Abstract
Background
Current right ventricular (RV) volume overload (VO) is established in adult mice. There are no neonatal mouse VO models and how VO affects postnatal RV development is largely unknown.
Methods and Results
Neonatal VO was induced by the fistula between abdominal aorta and inferior vena cava on postnatal day 7 and confirmed by abdominal ultrasound, echocardiography, and hematoxylin and eosin staining. The RNA‐sequencing results showed that the top 5 most enriched gene ontology terms in normal RV development were energy derivation by oxidation of organic compounds, generation of precursor metabolites and energy, cellular respiration, striated muscle tissue development, and muscle organ development. Under the influence of VO, the top 5 most enriched gene ontology terms were angiogenesis, regulation of cytoskeleton organization, regulation of vasculature development, regulation of mitotic cell cycle, and regulation of the actin filament‐based process. The top 3 enriched signaling pathways for the normal RV development were PPAR signaling pathway, citrate cycle (Tricarboxylic acid cycle), and fatty acid degradation. VO changed the signaling pathways to focal adhesion, the PI3K‐Akt signaling pathway, and pathways in cancer. The RNA sequencing results were confirmed by the examination of the markers of metabolic and cardiac muscle maturation and the markers of cell cycle and angiogenesis.
Conclusions
A neonatal mouse VO model was successfully established, and the main processes of postnatal RV development were metabolic and cardiac muscle maturation, and VO changed that to angiogenesis and cell cycle regulation.
Keywords: cardiomyocyte, proliferation, right ventricle, RNA sequencing, volume overload
Subject Categories: Animal Models of Human Disease, Basic Science Research, Myocardial Biology, Myocardial Regeneration, Pathophysiology
Nonstandard Abbreviations and Acronyms
- AVF
abdominal aorta and inferior vena cava fistula
- GO
gene ontology
- RVCM
right ventricular cardiomyocyte
- VO
volume overload
Clinical Perspective
What Is New?
A neonatal volume overload mouse model was successfully established. For the first time, postnatal right ventricular developmental transcriptome and how they were changed by volume overload were revealed.
What Are the Clinical Implications?
A new platform for the study of volume overload during postnatal right ventricular development was provided.
Developmental right ventricular responses to volume overload are different from that of adult mature right ventricles, highlighting the necessity of different management strategies between children with congenital heart disease and adults with congenital heart disease.
The right ventricle (RV) is an extremely versatile pumping chamber in children with congenital heart disease (CHD). It may be used not only as a pulmonary ventricle but as a pivotal systemic ventricle as well (for example, hypoplastic left heart syndrome and transposition of the great arteries after an atrial inversion operation).1, 2, 3, 4 RV volume overload (VO), which is very common in children with CHD, may cause severe consequences. For example, in atrial septal defect, RV VO will cause "paradoxical" motion of the interventricular septum, which has a deleterious impact on left ventricle (LV) filling and systemic output.5 Assessment of RV performance in the presence of volume loading is important to follow‐up after repair of neonatal tetralogy of Fallot.6 It has been noted that RV VO caused severe RV enlargement and dysfunction after neonatal repair of severe tetralogy of Fallot.7 Thus, understanding how VO contributes to postnatal RV development is critical to treat children with CHD.
Unfortunately, owing to the lack of a neonatal mouse VO model, little is known about the neonatal RV response to VO, let alone how VO influences postnatal RV development.8, 9 At the adult stage, RV responds to VO with fibrosis and hypertrophy.10, 11 However, how neonatal RV responds to VO is unknown. Because of the huge differences between the neonatal heart and the adult heart, the results obtained from adult hearts cannot be applied directly to neonatal hearts.12, 13, 14, 15 For example, adult hearts respond to pressure overload with fibrosis and hypertrophy,16, 17 but neonatal hearts respond to pressure overload with proliferation.18, 19 To establish a neonatal RV mouse model is necessary to understand how VO contributes to postnatal RV development.
It is well acknowledged that from postnatal day 1 (P1) to postnatal day 7 (P7) that mouse cardiomyocytes have a strong proliferative and regenerative potential.13, 20 At this stage, mouse cardiomyocytes are immature and use glycolysis as their primary energy source.21, 22 At P7, mouse cardiomyocytes begin the maturation process. At P21, the cardiomyocytes are fully mature and use oxidative phosphorylation as the primary energy source. From P7 to P21, according to the data obtained from the LV, it is believed that the developmental manner of RV is metabolic and cardiac muscle maturation; therefore, there is an increase in cell size, mitochondria and T‐tubules, and calcium handling abilities.23, 24, 25 Recently Talman et al combined transcriptomics with untargeted proteomic and global metabolic analyses to reveal the molecular atlas of postnatal mouse heart development, highlighting the metabolic maturation process of postnatal mouse heart development.26 However, owing to the huge differences between LV and RV, it is better to observe RV independently.27 To fully understand the postnatal RV developmental process and how VO alters the process, in this study, a neonatal mouse RV VO model is constructed at P7 and followed up with a model at P21.
Methods
Data generated in this study are available from the corresponding author upon reasonable request. All of the RNA sequencing data have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo) with accession number GSE157396.
All primers, reagents, and antibodies information were provided in Tables S1 through S3.
Animal Experiments
C57/BL6 mice were purchased from Xipu'er‐bikai Experimental Animal Co., Ltd (Shanghai, China). At postnatal day 7 (P7), the neonates males were randomized into 2 groups—an experimental group (VO group) and a control group (sham group)—who underwent the same procedure except for the puncture step. The fistula surgery protocol was modified according to previous publication.28 Briefly, the neonatal mice were anesthetized using 4% isoflurane. Under general anesthesia, a midline laparotomy was made, and the abdominal aorta (AA) and inferior vena cava (IVC) were exposed. A 0.07‐mm diameter needle was used to puncture fistula from AA into the IVC. The surrounding connective tissue was used for hemostatic compression for 2 minutes. We closed the abdominal wall and the pain was relieved with local lidocaine treatment, and the pups were returned to their mother. We have provided a surgical video of RV VO model in the supplemental materials to enable others to learn the technique. At the 3 time points: P14, P21, and 12 months, 6 mice from each group were euthanized, the mice were anesthetized with 1.5% isoflurane and then euthanized to obtain the hearts, which were washed with cold PBS and stored at −80°C for further analysis. All of the procedures conformed to the principles outlined in the Declaration of Helsinki and were approved by the Animal Welfare and Human Studies Committee at Shanghai Children's Medical Center (Institutional Review Board number:SCMCIRB‐Y2020094).
Abdominal Ultrasound and Echocardiography
At P14, mice were anesthetized with isoflurane and allowed to breathe spontaneously through a nasal cone (isoflurane/oxygen: 1.5%–2.0% maintenance). The fistula between AA and IVC (AVF) and pulmonary artery (PA) flow were analyzed with a Vevo 2100 imaging system (Visual Sonics, Toronto, Ontario, Canada). For confirmation of an AVF, the waveform in the IVC was recorded using pulse‐wave mode. To confirm the VO, the velocity time integral (VTI) of the PA (PA‐VTI) blood flow and the PA‐velocity were calculated from the mean of 3 consecutive measurements using 2‐dimensional and pulse doppler echocardiography.
