Abstract
Interaction with DNA is essential for the tumor suppressor functions of p53. We now show, for the first time, that the interaction of p53 with DNA can be stabilized by small molecules, such as ADP and dADP. Our results also indicate an ATP/ADP molecular switch mechanism which determines the off-on states for p53-DNA binding. This ATP/ADP molecular switch requires dimer-dimer interaction of the p53 tetramer. Dissociation of p53-DNA complexes by ATP is independent of ATP hydrolysis. Low-level ATPase activity is nonetheless associated with ATP-p53 interaction and may serve to regenerate ADP-p53, thus recycling the high-affinity DNA binding form of p53. The ATP/ADP regulatory mechanism applies to two distinct types of p53 interaction with DNA, namely, sequence-specific DNA binding (via the core domain of the p53 protein) and binding to sites of DNA damage (via the C-terminal domain). Further studies indicate that ADP not only stabilizes p53-DNA complexes but also renders the complexes susceptible to dissociation by specific p53 binding proteins. We propose a model in which the DNA binding functions of p53 are regulated by an ATP/ADP molecular switch, and we suggest that this mechanism may function during the cellular response to DNA damage.
The p53 tumor suppressor plays a central role in the cellular response to DNA damage and blocks the proliferation of cells which have undergone genomic damage. Exposure of cells to genotoxic stress activates p53 as a transcription factor capable of regulating a wide range of downstream genes involved in G1 arrest, in DNA repair, and in apoptosis (for recent reviews, see references 13, 20, and 26). The p53 protein has two separate domains involved in DNA binding. The central core domain (residues 98 to 303) is responsible for binding to sequence-specific DNA elements located near promoters of downstream target genes (3, 39, 49). p53 can also form stable complexes with “nonspecific” DNA targets, including mismatched DNA (or lesion DNA [L-DNA]), double-strand breaks, single-stranded DNA (ssDNA), and Holliday junction structures (1, 16, 22, 23, 36, 37, 40). Interaction with abnormal DNA involves the carboxyl-terminal domain of p53 (residues 363 to 392), and the p53-DNA complexes may serve to recruit other proteins which function in DNA repair. Interaction with sites of DNA damage may also contribute to the activation of p53 by inducing proteolytic cleavage with removal of negative regulatory domains from the protein (38).
Treatment of cells with inhibitors of nucleotide biosynthesis can also activate a p53 response with induction of G1 arrest (27; reviewed in reference 19). This suggests that p53 can respond to altered levels of nucleotides within cells. The mechanism of p53 activation under such conditions is unknown. One possibility is that limiting levels of nucleoside triphosphates (or their precursors) lead to abnormal DNA and/or RNA within the cell, thus indirectly activating a p53 response. Another possibility is that p53 directly interacts with ribonucleotides and, in nondamaged cells, this contributes to the normal cellular function(s) of p53. Indeed, there is some evidence that p53 may play a role in the maintenance of cellular nucleotide pools. p53 was identified as a possible regulator of guanine synthesis at the step of IMP conversion to XMP (42). A link with adenosine metabolism is also indicated since a functional p53 response element is located in the first intron of the adenosine deaminase gene (21). In addition, a direct interaction between p53 and nucleotides is possible, and p53 protein binds ATP at its C terminus (4) and ATP facilitates the release of p53 from sites of DNA damage (34, 38).
