 48 predicted fragments were observed (100%); 46 were within 1 bp of their predicted size, and 2 were within 2 bp of their predicted size. In one experiment, 48 of 48 (100%) predicted fragments were observed, in the second experiment 47 of 48 (98%) predicted fragments were observed, and in the third experiment 44 of 48 (92%) predicted fragments were observed. Five unpredicted fragments were recorded (one to three experiments each). For these three experiments, the mean accuracy of the FAFLP assay, calculated by averaging the percentage of predicted fragments occurring, is therefore 97%.
FIG. 1.
Sizes (in base pairs) and approximate locations of FAFLP assay-predicted fragments in the E. coli K-12 MG1655 genome obtained with primer set MseI+TA–EcoRI+0. The genes that the fragments are predicted to have been amplified from are also indicated. θ, an uncharacterized locus in E. coli bearing sequence similarity to characterized loci in other bacteria; ϕ, no matches were found, indicating a noncoding region; ∗, hypothetical, as yet uncharacterized genes.
Interstrain typing of the EcoR collection by the FAFLP assay.
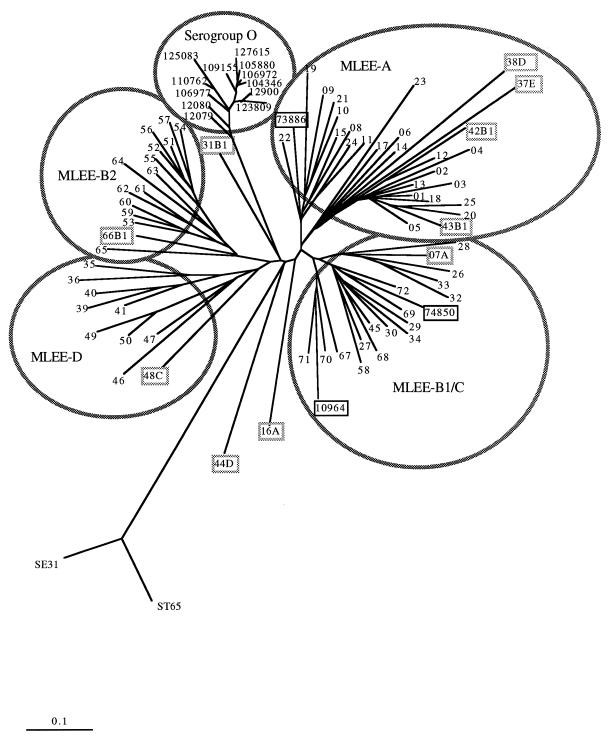
No strains from the EcoR collection gave the same profile by the FAFLP assay. An average of 46 fragments of between 100 and 500 bp were generated by the FAFLP assay. For each strain the sized fragments were scored on a spreadsheet as present or absent. A program that uses the coefficient of Nei and Li (18) was used to calculate pairwise similarities between the strains from a spreadsheet, creating an 89-by-89 lower triangular distance matrix of pairwise comparisons. This was used as input for the Fitch tree-building programs, and that output was then used for Drawtree in the PHYLIP suite of programs (7) to generate a distance tree with data from the FAFLP assay (Fig. 2). This tree shows a close correlation with the MLEE groupings of the EcoR collection (circled in Fig. 2). The nine EcoR strains that did not cluster with their original MLEE groupings in Fig. 2 are boxed in grey. All but one strain (EcoR31) of MLEE group B2 (MLEE-B2) clustered together on a unique branch. EcoR31 clustered apart from the other groups. All but two strains (EcoR16 and EcoR07) of MLEE group A (MLEE-A) clustered together on a single branch. EcoR16 clustered apart from the other groups. EcoR07 grouped with MLEE group B1/C (MLEE-B1/C). Strains of MLEE-B1 and MLEE-C were indistinguishable by the FAFLP assay and clustered together. Five other strains grouped with different MLEE types when they were analyzed by the FAFLP assay: strain 12784-37 (MLEE-E) and strains 12789-42 and 12790-43 (MLEE-B1) all clustered with the MLEE-A strains, strain 12795-48 (MLEE-C) clustered with the MLEE-D strains and strain 66 (MLEE-B1) clustered with the MLEE-B2 strains. An outlier branch grouped two control strains of Salmonella enterica as distantly related. Different tree-building methods (the parsimony and neighbor-joining methods; PAUP, version 4.0) were also used and generated trees with similar topologies (data not shown).