Histology
Mouse hearts were fixed in 10% paraformaldehyde overnight at room temperature, then dehydrated in an ethanol series, embedded in paraffin, and sectioned into 6‐µm slices. Hematoxylin and eosin staining were performed according to the manufacturer's instructions.
Total RNA Preparation and Real‐Time Quantitative Polymerase Chain Reaction Analysis
For mRNA quantification, mRNA was extracted from the RV free wall and purified using a PureLink RNA Micro Scale Kit (Catalog No. 12183016; Life Technologies, Carlsbad, CA). Reverse transcriptase polymerase chain reaction (PCR) was performed using the PrimeScript reagent kit (Takara Bio, Kusatsu, Japan). Quantitative real‐time PCR was performed using SYBR Green Power Premix Kits (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Quantitative real‐time PCR was performed with a 7900 Fast Real‐Time PCR System (Applied Biosystems), and the following conditions were used: 1 cycle at 95°C for 10 seconds, followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. The primers were obtained from Generay Biotech Co., Ltd (Shanghai, China). The relative fold change was then calculated using the ΔΔCT method.
Library Preparation for Transcriptome Sequencing
A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer's recommendations. Index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly‐T oligo‐attached magnetic beads. Fragmentation was performed using divalent cations under elevated temperature in NEB Next First Strand Synthesis Reaction Buffer (5X). First‐strand cDNA was synthesized using random hexamer primers and M‐MuLV Reverse Transcriptase (RNase H –). Second‐strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of the 3' ends of the DNA fragments, NEBNext Adaptors with hairpin loop structures were ligated to prepare for hybridization. To select cDNA fragments of preferentially 250–300 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, MA). Then 3 μL USER Enzyme (NEB, USA) was used with size‐selected, adaptor‐ligated cDNA at 37°C for 15 minutes followed by 5 minutes at 95°C. Then, PCR was performed with Phusion High‐Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. Finally, PCR products were purified (AMPure XP system), and library quality was assessed on an Agilent Bioanalyzer 2100 system.
Clustering and Sequencing
The clustering of the index‐coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3‐cBot‐HS (Illumina) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform, and 150 bp paired‐end reads were generated.
Quality Control, Read Mapping, and Quantification of Gene Expression Levels
Raw data (raw reads) in fastq format were first processed through in‐house Perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapters, reads containing poly‐N, and low quality reads from raw data. At the same time, Qphred ≥ 20, Qphred ≥ 30, and Guanine Cytosine content of the clean data were calculated. All of the downstream analyses were based on clean, high‐quality data.
Reference genome and gene model annotation files were downloaded from the genome website directly. The index of the reference genome was built using Hisat2 v2.0.5, and paired‐end clean reads were also aligned to the reference genome using Hisat2 v2.0.5.
The expected number of Fragments Per Kilobase of transcript sequence per Million base pairs sequenced considers the effect of sequencing depth and gene length for the reads counted at the same time. FeatureCounts v1.5.0‐p3 was used to count the number of reads mapped to each gene. Then, Fragments Per Kilobase of transcript sequence per Million of each gene was calculated based on the length of the gene and read counts mapped to each gene.
Differential Expression Analysis and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis of Differentially Expressed Genes
Differential expression analysis was performed using the DESeq2 R package (1.16.1). The resulting P values were adjusted using Benjamini and Hochberg approach for controlling the false discovery rate. Genes with an adjusted P value of <0.05 found by DESeq2 were considered differentially expressed.
Gene ontology (GO) enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package, in which gene length bias was corrected. GO terms with corrected P values under 0.05 were considered to be significantly enriched by differentially expressed genes.
The clusterProfiler R package was used to test the statistical enrichment of differentially expressed genes in Kyoto Encyclopedia of Genes and Genomes pathways.
Immunofluorescence
Slides were washed 3 times with PBS, fixed with 4% paraformaldehyde for 10 minutes, permeated with 0.5% Triton X‐100 for 15 minutes, blocked with 10% donkey serum for 30 minutes, and stained with primary antibodies overnight at 4°C. After washing the slides 3 more times, we incubated the sections or cells with secondary antibodies and 4',6‐diamidino‐2‐phenylindole for 30 minutes. Three researchers who were masked to sample identity quantified cellular Ki67, pHH3 (phospho‐histone H3), and aurora B via either manual counting or digital thresholding. This included image segmentation and creation of a binary image from a grayscale. We analyzed the converted binary images using ImageJ software (National Institutes of Health, Bethesda, Maryland, USA; Laboratory for Optical and Computational Instrumentation, University of Wisconsin, Madison, Wisconsin, USA).
Cardiomyocyte Isolation From RV Free Wall and Calcium Imaging
Cardiomyocytes were isolated with a Langendorff perfusion system as described previously.29 After perfusion, only the RV free wall was removed and cardiomyocytes from the RV were used for calcium image and immunostaining. Calcium imaging was performed according to a previous publication.29 Briefly, before contractility and calcium analyses, calcium was re‐introduced into isolated cardiomyocytes by treating cells with a series of 10‐mL 2,3‐Butanedione monoxime‐free perfusion buffers containing 100 nmol/L, 400 nmol/L, 900 nmol/L, and 1.2 µmol/L CaCl2. At each step, cardiomyocytes were settled down by gravity for 10 minutes at room temperature before cells were transferred to the next buffer with a higher calcium concentration. Cardiomyocytes were loaded with 5‐µmol/L Rhod‐4 for 20 minutes. The cells were next washed with normal Tyrode solution (NaCl, 140 mmol/L; KCl, 4 mmol/L; MgCl2, 1 mmol/L; CaCl2, 1.8 mmol/L; glucose, 10 mmol/L; and HEPES, 5 mmol/L, pH=7.4, adjusted with NaOH) for 20 minutes. The cells were next settled in a laminin‐coated glass‐bottom flow chamber at 30°C for 10 minutes and electrically stimulated at 1 Hz to produce steady‐state conditions. Calcium signals were next acquired through confocal line scanning using a ×60 objective. Line scan was positioned along the long axis of the cell in the cytosol, avoiding the nuclear area. Calcium signal was quantified manually using ImageJ.30
Statistical Analysis
Continuous data, including mRNA expression, protein expression, and number of Ki67/pHH3/aurora B‐positive cells, were expressed as means±SD. Differences were tested with Student t‐test if the data were normally distributed; otherwise, they were tested with the rank sum test. P<0.05 were considered to be statistically significant. Statistical analyses were performed using SAS software version 9.2 (SAS Institute Inc., Cary, NC).