In the present study, we have examined the effects of nucleotides on p53-DNA interactions in more detail by using murine and human p53s and specific and nonspecific DNA targets. The experimental model used p53-DNA complexes that were formed in vitro and incubated with different nucleotides. We observed that ATP, dATP, GTP, and dGTP facilitated the release of p53 from both sequence-specific and nonspecific DNA targets but, importantly, did not interfere with p53 binding to the DNA. In contrast, ADP and dADP stabilized p53-DNA complexes, and we demonstrated that tetramerization of p53 was required for this effect. Further experiments showed that p53, purified from a baculovirus expression system, was associated with Mg2+-dependent ATPase and GTPase activities: however, hydrolysis was not required for the release of p53 from DNA. The characteristics of the system bear a striking resemblance to the human mismatch recognition complex hMSH2-hMSH6, which functions as an ATP/ADP-dependent molecular switch. Thus, the hMSH2-hMSH6 complex binds mismatched DNA in the ADP-bound form (on) but not in the ATP-bound form (off) (14; reviewed in reference 10). Our results indicate that DNA binding by p53 is also on when bound to ADP and off when bound to ATP. Moreover, we also show that ADP-p53 can be dissociated from DNA by specific protein-protein interactions, raising the possibility that ADP-p53 functions as a molecular matchmaker for recruiting protein complexes to specific sites on DNA, followed by dissociation of p53 from the complex.
MATERIALS AND METHODS
Construction of p53 derivatives and mutants.
Wild-type murine p53 cDNA was used as a template to produce the truncated p53 derivatives. PCR primers were designed to generate cDNAs 50ΔN (residues 24 to 392), 50ΔC (residues 1 to 363), and 40ΔNC (residues 67 to 363) and C-terminal peptide (residues 323 to 363). All generated PCR fragments were His tagged at the amino terminus and cloned under the T7 promoter into the pBluescript II SK(+) vector. Plasmids containing p53 cDNAs coding for R273H, R175H, and M340Q/L344R mutants were kindly made available by Trevor Mee. All cloned cDNAs were verified by DNA sequencing.
p53 produced in vitro.
Transcription and translation were carried out as described previously (33) with plasmids encoding wild-type human p53, murine p53, or its derivatives under the SP6 or T7 promoters. The produced proteins were checked by immunoprecipitation with anti-p53 monoclonal antibodies PAb248, PAb246, PAb1620, PAb240, and PAb421 as described previously (33).
Expression, purification, and analysis of baculovirus-produced p53.
Recombinant baculovirus coding for His-tagged murine p53 was expressed in Sf9 insect cells as described previously (34) and p53 was purified as described previously (38). The final buffer used for the purified p53 contained 50 mM NaCl, 10 mM Tris-HCl (pH 7.0), and 5 mM dithiothreitol (DTT). p53 protein was quantitated by the Bradford assay (compared to a bovine serum albumin standard [Boehringer Mannheim]), and its purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by either Coomassie or silver staining. A single band corresponding to p53 was observed in all cases (for an example, see Fig. 4A). For immunoblotting, protein samples were separated by SDS-PAGE (15% polyacrylamide) and probed with PAb240 anti-p53 monoclonal antibody as described previously (38). Detection was performed with the Boehringer Mannheim chemiluminescence blotting kit.
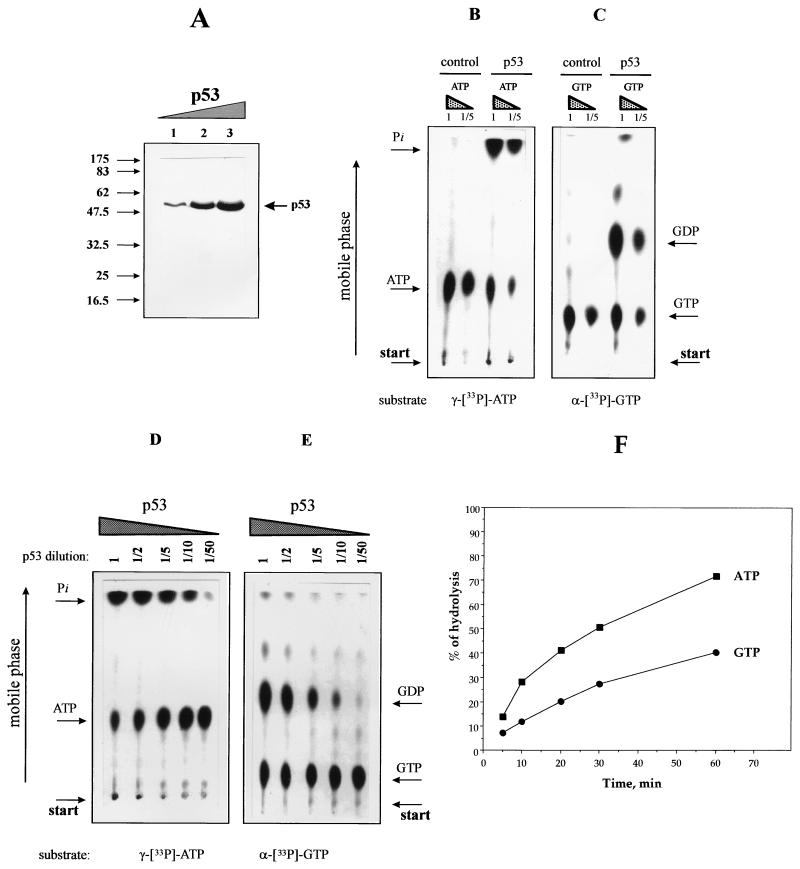
FIG. 4.
