Abstract
BACKGROUND
Crohn’s disease (CD) is an incurable intestinal disorder with unclear etiology and pathogenesis. Currently, there is a lack of specific biomarkers and drug targets for CD in clinical practice. It is essential to identify the precise pathophysiological mechanism of CD and investigate new therapeutic targets.
AIM
To explore a new biomarker and therapeutic target for CD and verify its role in the CD pathological mechanism.
METHODS
Proteomics was performed to quantify the protein profile in the plasma of 20 CD patients and 20 matched healthy controls. Hub genes among the selected differentially expressed proteins (DEPs) were detected via the MCODE plugin in Cytoscape software. The expression level of one hub gene with an immunoregulatory role that interested us was verified in the inflamed intestinal tissues of 20 CD patients by immunohistochemical analysis. After that, the effects of the selected hub gene on the intestinal inflammation of CD were identified in a CD cell model by examining the levels of proinflammatory cytokines by enzyme-linked immunosorbent assays and the expression of the NF-κB signalling pathway by quantitative real-time PCR analysis and Western blot assays.
RESULTS
Thirty-five DEPs were selected from 393 credible proteins identified by proteomic analysis. Among the DEPs, fibrinogen-like protein 1 (FGL1), which attracted our attention due to its function in the regulation of the immune response, had 1.722-fold higher expression in the plasma of CD patients and was identified as a hub gene by MCODE. Furthermore, the expression of FGL1 in the intestinal mucosal and epithelial tissues of CD patients was also upregulated (P < 0.05). In vitro, the mRNA levels of FGL1 and NF-κB; the protein expression levels of FGL1, IKKα, IKKβ, p-IKKα/β, p-IκBα, and p-p65; and the concentrations of the proinflammatory cytokines IL-1β, IL-6, IL-17, and TNF-α were increased (P < 0.05) after stimulation with lipopolysaccharide, which were reversed by knockdown of FGL1 with siRNA transfection (P < 0.05). Conversely, FGL1 overexpression enhanced the abovementioned results (P < 0.05).
CONCLUSION
FGL1 can induce intestinal inflammation by activating the canonical NF-κB signalling pathway, and it may be considered a potential biomarker and therapeutic target for CD.
Keywords: Crohn’s disease, Fibrinogen-like protein 1, Proteomics, NF-κB pathway
Core Tip: In this study, fibrinogen-like protein 1 (FGL1) was identified to be significantly upregulated in the plasma and intestinal mucosa of Crohn’s disease (CD) patients. In vitro, silencing FGL1 downregulated the levels of the proinflammatory cytokines IL-1β, IL-6, IL-17, and TNF-α. Furthermore, FGL1 knockdown suppressed the mRNA expression of NF-κB and the protein levels of IKKα, IKKβ, p-IKKα/β, p-IκBα, and p-p65. These results could be reversed by the overexpression of FGL1. Taken together, these data suggest that FGL1 may induce intestinal inflammation by activating the canonical NF-κB signalling pathway and has the potential to be a therapeutic target for CD.
INTRODUCTION
Crohn’s disease (CD) is a chronic, idiopathic intestinal inflammatory disease affecting any segment of the gastrointestinal tract. Although CD is believed to be a result of an imbalanced interaction among genetic susceptibility, environmental factors, the intestinal microflora, and the immune system, the precise pathogenesis is still not entirely clear[1]. Consequently, CD remains incurable even though great advancement has been achieved in medical therapy. Symptoms evolving in a relapsing and remitting manner indicate that CD has a progressive disease course that may induce complications, such as abscess, fistula, and stricture development. Eventually, up to 70% of CD patients require at least one intestinal surgery over their lifetime[2]. Targeted therapy is anticipated to change the natural course of CD, and even to cure it.
Currently, anti-tumor necrosis factor (TNF) agents (infliximab, adalimumab, and certolizumab pegol) are the most potent drugs for inducing and maintaining remission of CD. Unfortunately, anti-TNF treatment failure is common. Primary non-response occurs in 21.9% of infliximab-treated CD patients and 26.8% of adalimumab-treated patients[3]. More than 60% of patients treated with infliximab or adalimumab do not achieve deep remission[3]. These data indicate that increased TNF-α levels may be the result of an immunoinflammatory response instead of the cause. Vedolizumab blocking the α4β7 integrin can induce endoscopic remission in approximately one-third of CD patients at week 52[4]. The decreased long-term efficacy of biologic medications makes it urgent to investigate new therapeutic targets for CD.
Omics techniques, including genomics, metabolomics, and proteomics, have been applied to explore potential biomarkers and targets for CD in recent years. It is widely known that cellular function and biological behaviour are primarily regulated by proteins. The protein domain is likely the most ubiquitously affected in disease development, treatment response, and physical recovery. Hence, it is promising to reveal the crucial changes in CD pathogenesis and discover novel drug targets by proteomics directly profiling protein expression. Proteomic techniques are classified into three major stages: Discovery, verification, and validation. Currently, the application of proteomics in CD remains in the initial discovery phase[5].