Results
RV VO is Significantly Increased in AVF Mice
To investigate how VO contributes to postnatal RV development, first, we constructed an AVF in P7 mice (Figure 1A and Video S1). The results showed that, without AVF, there is no pulsatile blood flow in the IVC, while there is pulsatile blood flow in the aorta, with peak flow velocity up to 400 mm/s (Figure 1B and 1C). After AVF, clear flow appeared at the puncture point (Figure 1D), with peak flow velocity up to 450 mm/s at puncture point (Figure 1E), and a reverse pulsatile blood flow appeared in the IVC, with peak flow velocity up to 200 mm/s.
Figure 1. Establishment of the abdominal aorta and inferior vena cava fistula.
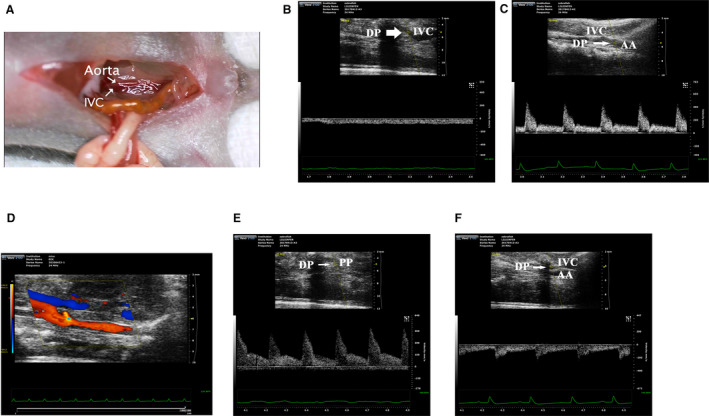
A, Schematic diagram of abdominal aorta and inferior vena cava fistula model. B, Under normal circumstances, the inferior vena cava has no pulsatile blood flow, and the detection point is located in the inferior vena cava. C, Under normal circumstances, there are pulsatile blood flow in abdominal aorta, with a peak blood flow velocity of 400 mm/s. D, The representative image of blood flow through the puncture point. E, The representative image of pulsating blood flow at the puncture point, with a peak blood flow velocity of 450 mm/s, which is higher than that of normal aortic blood flow. F, After puncture, the pulsatile blood flow appeared in the inferior vena cava, with a peak blood flow velocity of 200 mm/s. AA indicates abdominal aorta; DP, detection point; IVC, inferior vena cava; and PP, puncture point.
Seven days after AVF, at P14, volume load in the RV was significantly increased, indicated by the PA‐velocity and PA‐VTI. The PA‐ velocity in the sham and VO groups were 478.70±65.24 mm/s and 606.20±38.01 mm/s, respectively (P=0.002, n=6, Figure 2A and 2B), and the PA‐VTI in the sham and VO groups were 19.88±3.25 mm and 26.3±2.8 mm, respectively (P=0.0045, n=6, Figure 2A and 2B). Although PA diameter (D) changed with age, there were no significant differences between VO and sham groups at P14 or P21 (Table S4). According to the formula:,31 it is reasonable to use PA‐VTI as a surrogate of RV stroke volume. Twelve months after AVF, RV failure appeared, indicating by reduced RV stroke volume (Figure S1), with RV free wall thickness significantly increasing and ventricular septum moving toward to LV (Figure 2C and 2D). The survival rate (the number of mice in whom the fistula did not close as detected by echo) was ≈90% and the fistula close rate was 10% (60 mice in total).
Figure 2. Volume overload (VO) increased in the abdominal aorta and inferior vena cava fistula model.
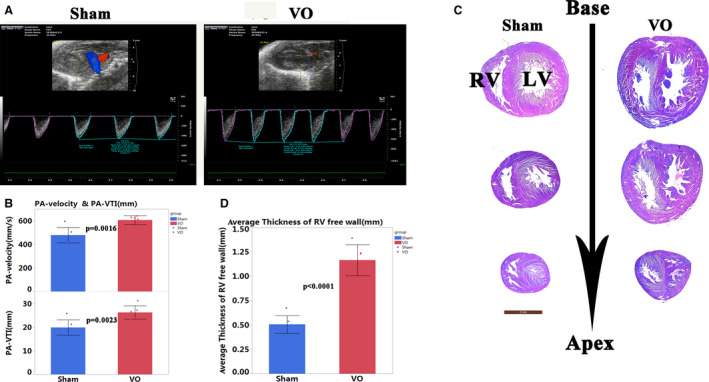
A, The representative echo image showed that 2 weeks after puncture, the pulmonary artery velocity and velocity time integral in the VO group increased. B, Quantitative statistics of pulmonary artery‐velocity and velocity time integral in sham and VO group, n=6, Student t‐test. C, Hematoxylin and eosin staining showed that 12 months after of puncture, the free wall of the right ventricle was thickened in the VO group, and the ventricular septum shifted to the left. D, Quantification of the thickness of the free wall of the right ventricle, n=6, Student t‐test. LV indicates left ventricle; PA, pulmonary artery; RV, right ventricle; VO, volume overload; and VTI, velocity time integral.
VO Changes the Transcriptome of Postnatal RV Development
To investigate how VO changes gene expression in postnatal RV development from P7 to P21, we selected the RVs of the hearts from AVF and sham‐operated mice at P14 and P21 and performed RNA sequencing. Our results showed that during normal postnatal RV development, there were 3012 differentially expressed genes between from P21 to P14, among which 1405 were downregulated and 1607 were upregulated (Figure 3A). The VO increased this number to 3470, among which 1732 were downregulated and 1738 were upregulated (Figure 3A). The heatmap results showed that the individual mice in the same group were similar to each other but differed noticeably from the mice in the other group (Figure 3B). The Venn diagram shows that the P21_sham group shared 11 459 genes with the P14_sham group, 451 genes were only expressed in the P14_sham group, and 304 genes were only expressed in the P21_sham group. The P21_VO group shared 11 523 genes with P14_VO group, 403 genes were only expressed in the P14_VO group, and 451 genes were only expressed in the P21_VO group (Figure 3C). These results indicate that VO changes the transcriptome of postnatal RV development.
Figure 3. The transcriptomic changes of postnatal right ventricular development.