p53 is able to hydrolyze ATP and GTP. (A) Recombinant p53 was purified, and increasing amounts of purified protein were separated by PAGE (15% polyacrylamide) and stained with Coomassie blue. Lanes: 1, 0.5 μg of recombinant p53; 2, 4 μg; 3, 8 μg. The positions of molecular weight standards are indicated in thousands. (B and C) Equal amounts of purified p53 were incubated in the presence of [γ-33P]ATP (B) or [α-33P]GTP (C). Samples were incubated for 60 min at 37°C and analyzed by TLC (see Materials and Methods). The amount of labelled ATP (GTP) was 5 pM or, for a 1:5 dilution, 1 pM. The amount of the p53 added, when indicated, was approximately 5 pM (calculated for the tetrameric protein). The negative control was an equivalent aliquot of cell lysate from cells infected with wild-type virus (see the text for details). (D and E) Experiment where equal amounts (1 pM) of [γ-33P]ATP (D) or [α-33P]GTP (E) were incubated in the presence of decreasing concentrations of purified p53, as indicated. Approximately 5 pM p53 was used as the initial concentration (lane 1), providing a protein-to-substrate ratio of 5:1 (for tetrameric protein). Arrows indicate the positions of radiolabelled substrates and products. (F) Time course analysis of the efficiency of nucleoside triphosphate hydrolysis by p53. Equal amounts of p53 were incubated in the presence of radiolabelled nucleotides (protein-to-substrate ratio, 5:1). After the indicated times, aliquots of the reaction products were loaded onto a TLC plate and the products were separated by TLC and quantitated by scintillation counting (see Materials and Methods).
Oligonucleotides for binding assays.
The following biotinylated oligonucleotides were used: p53-consensus (CON; 20 bases, biotinylated at the 5′ end) (5′-GGACATGCCCGGGCATGTCC-3′) (12) and L-DNA (5′-GGCTCGAAC CCGTTCTCGGAGCACCCCTGCCCCAGCCCAACCGCTTTGGCCGCCG CCCAGCC-3′) (62 bases) (22), where triple cytosine lesions are underlined. The oligonucleotides were annealed as follows: CON to itself and L-DNA oligonucleotide to the reverse sequence 5′-GGCTGGGCGGCGGCCAAAGCGGTTCTGCAGTGCTCCGAGAACGGGTTCGAGCC-3′ (53 bases). The latter was used also as an ssDNA template for binding assays. For NL-DNA (dsDNA with blunt double-strand ends), we used oligonucleotides with the same sequence as L-DNA but missing the triple cytosine lesions.
DNA binding and release assay with magnetic beads.