In the present study, we applied proteomics to identify differentially expressed proteins (DEPs) in the plasma of CD patients in an attempt to discover a potential biomarker and therapeutic target for CD. Our data showed that fibrinogen-like protein 1 (FGL1) was significantly upregulated in the plasma of CD patients. FGL1, also known as hepassocin or hepatocyte-derived fibrinogen-related protein 1 (HFREP1), is a hepatocyte-secreted protein that belongs to the fibrinogen family[6]. However, FGL1 lacks a platelet-binding site, a cross-linking region, and a thrombin-sensitive site, which are crucial for fibrin clot formation. Several studies have demonstrated that FGL1 can regulate immune systems to induce inflammatory response and tumor immune evasion[7,8]. To date, whether FGL1 is correlated with the development of CD remains unclear. Therefore, we further verified the expression of FGL1 in intestinal tissues of CD patients and validated its crucial role in the pathogenesis of CD in vitro.
MATERIALS AND METHODS
Clinical samples
Plasma samples were collected from 20 treatment-naive patients with CD and 20 age- and sex-matched healthy individuals between July 2017 and August 2018. The protein profiles in the plasma were analysed by tandem mass tag (TMT)-based quantitative proteomics. Paraffin-embedded mucosal biopsy specimens from an additional 20 treatment-naive patients with active CD and 20 matched healthy individuals undergoing colonoscopy screening were obtained for immunohistochemical examination. The protocols of this study were approved by the ethics committee of the Affiliated Hospital of Nanjing University of Chinese Medicine (2018NL-171-02). All patients provided informed consent.
TMT-based quantitative proteomics
Plasma samples were homogenized in sodium dodecyl sulfate (SDS) lysis buffer. Centrifugation was performed to collect the supernatant. Total protein concentrations were quantified using a bicinchoninic acid (BCA) assay (Thermo Scientific, United States). Protein extracts were reduced with reducing buffer (10 mmol/L dithiothreitol, 8 mol/L urea, and 100 mmol/L tetraethylammonium bromide (TEAB), pH 8.0) at 60 °C for 1 h. All samples were alkylated with iodoacetamide for 40 min at room temperature in the dark. After centrifugation, the protein pellets were digested with TEAB (100 mmol/L) and sequencing-grade trypsin (1 μg/μL) at 37 °C for 12 h.
For TMT labelling, 100 μL of protein sample was incubated with a mixed solution of 41 μL of TMT labelling reagent (Thermo Fisher Scientific, United States) and 41 μL of anhydrous acetonitrile for 1 h at room temperature. The reaction was terminated with 8 μL of 5% hydroxylamine. The samples from the CD patients were labelled with TMT-130 and TMT-131, while those from the healthy controls were labelled with TMT-126 and TMT-127.
The TMT-labelled peptides were eluted by using an Agilent Zorbax Extend-C18 column (2.1 mm × 150 mm, 5 μm) and fractionated with a high-performance liquid chromatography (HPLC) system at a flow rate of 300 μL/min. The elution gradient was set to 98%, 95%, 75%, 60%, and 10%. The collected peptides were loaded on a reverse-phase trap column (C18, 100 μm × 20 mm, Thermo Fisher Scientific, United States) and enriched on an analysis column (C18, 75 μm × 150 mm, Thermo Fisher Scientific, United States) following redissolution in nano-HPLC buffer (HPLC water containing 0.1% formic acid). The flow rate was 300 nL/min, and the linear elution gradient was set as 5%, 30%, 50%, and 100%.
For mass spectrometry (MS) survey scans, the ion spray voltage, interface heating temperature, MS resolution, and ion population were set to 1,800 V, 250 °C, 70000, and 1 × 106, respectively. The precursor ion was acquired at 300-1600 m/z. A maximum of 10 precursors were selected for higher-energy collisional dissociation with analysis in an LTQ Orbitrap Velos Pro (Thermo Fisher Scientific, United States), and the normal chemical energy was 32%. For MS/MS detection, the tandem MS resolution, ion population, ion maximum injection time, and dynamic exclusion time were set to 17500, 2 × 105, 80 ms, and 30 s, respectively.
Quantitative proteomic analysis
The raw proteomic data were analysed using Proteome Discoverer software (version 2.2, Thermo Fisher Scientific, United States) and searched against the UniProtKB database (Hunam, 2015-09, 88473 sequences). Andromeda was used as the search engine with the following parameters: (1) Homo sapiens taxonomy; (2) Q Exactive plus as instrument type; (3) Trypsin as the proteolytic enzyme, with two missed cleavages allowed; (4) TMT 6 plex and cysteine carbamidomethylations as fixed modifications; (5) Oxidation of methionine as a variable modification; (6) 20 ppm as the MS tolerance; and (7) Seven amino acids as minimum cut-off for peptide length. A false discovery rate (FDR) of less than 1% was set to refine the results.
For quantitative analysis, the TMT reporter ion intensity of each protein was analysed using Proteome Discoverer software. Proteins with empty values were discarded. Student’s t test was performed to examine the difference in each protein between the two groups with Perseus software. Proteins with a fold change > 1.5 or < 0.67 and a P value < 0.05 were considered to be DEPs.
Bioinformatics analysis
Genes of DEPs were visualized in Cytoscape software (version 3.7.2), in which the MCODE plugin was used to select significant modules for identification of hub genes. Subsequently, the hub genes were input into the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) to construct a protein-protein interaction (PPI) network. The Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/) was applied for gene ontology (GO) enrichment analysis. Reactome pathway analysis (https://www.reactome.org/) was performed for pathway enrichment analysis.