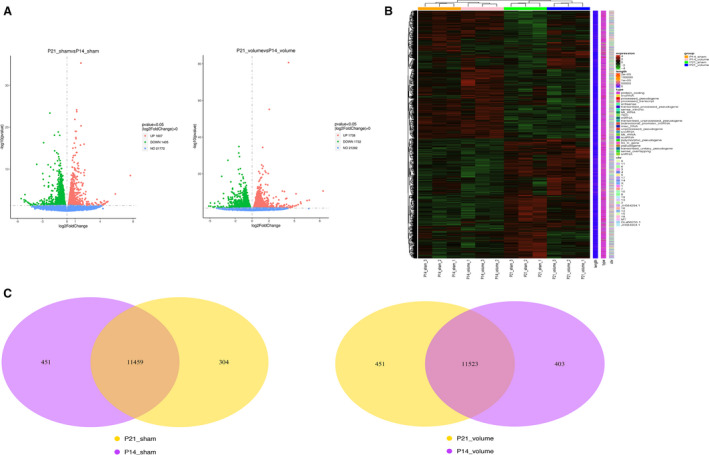
A, Volcano map of differentially expressed genes of postnatal right ventricular development in the normal condition (postnatal day 21 sham vs postnatal day 14 sham) and under the influence of volume overload (postnatal day 21 volume overload and postnatal day 14 volume overload). B, Cluster analysis of differentially expressed genes. Every group had 3 mice. The redder the color, the higher the expression level; the bluer, the lower. The clusters of genes in each group are quite different from each other but similar in the same group. C, Venn diagram of differentially expressed genes. P14 indicates postnatal day 14; and P21, postnatal day 21.
Processes of Normal Postnatal RV Development Were Primarily Associated With Metabolic and Cardiac Muscle Maturation, as Indicated by the GO Analysis
To understand normal postnatal RV development from P14 to P21, the differentially expressed genes in the sham groups were analyzed using GO analysis. The results showed that the top 10 most enriched GO terms of biological process were energy derivation by oxidation of organic compounds, generation of precursor metabolites and energy, cellular respiration, striated muscle tissue development, muscle organ development, muscle tissue development, sterol metabolic process, cholesterol metabolic process, cofactor metabolic process, and lipid homeostasis (Figure 4A and 4B). The top 10 most enriched GO terms of the cellular component were the mitochondrial inner membrane, organelle inner membrane, mitochondrial matrix, mitochondrial protein complex, myelin sheath, myofibrils, extracellular matrix, contractile fiber components, contractile fibers, and sarcomeres (Figure 4A and 4B). The top 10 most enriched GO terms of molecular function were cofactor binding, extracellular matrix structural constituent, coenzyme binding, structural molecule activity, O‐acyltransferase activity, alpha‐actinin binding, oxidoreductase activity, acting on the aldehyde or oxo group of donors, anion transmembrane transporter activity, unfolded protein binding, and active transmembrane transporter activity (Figure 4A and 4B). These results suggested that the processes of normal postnatal RV development were primarily associated with metabolic and cardiac muscle maturation, similar to that of the LV.
Figure 4. The processes of normal postnatal right ventricular development were primarily associated with metabolic and cardiac muscle maturation, as indicated by the gene ontology (GO) analysis.
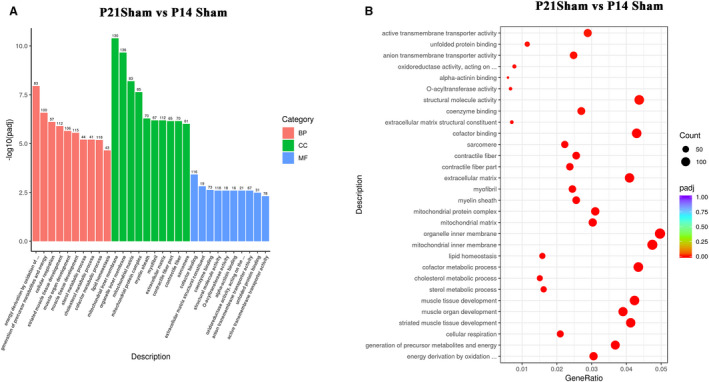
A, From the results of the GO enrichment analysis, the most significant 10 terms are displayed. The abscissa is the GO term, and the ordinate is the significance level of GO term enrichment. The higher the value, the more significant the result, and the different colors represent 3 different GO subclasses: biological process, cellular component, and molecular function. B, From the results of the GO enrichment analysis, we selected the most significant 30 terms to draw scatter plots for display. The abscissa is the ratio of the number of differentially expressed genes on the GO term to the total number of differentially expressed genes, the ordinate is GO term, the size of the dots represents the number of genes annotated to the GO term, and the color from red to purple represents the significance level of GO term enrichment. BP indicates biological process; CC, cellular component; and MF, molecular function; P14, postnatal day 14; P21, postnatal day 2; and VO, volume overload.
Process of Postnatal RV Development Under the Influence of VO Were Partially Changed to Angiogenesis and Cell Cycle Regulation, as Indicated by the GO Analysis
To understand how the VO changed the postnatal RV development from P14 to P21, the differentially expressed genes in the VO groups were analyzed using a GO analysis. The results showed that the top 10 most enriched GO terms of the biological process were angiogenesis, regulation of cytoskeleton organization, regulation of vasculature development, regulation of mitotic cell cycle, regulation of actin filament‐based process, regulation of angiogenesis, regulation of cell morphogenesis, regulation of cell morphogenesis involved in differentiation, monocarboxylic acid metabolic process, and regulation of cell‐substrate adhesion (Figure 5A and 5B). The 10 most enriched GO terms of the cellular component were sarcolemma, extracellular matrix, actin cytoskeleton, extracellular matrix component, proteinaceous extracellular matrix, cell leading edge, cell cortex, contractile fiber part, contractile fiber, and myofibril (Figure 5A and 5B). The top 10 most enriched GO terms of the molecular function were cell adhesion molecule binding, actin binding, cadherin binding, actin filament binding, Rho GTPase binding, integrin binding, ubiquitin protein ligase binding, ubiquitin‐like protein ligase binding, tubulin binding, and protein heterodimerization activity (Figure 5A and 5B). These results suggested that the process of postnatal RV development was partially changed by the VO. During the change process, angiogenesis and cell cycle regulation played a role.
Figure 5. The processes of postnatal right ventricular development partially switched to angiogenesis and cell cycle regulation, as indicated by gene ontology (GO) analysis.
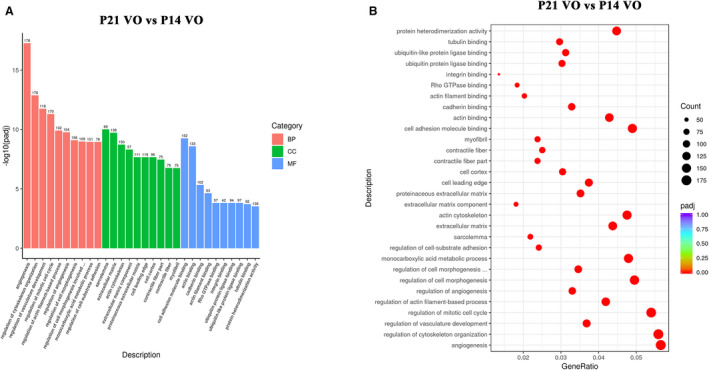
A, From the results of the GO enrichment analysis, the most significant 10 terms are displayed. The abscissa is the GO term, and the ordinate is the significance level of the GO term enrichment. The higher the value, the more significant the result, and the different colors represent 3 different GO subclasses: biological process, cellular component, and molecular function. B, From the results of the GO enrichment analysis, the most significant 30 terms were selected to draw scatter plots for display. The abscissa is the ratio of the number of differentially expressed genes on the GO term to the total number of differentially expressed genes, the ordinate is GO term, the size of the dots represent the number of genes annotated to the GO term, and the colors from red to purple represent the significance level of the GO term enrichment. P14 indicates postnatal day 14; BP, biological process; CC, cellular component; MF, molecular function; and P21, postnatal day 2.