Streptavidin-coated magnetic beads (M-280 Dynabeads; Dynal) were used to harvest biotinylated DNA-protein complexes. dsDNA or ssDNA oligonucleotides were bound to the beads (typically 75 pmol of oligonucleotide per 40 μl of beads for each reaction) in Tris-EDTA (pH 7.5)–1 M NaCl (TE-NaCl) for 15 min at 20°C and washed twice with 400 μl of TE-NaCl and twice with 400 μl of DNA binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1% NP-40, 10% glycerol, 5 mM DTT) to remove nonbound DNA. The supernatant was replaced with fresh 50 μl of DNA binding buffer containing either 25 pmol of purified p53 (in buffer [50 mM NaCl, 10 mM Tris-HCl at pH 7.0, 5 mM DTT]) or a 10-μl aliquot of an in vitro translation reaction mixture. Typically, 40 μl of beads was used in a total reaction volume of 50 μl. After a 20-min incubation at room temperature, the p53-DNA complexes were collected on a magnetic harvester, washed four times with 400 μl of DNA binding buffer to remove all free p53, resuspended in 50 μl of DNA binding buffer, and incubated under the conditions of the experiment. When nucleotides were present, the final concentration of each was 5 mM. After addition of the nucleotide, samples were incubated for 10 min at 37°C (unless otherwise stated). The ATP-regenerating system, when present, contained 4mM ATP, 0.025 U of creatine phosphokinase per ml, 5 mM phosphate creatine, and 0.25 mg of bovine serum albumin per ml. For experiments studying the effect of magnesium, the final concentration of MgCl2 was 5 mM. No difference in the efficiency of initial p53 binding to DNA was observed in the presence or absence of Mg2+ or in the presence of an ATP-regenerating system.
After incubation, released and DNA-bound p53 fractions (see Fig. 1A) were analyzed by SDS-PAGE (15% polyacrylamide) and immunoblotted (for baculovirus-produced protein). For in vitro-produced p53 proteins, equal aliquots were taken from bound and released fractions and analyzed by scintillation counting and SDS-PAGE (15% polyacrylamide) followed by autoradiography.
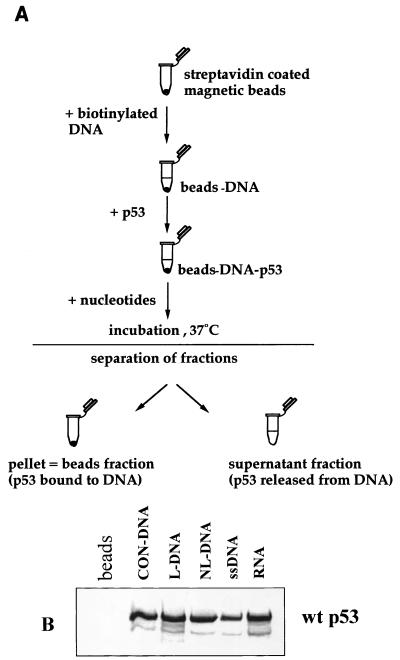
FIG. 1.
Scheme of the DNA binding and release assay. (A) Streptavidin-coated magnetic beads were used to bind biotinylated oligonucleotide targets. After washing, the DNA-bound beads were incubated with p53. Bound p53-DNA complexes on the beads were washed (to remove nonbound p53) and incubated in fresh buffer, with or without nucleotides, for 10 min at 37°C (or longer for time course experiments). Subsequently the released p53 (in solution) and DNA-bound p53 were analyzed by scintillation counting (for radiolabelled p53) and PAGE (15% polyacrylamide) followed by immunoblotting or autoradiography. (B) Binding of 35S-labelled p53 to magnetic beads coated with different oligonucleotides (detailed in Materials and Methods). An aliquot of beads without DNA was included as negative control (beads lane). Other lanes: CON-DNA, beads prebound with a consensus sequence-specific p53 DNA binding site; L-DNA, dsDNA containing triple insertion-deletion lesion; NL-DNA, dsDNA with blunt ends (the bottom strand of NL-DNA alone was used as ssDNA target) (see Materials and Methods). RNA was obtained by in vitro transcription from p53 cDNA under the T7 promoter by using biotinylated UTP. The faster-migrating band (below p53) represents a truncated p53 product that is routinely observed for in vitro-translated p53. wt, wild type.
Hydrolysis assay and TLC.
