Abstract
Background
Circadian rhythm disorders, often seen in modern lifestyles, are a major social health concern. The aim of this study was to examine whether circadian rhythm disorders would influence angiogenesis and blood perfusion recovery in a mouse model of hind limb ischemia.
Methods and Results
A jet‐lag model was established in C57BL/6J mice using a light‐controlled isolation box. Control mice were kept at a light/dark 12:12 (12‐hour light and 12‐hour dark) condition. Concentrations of plasma vascular endothelial growth factor and circulating endothelial progenitor cells in control mice formed a circadian rhythm, which was diminished in the jet‐lag model (P<0.05). The jet‐lag condition deteriorated tissue capillary formation (P<0.001) and tissue blood perfusion recovery (P<0.01) in hind limb ischemia, which was associated with downregulation of vascular endothelial growth factor expression in local ischemic tissue and in the plasma. Although the expression of clock genes (ie, Clock, Bmal1, and Cry) in local tissues was upregulated after ischemic injury, the expression levels of cryptochrome (Cry) 1 and Cry2 were inhibited by the jet‐lag condition. Next, Cry1 and Cry2 double‐knockout mice were examined for blood perfusion recoveries and a reparative angiogenesis. Cry1 and Cry2 double‐knockout mice revealed suppressed capillary density (P<0.001) and suppressed tissue blood perfusion recovery (P<0.05) in the hind limb ischemia model. Moreover, knockdown of CRY1/2 in human umbilical vein endothelial cells was accompanied by increased expression of WEE1 and decreased expression of HOXC5. This was associated with decreased proliferative capacity, migration ability, and tube formation ability of human umbilical vein endothelial cells, respectively, leading to impairment of angiogenesis.
Conclusions
Our data suggest that circadian rhythm disorder deteriorates reparative ischemia‐induced angiogenesis and that maintenance of circadian rhythm plays an important role in angiogenesis.
Keywords: angiogenesis, circadian rhythm, cryptochrome, endothelial progenitor cell, hind limb ischemia, vascular endothelial growth factor
Subject Categories: Angiogenesis, Ischemia, Vascular Biology
Nonstandard Abbreviations and Acronyms
- Cry
cryptochrome
- EPC
endothelial progenitor cell
- FBS
fetal bovine serum
- HLI
hind limb ischemia
- HUVEC
human umbilical vein endothelial cell
- POD
postoperative day
- SDF‐1
stromal cell–derived factor‐1
- VEGFR
vascular endothelial growth factor receptor
Clinical Perspective
What Is New?
Circulating vascular endothelial growth factor and endothelial progenitor cell in physiological condition display the rhythm peaking at daytime (ie, resting period for mouse), which were diminished by constant jet‐lag condition.
Cryptochrome 1/2 deficiency mice were impaired in blood perfusion recovery in a unilateral lower limb ischemia model and displayed reduced capillary density compared with wild‐type mice.
What Are the Clinical Implications?
Environment‐induced constant circadian rhythm disorder has an adverse effect on reparative angiogenesis in a hind limb ischemia model.
Circadian rhythm is an endogenous diurnal rhythm that induces oscillations in organisms and generates periodicity even when there is no time information from the outside environment.1 It is a fundamental function that has been acquired by life on the Earth, from plants, cyanobacteria, Drosophila, fish, and birds to mammals and higher primates, and is responsible for adaptation to the environmental changes during the rotation cycle.1 Conversely, circadian rhythm disorders in humans, resulting from modern society lifestyles changes, such as shift work, jet lag, or nighttime mobile telephone use, have emerged as major social health problems.2, 3, 4 For instance, many epidemiological studies have reported that circadian rhythm disorders correlate with an increased risk of obesity, diabetes mellitus, hypertension, arrhythmia, and ischemic heart disease.5, 6, 7, 8, 9, 10, 11 However, there is still a large knowledge gap between the precise causal mechanism underlying the pathogenesis of circadian rhythm disorders, especially in the setting of reparative angiogenesis.12
Since the discovery of clock genes, an accumulation of evidence has revealed that the core loop (formed by CLOCK, BMAL1, Period [PER], and cryptochrome [CRY]) and the auxiliary interlocking loop (formed by Rev‐Erb and ROR) regulate the transcriptional activity of the core loop by dynamically forming the spontaneous circadian rhythm and are involved in various physiological phenomena as clock outputs.13 In cardiomyocytes, vascular endothelial cells, and vascular smooth muscle cells, which are the main constituent cells of the cardiovascular system, 5% to 10% of genes have a cycle of gene expression that is regulated in a 24‐hour rhythm,14 and at least part of these genes is thought to be regulated by peripheral clock genes in each cell. In the field of cardiovascular disease (CVD), there are observation clinical studies about the relationship between some CVDs and the timing of frequent onset. However, research into the genetic and molecular mechanisms of circadian rhythm–regulated angiogenesis is limited. Accordingly, we investigated the effects of circadian rhythm disorder on reparative angiogenesis and its potential molecular mechanisms in the setting of hind limb ischemia (HLI) using both an environment‐induced and a genetically induced circadian rhythm disruption in a mouse model.
Methods
The data, analytic methods, and study materials of this study are available from the corresponding author on reasonable request.
Animal Care
All procedures of animal care and use in this study were approved by the Animal Ethics Review Board of Nagoya University School of Medicine. Our study conformed to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes or the National Institutes of Health Guide for the Care and Use of Laboratory Animals. We used the only male mice in present studies, as described in the ATVB Council Statement for considering sex difference as a biological variable.15 Male C57BL/6J mice (age, 8 weeks) were purchased from Charles River Laboratories Japan, Inc (Kanagawa, Japan). Cry1 and Cry2 deficiency mice were kindly provided by Dr Takeshi Todo (Osaka University). In this study, we use Cry1 and Cry2 double‐heterozygous knockout (Cry1/2‐DKO) mice.16 A new set of wild‐type HLI mice was repeated for the DKO experiments. Wild‐type littermates were used as controls for the DKO experiments. Mice (male, aged 8–10 weeks) were randomly assigned to the experimental groups. All the mice were anesthetized with medetomidine hydrochloride (0.3 mg/kg), midazolam (4 mg/kg), and butorphanol tartrate (5 mg/kg) before the surgical procedure as well as during laser Doppler measurements of limb blood perfusion. We used a 26‐gauge needle to draw about 500 to 1000 μL of blood from the heart cavity of mice by slow aspiration, taking care not to destroy blood cells, and used it for the following experiments. Cervical dislocation was used for the animal euthanasia method.
Mouse Jet‐Lag Model
Male C57BL/6J mice (aged 8 weeks) were set in a home cage in a light‐shield mouse‐housing box (MELQUEST, No. MBX‐002). The mice were acclimatized to the environment in Nagoya University Graduate School of Medicine for 1 week (light‐dark [LD] condition with 9:00 [zeitgeber time 0: ZT0]–21:00 [ZT12] light period) before being housed under the control condition and advance time shift condition (8‐hour phase advance once every 4 days) with LED lights on and off that are adjusted by an optional timer (MELQUEST, No. LCT‐8).17 The animals were allowed ad libitum access to food and water and were regularly subjected to observation. After 12 days of housing in control or jet‐lag environment, a unilateral lower limb ischemia model was created in these mice as described below. Thereafter, the control group was housed under normal condition, and the jet‐lag group was continued to be housed in the jet‐lag model.
Enzyme‐Linked Immunosorbent Assay
Concentrations of vascular endothelial growth factor (VEGF) and stromal cell–derived factor‐1 (SDF‐1) α proteins in murine blood plasma samples were determined by ELISA kits (mouse VEGF ELISA kit, R&D Systems, and mouse CXCL12/SDF‐1α Quantikine ELISA kit, respectively), according to manufacturer's instructions.18 Plasma levels of VEGF and SDF‐1α in sham operation and at day 7 after HLI surgery were measured every 6 hours (6:00, 12:00, 18:00, and 24:00) in both the control and jet‐lag groups.