Pathways of the Postnatal RV Developmental Trajectory Changed by VO
Kyoto Encyclopedia of Genes and Genomes pathway analysis was applied to find out what types of pathways regulating normal RV development were changed by VO. The results indicated that the top 20 enriched pathways in the normal RV development were the PPAR signaling pathway, citrate cycle (Tricarboxylic acid cycle), fatty acid degradation, valine, leucine and isoleucine degradation, propanoate metabolism, steroid biosynthesis, carbon metabolism, cardiac muscle contraction, fatty acid metabolism, cholesterol metabolism, thermogenesis, fatty acid elongation, aminoacyl‐tRNA biosynthesis, pyruvate metabolism, protein digestion and absorption, AMPK signaling pathway, Alzheimer disease, butanoate metabolism, oxidative phosphorylation, and alanine, aspartate, and glutamate metabolism (Figure 6A and 6C). These results further confirmed that metabolic and cardiac muscle maturation were the primary processes of postnatal RV development, similar to that of the LV.23, 24, 25
Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the differentially expressed genes of normal postnatal right ventricular (RV) development and the postnatal RV development under the influence of volume overload.
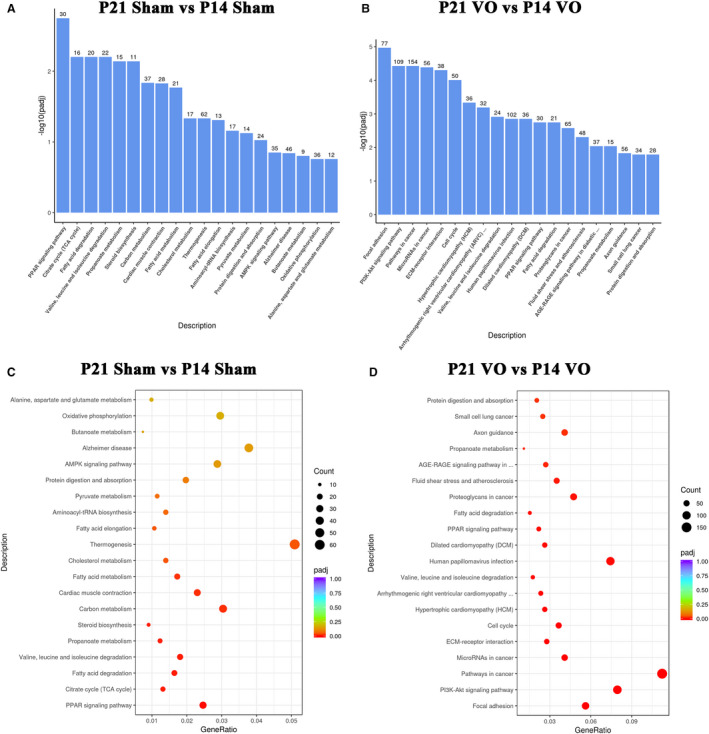
A, The KEGG pathways of normal RV development. The most significant 20 KEGG pathways from the KEGG enrichment results are displayed. The abscissa is the KEGG pathway, and the ordinate is the significance level of pathway enrichment. The higher the value, the greater the significance. B, The KEGG pathway of postnatal RV development under the influence of volume overload. The most significant 20 KEGG pathways from the KEGG enrichment results are displayed. The abscissa is the KEGG pathway, and the ordinate is the significance level of pathway enrichment. The higher the value, the greater the significance. C, The KEGG pathways of normal RV development. From the KEGG enrichment results, the most significant 20 KEGG pathways were selected for the scatter plots. The abscissa is the ratio of the number of differentially expressed genes on the KEGG pathway to the total number of differentially expressed genes, the ordinate is the KEGG pathway, the size of the dots represents the number of genes annotated to the KEGG pathway, and the color from red to purple the represents significance level of KEGG pathway enrichment. D, The KEGG pathways of postnatal RV development under the influence of volume overload. From the KEGG enrichment results, the most significant 20 KEGG pathways were selected for the scatter plots. The abscissa is the ratio of the number of differentially expressed genes on the KEGG pathway to the total number of differentially expressed genes, the ordinate is the KEGG pathway, the size of the dots represents the number of genes annotated to the KEGG pathway, and the color from red to purple the represents significance level of KEGG pathway enrichment. P14 indicates postnatal day 14; P21, postnatal day 2; and VO, volume overload.
Under the influence of VO, the top 20 enriched pathways were focal adhesion, PI3K‐Akt signaling pathway, pathways in cancer, microRNAs in cancer, extracellular matrix‐receptor interaction, cell cycle, hypertrophic cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, valine, leucine and isoleucine degradation, human papillomavirus infection, dilated cardiomyopathy, PPAR signaling pathway, fatty acid degradation, proteoglycans in cancer, fluid shear stress and atherosclerosis, advanced glycation end products and their receptors signaling pathway in diabetic complications, propanoate metabolism, axon guidance, small cell lung cancer, and protein digestion and absorption (Figure 6B and 6D). These results further confirmed that the primary process of postnatal RV development was changed by the VO, in which angiogenesis and cell cycle regulation played a role.
Verification of RNA‐Sequencing Results by Examination of Metabolic and Cardiac Muscle Maturation and Cell Cycle Markers
To confirm the RNA‐sequencing results, top 20‐fold changed genes were verified by quantitative real‐time PCR (Figures 7A and 8A).
Figure 7. The processes of normal postnatal right ventricular (RV) development are primarily associated with metabolic and cardiac muscle maturation.