Immunohistochemical staining
Immunohistochemical staining was implemented to detect the expression of FGL1 in inflamed intestinal tissues of CD patients and normal intestinal biopsies. Mucosal biopsy specimens were fixed in 10% neutral formalin for 24 h. Afterwards, they were embedded in paraffin and cut into 5 μm sections. The sections were deparaffinized and rehydrated and then incubated in citrate buffer (pH 6.0) for antigen retrieval. After endogenous peroxidase activity was quenched with 3% hydrogen peroxide, the samples were incubated in 1% bovine serum albumin (BSA) to block non-specific immunoglobulin binding. Subsequently, the slides were incubated with an anti-FGL1 antibody (1:200 dilution, 16000-1-AP, Proteintech, United States) at 4 °C overnight. Following washing with phosphate buffered saline (PBS), the slides were incubated with a secondary IgG antibody (1:1000 dilution, ab6721, Abcam, United Kingdom) at room temperature for 1 h, counterstained with haematoxylin, and stained with a diaminobenzidine kit (DAB, Beyotime, China). All the sections were visualized under a light microscope (Nikon 80i, Japan). ImageJ software (version 1.52) was used to calculate the integrated optical density (IOD) values.
Cell culture and treatment
The human colonic adenoma cell line HT-29 (ATCC, United States) was cultured with Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum, 100 μg/mL penicillin, and 100 U/mL streptomycin at 37 °C with 5% CO2. The HT-29 cells were stimulated with 100 ng/mL lipopolysaccharide (LPS, Sigma, United States) to establish a cell model of intestinal inflammation. To uncover the impact of FGL1 on intestinal inflammation, the HT-29 cells were transfected with FGL1 siRNA and plasmid DNA (Nanjing KeyGen Biotech Co., Ltd., China) before stimulation with LPS. The transfection efficiency was determined by examining the mRNA expression of FGL1.
Quantitative real-time PCR analysis
Total RNA in HT-29 cells was extracted using a TRIzol reagent kit (Takara, Japan) according to the manufacturer’s instructions. A PrimeScript RT reagent kit (Takara, Japan) was used for reverse transcription of the extracted RNA into cDNA. Quantitative real-time PCR (qRT-PCR) was conducted to detect the mRNA expression of FGL1 and NF-κB by using a SYBR green kit (Takara, Japan). The housekeeping gene β-actin was used for normalization to an endogenous reference. The relative gene expression was evaluated by using the 2−ΔΔCt method. The sequences of the PCR primers are as follows: FGL1-forward: 5’-ATGGCAAAGGTGTTCAGTTTCA-3’, reverse: 5’-ACAATCTGCATACTGCCTCTTG-3’; NF-κB-forward: 5’-GAAGCACGAATGACAGAGGC-3’, reverse: 5’-GCTTGGCGGATTAGCTCTTTT-3’; and β-actin-forward: 5’-CATGTACGTTGCTATCCAGGC-3’, reverse: 5’-CTCCTTAATGTCACGCACGAT-3’.
Enzymelinked immunosorbent assay
The levels of the proinflammatory cytokines IL-1β, IL-6, IL-17, and TNF-α (Sigma, United States) in the culture medium collected after 48 h were examined by enzyme linked immunosorbent assay (ELISA) according to the manufacturer’s protocol.
Western blot assay
Cells lysed with radioimmunoprecipitation assay (RIPA) lysis buffer were centrifuged at 12000 g for 20 min at 4 °C. Protein concentrations in the collected supernatant were quantified with a BCA assay kit. After equal amounts of protein (20 μg/well) were loaded and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), they were transferred onto polyvinylidene difluoride (PVDF) membranes and incubated in 5% BSA for 1 h at room temperature. The membranes were washed with Tris-borate saline containing 0.1% Tween-20 (TBST) and were incubated with primary antibodies against FGL1 (1:1000 dilution, 16000-1-AP, Proteintech, United States), IKKα (1:1000 dilution, ab32041, Abcam, United Kingdom), IKKβ (1:1000 dilution, ab32135, Abcam), p-IKKα/β (1:1000 dilution, ab194528, Abcam), IκBα (1:1000 dilution, ab32518, Abcam), p-IκBα (1:1000 dilution, ab133462, Abcam), NF-κB (p65, 1:1000 dilution, ab32536, Abcam), p-p65 (1:1000 dilution, ab76302, Abcam) and β-actin (1:1000 dilution, 20536-1-AP, Proteintech) at 4 °C overnight. The membranes were washed with TBST again and incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. The blots were imaged using enhanced chemiluminescence (ECL). ImageJ software was used to calculate the protein signal grey values.
Statistical analysis
All data were statistically analysed with SPSS 22.0 (SPSS Inc., United States). Continuous variables with a normal distribution are summarized using the mean ± SD, which in a skewness distribution are expressed as the median with range. The Mann-Whitney U test, Student’s t tests, and chi-square test were performed to compare numerical variables and categorical variables as appropriate. One-way analysis of variance was used for multi-group comparisons. A two-sided P value < 0.05 was considered statistically significant.
RESULTS
Patients’ characteristics
Twenty treatment-naive CD patients and 20 healthy controls were recruited for plasma proteomic analysis. The diagnostic criteria for CD referred to the clinical guidelines of the American College of Gastroenterology (ACG)[9]. Thirteen males and seven females with a median age of 20.5 (14-43) years were included in the CD group, and eleven males and nine females with a median age of 24.5 (18-46) years were included in the normal control group. Baseline demographic characteristics were comparable between the two groups (P > 0.05).
Colonoscopic biopsy specimens from an additional 20 treatment-naive patients with active CD and 20 healthy controls were used for immunohistochemical staining. There was no significant difference in sex distribution, age, or biopsy site between the two groups (P > 0.05). The baseline clinical characteristics of patients for plasma proteomic detection and immunohistochemical analysis are presented in Tables 1 and 2, respectively.