Flow Cytometry
Blood samples were collected every 6 hours (6:00, 12:00, 18:00, and 24:00). Endothelial progenitor cell (EPC) populations in blood were analyzed using fluorescence‐activated cell sorting with fluorescein isothiocyanate–labeled anti‐CD34 monoclonal antibody (1:80, BD Bioscience) and phycoerythrin‐labeled anti‐flk1 monoclonal antibody (1:100, BD Bioscience).19 Isotype‐matched IgGs were used for negative control. Immunofluorescence‐labeled cells were analyzed by fluorescence‐activated cell sorting and Cell Quest Software, counting 10 000 events per sample.20
Mouse Model of HLI
Unilateral HLI was induced in mice under anesthesia. Left femoral artery ligation and total excision of the branches were performed as previously described.20, 21
Blood Perfusion Assessment
The laser Doppler perfusion imaging system (Moor Instruments, Devon, UK) was used to record blood flow changes before and after surgery on postoperative days (PODs) 0, 7, 14, 21, and 28 around 10 am.20 Briefly, the animals were kept in a dorsal position after intraperitoneal anesthesia with their hind limbs slightly fixed and excess hairs removed before imaging. Room temperature was stabilized at 25 °C, and mouse body temperatures were kept at 37 °C by using a hotplate. To account for variables, including light and temperature, calculated perfusion was expressed as a ratio of surgical (ischemic)/nonsurgical (nonischemic) limb.20 Individuals with limb amputations were excluded from this analysis because it was not possible to evaluate blood flow to the target limb using laser Doppler.
Severity of Ischemic Limb Status
States of ischemic limbs were divided into 3 groups: limb loss, toe necrosis, and limb salvage. Limb ischemia severity was compared between the control and jet‐lag groups.
Immunohistochemical Staining and Capillary Density Analysis
The frozen sections were collected from ischemic hind limb muscles at POD 28. Sections at 8‐μm thick were fixed in 4% paraformaldehyde, washed twice with PBS, and blocked using 0.5% BSA at room temperature for 1 hour. Sections were then incubated with primary antibody CD31 monoclonal antibody (1:1000, BD Pharmingen) at 4 °C overnight, followed by incubation with secondary antibody Alexa Fluor 594–conjugated anti‐rat antibody (1:1000, Thermo Fisher Scientific) at room temperature for 1 hour. The nuclei were identified with 4′,6‐diamidino‐2‐phenylindole (1:1000, DOJINDO). Images were visualized on a BZ‐X710 fluorescent microscope (KEYENCE, Japan). Results were counted as the average positive cells per field.18, 21, 22
Isolation of RNA and Real‐Time Reverse Transcription–Polymerase Chain Reaction
RNA was isolated using the RNeasy Micro Kit, according to the manufacturer's instructions (QIAGEN). Reverse transcription was performed with 1‐μg total RNA in a standard manner with ReverTra Ace qPCR RT Master Mix (TOYOBO) supplemented with DNase treatment.18 Real‐time reverse transcription–polymerase chain reaction analysis of the VEGF, SDF‐1, Clock, Bmal1, Cry1, Cry2, Per2, VEGF receptor (VEGFR) 1, VEGFR2, WEE1, HOXC5, and GAPDH23, 24, 25 mRNAs was performed on C1000 Thermal Cycler (BIO‐RAD) using SYBR Green I and the following conditions: 95 °C for 10 minutes, followed by 40 cycles at 95 °C for 15 seconds and 60 °C for 45 seconds.18 The forward primer for mouse VEGF was as follows: 5′‐AGCACAGCAGATGTGAATGC‐3′; the reverse primer: 5′‐AATGCTTTCTCCGCTCTGAA‐3′. The forward primer for mouse SDF‐1 was as follows: 5′‐TGCATCTGGATAGGGAAAGG‐3′; the reverse primer: 5′‐ATCAGGCAATGAACCAGAGG‐3′. The forward primer for mouse HIF1 was as follows: 5′‐CCTGCACTGAATCAAGAGGTTGC‐3′; the reverse primer: 5′‐CCATCAGAAGGACTTGCTGGCT‐3′. The forward primer for mouse Wee1 was as follows: 5′‐GAAACAAGACCTGCCAAAAGAA‐3′; the reverse primer: 5′‐GCATCCATCTAACCTCTTCACAC‐3′. The forward primer for mouse HOXC5 was as follows: 5′‐CATGCCTGTTTGCTGTCATC‐3′; the reverse primer: 5′‐CATTGTGGAAGGCTGGAGAG‐3′. The forward primer for mouse CXCR4 was as follows: 5′‐TCAGTGGCTGACCTCCTCTT‐3′; the reverse primer: 5′‐CTTGGCCTTTGACTGTTGGT‐3′. The forward primer for mouse Clock was as follows: 5′‐AAGATTCTGGGTCTGACAAT‐3′; the reverse primer: 5′‐TTGCAGCTTGAGACATCGCT‐3′. The forward primer for human Clock was as follows: 5′‐AAGTTAGGGCTGAAAGACGACG‐3′; the reverse primer: 5′‐GAACTCCGAGAAGAGGCAGAAG‐3′. The forward primer for mouse Bmal1 was as follows: 5′‐TGACCCTCATGGAAGGTTAGAA‐3′; the reverse primer: 5′‐GGACATTGCATTGCATGTTGG‐3′. The forward primer for human Bmal1 was as follows: 5′‐CCACCAATCCATACACAGAAGC‐3′; the reverse primer: 5′‐TCCCTCGGTCACATCCTACG‐3′. The forward primer for mouse Cry1 was as follows: 5′‐CACTGGTTCCGAAAGGGACTC‐3′; the reverse primer: 5′‐CTGAAGCAAAAATCGCCACCT‐3′. The forward primer for human Cry1 was as follows: 5′‐TTGGAAAGGAACGAGACGCA‐3′; the reverse primer: 5′‐GCGGTTGTCCACCATTGAGT‐3′. The forward primer for mouse Cry2 was as follows: 5′‐GGGACTCTGTCTATTGGCATCTG‐3′; the reverse primer: 5′‐GTCACTCTAGCCCGCTTGGT‐3′. The forward primer for human Cry2 was as follows: 5′‐GTGTTCCCAAGGCTGGTTCTCC‐3′; the reverse primer: 5′‐GTAGGTCTCGTCGTGGTTCTCC‐3′. The forward primer for mouse Per2 was as follows: 5′‐ACACCACCCCTTACAAGCTTC‐3′; the reverse primer: 5′‐CGCTGGATGATGTCTGGCTC‐3′. The forward primer for human Per2 was as follows: 5′‐GCAGGTGAAAGCCAATGAAG‐3′; the reverse primer: 5′‐CACCGCAAACATATCGGCAT‐3′. The forward primer for human VEGFR1 was as follows: 5′‐CCTCACTGCCACTCTAATTGTC‐3′; the reverse primer: 5′‐ACAGTTTCAGGTCCTCTCCTT‐3′. The forward primer for human VEGFR2 was as follows: 5′‐CTCATGTCTGAACTCAAGATCC‐3′; the reverse primer: 5′‐CCAGAATCCTCTTCCATGCTCA‐3′. The forward primer for human WEE1 was as follows: 5′‐ACTGATAGAATCCAGTTTGC‐3′; the reverse primer: 5′‐ATGCAATGCCTACAAAGTGC‐3′. The forward primer for human HOXC5 was as follows: 5′‐AGGTGCAGGCATCCAGGTACT‐3′; the reverse primer: 5′‐GGGTTGGCAGCCATGTCTAC‐3′. The forward primer for mouse GAPDH was as follows: 5′‐ACCCAGAAGACTGTGGATGG‐3′; the reverse primer: 5′‐CACATTGGGGGTAGGAACAC‐3′. The forward primer for human GAPDH was as follows: 5′‐AGGTGCAGGCATCCAGGTACT‐3′; the reverse primer: 5′‐GGCCATCCACAGTCTTCTCAG‐3′.
Cell Culture
Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza. When cultured HUVECs reached ≈90% confluent, monolayers of cells were transfected with siRNA against: both Cry1 (100 pmol/L, Ambion) and Cry2 (100 pmol/L, Ambion) or scrambled sequence (negative control, 200 pmol/L, Ambion).26 Transfections were performed with Lipofectamine 2000 (Thermo Fisher Scientific) by incubating the HUVECs with the respective siRNA for 1 day.26 Afterward, medium containing the transfecting siRNA agent was removed from the cell monolayers, washed twice with cold PBS, and replenished with fresh EBM‐2 with 0.5% fetal bovine serum (FBS). For another experiment, HUVECs were cultured with 0.1% dimethyl sulfoxide (vehicle group) or 40 μmol/L of KL001 (funakoshi) in EBM‐2 with 0.5% FBS for 16 hours.