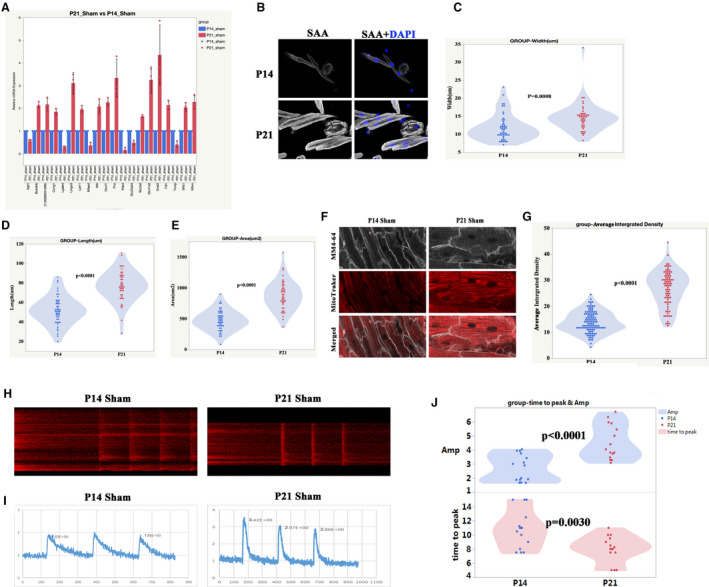
A, The top 20‐fold change genes between postnatal day 21 (P21) sham and postnatal day 14 (P14) sham were selected for verification by quantitative real‐time polymerase chain reaction. P<0.05, n=3 mice, Mann‐Whitney test. B, Representative isolated cardiomyocytes from P14 and P21 RV. Sarcomeric α‐actinin (SAA, white), 4',6‐diamidino‐2‐phenylindole (blue). C, Quantification of cell width from P14 and P21 RV, Student t test. D, Quantification of cell length from P14 and P21 RV. E, Quantification of cell area from P14 and P21 RV. F, Representative of T‐tubule and mitochondria from P14 and P21 RV. MM4‐64 (T‐tubule, white), mitochondria (red). G, Quantification of mitochondria intensity from P14 and P21 RV, Student t‐test. H, Representative of the calcium transient image of cardiomyocytes from P14 and P21 RV, Student t test. I, Plot representative of the calcium transient image of cardiomyocytes from P14 and P21 RV. J, Quantification of the calcium transient parameters (amplitude (Amp) and time to peak), Student t‐test. P14, postnatal day 14; and P21, postnatal day 2.
Figure 8. The processes of postnatal right ventricular development partially switched to cell cycle regulation and angiogenesis.
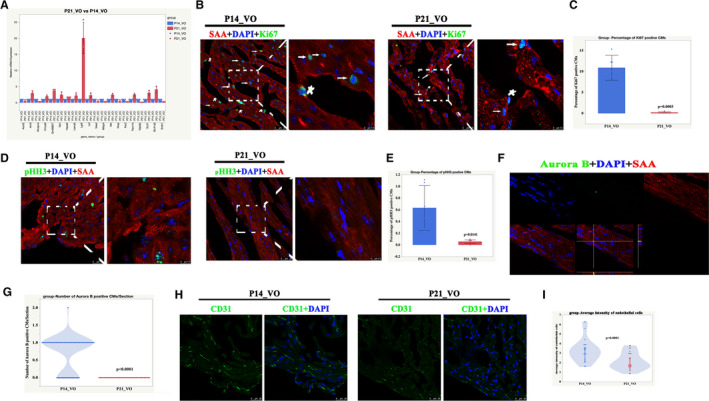
A, The top 20‐fold change genes between postnatal day 21 volume overload and postnatal day 14 volume overload were selected for verification by quantitative real‐time polymerase chain reaction. P<0.05, n=3 mice, Mann‐Whitney test. B, Representative Ki67‐positive cardiomyocytes. Sarcomeric α‐actinin (red), 4',6‐diamidino‐2‐phenylindole (blue), Ki67 (green). Star indicates Ki67‐positive non‐cardiomyocytes; arrow indicates Ki67‐positive cardiomyocytes. C, Quantification of Ki67‐positive cardiomyocytes, n=6 mice, Student t‐test. D, Representative pHH3 (phospho‐histone H3)‐positive cardiomyocytes from postnatal day 14 volume overload and postnatal day 21 volume overload. Sarcomeric α‐actinin (red), 4',6‐diamidino‐2‐phenylindole (blue), pHH3 (green). E, Quantification of pHH3‐positive cardiomyocytes, n=6 mice, Student t‐test. F, Representative aurora B‐positive cardiomyocytes from postnatal day 14 volume overload and postnatal day 21 volume overload . Sarcomeric α‐actinin (red), diamidino‐2‐phenylindole (blue), aurora B (green). G, Quantification of aurora B‐positive cardiomyocytes, n=60 sections from 6 mice, Student t‐test. H, Representative CD31‐positive cells. I, Quantification of CD31‐positive cells, n=30 sections from 6 mice, Student t‐test. DAPI indicates diamidino‐2‐phenylindole; P14, postnatal day 14; P21, postnatal day 2; SAA, sarcomeric α‐actinin; and VO, volume overload.
During normal RV development, among the top 20‐fold changed genes, 7 of them (Slc41a3, Whrn, Lpin1, Wfs1, Slc25a34, Ttll1, and Slc2a4) were closely related to muscle development, and 8 of them (Oxct1, Lgals4, Pcx, Mfap4, Bckdhb, Pdk4, Mtr, and Txnip) were associated with energy metabolism (Figure 7A). The metabolic and cardiac muscle maturation markers, such as cardiomyocyte size, mitochondria, and T‐tubule intensity, and calcium handling ability were further analyzed. The results showed that the width, length, and area of cardiomyocytes at P21 were significantly increased compared with those at P14 (Figure 7B through 7E). The mitochondria and T‐tubule intensity were increased in the RV at P21 (Figure 7F and 7G). Accordingly, the amplitude of calcium transient of cardiomyocytes from P21 RV was increased while the time‐to‐peak of the calcium transient of cardiomyocytes from P21 RV was reduced (Figure 7H through 7J).
In RV development under the influence of VO, among the top 20‐fold changed genes, 2 of them (Lgr6 and Rab6b) were closely related to the cell cycle regulation (Figure 8A). The cell cycle and intensity of endothelial cells were further examined to confirm the above results. The results showed that cell cycle markers, including Ki67/pHH3/aurora B, increased at P14 but was reduced at P21 (Figure 8B through 8G). The intensity of the endothelial cells increased at P14 but was reduced at P21 (Figure 8H and 8I). These results confirmed that the VO changed the postnatal RV development, in which angiogenesis and cell cycle regulation played a role.
Discussion
The crucial role of RV in determining functional status and prognosis in CHDs has been well recognized in recent years.4, 6 The level of RV exposure to VO and RV intrinsic contractile function largely determine the RV adaptation, which are associated with distinct clinical courses and therapeutic approaches.6, 7 VO can persist throughout the patient's lifetime even if most CHD are repaired at a young age, which means that CHD RV development can be affected by VO.7 Improving our understanding of RV adaption at a young age may ultimately result in improved outcomes. However, to the best of our knowledge, there were no neonatal VO mouse models, so determining how VO modify RV remodeling at a young age is largely unknown.9, 10 This study was the first to introduce a neonatal VO mouse model, providing a platform for studying the effect of VO on the neonatal RV. Compared with adult VO models,32 the neonatal VO model is unique. Although there is increased RV cardiac output early after the surgery and decreased cardiac output later on in both models, neonatal mice took longer to develop heart failure (reduced cardiac output, Figure S1). This may be because the PA is under development and the diameter of the PA is expanding in the neonatal mouse model. In addition, the response to VO also differed between adult and neonatal RV. Adult RV responds as early hypertrophy and late fibrosis,32 while neonatal RV responds as cell cycle regulation and angiogenesis (Figures 5 and 8). Further research is required to determine the underlying mechanism and whether a similar phenomenon takes place in human beings.