Purified p53 (5 pM) was added to the reaction mixture containing [33P]ATP, [33P]dATP, or [33P]GTP. The reactions were typically carried out in a total volume of 21 μl, comprising 10 μl of protein sample, 10 μl of the reaction buffer (50 mM NaCl, 10 mM Tris-HCl [pH 7.0]), and 1 μl of radiolabelled nucleoside triphosphate either neat (5 pM) or diluted 1:5 (1 pM). When magnesium was present, the final concentration was 5 mM MgCl2. All reaction mixtures were incubated at 37°C for 1 h (except the time course samples). Aliquots (4 μl) from each reaction mixture were analyzed by thin-layer chromatography (TLC) on PEI cellulose F plates (Merck) with 0.3 M sodium phosphate buffer (pH 3.5) as the mobile phase. After the chromatography, the TLC plates were air dried and analyzed by autoradiography. The autoradiography image was used to locate the zones of the TLC plate corresponding to the initial substrates and products of hydrolysis, these zones were cut out, and radioactivity was quantitated by scintillation counting.
Gel mobility shift assay.
Equal aliquots of in vitro-translated p53 were incubated with 5 ng of 32P-5′-end-labelled self-annealed double-stranded CON-DNA, in DNA binding buffer in a final volume of 50 μl. After 20 min of incubation at a room temperature, appropriate amounts of adenosine nucleotides were added (see Fig. 6) and incubation was continued for another 10 min. Where stated, 20 μl of hybridoma supernatant of anti-p53 monoclonal antibody was added and the mixture was incubated for another 5 min. If the antibody was not added, the final volume of the reaction mixture was adjusted by addition of 20 μl of DNA binding buffer. Complexes were loaded onto a native 4% polyacrylamide gel in TBE (containing 1 mM EDTA) and electrophoresed for 4 h at 120 V with water cooling (8 to 12°C). The gels were exposed for autoradiography at −70°C with aluminum foil placed between the gel and the X-ray film to cut out the signal from the [35S]methionine.
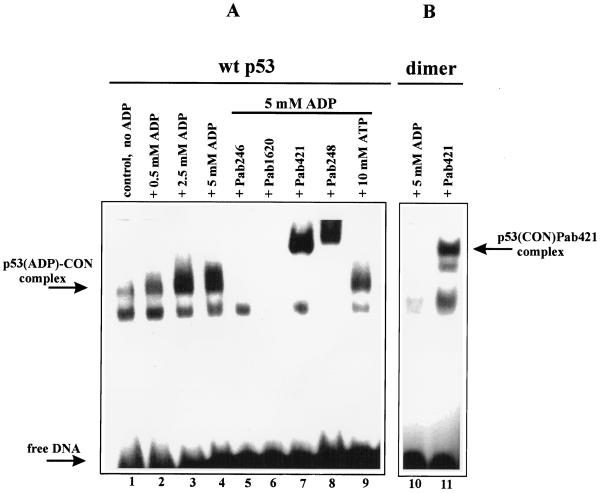
FIG. 6.
Stabilization of the p53-CON-DNA complex by ADP and comparison of wild-type tetrameric p53 with dimeric p53. (A) Gel mobility shift assay of wild-type (wt) p53-CON-DNA complexes in the presence of increasing amounts of ADP (lanes 1 to 4, as indicated) or with a constant amount of ADP (5 mM; lanes 5 to 9). Where relevant, the addition of different monoclonal antibodies or 10 mM ATP is indicated. Upper bands in lanes with PAb421 and PAb248 represent antibody supershifts of the p53-DNA complexes. (B) Gel mobility shift assay of dimeric p53 complexed with CON-DNA in the presence of 5 mM ADP (lane 10) or PAb421 (lane 11). Results obtained in the absence of ADP and PAb421 are equivalent to those obtained in the presence of ADP. The faster-migrating band, below the p53-DNA complexes, is derived from a component present in the rabbit reticulocyte lysate. It is routinely observed in negative control translations (without mRNA [not shown]) and is reduced by addition of certain hybridoma supernatants, including PAb1620.