Tube Formation Assay
HUVECs were cultured in the complete medium, and were changed to EBM‐2 with 0.5% FBS medium as a serum starvation overnight. After overnight starvation, the cells were transferred onto a 6‐well plate coated with Matrigel (Becton Dickinson, Bedford, MA) at 2×104 cells/well in EBM‐2 with 0.5% FBS medium.22 Cells were incubated at 37 °C for 6 hours, and morphological changes of HUVECs were observed and photographed in 3 different fields per well under a microscope. Inhibition of tube formation was assessed by the measurement of total tube length formed in each photograph using the Image J software (version 1.51).22
Proliferation Assay
Cells were incubated in a 96‐well tissue culture plate with the WST‐1 reagent for 6 hours.18, 22 We then evaluated cell proliferation using the Premix WST‐1 Cell Proliferation Assay System (TAKARA Bio Inc), according to the manufacturer's instructions, with a scanning multiwell spectrophotometer (440 nm; reference, 600 nm).18, 22 The measured absorbance directly correlates to the number of viable cells.
Migration Assay
Cell migration assay was performed using 3‐mm pore size Costar transwell migration chambers (Corning Inc) in 24‐well plates.18, 21 Then, 4×104 cells/100 μL in EBM‐2 with 0.5% FBS were plated on the top of the chamber membrane and incubated for 8 hours.18, 21 Cells were then fixed with 3% paraformaldehyde at room temperature for 15 minutes and stained with 0.1% 4′,6‐diamidino‐2‐phenylindole for 3 minutes. The stain was rinsed off thoroughly with PBS. Cells remaining on the top of the migration chamber were removed by gently swabbing using a cotton tip, and stained cells adhering to the bottom of the chamber membrane were counted. The cell numbers of 5 random fields at the magnification of 200 for each migration chamber membrane were counted, and the average number was recorded.
Statistical Analysis
Results are expressed as mean±SEM. Shapiro‐Wilk normality test was performed to evaluate data distribution. Homogeneity of variance was evaluated by F test. Normally distributed data with 1 variable were analyzed by the unpaired Student t test to evaluate the statistical significance between the 2 groups; 1‐way ANOVA along with the Tukey post hoc test was used for ≥3 groups. We also used a 2‐way repeated‐measures ANOVA (Bonferroni) to assess the changes over time. Nonnormally distributed data were analyzed by 2‐tailed Mann‐Whitney U test between 2 groups and Kruskal‐Wallis H test with post hoc Dunn test among 3 groups. GraphPad Prism software version 8.0 (GraphPad Software Inc) was used. Values of P<0.05 were considered statistically significant.27
Results
Relationship Between Circadian Rhythm and Circulating VEGF, SDF‐1, and EPCs With or Without Daily Jet Lag
Circadian fluctuations in plasma VEGF levels were observed in normally housed control mice peaking at around noon (ie, resting period for mouse). In contrast, the circadian variation in plasma VEGF concentrations disappeared in the jet‐lag group. In addition, plasma VEGF levels were consistently decreased in the jet‐lag group compared with the control group (Figure 1A). Figure 2B demonstrates that circadian fluctuations of SDF‐1 levels peaked at around noon in a similar manner as the kinetics for plasma VEGF levels in normally housed control mice. However, the jet‐lag intervention merely shifted the peak in plasma SDF‐1 levels. Next, the number of mobilized EPCs in the peripheral blood peaked at daytime (ie, resting period for mice) in normally housed mice. Conversely, the intraday variability of circulating EPCs disappeared in the jet‐lag model, and the number of EPCs in the peripheral blood decreased compared with the control group (Figure 1C and 1D).
Figure 1. Circadian kinetics of circulating vascular endothelial growth factor (VEGF), stromal cell–derived factor‐1 (SDF‐1), and endothelial progenitor cells in mice.
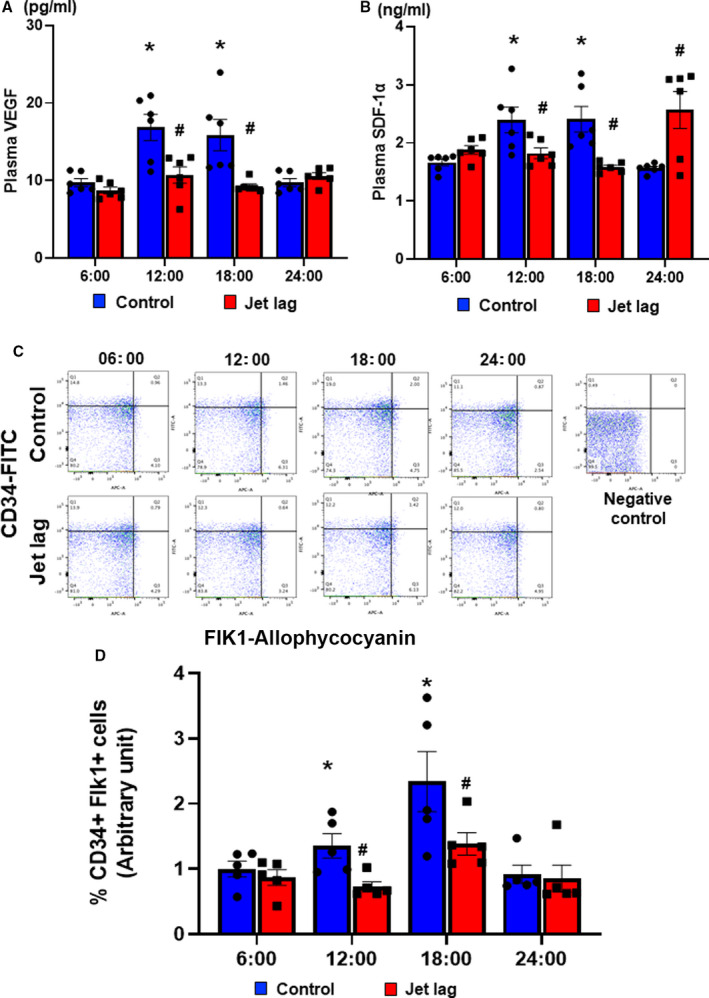
A, Plasma level of VEGF concentration under control or jet‐lagged condition for 13 days at 6:00, 12:00, 18:00, and 24:00 detected by ELISA kit. Values are mean±SEM (n=8 for each). *P<0.05 vs 6:00, # P<0.05 vs at the same time of control, by 1‐way ANOVA and Tukey post hoc tests. B, Plasma level of SDF‐1α concentration under control or jet‐lagged condition for 13 days at 6:00, 12:00, 18:00, and 24:00 detected by ELISA kit. Values are mean±SEM (n=8 for each). *P<0.05 vs 6:00, # P<0.05 vs at the same time of control, by 1‐way ANOVA and Tukey post hoc tests. Representative images of circulating CD34‐ and Flik1‐positive cells by fluorescence‐activated cell sorting analysis (C) and the quantification of CD34 and Flik1 double‐positive cell ratio (D) at 6:00, 12:00, 18:00, and 24:00. Data are mean±SEM (n=5). *P<0.05 vs 6:00, # P<0.05 vs at the same time of control, by 1‐way ANOVA and Tukey post hoc tests. FITC indicates fluorescein isothiocyanate.
Figure 2. Constant jet lag blocked angiogenesis and blood perfusion recovery in hind limb ischemia (HLI), following the downregulation of vascular endothelial growth factor (VEGF) and stromal cell–derived factor‐1 (SDF‐1) expression.