Moreover, this was the first time that postnatal RV development and how the VO changed the RV developmental track were revealed. Previously, Talman et al showed the molecular atlas of postnatal mouse ventricle development by the combination of transcriptomics, proteomic and metabolic analyses, highlighting the importance of metabolic pathways in the cell cycle regulation.26 Our RV results were similar to the overall ventricle results with respect to normal RV development, such as the upregulation of carbon metabolism, fatty acid metabolism, and pyruvate metabolism (Figure 6A). However some enriched pathways appeared solely in RV, such as the PPAR signaling pathway and Tricarboxylic acid cycle (TCA cycle) (Figure 6A), highlighting the uniqueness of RV. In addition, although it is well acknowledged that from P7 to P21, the mouse heart is undergoing a maturation process, this is the first time that the isolated RV maturation process was revealed. VO interrupted the metabolic and cardiac muscle maturation process of normal RV development, which was partially switched to angiogenesis and cell cycle regulation. Regarding cell cycle regulation, another interesting finding was that previously it has been believed that the cardiomyocyte of a P7 mouse was exited from the cell cycle, and hypertrophic growth was its main developmental manner. This current study challenges this traditional notion, pointing that in some pathological conditions, cardiomyocytes are able to reenter cell cycle activities. Therefore, this study provides another platform for heart regeneration research, allowing people to study how to make cardiomyocytes reenter into the cell cycle. This current study also showed a different response to VO between neonatal and adult RVs, highlighting the importance of different management strategies for childhood and adult CHD.
One limitation of this study is that it only observed the RV. The VF increased VO in the RV, and it may increase VO in LV correspondingly. Will there be corresponding changes in the LV? Because the RV is different from the LV in its anatomic, electrical, and cellular configuration,27 further research is required to analyze the LV independently. Another limitation is that how the RV responded to the VO from P1 to P7, the cardiomyocyte proliferative stage, is still unknown. The successful establishment of a P1 RV VO mouse model will help answer this question. The third limitation is that longstanding increased VO may subject the RV to pressure load, which can confound the study results. We found that the effect of VO on the RV persisted for 2 months before being dominated by the pressure overload, as evidenced by decreased RV stroke volume and elevated PA pressure at 3 months (Figure S1). Therefore, elevated afterload or pressure overload may have only a trivial effect or even no effect on RV during early stages, such as P14 and P21.
In summary, the current study successfully established a neonatal RV VO mouse model and revealed how the neonatal RV responds to VO. P1 RV VO and LV VO mouse models are required to fully understand how the VO will affect postnatal heart development.
Sources of Funding
This work was supported by National Key R&D Program of China (No. 2019YFA0110401), Science and Technology Innovation Action Plan of Shanghai—Experimental Animal Research (No. 201409005900), National Natural Science Foundation of China (No. 81800285), the Foundation of Pudong Science and Technology Development (No. PKJ2019‐Y12), the Clinical Science and Technology Innovation Project of Shanghai Shenkang Hospital Development Center (No. SHDC12017X08), and the Program for Outstanding Medical Academic Leader in Shanghai (Zhang).
Disclosures
None.
Supporting information
Tables S1–S4
Figure S1
Video S1
(J Am Heart Assoc. 2021;10:e020854. DOI: 10.1161/JAHA.121.020854.)
Supplementary Materials for this article are available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.121.020854
For Sources of Funding and Disclosures, see page 13.
Contributor Information
Fen Li, Email: lifen@scmc.com.cn.
Lincai Ye, Email: ylc717@163.com.
References
- 1.Latus H, Nassar MS, Wong J, Hachmann P, Bellsham‐Revell H, Hussain T, Apitz C, Salih C, Austin C, Anderson D, et al. Ventricular function and vascular dimensions after Norwood and hybrid palliation of hypoplastic left heart syndrome. Heart. 2018;104:244–252. DOI: 10.1136/heartjnl-2017-311532. [DOI] [PubMed] [Google Scholar]
- 2.Borrelli N, Di Salvo G, Sabatino J, Ibrahim A, Avesani M, Sirico D, Josen M, Penco M, Fraisse A, Michielon G, et al. Serial changes in longitudinal strain are associated with outcome in children with hypoplastic left heart syndrome. Int J Cardiol. 2020;317:56–62. DOI: 10.1016/j.ijcard.2020.03.085. [DOI] [PubMed] [Google Scholar]
- 3.Warnes CA. Transposition of the great arteries. Circulation. 2006;114:2699–2709. DOI: 10.1161/CIRCULATIONAHA.105.592352. [DOI] [PubMed] [Google Scholar]
- 4.Broberg CS, Valente AM, Huang J, Burchill LJ, Holt J, Van Woerkom R, Powell AJ, Pantely GA, Jerosch‐Herold M. Myocardial fibrosis and its relation to adverse outcome in transposition of the great arteries with a systemic right ventricle. Int J Cardiol. 2018;271:60–65. DOI: 10.1016/j.ijcard.2018.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feneley M, Gavaghan T. Paradoxical and pseudoparadoxical interventricular septal motion in patients with right ventricular volume overload. Circulation. 1986;74:230–238. DOI: 10.1161/01.CIR.74.2.230. [DOI] [PubMed] [Google Scholar]
- 6.Egbe AC, Najam M, Banala K, Vojjinni R, Faizan F, Ammash NM, Khalil F, Mathew J, Angirekula M, Connolly HM. Usefulness of right ventricular volumetric and afterload indices for risk stratification in patients with tetralogy of Fallot. Am J Cardiol. 2019;124:1293–1297. DOI: 10.1016/j.amjcard.2019.07.025. [DOI] [PubMed] [Google Scholar]
- 7.Rommel K‐P, Besler C, Noack T, Blazek S, von Roeder M , Fengler K, Ender J, Gutberlet M, Desch S, Borger MA, et al. Physiological and clinical consequences of right ventricular volume overload reduction after transcatheter treatment for tricuspid regurgitation. JACC Cardiovasc Interv. 2019;12:1423–1434. DOI: 10.1016/j.jcin.2019.02.042. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg MK, Reddy S. Right ventricular failure in congenital heart disease. Curr Opin Pediatr. 2019;31:604–610. DOI: 10.1097/MOP.0000000000000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karamlou T, Giraud GD, McKeogh D, Jonker SS, Shen I, Ungerleider RM, Thornburg KL. Right ventricular remodeling in response to volume overload in fetal sheep. Am J Physiol Heart Circ Physiol. 2019;316:H985–H991. DOI: 10.1152/ajpheart.00439.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanz J, Sánchez‐Quintana D, Bossone E, Bogaard HJ, Naeije R. Anatomy, function, and dysfunction of the right ventricle: JACC state‐of‐the‐art review. J Am Coll Cardiol. 2019;73:1463–1482. DOI: 10.1016/j.jacc.2018.12.076. [DOI] [PubMed] [Google Scholar]