Table 1.
Clinical characteristics of patients for plasma proteomic analysis
|
Item
|
Crohn’s disease (n = 20)
|
Normal control (n = 20)
|
P
value
|
| Sex | |||
| Male | 13 | 11 | 0.519 |
| Female | 7 | 9 | |
| Median age (range), yr | 20.5 (14-43) | 24.5 (18-46) | 0.069 |
| Disease location in the endoscopy | |||
| Ileum | 6 | N/A | |
| Colon | 6 | N/A | |
| Ileocolon | 8 | N/A |
N/A: Not applicable.
Table 2.
Clinical characteristics of patients for immunohistochemical assay
|
Item
|
Crohn’s disease (n = 20)
|
Normal control (n = 20)
|
P
value
|
| Sex | |||
| Male | 15 | 13 | 0.731 |
| Female | 5 | 7 | |
| Age (mean ± SD), yr | 27.1 ± 7.9 | 25.6 ± 4.5 | 0.465 |
| Biopsy site | |||
| Terminal ileum | 8 | 5 | 0.832 |
| Ascending colon | 2 | 1 | |
| Transverse colon | 2 | 2 | |
| Descending colon | 2 | 4 | |
| Sigmoid colon | 4 | 6 | |
| Rectum | 2 | 2 |
SD: Standard deviation.
FGL1 is significantly upregulated in plasma proteomic analysis
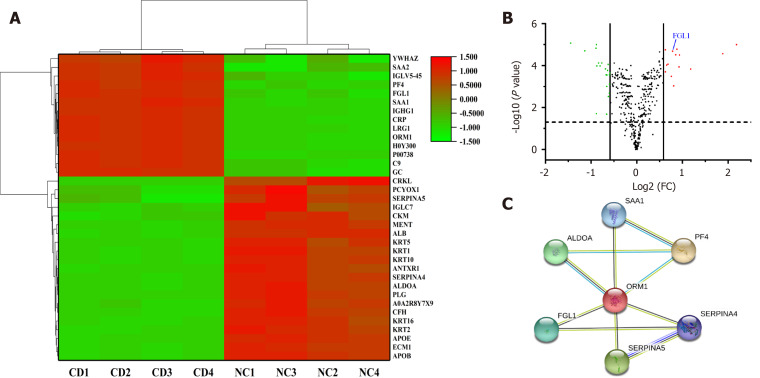
Plasma samples in each group were randomly divided into four subclusters. A total of 393 credible proteins were identified by proteomic analysis, among which 35 had differential expression between the two groups (Figure 1A). Among the DEPs, FGL1 attracted our attention because of its function in the regulation of the immune response. The expression level of FGL1 in the plasma of CD patients was 1.722-fold greater than that in healthy people (Figure 1B). Three MCODE modules were established to screen hub genes via Cytoscape software. As FGL1 was contained in the 3rd module, the genes in this module were used for further bioinformatics analysis. Figure 1C shows the PPI network of the genes. GO enrichment analysis showed that the genes were involved in the biological processes of platelet degranulation, acute phase response, platelet activation, and negative regulation of endopeptidase activity, and the molecular functions of heparin binding and serine-type endopeptidase inhibitor activity. Reactome pathway analysis demonstrated that the genes were related to the common pathway of fibrin clot formation and the pathways of platelet degranulation, peptide ligand-binding receptors, haemostasis, G alpha (i) signalling events, and innate immune system.
Figure 1.
Fibrinogen-like protein 1 expression in the plasma of Crohn’s disease patients. A: Heat map showing 35 differentially expressed proteins between Crohn’s disease (CD) patients and healthy individuals, among which fibrinogen-like protein 1 (FGL1) expression was upregulated in the CD group; B: The FGL1 expression level in the plasma of CD patients was 1.722-fold greater than that in healthy people; C: Protein-protein interaction network of an MCODE module containing FGL1 as a hub gene. CD: Crohn’s disease; NC: Normal control; FGL1: Fibrinogen-like protein 1.
FGL1 expression is increased in intestinal tissues of CD patients
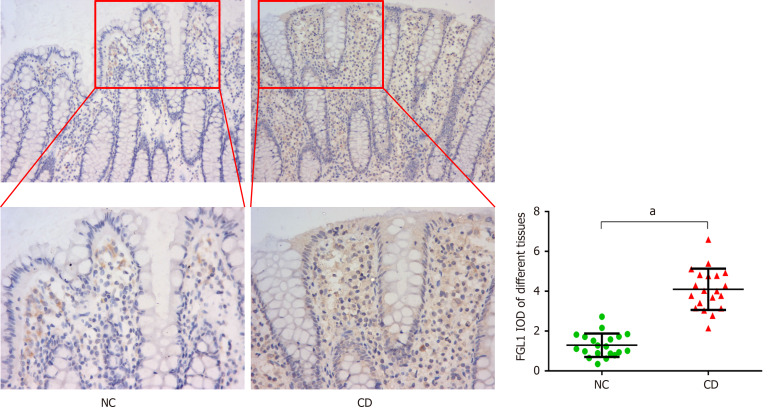
Immunohistochemical analysis was performed to verify the expression of FGL1 in the intestinal tissues of CD patients. The results demonstrated that the FGL1 levels in the intestinal mucosal and epithelial tissues were higher than those in the normal intestinal tissues (P < 0.01, Figure 2).
Figure 2.