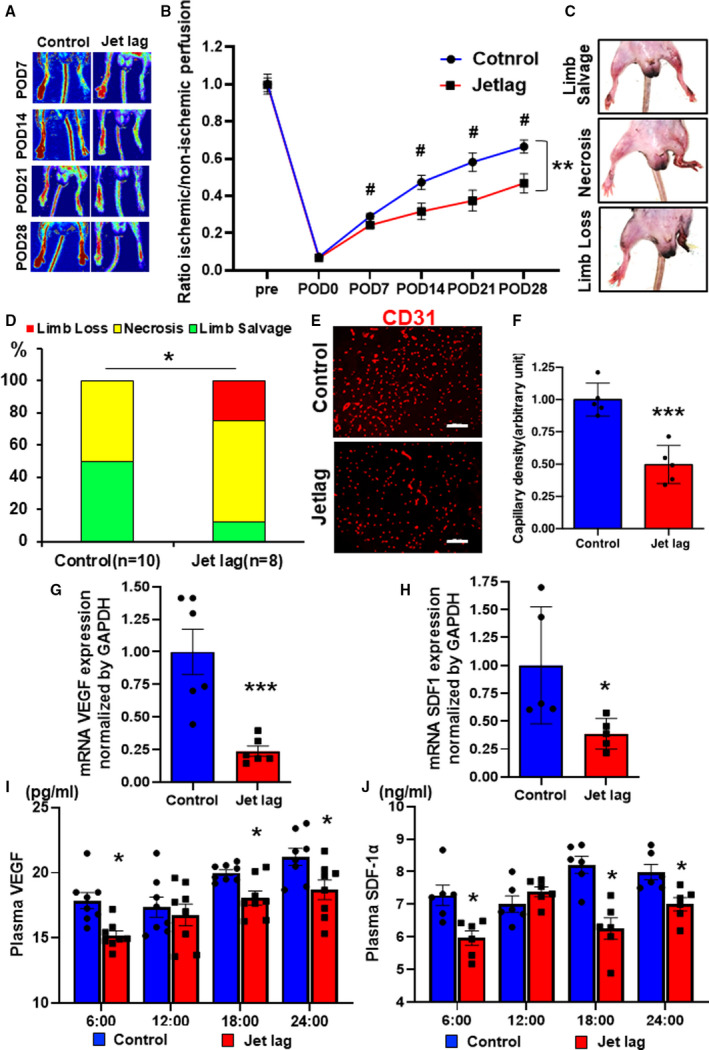
Representative laser Doppler perfusion imaging (LDPI) A, and summary of the LDPI ratio (ischemic/nonischemic) B, in the setting of blood perfusion recovery under control or constant jet‐lagged condition. Data are mean±SEM (n=8). **P<0.01, by 2‐way ANOVA and Bonferroni post hoc tests. # P<0.05 vs at the same time of control, by 1‐way ANOVA and Tukey post hoc tests. C, Representative images of limb loss, necrosis, and limb salvage in HLI mice. D, Quantitative analysis of limb ischemia severity for each group in an ischemic limb at day 28. Data are mean±SEM (n=10 for the control group, n=8 for the jet lag group). *P<0.05, by 2‐way ANOVA and Bonferroni post hoc tests. Immunostaining with anti‐CD31 (red) in the control or jet‐lag group E, and quantitative analysis of CD31‐positive cells for each group F, in an ischemic limb at day 28. Bar=100 μm. Data are mean±SEM (n=5). ***P<0.001 vs control, by unpaired Student t test. G and H, The expressions of mRNA VEGF and SDF‐1 in HLI muscles of the control or jet‐lag group. Data are mean±SEM (n=6). *P<0.05, ***P<0.001 vs control, by unpaired Student t test. I and J, Plasma level of VEGF and SDF‐1α concentration in HLI mice of the control or jet‐lag group at 6:00, 12:00, 18:00, and 24:00 detected by ELISA kit. Values are mean±SEM (n=8). *P<0.05 vs 6:00 vs at the same time of control, by 1‐way ANOVA and Tukey post hoc tests. POD indicates postoperative day.
Continuous Jet Lag Inhibits Angiogenesis and Blood Perfusion Recovery in a Murine Model of HLI
Jet lag did not change the food and water consumption of the experimental mouse (Figure S1). Laser Doppler measurement of the blood flow in the lower limbs revealed that the jet‐lag group had poorer blood flow recovery compared with the control group (Figure 2A and 2B). Moreover, on day 28 of the limb ischemia, the leg/toe necrosis and prolapses were more severe in the jet‐lag model than in the control group (Figure 2C and 2D). Furthermore, when ischemic skeletal muscle tissues were harvested and the capillary density was assessed at day 28 of limb ischemia, the capillary density was lower in the jet‐lag group than in the control group (Figure 2E and 2F). Both VEGF and SDF‐1 mRNA were significantly downregulated in the jet‐lag group compared with the control group (Figure 2G and 2H). In addition, Figure 2I and 2J show that plasma VEGF and SDF‐1 levels were also lower in the jet‐lag group than in the control group. Furthermore, circulating EPCs after limb ischemia were suppressed in the jet‐lag group compared with the control group (Figure S2). Taken together, these results indicate that environment‐induced circadian rhythm disturbance decreases VEGF and SDF‐1 expression in both local ischemic tissues and plasma, resulting in reduced angiogenic response and impaired blood flow recovery.
Time Course of Clock Gene Expression After Ischemic Injury In Vivo and Impact of Cryptochrome Deficiency on Blood Perfusion Recovery and Angiogenesis in an HLI Model
Next, the time courses of core‐loop forming clock gene expression, mRNA of Clock, Bmal, Period (Per), and Cry, were investigated in the setting of blood perfusion recovery in ischemic tissues to elucidate the molecular mechanisms in terms of a peripheral clock on reparative angiogenesis. Figure 3A through 3C revealed that the expression of Cry1, Cry2, and Clock in local tissues were upregulated, peaking around POD7 after ischemic injury. The expression of Bmal1 was upregulated, peaking around POD3 in ischemic hind limbs (Figure 3D). In contrast, the expression of Per2 was downregulated at POD5, as shown in Figure 3E. Interestingly, in the jet‐lag group, the expression levels of Cry1 and Cry2, Clock, and Bmal1 in ischemic tissues were down‐regulated at POD7 (Figure 3F through 3I) compared with the control group. On the other hand, the expression of Per2 was upregulated, contrary to other clock genes at POD5 (Figure 3J). Subsequently, we examined whether Cry deficiency would deteriorate reparative angiogenesis in HLI. Figure 4A and 4B revealed that Cry1/2‐DKO mice demonstrated worse blood perfusion recovery compared with control wild‐type mice. In addition, Cry1/2 deficiency suppressed capillary density at POD28 in the ischemic limb (Figure 4C and 4D). Furthermore, the expressions of VEGF and SDF‐1 in the ischemic skeletal muscle were downregulated in the CRY1/2 mice following HLI (Figure S3A and S3B).
Figure 3. The time series expression of peripheral clock genes in ischemic muscles and the impact of jet‐lag condition.
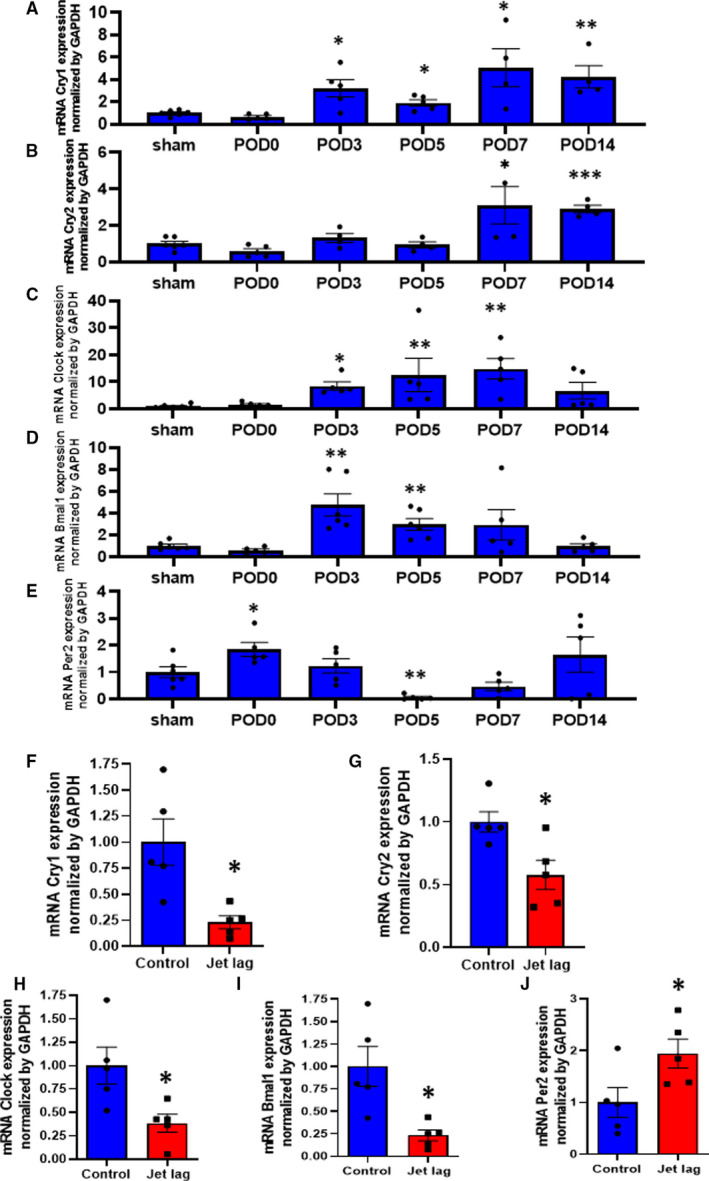
A through E, The time series of mRNA levels of cryptochrome (Cry) 1, Cry2, Clock, Bmal1, and Period 2 (Per2) in the samples collected from sham or ischemic muscles at post–hind limb ischemia day 0, 3, 5, 7, and 14. Data are mean±SEM. *P<0.05, **P<0.01, and ***P<0.001 vs sham, by 2‐way ANOVA and Bonferroni post hoc tests. F through J, The expressions of Cry1, Cry2, Clock, Bmal1, and Per2 in ischemic muscles at day 7 of postoperation in the control or jet‐lag group. Data are mean±SEM (n=5). *P<0.05, **P<0.01 vs control, by unpaired Student t test. POD indicates postoperative day.