RESULTS
Previously we have shown that addition of ATP induces the release of p53 from mismatched DNA and ssDNA (34, 38). We have now studied in more detail the effects of ATP and other nucleotides on the interaction of p53 with DNA, by using both sequence-specific and nonspecific DNA targets. The results presented are for sequence-specific DNA (CON-DNA [12]) and mismatched DNA targets (22). Other nonspecific DNA targets included ssDNA and dsDNA with double-strand breaks (see Materials and Methods for details).
ATP and ADP have opposing effects on the stability of p53-DNA complexes.
Radiolabelled p53 protein was obtained by in vitro translation with rabbit reticulocyte lysate (see Materials and Methods). The experimental assay used p53-DNA comlexes formed in vitro with biotinylated oligonucleotides bound to streptavidin-coated magnetic beads (Fig. 1A). p53-DNA beads were incubated at 37°C in the presence of different nucleotides. Both supernatant (released p53) and beads (DNA-bound p53) were analyzed by SDS-PAGE (15% polyacrylamide) plus autoradiography. Examples of p53 binding to different oligonucleotide targets are shown in Fig. 1B. In general, the observed binding efficiency was 25 to 30% of the total input radiolabelled p53. The lower efficiency of binding to ssDNA (Fig. 1B) is consistent with previously published results from other laboratories (23, 37). Control incubations demonstrated that there was no binding of radiolabelled p53 to the beads alone (i.e., in the absence of oligonucleotide [Fig. 1B, beads lane]).
Further controls showed that only a small amount of protein, 5% of the total for CON-DNA and approximately 10% for L-DNA, was released from the preformed p53-DNA complexes during 10 min of incubation at 37°C. The effect of different nucleotides on the stability of the p53-DNA complexes was next examined. When the nucleoside triphosphates ATP or GTP (and dATP or dGTP) were added, the release of p53 was enhanced more than threefold, with 17 to 20% release from CON-DNA and 32 to 34% from the L-DNA target (represented graphically in Fig. 2A, with actual examples shown in Fig. 2B and C). Only a marginal effect was observed with the pyrimidine triphosphates CTP and UTP (Fig. 2A), indicating that the release of p53 from DNA is preferentially induced by purine-based triphosphates.
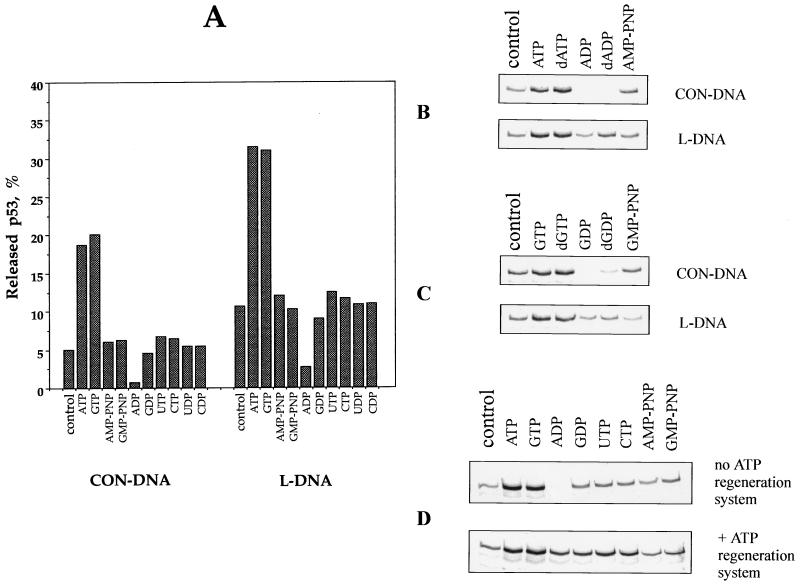
FIG. 2.