Fibrinogen-like protein 1 expression, as determined by immunohistochemical analysis (× 200, × 400), was increased in the intestinal mucosal and epithelial tissues of Crohn’s disease patients. aP < 0.01 vs normal control group. NC: Normal control group; CD: Crohn’s disease group; FGL1: Fibrinogen-like protein 1.
FGL1 mediates the expression of proinflammatory cytokines in intestinal epithelial cells
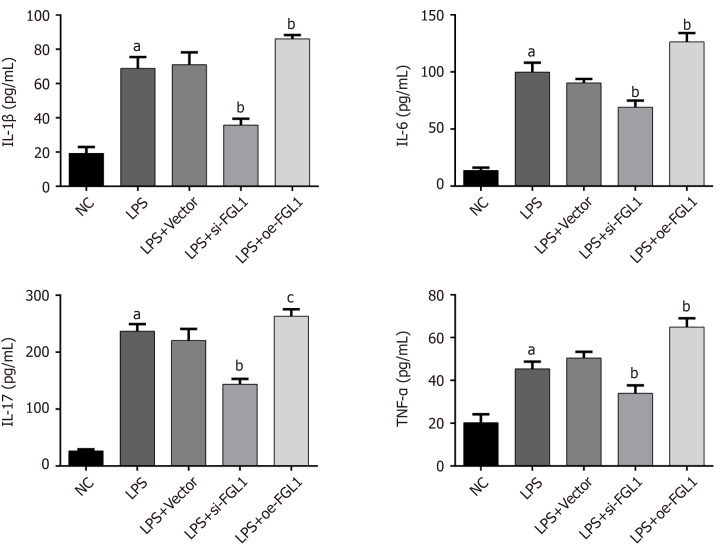
To investigate the regulation of intestinal inflammation by FGL1, proinflammatory cytokines were detected by ELISA after FGL1 siRNA and plasmids were transfected into HT-29 cells. After LPS stimulation, the IL-1β, IL-6, IL-17, and TNF-α levels were significantly upregulated. FGL1 knockdown reversed the expression of IL-1β, IL-6, IL-17, and TNF-α, while overexpression of FGL1 elevated the levels of the four proinflammatory cytokines (P < 0.05, Figure 3).
Figure 3.
Expression of proinflammatory cytokines in different groups, as determined by enzyme linked immunosorbent assay. aP < 0.01 vs normal control group; bP < 0.01, cP < 0.05 vs lipopolysaccharide. NC: Normal control group; LPS: Crohn’s disease (CD) model induced by lipopolysaccharide (LPS); LPS+Vector: Transfected with an empty vector based on the CD model; LPS+si-FGL1: Transfected with FGL1 siRNA based on the CD model; LPS+oe-FGL1: Transfected with the FGL1 plasmid based on the CD model; FGL1: Fibrinogen-like protein 1.
FGL1 activates the NF-κB signalling pathway
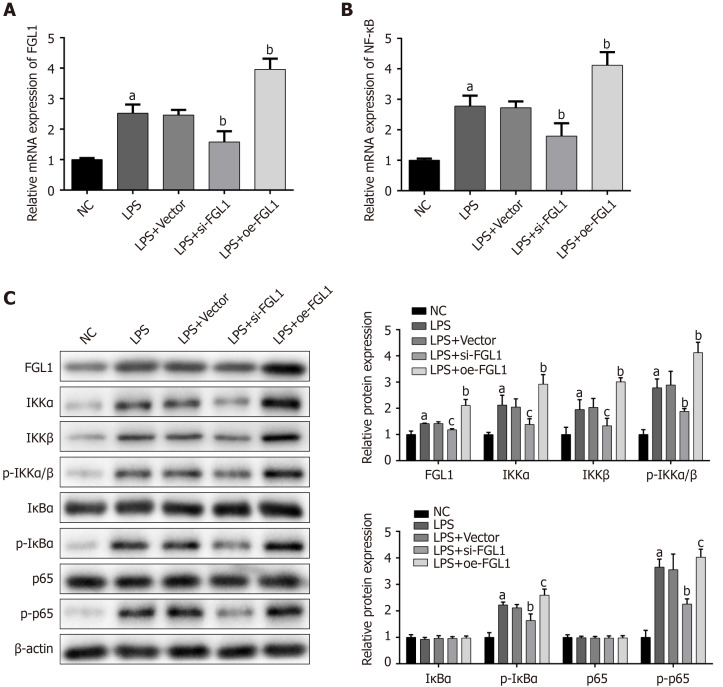
Given that NF-κB plays a fundamental role in the intestinal inflammation of CD, the modulation of the NF-κB signalling pathway by FGL1 was investigated. The mRNA expression levels of FGL1 and NF-κB were increased by LPS stimulation. After intervention with FGL1 siRNA, the mRNA expression levels of FGL1 and NF-κB were both downregulated, while the mRNA levels were enhanced following FGL1 overexpression with plasmid transfection (P < 0.01, Figure 4A and B).
Figure 4.
Impact of fibrinogen-like protein 1 on the NF-κB signalling pathway. A and B: Effect of knockdown or overexpression of fibrinogen-like protein 1 (FGL1) on the mRNA expression of FGL1 and NF-κB, as determined by quantitative real-time PCR assay; C: Effect of FGL1 on the protein expression of related proteins in the canonical NF-κB signalling pathway, as determined by Western blot assay. aP < 0.01 vs NC; bP < 0.01, cP < 0.05 vs LPS. NC: Normal control group; LPS: Crohn’s disease (CD) model induced by LPS; LPS+Vector: Transfected with an empty vector based on the CD model; LPS+si-FGL1: Transfected with FGL1 siRNA based on the CD model; LPS+oe-FGL1: Transfected with the FGL1 plasmid based on the CD model; LPS: Lipopolysaccharide; FGL1: Fibrinogen-like protein 1.