Figure 4. Deficiency of cryptochrome (Cry) 1 and Cry2 genes inhibited angiogenesis and blood perfusion recovery in hind limb ischemia (HLI).
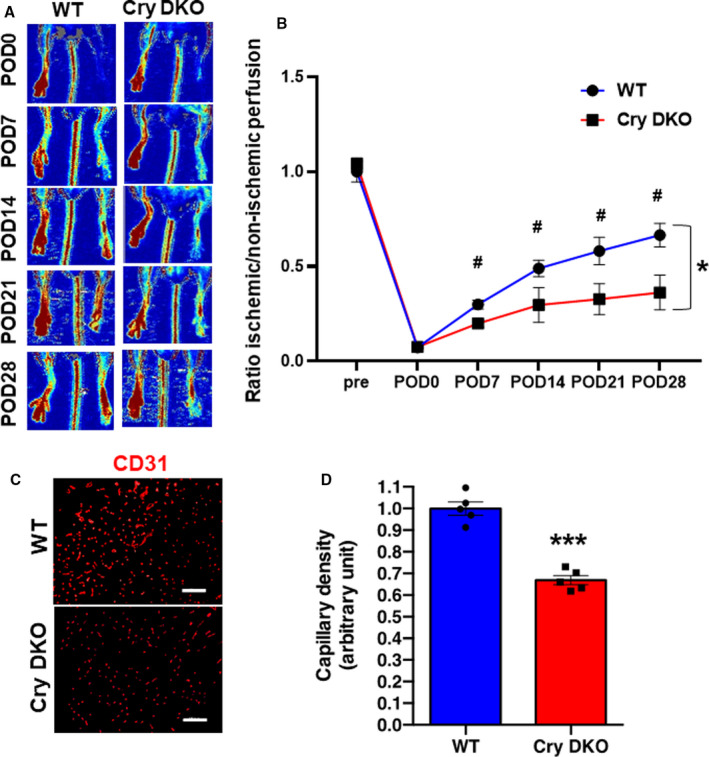
Representative images A, and summary of laser Doppler perfusion imaging ratio (ischemic/nonischemic) B, in wild‐type (WT) or Cry double‐knockout (DKO) mice. Data are mean±SEM (n=8). *P<0.05 vs WT analyzed using 2‐way ANOVA and Bonferroni post hoc tests. # P<0.05 vs at the same time of WT, by 1‐way ANOVA and Tukey post hoc tests. C, Immunostaining with anti‐CD31 (red) to detect blood endothelial cells. Bar=100 μm. D, Quantitative analysis of CD31‐positive cells in HLI in each group. Data are mean±SEM (n=5). ***P<0.001 vs WT, by unpaired Student t test. POD indicates postoperative day.
Cryptochrome Promotes the Proliferation, Migration, and Tube Formation of Endothelial Cells via Modulating WEE1 and HOXC5 Expression
Finally, to gain insight into the potential molecular mechanism, the role of Cry in endothelial cells on angiogenesis was investigated by loss‐of‐function studies. Knockdown of CRY1 and CRY2 in HUVECs did not influence the expression of CLOCK or BMAL1 (Figure S4A and S4B). Interestingly, PER, which is known to form a dimer with CRY, was upregulated in response to CRY1/2 double knockdown that could be considered a compensatory response (Figure S4C). The effects on angiogenesis were next evaluated in terms of proliferation, migration, and tube formation ability of endothelial cells as a functional analysis experiment. Analyses revealed that the CRY1/2 knockdown group inhibited the proliferation of vascular endothelial cells (Figure 5A). In addition, the migration ability of HUVECs with CRY1/2 knockdown, as evaluated by Boyden chamber experiments, demonstrated that the migration ability of HUVECs with CRY1/2 knockdown was inhibited (Figure 5B and 5C). Furthermore, when the tubular morphological features were evaluated by Matrigel experiments, it was found that CRY1/2 knockdown‐treated HUVECs inhibited the tubular morphological features (Figure 5D and 5E). Taken together, CRY1/2 in HUVECs plays an important role in angiogenesis.
Figure 5. Suppression of cryptochrome (Cry) 1 and Cry2 has the inhibitory effects on angiogenesis in human umbilical vein endothelial cells (HUVECs).
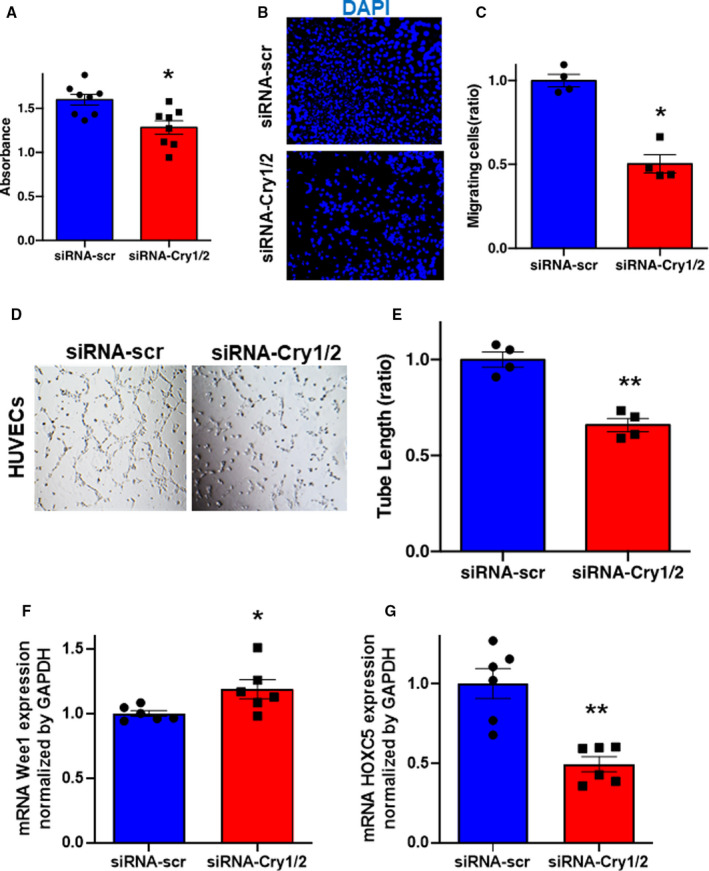
A, WST‐1 proliferation assay of HUVECs in the response to the treatment with siRNA‐scr or siRNA‐Cry1/2. B and C, Representative images of migrating cells stained by 4′,6‐diamidino‐2‐phenylindole and its quantitative analysis. Data are mean±SEM. *P<0.05 vs siRNA‐scr, analyzed by 2‐tailed Mann‐Whitney U tests. Bar=300 μm. Representative images of tube formation of HUVECs (D) and quantitative analysis by tube length (E) in each group. Data are mean±SEM (n=4). **P<0.01 vs siRNA‐scr, analyzed by unpaired Student t test. Bar=1 mm. F and G, The expressions of mRNA Wee1 and HOXC5 in HUVECs treated with siRNA‐scr or siRNA‐Cry1/2 by quantitative polymerase chain reaction. Data are mean±SEM. *P<0.05, **P<0.01, vs siRNA‐scr, by unpaired Student t test.