FIG. 2.
Distance tree of the EcoR collection of strains and 15 serogroup O157 strains obtained by the FAFLP assay. EcoR strains are labelled 01 to 72. The tree was generated by using a matrix of pairwise distances calculated for all strains with the coefficient of Nei and Li (Dice) (18). The MLEE groupings of the EcoR collection and the O157 VTEC group are circled. The VT-negative strains of serotypes O157:H8 (strain 74850), O157:H19 (strain 10964), and O157:H42 (strain 73886) that did not cluster with the O157 VTEC group are boxed in black. The nine EcoR strains that did not cluster with their original MLEE groupings are boxed in grey and are followed by their MLEE group designation.
Subtyping of serogroup O157 strains by the FAFLP assay.
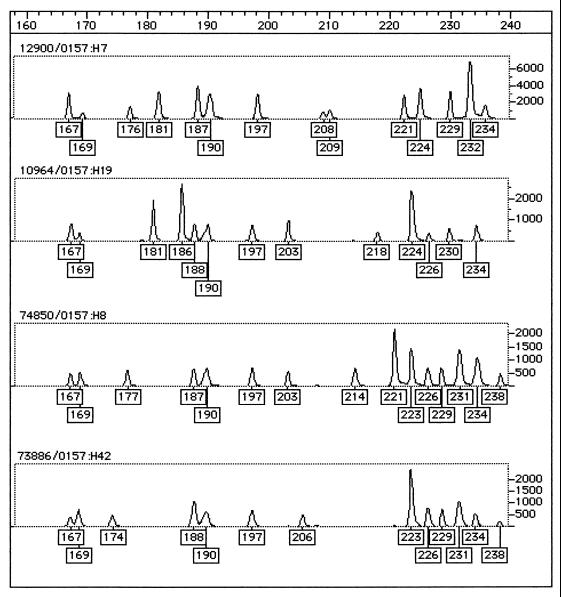
All strains of serogroup O157 had distinct profiles by the FAFLP assay. The 11 O157 VTEC and the VT-negative variant (serotype O157:H7 or O157:H−) strains grouped together on a distinct outlier branch. The VT-negative strains of serotypes O157:H8 (strain 74850) and O157:H19 (strain NCTC10964) clustered with the MLEE-B1/C group. O157:H42 (strain 73886) clustered with MLEE-A. An example of a Genotyper output for strains representing the diversity of O157 by the FAFLP assay is shown in Fig. 3. The VT-negative strains that did not group with the serogroup O cluster are shown boxed in black in Fig. 2.
FIG. 3.
Genotyper FAFLP assay output for four O157 serotypes representing the diversity of O157: O157:H7 (strain 12900), O157:H19 (strain 10964), O157:H8 (strain 74850), and O157:H42 (strain 73886). The boxed numbers under the peaks of the traces are the fragment sizes assigned by Genotyper after comparison with the standard curve generated with the internal size standard.
DISCUSSION
Our results indicate that experimental FAFLP analysis of E. coli K-12 MG1655 with the selective primers MseI+TA and EcoRI+0 produced 97% of the fragments (±1 bp) in the 100- to 500-bp size range predicted from computer analysis of the K-12 MG1655 genome sequence. This sizing precision within the 100- to 500-bp range was achieved by having a differentially labelled size standard in each lane so that the size of every fragment obtained by the FAFLP assay was determined by comparison with an accurate standard curve created individually for the lane in which the reaction was run. This precision allowed each strain to be assigned a unique genotype. Computer modeling with enzyme combinations other than MseI and EcoRI (unpublished data) showed that the choice of restriction enzymes is an important determinant of the discriminatory power of FAFLP analysis. Hence, for each bacterial species the best combination of restriction enzymes and selective primers should be modeled from the whole genome sequence once this information is available. In addition to its high resolving power, AFLP offers greater throughput than other molecular methods for bacterial strain typing, since it does not depend on time-consuming or labor-intensive steps such as Southern blotting or careful cell lysis in agarose.