The exact mechanism by which FGL1 regulates the NF-κB signalling pathway was revealed by Western blot assay. The protein levels of FGL1, IKKα, IKKβ, p-IKKα/β, p-IκBα, and p-p65 were upregulated in HT-29 cells stimulated with LPS (P < 0.05). FGL1 gene knockdown inhibited the protein expression of FGL1 and downregulated the protein expression of IKKα, IKKβ, p-IKKα/β, p-IκBα, and p-p65 (P < 0.05). Conversely, the overexpression of the FGL1 gene enhanced the protein expression of FGL1, IKKα, IKKβ, p-IKKα/β, p-IκBα, and p-p65 (P < 0.05, Figure 4C).
DISCUSSION
In the present study, we took advantage of proteomics for a large-scale screen of DEPs between the plasma of CD patients and healthy people. The expression of FGL1, a hub gene among the DEPs, was increased in the plasma of CD patients, which was verified in intestinal mucosal and epithelial tissues. Furthermore, FGL1 was validated to exacerbate the inflammatory response in intestinal epithelial cells by activating the NF-κB signalling pathway.
At present, knowledge about the etiology and pathogenesis of CD is limited, which makes it incurable. Hence, it is essential to detect potential drug targets for CD. As proteins are directly involved in nearly all pathophysiological processes, proteomics has become a hotspot tool for the discovery of novel biomarkers or therapeutic targets for CD, differential diagnosis between CD and ulcerative colitis, and disease stratification by examining and quantifying thousands of proteins encoded by the genome in a holistic manner[10-13]. To our knowledge, the present study revealed for the first time by proteomics that FGL1 may be a key contributor to CD onset and progression.
Accumulating evidence has demonstrated that FGL1 plays a prominent role in the pathogenesis of various diseases, including hepatocellular carcinoma, gastric cancer, lung cancer, diabetes mellitus, and obesity[14-19]. Additionally, FGL1 is also considered a potential biomarker and drug target in certain inflammatory conditions. FGL1 may promote liver injury-induced inflammation via the IL-6/STAT3 signalling pathway[20]. Proteomics revealed that FGL1 is a specific biomarker for predicting the progression of rheumatoid arthritis[7]. This finding displays a fundamental role of FGL1 in regulating immune-mediated inflammation.
Pierre and colleagues have demonstrated that CD relapse is correlated with the innate immune response of the liver[21]. Given that FGL1 is a liver-derived protein and is involved in the innate immune system pathway, we hypothesize that FGL1 may influence the pathophysiology of CD based on the evidence of increased expression of FGL1 in the plasma and intestinal tissues of CD patients. Although FGL1 has been demonstrated to be a potent target for cancer immunotherapy, its precise role in CD therapy is unknown[8]. To unravel the mystery, a cell experiment was designed in the current study that focused on the FGL1-mediated regulation of signalling by NF-κB, an important proinflammatory transcription factor for inflammatory disorders.
Activation of NF-κB plays a central role in the induction and exacerbation of the intestinal inflammatory response of CD patients[22]. The NF-κB family consists of p65 (RELA), RELB, c-REL, p50/p105 (NF-κB1), and p52/p100 (NF-κB2). Activated p65 can translocate into the nucleus to upregulate the transcriptional expression of proinflammatory cytokines. In the present study, the mRNA and protein levels of FGL1 and NF-κB and the concentrations of IL-1β, IL-6, IL-17, and TNF-α were markedly upregulated in HT-29 cells stimulated with LPS, and these effects were reversed by depleting FGL1 with specific siRNA. Correspondingly, the expression of NF-κB and the four proinflammatory cytokines was enhanced following overexpression of FGL1. These results indicate that FGL1 may promote the intestinal inflammatory response by activating NF-κB signalling. The canonical pathway of NF-κB activation involves the IKK complex, consisting of NEMO, IKKα, and IKKβ, and the IκB protein family, including IκBα, IκBβ, and IκBe. After stimulation, IKKα and IKKβ activation promotes phosphorylation of IκBα. Degradation of phosphorylated IκBα releases the p65-p50 dimer for nuclear translocation[23]. In this study, the protein expression levels of IKKα, IKKβ, p-IKKα/β, p-IκBα, and p-p65 were decreased after knockdown of FGL1 compared to those in the cell model, and the inverse effect was verified by overexpression of FGL1. Therefore, FGL1 may induce intestinal inflammation by activating the canonical NF-κB pathway.
CONCLUSION
In summary, we found for the first time that the expression of FGL1 is considerably upregulated in the plasma and intestinal mucosal and epithelial tissues of CD patients. FGL1 might induce intestinal inflammation by activating the canonical NF-κB signalling pathway to stimulate the secretion of proinflammatory cytokines, such as IL-1β, IL-6, IL-17, and TNF-α. Hence, FGL1 may be considered a potential biomarker and therapeutic target for CD. However, given the exploratory design of our study, the precise role of FGL1 in the pathogenesis of CD needs to be deeply investigated and further validated.
ARTICLE HIGHLIGHTS
Research background
Currently, the etiology and pathogenesis of Crohn’s disease (CD) are not completely known, which makes it incurable. It is urgent to reveal the pathophysiological mechanism of CD and investigate new therapeutic targets.