To gain insight into the clock control gene by CRY, quantitative polymerase chain reaction was performed on CRY1/2 knockdown‐treated HUVECs. Although the expression levels of VEGFR1 and VEGFR2 were not changed (Figure S5A and S5B), the expression of WEE1, a mitotic cycle stopper, was upregulated and the expression of HOXC5, a gene involved in a migratory capacity of endothelial cells, was downregulated within CRY1/2 knockdown HUVECs (Figure 5F and 5G). Conversely, treatment with KL001, which is a stabilizer of CRY1 and CRY2, promoted proliferation, migration, and tubular formation of HUVECs (Figure 6A through 6E). Moreover, the expression of WEE1 was downregulated (Figure 6F), and the expression of HOXC5 was upregulated, by KL001 (Figure 6G).
Figure 6. Stabilization of cryptochrome (Cry) 1 and Cry2 promotes angiogenesis in human umbilical vein endothelial cells (HUVECs).
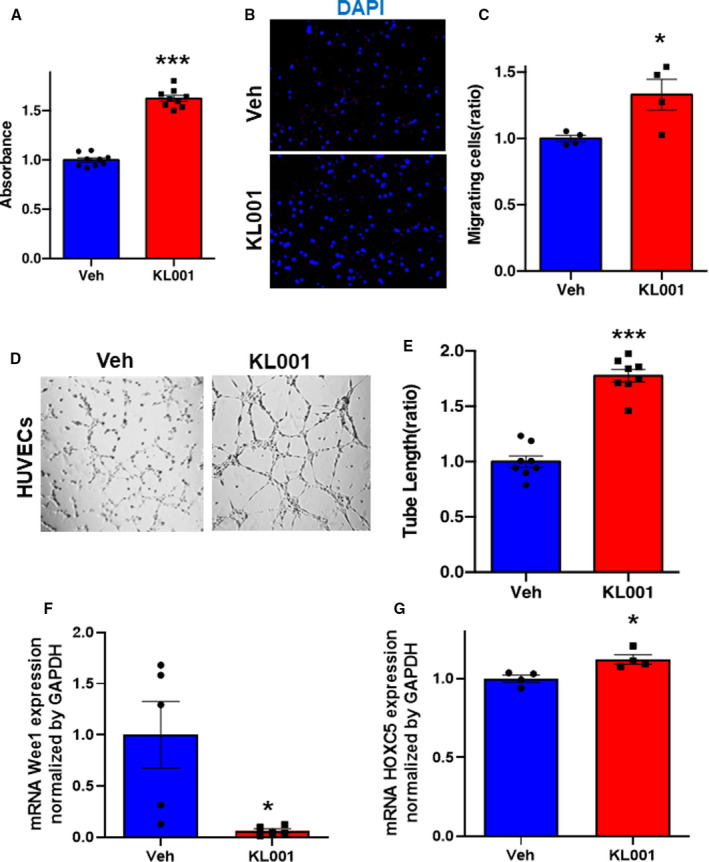
A, WST‐1 proliferation assay of HUVECs in the response to the treatment with vehicle (Veh) or KL001 (40 μmol/L). Data are mean±SEM (n=8). ***P<0.001 vs Veh, analyzed by unpaired Student t test. B and C, Representative images of migrating cells stained by 4′,6‐diamidino‐2‐phenylindole (DAPI) and its quantitative analysis. Data are mean±SEM (n=4). *P<0.05 vs Veh, analyzed by unpaired Student t test. Bar=300 μm. Representative images of tube formation of HUVECs (D) and quantitative analysis by tube length (E) in each group. Data are mean±SEM. ***P<0.001 vs Veh, analyzed by unpaired Student t test. Bar= 1 mm. F and G, The expressions of mRNA Wee1 and HOXC5 in HUVECs treated with vehicle or KL001 (40 μmol/L) by quantitative polymerase chain reaction. Data are mean±SEM. *P<0.05 vs Veh, by unpaired Student t test.
Discussion
The major findings of the present study are as follows: (1) plasma VEGF levels display circadian rhythm peaking at around noon in mice, which is diminished by circadian rhythm disturbance by a jet‐lag model; (2) circulating EPCs show diurnal variation in mice, which is canceled by jet‐lag model‐induced circadian rhythm disruption; (3) constant jet lag inhibits reparative angiogenesis as well as blood perfusion recovery in a mouse model of HLI; (4) deficiency of Cry1 and Cry2 genes inhibits angiogenesis and blood perfusion recovery in a mouse model of HLI; and (5) cryptochrome in endothelial cells promotes angiogenesis in terms of the proliferation, migration, and tube formation. The summary of the adverse effect of circadian rhythm disorder on reparative angiogenesis in HLI is shown in Figure S6.
The number of patients with ischemic CVD is increasing worldwide, especially in developed countries, and interventions against these diseases are one of the most important medical issues as they cannot only prolong life expectancy but also improve quality of life.28 The disturbance of regular circadian rhythms attributable to modern social lifestyle, such as shift work and jet lag, has recently become a hot topic of modern health concerns,2, 3, 4 and observational studies have suggested an association between circadian rhythm disorders and the risk of cardiovascular events.29 However, most of the reports on circadian rhythm disorders and the cardiovascular systems have come from epidemiological studies, and their molecular mechanisms are not fully understood.
VEGF is one of the key growth factors to augment postnatal angiogenesis and vasculogenesis.30, 31 Previous studies reported that tumor‐related VEGF expression in mice displayed a circadian rhythm,32, 33 and another study demonstrated that Per2‐deficient mice showed less VEGF expression in a myocardial infarction model.34 However, it is not known if a circulating VEGF level displays a 24‐hour circadian rhythm under physiological conditions or if a disruption in circadian rhythms disturbs the circulating level of VEGF. Bone marrow–derived EPCs circulate in the peripheral blood and are incorporated into newly formed blood vessels, contributing to vasculogenesis.35 Thus, circulating EPCs play an important role in vascular repair/regeneration during blood perfusion recovery at ischemic tissues. Circadian changes of circulating EPCs in blood have been reported to be disturbed in non–dipper‐type hypertension36 and type 2 diabetes mellitus.37 In addition, in an animal model, mobilization of EPCs in blood was inhibited by Per2 knockout.19, 34, 38 However, there is not any evidence on the impact of circadian rhythm disorder on the kinetics of circulating EPCs in 24‐hour intraday variation to date.39 Herein, our data unveiled the evidence that circulating VEGF and EPCs in physiological conditions display circadian rhythm peaking at daytime (ie, resting period for mouse), which were diminished by a constant jet‐lag condition. Moreover, the present study also revealed that the environment‐induced constant circadian rhythm disorder has an adverse effect on reparative angiogenesis in a mouse model of HLI.
The suprachiasmatic nucleus in the hypothalamus is the main body of the biological clock, and the clock cells in this region contain clock genes and keep time at a rhythm of ≈24 hours.40 However, each cell in the peripheral tissues also individually contains clock genes, which are coordinated with the central clock and form a 24‐hour cycle of gene expression at the cellular level.40 As a central oscillation mechanism for clock genes, the core loops formed by CLOCK, BMAL1, PER, and CRY and auxiliary interlocking loops formed by Rev‐Erb and ROR control the transcriptional activity of the core loops by dynamically forming spontaneous circadian rhythms and are involved in various physiological phenomena as clock outputs.41 The molecules act as transcriptional factors, with CLOCK and BMAL forming heterodimers and regulating gene expression positively.42 In contrast, heterodimers of PER and CRY regulate gene expression negatively,43 creating an oscillation of gene expression in the cell over a period of 24 hours in an exquisite balance.44, 45 In the present study, we focused on CRY among the 4 major clock genes.46 It has been reported in humans that CRY1 mutation develops sleep phase regression syndrome,47 whereas CRY2 mutation exhibits a phenotype of sleep phase advance syndrome.48 Another article reported that double knockout of Cry1 and Cry2 in mice results in strongly impaired circadian rhythm.16 Herein, we observed that Cry1/2 DKO mice were impaired in blood perfusion recovery in a unilateral lower limb ischemia model and displayed reduced capillary density compared with wild‐type mice.