Large-scale application of MLEE to E. coli and other bacterial genera has elucidated the principal features of the genetic structures of bacterial populations and species. The MLEE-derived framework has permitted analysis of the distribution of serotypes and biotypes, as well as of mobile genetic elements (22). For bacteria, MLEE is the most extensively documented methodology to which statistical population genetics has been applied (22). The EcoR collection is a set of 72 strains from humans and 16 other mammalian species and was selected to be broadly representative of the enzyme genotypic diversity in E. coli as a whole (19). Members of the collection were defined by electrophoretic analysis of 35 enzymes (22). Among the electrophoretic types (ETs) of the collection, the mean allelic diversity varied from 0 for the citrate synthase locus (monomorphic) to 0.82 for the β-galactosidase locus (12 alleles were detected). Six phylogenetic groups designated A, B1, B2, C, D, and E were identified. Figure 2 shows that the groupings obtained by the FAFLP assay correlate with these MLEE data. For example, the 23 ETs of MLEE-A all have the same root except strain EcoR07 and EcoR16 (07A and 16A, respectively, in Fig. 2). EcoR16, a leopard strain, stands apart from other strains by the FAFLP assay. Strains in MLEE-B1 and MLEE-C generally cluster together, with three exceptions: strains EcoR42 and EcoR43 (MLEE-B1) cluster with the MLEE-A strains, strains EcoR31 clusters apart from the other groups, and strain EcoR48 (MLEE-C) is placed among the MLEE-D strains. Strain EcoR48 is thought to be defective in methyl-directed mismatch repair and hence may be a mutator for which horizontal transfer of genes from similar or disparate species takes place at a much higher rate, and this is also the case for some O157 strains (4, 16). The 15 MLEE-B2 strains (10 ETs) were grouped by the FAFLP assay as a tight cluster with the inclusion of EcoR66 (MLEE-B2). Strain EcoR44 (MLEE-D) clustered away from the other groups. Interestingly, strains EcoR16, EcoR31, and EcoR44 did not cluster with any of the groups and are the only strains in this collection to be isolated from the cat family (leopard and cougar). The FAFLP assay apparently resolves differences between strains that are clonal by MLEE.
The serogroup O157 strains fell into three groups, one of which contained all of the O157 VTEC strains and a VT-negative O157 VTEC derivative strain and clustered in a remote group apart from other E. coli strains. The different profiles of the strains in this group obtained by the FAFLP assay confirmed those obtained in a previous PFGE analysis which showed that all the isolates, including those belonging to the same phage type, are distinguishable (24, 28). The VT-negative serogroup O157 strains that grouped apart from the VTEC by the FAFLP assay were biochemically and antigenically different from O157 VTEC (27). The FAFLP assay thus confirms independent lines of evolution within the highly diverse O157 serogroup. The average number of fragments generated by the O157 VTEC strains was 40, whereas, on average, 47 fragments were generated by strains from the EcoR collection; moreover, the serogroup O157 strains produced more large fragments than the EcoR collection. This offers further proof of the unusual nature of serogroup O157, the genome of which is thought to be 20% larger than the K-12 genome (20).
We have previously demonstrated that the empirically derived FAFLP assay can provide high-resolution molecular epidemiological analysis for outbreaks of S. pyogenes infection (5). We suggest that the FAFLP assay conditions optimized for E. coli in the present study will provide a basis for genotyping of strains of this species. The data generated by the FAFLP assay are suitable for rapid electronic dissemination, manipulation, and interlaboratory comparison. They could be stored in national or international epidemiological databases for further analysis.
In summary, we have compared predicted and observed FAFLP assay data and determined the requirements for the precise sizing of individual genome fragments (and, therefore, the identification of individual strains) from a singly labelled AFLP reaction generating approximately 50 fragments with a size range of 100 to 500 bp. The predicted fragments of strain MG1655 generated by the FAFLP assay and documented in this study were suitable for the standardization and calibration of the FAFLP profiles of all E. coli strains examined. We suggest that a standardized molecular method such as this can be usefully applied in clinical microbiology, molecular epidemiology, and population genetics.
ACKNOWLEDGMENTS
We thank Philip Mortimer and Jon Clewley for valuable comments.