Research motivation
To explore a potential therapeutic target for CD and verify its role in the CD pathological mechanism.
Research objectives
In this study, we attempted to find a potential therapeutic target for CD and verify its role in the CD pathological mechanism in vitro.
Research methods
Proteomics was implemented to quantify the protein profile in the plasma of CD patients. Among the differentially expressed proteins, a hub gene that could regulate the immune response was selected for further study. The expression of the selected hub gene in the inflamed intestinal mucosa was verified by immunohistochemical staining. In vitro, the effects of the hub gene on the expression of proinflammatory cytokines and the NF-κB signalling pathway were evaluated by ELISA, qRT-PCR, and Western blot analysis.
Research results
Fibrinogen-like protein 1 (FGL1), as a hub gene of the differentially expressed proteins, was confirmed to be markedly upregulated in the plasma and intestinal mucosa of CD patients. Silencing FGL1 downregulated the levels of the proinflammatory cytokines IL-1β, IL-6, IL-17, and TNF-α. Furthermore, FGL1 knockdown repressed the mRNA expression of NF-κB and the protein levels of IKKα, IKKβ, p-IKKα/β, p-IκBα, and p-p65. Overexpression of FGL1 enhanced these results.
Research conclusions
FGL1 may promote intestinal inflammation modulated by the canonical NF-κB signalling pathway and has the potential to be a therapeutic target for CD.
Research perspectives
Our findings indicate a critical role of FGL1 in the onset and progression of CD, which may serve as a potential prognostic biomarker and therapeutic target for CD.
ACKNOWLEDGEMENT
The authors are deeply grateful to Fan Yang from the Faculty of Art and Science, St. George Campus, University of Toronto, for helping us analyze the data in the statistics of this study.
Footnotes
Institutional review board statement: The study was approved by the Ethics Committee of the Affiliated Hospital of Nanjing University of Chinese Medicine (2018NL-171-02).
Conflict-of-interest statement: To the best of our knowledge, no conflict of interest exists.
Manuscript source: Unsolicited manuscript
Peer-review started: March 5, 2021
First decision: April 5, 2021
Article in press: August 10, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kumar S S-Editor: Gong ZM L-Editor: Wang TQ P-Editor: Xing YX
Contributor Information
Xue-Liang Sun, First Clinical Medical College, The Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, Jiangsu Province, China; Department of Colorectal Surgery, Suzhou TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Suzhou 215000, Jiangsu Province, China.
Li-Chao Qiao, First Clinical Medical College, The Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, Jiangsu Province, China.
Jing Gong, First Clinical Medical College, The Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, Jiangsu Province, China.
Ke Wen, Department of Colorectal Surgery, Suzhou TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Suzhou 215000, Jiangsu Province, China.
Zhi-Zhong Xu, Department of Colorectal Surgery, Suzhou TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Suzhou 215000, Jiangsu Province, China.
Bo-Lin Yang, First Clinical Medical College, The Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, Jiangsu Province, China; Department of Colorectal Surgery, Jiangsu Province Hospital of Chinese Medicine, Nanjing 210029, Jiangsu Province, China. yfy0051@njucm.edu.cn.
Data sharing statement
No additional data are available.
References
- 1.Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn's disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 2.Patel KV, Darakhshan AA, Griffin N, Williams AB, Sanderson JD, Irving PM. Patient optimization for surgery relating to Crohn's disease. Nat Rev Gastroenterol Hepatol. 2016;13:707–719. doi: 10.1038/nrgastro.2016.158. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy NA, Heap GA, Green HD, Hamilton B, Bewshea C, Walker GJ, Thomas A, Nice R, Perry MH, Bouri S, Chanchlani N, Heerasing NM, Hendy P, Lin S, Gaya DR, Cummings JRF, Selinger CP, Lees CW, Hart AL, Parkes M, Sebastian S, Mansfield JC, Irving PM, Lindsay J, Russell RK, McDonald TJ, McGovern D, Goodhand JR, Ahmad T UK Inflammatory Bowel Disease Pharmacogenetics Study Group. Predictors of anti-TNF treatment failure in anti-TNF-naive patients with active luminal Crohn's disease: a prospective, multicentre, cohort study. Lancet Gastroenterol Hepatol. 2019;4:341–353. doi: 10.1016/S2468-1253(19)30012-3. [DOI] [PubMed] [Google Scholar]
- 4.Löwenberg M, Vermeire S, Mostafavi N, Hoentjen F, Franchimont D, Bossuyt P, Hindryckx P, Rispens T, de Vries A, van der Woude CJ, Berends S, Ambarus CA, Mathot R, Clasquin E, Baert F, D'Haens G. Vedolizumab Induces Endoscopic and Histologic Remission in Patients With Crohn's Disease. Gastroenterology. 2019;157:997–1006.e6. doi: 10.1053/j.gastro.2019.05.067. [DOI] [PubMed] [Google Scholar]