Nucleotide-induced release of 35S-labelled p53 from specific (CON-DNA) and nonspecific (L-DNA) targets. (A) In vitro-translated p53 was bound to either CON-DNA or L-DNA beads as indicated and incubated in the presence of different nucleotides (Fig. 1). DNA-bound p53 and released p53 were quantitated by scintillation counting. Bars represent the percentage of released p53 with respect to released plus bound p53. (B and C) Comparison of the release of p53 from specific and nonspecific DNA targets incubated with either adenosine or guanosine nucleotides. Equal aliquots of p53 bound to DNA-beads were incubated in the presence of adenosine nucleotides (B) or guanosine nucleotides (C). (D) Release of p53 from CON-DNA-beads after incubation with different nucleotides in the presence or absence of an ATP-regenerating system. p53 released from the DNA was analyzed by PAGE (15% polyacrylamide). The control consisted of p53 released in the absence of nucleotide addition.
ADP, on the other hand, appeared to block the dissociation of p53-DNA complexes, and dADP had a similar effect (Fig. 2A to C). This was unexpected and indicates that the ATP-induced release of p53 from the DNA described above was not the result of competition between DNA and nucleotides for the DNA binding surface of p53. To determine if the stabilizing effect of ADP is reversible, we incubated p53-DNA complexes in the presence of nucleoside tri- or diphosphates with or without an ATP regeneration system. As shown in Fig. 2D, addition of an ATP regeneration system partially restores p53 release from CON-DNA in the presence of ADP.
These overall results indicate that purine triphosphates enhance the dissociation of p53-DNA complexes whereas ADP stabilizes p53-DNA complexes. Similar results were obtained for recombinant p53, for human and murine p53 proteins, and for each of the DNA targets (listed in Materials and Methods). Thus, the observed ATP/ADP effect applies to two distinct types of p53 interaction with DNA, namely, sequence-specific DNA binding (via the core domain of the p53 protein) and binding to sites that imitate DNA damage (via the C-terminal domain).
Other proteins involved in the recognition and binding of sites of DNA damage are also influenced by ATP. However, in general, ATP stimulates protein-DNA binding (10, 41). For p53, we found the opposite: ATP released p53 from DNA, while ADP stabilized the protein-DNA complex.
One possible explanation for these observations is that ATP and GTP chelate divalent cations much more efficiently than ADP and GDP do, thereby causing differential effects on the structural stability of the DNA target. However, this can be excluded on a number of grounds. (i) No divalent cations were present in the incubation buffers (see Materials and Methods). Nucleotides were used at 5 mM (see Materials and Methods), and it is already established that EGTA (5 mM) has no effect on p53-DNA binding in vitro (reference 34 and unpublished observations). (ii) Chelation of divalent cations by the phosphates of 5′-adenylylimidodiphosphate (AMP-PNP) and 5′-guanylylimidodiphosphate (GMP-PNP) would be expected to give results similar to those for ATP and GTP: this was not observed. Instead, AMP-PNP and GMP-PNP had little, if any, effect on the release of p53 from DNA (Fig. 2; see also Discussion). (iii) If the observed effect is due to chelation by the base rings of the nucleotides, it follows that each of the purine nucleotides should give similar effects independent of the number of phosphate groups. This was not observed (Fig. 2). We therefore conclude that the opposing effects of ATP and ADP on the stability of p53-DNA complexes cannot simply be attributed to chelation of divalent cations.
Tetramerization is required for nucleotide-dependent regulation of the stability of p53-DNA complexes.