REFERENCES
- 1.Arbeit R, Arthur M, Dunn R, Cheung K, Selander R K, Goldstein R. Resolution of recent divergence among Escherichia coli from related lineages: the application of pulsed field gel electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingstown R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology I. New York, N.Y: John Wiley & Sons, Inc.; 1989. Preparation of genomic DNA from bacteria. [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cebula T A, Li B, Payne W L, Leclerc L E. ASM Conference on Small Genomes. Washington, D.C: American Society for Microbiology; 1998. Mutators among Escherichia coli and Salmonella enterica: adaptation and emergence of bacterial pathogens; p. 16. [Google Scholar]
- 5.Desai M, Tanna A, Wall R, Efstratiou A, George R, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J Clin Microbiol. 1998;36:3133–3137. doi: 10.1128/jcm.36.11.3133-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dijkshoorn L, Aucken H, Gerner-Smidt P, Janssen P, Kaufmann M E, Garaizar J, Ursing J, Pitt T L. Comparison of outbreak and nonoutbreak Acinetobacter baumanni strains by genotypic and phenotypic methods. J Clin Microbiol. 1996;34:1519–1525. doi: 10.1128/jcm.34.6.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 8.Fitzgerald C, Owen R J, Stanley J. A comprehensive ribotyping scheme for the heat stable serotypes of Campylobacter jejuni. J Clin Microbiol. 1996;34:265–269. doi: 10.1128/jcm.34.2.265-269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimont F, Grimont P A D. Ribosomal ribonucleic acid restriction patterns as potential taxonomic tools. Ann Inst Pasteur. 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 10.Hall L M, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huys G, Coopman R, Janssen P, Kersters K. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int J Syst Bacteriol. 1996;46:572–580. doi: 10.1099/00207713-46-2-572. [DOI] [PubMed] [Google Scholar]
- 12.Huys G, Kersters I, Coopman R, Janssen P, Kersters K. Genotypic diversity among Aeromonas isolates recovered from drinking water production plants as revealed by AFLP™ analysis. Syst Appl Microbiol. 1996;19:428–435. [Google Scholar]
- 13.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 14.Janssen P, Dijkshoorn L. High resolution DNA fingerprinting of Acinetobacter outbreak strains. FEMS Microbiol Lett. 1996;142:191–194. doi: 10.1111/j.1574-6968.1996.tb08429.x. [DOI] [PubMed] [Google Scholar]
- 15.Kiem P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 17.Meunier J-R, Grimont P A D. Factors affecting reproducibility of random amplified polymorphic DNA fingerprinting. Res Microbiol. 1993;144:373–379. doi: 10.1016/0923-2508(93)90194-7. [DOI] [PubMed] [Google Scholar]
- 18.Nei M, Li W-H. Mathematical model for studying genetic variations in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochman H, Whittam T S, Caugant D A, Selander R K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 20.Perna N T, Burland V, Plunkett III G, Gregor J, Mayhew G F, Rose D J, Shao Y, Blattner F R. ASM Conference on Small Genomes. Washington, D.C: American Society for Microbiology; 1998. Comparative genomics of E. coli K-12, O157:H7 and related entobacterial pathogens; p. 7. [Google Scholar]
- 21.Rafalski J A, Tingey S V, Williams J G K. RAPD markers—a new technology for genetic mapping and plant breeding. AgBiotech News Info. 1991;3:645–648. [Google Scholar]
- 22.Selander R K, Caugant D A, Whittham T S. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F C, editor. Escherichia coli and Salmonella typhimurium cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 23.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trevena W B, Hooper R S, Wray C, Willshaw G A, Cheasty T, Domingue G. Vero cytotoxin-producing Escherichia coli O157 associated with companion animals. Vet Rec. 1996;138:400. . (Letter.) [PubMed] [Google Scholar]
- 25.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kulper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7224. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willshaw G A, Scotland S M, Smith H R, Cheasty T, Thomas A, Rowe B. Hybridization of strains of Escherichia coli O157 with probes derived from the eaeA gene of enteropathogenic E. coli and the eaeA homolog from a vero cytotoxin-producing strain of E. coli O157. J Clin Microbiol. 1994;32:897–902. doi: 10.1128/jcm.32.4.897-902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willshaw G A, Smith H R, Cheasty T, Wall P G, Rowe B. Vero cytotoxin-producing Escherichia coli O157 outbreaks in England and Wales, 1995: phenotypic methods and genotypic subtyping. Emerg Infect Dis. 1997;3:561–565. doi: 10.3201/eid0304.970422. [DOI] [PMC free article] [PubMed] [Google Scholar]