- 5.Gisbert JP, Chaparro M. Clinical Usefulness of Proteomics in Inflammatory Bowel Disease: A Comprehensive Review. J Crohns Colitis. 2019;13:374–384. doi: 10.1093/ecco-jcc/jjy158. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto T, Gotoh M, Sasaki H, Terada M, Kitajima M, Hirohashi S. Molecular cloning and initial characterization of a novel fibrinogen-related gene, HFREP-1. Biochem Biophys Res Commun. 1993;193:681–687. doi: 10.1006/bbrc.1993.1678. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Guo Y, Lu L, Lu J, Ke M, Xu T, Lu Y, Chen W, Wang J, Kong D, Shen Q, Zhu Y, Tan W, Ji W, Zhou W. Fibrinogen-Like Protein 1 Is a Novel Biomarker for Predicting Disease Activity and Prognosis of Rheumatoid Arthritis. Front Immunol. 2020;11:579228. doi: 10.3389/fimmu.2020.579228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Sanmamed MF, Datar I, Su TT, Ji L, Sun J, Chen L, Chen Y, Zhu G, Yin W, Zheng L, Zhou T, Badri T, Yao S, Zhu S, Boto A, Sznol M, Melero I, Vignali DAA, Schalper K. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell. 2019;176:334–347.e12. doi: 10.1016/j.cell.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichtenstein GR, Loftus EV, Isaacs KL, Regueiro MD, Gerson LB, Sands BE. ACG Clinical Guideline: Management of Crohn's Disease in Adults. Am J Gastroenterol. 2018;113:481–517. doi: 10.1038/ajg.2018.27. [DOI] [PubMed] [Google Scholar]
- 10.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 11.Titz B, Gadaleta RM, Lo Sasso G, Elamin A, Ekroos K, Ivanov NV, Peitsch MC, Hoeng J. Proteomics and Lipidomics in Inflammatory Bowel Disease Research: From Mechanistic Insights to Biomarker Identification. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starr AE, Deeke SA, Ning Z, Chiang CK, Zhang X, Mottawea W, Singleton R, Benchimol EI, Wen M, Mack DR, Stintzi A, Figeys D. Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn's disease from UC. Gut. 2017;66:1573–1583. doi: 10.1136/gutjnl-2015-310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Townsend P, Zhang Q, Shapiro J, Webb-Robertson BJ, Bramer L, Schepmoes AA, Weitz KK, Mallette M, Moniz H, Bright R, Merrick M, Shah SA, Sands BE, Leleiko N. Serum Proteome Profiles in Stricturing Crohn's Disease: A Pilot Study. Inflamm Bowel Dis. 2015;21:1935–1941. doi: 10.1097/MIB.0000000000000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Wei W, Tang Q, Lu L, Luo Z, Li W, Lu Y, Pu J. Oxysophocarpine suppresses hepatocellular carcinoma growth and sensitizes the therapeutic blockade of anti-Lag-3 via reducing FGL1 expression. Cancer Med. 2020;9:7125–7136. doi: 10.1002/cam4.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun C, Gao W, Liu J, Cheng H, Hao J. FGL1 regulates acquired resistance to Gefitinib by inhibiting apoptosis in non-small cell lung cancer. Respir Res. 2020;21:210. doi: 10.1186/s12931-020-01477-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bie F, Wang G, Qu X, Wang Y, Huang C, Du J. Loss of FGL1 induces epithelialmesenchymal transition and angiogenesis in LKB1 mutant lung adenocarcinoma. Int J Oncol. 2019;55:697–707. doi: 10.3892/ijo.2019.4838. [DOI] [PubMed] [Google Scholar]
- 17.Wu HT, Ou HY, Hung HC, Su YC, Lu FH, Wu JS, Yang YC, Wu CL, Chang CJ. A novel hepatokine, HFREP1, plays a crucial role in the development of insulin resistance and type 2 diabetes. Diabetologia. 2016;59:1732–1742. doi: 10.1007/s00125-016-3991-7. [DOI] [PubMed] [Google Scholar]
- 18.Kang L, Li HY, Ou HY, Wu P, Wang SH, Chang CJ, Lin SY, Wu CL, Wu HT. Role of placental fibrinogen-like protein 1 in gestational diabetes. Transl Res. 2020;218:73–80. doi: 10.1016/j.trsl.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Wu HT, Chen SC, Fan KC, Kuo CH, Lin SY, Wang SH, Chang CJ, Li HY. Targeting fibrinogen-like protein 1 is a novel therapeutic strategy to combat obesity. FASEB J. 2020;34:2958–2967. doi: 10.1096/fj.201901925R. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Ukomadu C. Fibrinogen-like protein 1, a hepatocyte derived protein is an acute phase reactant. Biochem Biophys Res Commun. 2008;365:729–734. doi: 10.1016/j.bbrc.2007.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierre N, Baiwir D, Huynh-Thu VA, Mazzucchelli G, Smargiasso N, De Pauw E, Bouhnik Y, Laharie D, Colombel JF, Meuwis MA, Louis E GETAID (Groupe d'Etude Thérapeutique des Affections Inflammatoires du tube Digestif) Discovery of biomarker candidates associated with the risk of short-term and mid/long-term relapse after infliximab withdrawal in Crohn's patients: a proteomics-based study. Gut. 2020 doi: 10.1136/gutjnl-2020-322100. [DOI] [PubMed] [Google Scholar]
- 22.Atreya I, Atreya R, Neurath MF. NF-kappaB in inflammatory bowel disease. J Intern Med. 2008;263:591–596. doi: 10.1111/j.1365-2796.2008.01953.x. [DOI] [PubMed] [Google Scholar]
- 23.Rius-Pérez S, Pérez S, Martí-Andrés P, Monsalve M, Sastre J. Nuclear Factor Kappa B Signaling Complexes in Acute Inflammation. Antioxid Redox Signal. 2020;33:145–165. doi: 10.1089/ars.2019.7975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.