The expression of the G2/M inhibitor WEE1 is under circadian control via CLOCK/BMAL1‐responsive E‐box elements in the Wee1 gene promoter.49 Moreover, Cry deficiency in liver cells displays the continuous expression of Wee1.49 Systematic genome‐wide RNA interference screens found that HOXC5 is one of the potent molecules involved in the migratory activity of endothelial cells.50, 51 Our results from the loss‐of‐function study of the CRY1/2 in endothelial cells are accompanied by increased expression of WEE1 and decreased expression of HOXC5. More important, this was associated with decreased proliferative capacity and migration ability, respectively, leading to impairment of angiogenesis. Conversely, gain‐of‐function studies using KL00152, 53 revealed that the expression of WEE1 was increased and that of HOXC5 was decreased with the stability of CRY1 and CRY2, accompanied by increased proliferation capacity and migration ability of endothelial cells.
In summary, the present work suggests that cryptochrome could play an important role in reparative angiogenesis as a peripheral circadian rhythm regulator.
There are some limitations in the present study. It is still needed to validate whether the precise mechanisms underlying our findings would be through the regulation of peripheral clock genes in a direct manner, or at least in part through multiple factors in an indirect manner, because circadian rhythm disorder was reported to influence various pathophysiological reactions, such as metabolism, nerve system, hemodynamic rhythms, endocrine rhythms, and inflammation. In the future, studies aimed at exploring the mechanisms via the regulation of peripheral clock genes by an inducible Cry1/2 endothelial‐specific knockout mouse will be warranted. Nevertheless, our current information is important for the management of ischemic diseases and would be one of the therapeutic targets in clinical applications.
A past clinical study reported circadian variation in the onset of acute critical limb ischemia.54 Although the mechanisms underlying the rhythm formation still remain to be investigated, they showed that a circadian pattern of occurrence formed the peak in the morning and the bottom around midnight for human.54 Our data may support this observation with the evidence that endogenous angiogenic behavior in mammals would be active in a resting period, and suppressed in an active period. Despite the lack of evidence on the relationship between prognosis in peripheral artery disease and circadian rhythm disorder, studies demonstrated that shift work increased the risk for the onset of coronary heart disease.10 More important, the risk will be waning after cessation of shift work.10 Herein, our results suggested that continuous circadian rhythm disturbance impairs reparative angiogenesis and is associated with poor blood perfusion recovery. Taken together, these findings suggest that correcting circadian rhythm disorders should be recommended in terms of prevention of disease progression, especially in patients with ischemic CVD. Furthermore, on the basis of our new findings, interventions for the therapeutic timing with our chronobiological understanding in diurnal kinetics of angiogenesis and vascular formation, or the regulations of molecules (VEGF, EPC, and cryptochrome) involved in it, might lead to clinical applications. For example, chronotherapy based on our findings would be expected to improve therapeutic efficacy and safety with maximum efficiency and fewer adverse effects in therapeutic angiogenesis in ischemic diseases, or conversely in anti‐cancer therapy in the future.
In conclusion, circadian rhythm disorders reduced angiogenic activity in the ischemic tissue, suggesting that they may have adverse effects on CVD. Correction of circadian rhythm may be an important therapeutic target in patients with CVD.
Sources of Funding
This work was supported by a grant (No. 17H06745 to Dr Shimizu) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and a grant from Japan Foundation for Applied Enzymology (No. 14T020 to Dr Shimizu).
Disclosures
None.
Supporting information
Figures S1–S6
Acknowledgments
We are grateful to the staff from the Division of Experimental Animals at the Nagoya University School of Medicine for assisting with animal experiments. We thank Yoko Inoue for her technical assistance.
For Sources of Funding and Disclosures, see page 14.
References
- 1.Huang RC. The discoveries of molecular mechanisms for the circadian rhythm: the 2017 Nobel Prize in Physiology or Medicine. Biomed J. 2018;41:5–8. DOI: 10.1016/j.bj.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golombek DA, Casiraghi LP, Agostino PV, Paladino N, Duhart JM, Plano SA, Chiesa JJ. The times they're a‐changing: effects of circadian desynchronization on physiology and disease. J Physiol Paris. 2013;107:310–322. DOI: 10.1016/j.jphysparis.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Roenneberg T, Kantermann T, Juda M, Vetter C, Allebrandt KV. Light and the human circadian clock. Handb Exp Pharmacol. 2013;217:311–331. [DOI] [PubMed] [Google Scholar]
- 4.Wittmann M, Dinich J, Merrow M, Roenneberg T. Social jetlag: misalignment of biological and social time. Chronobiol Int. 2006;23:497–509. DOI: 10.1080/07420520500545979. [DOI] [PubMed] [Google Scholar]
- 5.Antunes LC, Levandovski R, Dantas G, Caumo W, Hidalgo MP. Obesity and shift work: chronobiological aspects. Nutr Res Rev. 2010;23:155–168. DOI: 10.1017/S0954422410000016. [DOI] [PubMed] [Google Scholar]
- 6.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. DOI: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris CJ, Purvis TE, Hu K, Scheer FA. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc Natl Acad Sci USA. 2016;113:E1402–E1411. DOI: 10.1073/pnas.1516953113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Amelsvoort LG, Schouten EG, Maan AC, Swenne CA, Kok FJ. Changes in frequency of premature complexes and heart rate variability related to shift work. Occup Environ Med. 2001;58:678–681. DOI: 10.1136/oem.58.10.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawachi I, Colditz GA, Stampfer MJ, Willett WC, Manson JE, Speizer FE, Hennekens CH. Prospective study of shift work and risk of coronary heart disease in women. Circulation. 1995;92:3178–3182. DOI: 10.1161/01.CIR.92.11.3178. [DOI] [PubMed] [Google Scholar]
- 10.Vetter C, Devore EE, Wegrzyn LR, Massa J, Speizer FE, Kawachi I, Rosner B, Stampfer MJ, Schernhammer ES. Association between rotating night shift work and risk of coronary heart disease among women. JAMA. 2016;315:1726–1734. DOI: 10.1001/jama.2016.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chellappa SL, Vujovic N, Williams JS, Scheer F. Impact of circadian disruption on cardiovascular function and disease. Trends Endocrinol Metab. 2019;30:767–779. DOI: 10.1016/j.tem.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams C, Blacker E, Burke W. Night shifts: circadian biology for public health. Nature. 2017;551:33. DOI: 10.1038/551033b. [DOI] [PubMed] [Google Scholar]
- 13.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. DOI: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 14.Kumaki Y, Ukai‐Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR. Analysis and synthesis of high‐amplitude Cis‐elements in the mammalian circadian clock. Proc Natl Acad Sci USA. 2008;105:14946–14951. DOI: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinet P, Milewicz DM, Cassis LA, Leeper NJ, Lu HS, Smith JD. Consideration of sex differences in design and reporting of experimental arterial pathology studies‐statement from ATVB council. Arterioscler Thromb Vasc Biol. 2018;38:292–303. DOI: 10.1161/ATVBAHA.117.309524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. DOI: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minami Y, Ohashi M, Hotta E, Hisatomi M, Okada N, Konishi E, Teramukai S, Inokawa H, Yagita K. Chronic inflammation in mice exposed to the long‐term un‐entrainable light–dark cycles. Sleep Biol Rhythms. 2018;16:63–68. DOI: 10.1007/s41105-017-0127-5. [DOI] [Google Scholar]