- 11.Hahn RT, Waxman AB, Denti P, Delhaas T. Anatomic relationship of the complex tricuspid valve, right ventricle, and pulmonary vasculature: a review. JAMA Cardiol. 2019;4:478–487. DOI: 10.1001/jamacardio.2019.0535. [DOI] [PubMed] [Google Scholar]
- 12.Lam NT, Sadek HA. Neonatal heart regeneration: comprehensive literature review. Circulation. 2018;138:412–423. DOI: 10.1161/CIRCULATIONAHA.118.033648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Cui M, Shah AM, Tan W, Liu N, Bassel‐Duby R, Olson EN. Cell‐type‐specific gene regulatory networks underlying murine neonatal heart regeneration at single‐cell resolution. Cell Rep. 2020;33:108472. DOI: 10.1016/j.celrep.2020.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Liang C, Yang KY, Huang X, Han MY, Li X, Chan VW, Chan KS, Liu D, Huang ZP, et al. Specific ablation of CD4+ T‐cells promotes heart regeneration in juvenile mice. Theranostics. 2020;10:8018–8035. DOI: 10.7150/thno.42943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadota S, Pabon L, Reinecke H, Murry CE. In vivo maturation of human induced pluripotent stem cell‐derived cardiomyocytes in neonatal and adult rat hearts. Stem Cell Reports. 2017;8:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lerchenmüller C, Rabolli CP, Yeri AS, Kitchen R, Salvador AM, Liu LX, Ziegler O, Danielson KM, Platt C, Shah R, et al. CITED4 protects against adverse remodeling in response to physiological and pathological stress. Circ Res. 2020;127:631–646. DOI: 10.1161/CIRCRESAHA.119.315881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards DA, Aronovitz MJ, Calamaras TD, Tam K, Martin GL, Liu P, Bowditch HK, Zhang P, Huggins GS, Blanton RM. Distinct phenotypes induced by three degrees of transverse aortic constriction in mice. Sci Rep. 2019;9:5844. DOI: 10.1038/s41598-019-42209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malek Mohammadi M, Abouissa A, Isyatul A, Xie Y, Cordero J, Shirvani A, Gigina A, Engelhardt M, Trogisch FA, Geffers R, et al. Induction of cardiomyocyte proliferation and angiogenesis protects neonatal mice from pressure overload‐associated maladaptation. JCI Insight. 2019;5:e128336. DOI: 10.1172/jci.insight.128336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye L, Wang S, Xiao Y, Jiang C, Huang Y, Chen H, Zhang H, Zhang H, Liu J, Xu Z, et al. Pressure overload greatly promotes neonatal right ventricular cardiomyocyte proliferation: a new model for the study of heart regeneration. J Am Heart Assoc. 2020;9:e015574. DOI: 10.1161/JAHA.119.015574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, De la Cruz E, Gu X, Balint L, Oxendine‐Burns M, Terrones T, Ma W, Kuo H‐H, Lantz C, Bansal T, et al. Lymphoangiocrine signals promote cardiac growth and repair. Nature. 2020;588:705–711. DOI: 10.1038/s41586-020-2998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardoso AC, Lam NT, Savla JJ, Nakada Y, Pereira AHM, Elnwasany A, Menendez‐Montes I, Ensley EL, Bezan Petric U, Sharma G, et al. Mitochondrial substrate utilization regulates cardiomyocyte cell cycle progression. Nat Metab. 2020;2:167–178. DOI: 10.1038/s42255-020-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardoso AC, Pereira AHM, Sadek HA. Mechanisms of neonatal heart regeneration. Curr Cardiol Rep. 2020;22:33. DOI: 10.1007/s11886-020-01282-5. [DOI] [PubMed] [Google Scholar]
- 23.Galdos FX, Guo Y, Paige SL, VanDusen NJ, Wu SM, Pu WT. Cardiac regeneration: lessons from development. Circ Res. 2017;120:941–959. DOI: 10.1161/CIRCRESAHA.116.309040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. DOI: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 25.Reynolds JO, Chiang DY, Wang W, Beavers DL, Dixit SS, Skapura DG, Landstrom AP, Song LS, Ackerman MJ, Wehrens XH. Junctophilin‐2 is necessary for T‐tubule maturation during mouse heart development. Cardiovasc Res. 2013;100:44–53. DOI: 10.1093/cvr/cvt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talman V, Teppo J, Pöhö P, Movahedi P, Vaikkinen A, Karhu ST, Trošt K, Suvitaival T, Heikkonen J, Pahikkala T, et al. Molecular atlas of postnatal mouse heart development. J Am Heart Assoc. 2018;7:e010378. DOI: 10.1161/JAHA.118.010378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakaletsis S, Meador WD, Mathur M, Sugerman GP, Jazwiec T, Malinowski M, Lejeune E, Timek TA, Rausch MK. Right ventricular myocardial mechanics: multi‐modal deformation, microstructure, modeling, and comparison to the left ventricle. Acta Biomater. 2021;123:154–166. DOI: 10.1016/j.actbio.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto K, Li X, Shu C, Miyata T, Dardik A. Technical aspects of the mouse aortocaval fistula. J Vis Exp. 2013;77:e50449. DOI: 10.3791/50449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Liu C, Bao M, Liu W, Nie Y, Lian H, Hu S. Optimized Langendorff perfusion system for cardiomyocyte isolation in adult mouse heart. J Cell Mol Med. 2020;24:14619–14625. DOI: 10.1111/jcmm.15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter A, Šarić T, Hescheler J, Papadopoulos S. Calcium imaging in pluripotent stem cell‐derived cardiac myocytes. Methods Mol Biol. 2016;1353:131–146. [DOI] [PubMed] [Google Scholar]
- 31.Quiñones MA, Otto CM, Stoddard M, Waggoner A, Zoghbi WA; Doppler quantification Task Force of the Nomenclature and Standards Committee of the American Society of Echocardiography . Recommendations for quantification of Doppler echocardiography: a report from the Doppler Quantification Task Force of the Nomenclature and Standards Committee of the American Society of Echocardiography. J Am Soc Echocardiogr. 2002;15:167–184. DOI: 10.1067/mje.2002.120202. [DOI] [PubMed] [Google Scholar]
- 32.Bossers GPL, Hagdorn QAJ, Ploegstra MJ, Borgdorff MAJ, Silljé HHW, Berger RMF, Bartelds B. Volume load‐induced right ventricular dysfunction in animal models: insights in a translational gap in congenital heart disease. Eur J Heart Fail. 2018;20:808–812. DOI: 10.1002/ejhf.931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S4
Figure S1
Video S1