- 18.Shimizu Y, Shibata R, Shintani S, Ishii M, Murohara T. Therapeutic lymphangiogenesis with implantation of adipose‐derived regenerative cells. J Am Heart Assoc. 2012;1:e000877. DOI: 10.1161/JAHA.112.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun YY, Bai WW, Wang B, Lu XT, Xing YF, Cheng W, Liu XQ, Zhao YX. Period 2 is essential to maintain early endothelial progenitor cell function in vitro and angiogenesis after myocardial infarction in mice. J Cell Mol Med. 2014;18:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo K, Shintani S, Shibata R, Murakami H, Murakami R, Imaizumi M, Kitagawa Y, Murohara T. Implantation of adipose‐derived regenerative cells enhances ischemia‐induced angiogenesis. Arterioscler Thromb Vasc Biol. 2009;29:61–66. DOI: 10.1161/ATVBAHA.108.166496. [DOI] [PubMed] [Google Scholar]
- 21.Hao C, Shintani S, Shimizu Y, Kondo K, Ishii M, Wu H, Murohara T. Therapeutic angiogenesis by autologous adipose‐derived regenerative cells: comparison with bone marrow mononuclear cells. Am J Physiol Heart Circ Physiol. 2014;307:H869–H879. DOI: 10.1152/ajpheart.00310.2014. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu Y, Shibata R, Ishii M, Ohashi K, Kambara T, Uemura Y, Yuasa D, Kataoka Y, Kihara S, Murohara T, et al. Adiponectin‐mediated modulation of lymphatic vessel formation and lymphedema. J Am Heart Assoc. 2013;2:e000438. DOI: 10.1161/JAHA.113.000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adeola F. Normalization of gene expression by quantitative RT‐PCR in human cell line: comparison of 12 endogenous reference genes. Ethiop J Health Sci. 2018;28:741–748. DOI: 10.4314/ejhs.v28i6.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakao R, Okauchi H, Hashimoto C, Wada N, Oishi K. Determination of reference genes that are independent of feeding rhythms for circadian studies of mouse metabolic tissues. Mol Genet Metab. 2017;121:190–197. DOI: 10.1016/j.ymgme.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Yang W, Kang X, Liu J, Li H, Ma Z, Jin X, Qian Z, Xie T, Qin N, Feng D, et al. Clock gene Bmal1 modulates human cartilage gene expression by crosstalk with Sirt1. Endocrinology. 2016;157:3096–3107. DOI: 10.1210/en.2015-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu Y, Lambert JP, Nicholson CK, Kim JJ, Wolfson DW, Cho HC, Husain A, Naqvi N, Chin LS, Li L, et al. DJ‐1 protects the heart against ischemia‐reperfusion injury by regulating mitochondrial fission. J Mol Cell Cardiol. 2016;97:56–66. DOI: 10.1016/j.yjmcc.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu Y, Polavarapu R, Eskla KL, Pantner Y, Nicholson CK, Ishii M, Brunnhoelzl D, Mauria R, Husain A, Naqvi N, et al. Impact of lymphangiogenesis on cardiac remodeling after ischemia and reperfusion injury. J Am Heart Assoc. 2018;7:e009565. DOI: 10.1161/JAHA.118.009565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenland P, Peterson ED, Gaziano JM. Progress against cardiovascular disease: putting the pieces together. JAMA. 2014;312:1979–1980. DOI: 10.1001/jama.2014.15554. [DOI] [PubMed] [Google Scholar]
- 29.Mohd Azmi NAS, Juliana N, Mohd Fahmi Teng NI, Azmani S, Das S, Effendy N. Consequences of circadian disruption in shift workers on chrononutrition and their psychosocial well‐being. Int J Environ Res Public Health. 2020;17:2043. DOI: 10.3390/ijerph17062043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibuya M. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J Biochem. 2013;153:13–19. DOI: 10.1093/jb/mvs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalka C, Asahara T, Krone W, Isner JM. Angiogenesis and vasculogenesis: therapeutic strategies for stimulation of postnatal neovascularization [in German]. Herz. 2000;25:611–622. [DOI] [PubMed] [Google Scholar]
- 32.Koyanagi S, Kuramoto Y, Nakagawa H, Aramaki H, Ohdo S, Soeda S, Shimeno H. A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 2003;63:7277–7283. [PubMed] [Google Scholar]
- 33.Andrini LB, Garcia MN, Inda AM, Errecalde AL. Study of DNA synthesis and mitotic activity of hepatocytes and its relation to angiogenesis in hepatectomised tumour bearing mice. Cell Biol Int. 2013;37:1233–1237. DOI: 10.1002/cbin.10159. [DOI] [PubMed] [Google Scholar]
- 34.Wang CY, Wen MS, Wang HW, Hsieh IC, Li Y, Liu PY, Lin FC, Liao JK. Increased vascular senescence and impaired endothelial progenitor cell function mediated by mutation of circadian gene Per2. Circulation. 2008;118:2166–2173. DOI: 10.1161/CIRCULATIONAHA.108.790469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. DOI: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Kim NH, Kim YK, Yoo JH, Shin SN, Ko JS, Kim YK, Rhee SJ, Yun KH, Lee EM, et al. The number of endothelial progenitor cells is decreased in patients with non‐dipper hypertension. Korean Circ J. 2012;42:329–334. DOI: 10.4070/kcj.2012.42.5.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busik JV, Tikhonenko M, Bhatwadekar A, Opreanu M, Yakubova N, Caballero S, Player D, Nakagawa T, Afzal A, Kielczewski J, et al. Diabetic retinopathy is associated with bone marrow neuropathy and a depressed peripheral clock. J Exp Med. 2009;206:2897–2906. DOI: 10.1084/jem.20090889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staff PO. Correction: Period2 deficiency blunts hypoxia‐induced mobilization and function of endothelial progenitor cells. PLoS One. 2015;10:e0119196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al Mheid I, Corrigan F, Shirazi F, Veledar E, Li Q, Alexander WR, Taylor WR, Waller EK, Quyyumi AA. Circadian variation in vascular function and regenerative capacity in healthy humans. J Am Heart Assoc. 2014;3:e000845. DOI: 10.1161/JAHA.114.000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atger F, Mauvoisin D, Weger B, Gobet C, Gachon F. Regulation of mammalian physiology by interconnected circadian and feeding rhythms. Front Endocrinol (Lausanne). 2017;8:42. DOI: 10.3389/fendo.2017.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojetin DJ, Burris TP. REV‐ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13:197–216. DOI: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. DOI: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 43.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. DOI: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 44.Hamilton EE, Kay SA. SnapShot: circadian clock proteins. Cell. 2008;135:368–368.e1. DOI: 10.1016/j.cell.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 45.Bell‐Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. DOI: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279:34079–34082. DOI: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 47.Patke A, Murphy PJ, Onat OE, Krieger AC, Ozcelik T, Campbell SS, Young MW. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell. 2017;169:203–215.e13. DOI: 10.1016/j.cell.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirano A, Shi G, Jones CR, Lipzen A, Pennacchio LA, Xu Y, Hallows WC, McMahon T, Yamazaki M, Ptacek LJ, et al. A Cryptochrome 2 mutation yields advanced sleep phase in humans. Elife. 2016;5:e16695. DOI: 10.7554/eLife.16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. DOI: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 50.Williams SP, Odell AF, Karnezis T, Farnsworth RH, Gould CM, Li J, Paquet‐Fifield S, Harris NC, Walter A, Gregory JL, et al. Genome‐wide functional analysis reveals central signaling regulators of lymphatic endothelial cell migration and remodeling. Sci Signal. 2017;10:eaal2987. DOI: 10.1126/scisignal.aal2987. [DOI] [PubMed] [Google Scholar]
- 51.Williams SP, Gould CM, Nowell CJ, Karnezis T, Achen MG, Simpson KJ, Stacker SA. Systematic high‐content genome‐wide RNAi screens of endothelial cell migration and morphology. Sci Data. 2017;4:170009. DOI: 10.1038/sdata.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oshima T, Yamanaka I, Kumar A, Yamaguchi J, Nishiwaki‐Ohkawa T, Muto K, Kawamura R, Hirota T, Yagita K, Irle S, et al. C‐H activation generates period‐shortening molecules that target cryptochrome in the mammalian circadian clock. Angew Chem Int Ed Engl. 2015;54:7193–7197. DOI: 10.1002/anie.201502942. [DOI] [PubMed] [Google Scholar]
- 53.Lee JW, Hirota T, Kumar A, Kim NJ, Irle S, Kay SA. Development of small‐molecule Cryptochrome stabilizer derivatives as modulators of the circadian clock. ChemMedChem. 2015;10:1489–1497. DOI: 10.1002/cmdc.201500260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manfredini R, Gallerani M, Portaluppi F, Salmi R, Zamboni P, Fersini C. Circadian variation in the onset of acute critical limb ischemia. Thromb Res. 1998;92:163–169. DOI: 10.1016/S0049-3848(98)00127-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S6