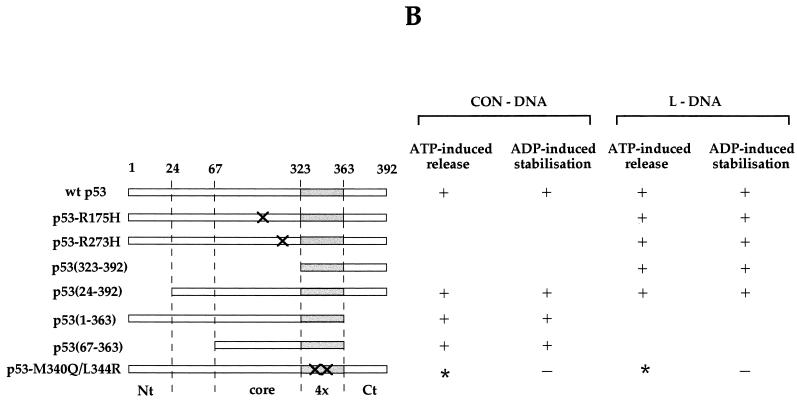
Our next experiments were designed to identify the domain(s) of p53 required for ATP/ADP-dependent regulation of p53-DNA interaction. For this purpose, we compared the following series of p53 constructs: Arg175His, Arg273His, a C-terminal peptide (residues 323 to 392), p53 ΔN50 (residues 24 to 392), p53 ΔC50 (residues 1 to 363), and p53 40ΔNC (residues 67 to 363). Throughout this series of p53 mutants, the oligomerization domain was intact. To ask if modification of the oligomerization domain can affect ATP-induced dissociation and/or ADP stabilization of p53-DNA complexes, we also included a double mutant, M340Q/L344R, which is dimeric but retains “wild-type” p53 immunological conformation, reactive with PAb1620 and nonreactive with PAb240 (8, 31).
Each of the above p53 mutants was tested with CON-DNA and/or mismatched DNA, bearing in mind that mutants lacking an intact core domain do not bind CON-DNA and those lacking the C terminus do not bind mismatched DNA. All, with the exception of the dimeric p53 mutant, were similar to wild-type p53 in that ATP induced the release of the p53 protein from DNA and, conversely, ADP stabilized the p53-DNA complex (Fig. 3A, summarized in Fig. 3B). With dimeric p53, however, ADP failed to stabilize the protein-DNA complexes (Fig. 3A). Dimeric p53-DNA complexes are more labile than tetrameric wild-type p53-DNA (reference 30 and observations in this study). This explains the higher level of p53 dimer released from DNA in the control incubation, and ATP did not induce any additional release under these conditions (Fig. 3A).
FIG. 3.
Release of p53 from DNA: effects of nucleotides on different p53 mutants. (A) Nucleotide-induced release of 35S-labelled p53 mutants from DNA targets (see the text for details). Equal aliquots for each p53 mutant were bound to DNA-beads and were incubated in the presence of different nucleotides as detailed in the legend to Fig. 1 and Materials and Methods. No nucleotide was added in the control lane. (B) Summary of results obtained from four independent experiments with each p53 protein. The major structural domains of p53 are also indicated. Nt, N terminus; Ct, C terminus; 4×, tetramerization domain. Sites of point mutations are indicated by crosses. ∗, dimeric p53 forms DNA complexes with a shorter half-life than that of wild-type p53 (see the text).
Taken together, these results indicate that tetramerization is necessary for the observed off-on effects of ATP/ADP on the stability of p53-DNA complexes. We suggest (i) that certain dimer-dimer interactions of the p53 tetramer are important for regulating the stability of p53-DNA complexes and (ii) that ATP/ADP may destabilize or stabilize p53-DNA complexes at the level of dimer-dimer interactions. It is interesting that naturally occurring oligomerization mutants of p53 are defective for function and are linked with cancer predisposition in humans (9, 28). Our present results raise the possibility that this defective functioning in vivo involves the loss of ADP/ATP regulation of p53-DNA interactions.
p53 can hydrolyze ATP and GTP.
Having demonstrated that ATP and GTP induce the release of p53 from p53-DNA complexes (Fig. 2), we wished to determine whether p53 has the capacity to hydrolyze ATP and/or GTP. For this purpose, we used nucleoside triphosphates radiolabelled at the γ-phosphate position ([γ-33P]ATP) or the α-phosphate position ([α-33P]GTP) and p53 purified from a baculovirus expression system (Fig. 4A) (see Materials and Methods). Radiolabelled ATP or GTP was incubated with p53 for 60 min at 37°C. The release of radiolabel was determined by TLC and autoradiography. In all cases, we observed hydrolysis of the nucleoside triphosphates, with the release of radiolabelled Pi for [γ-33P]ATP (Fig. 4B) and radiolabelled GDP for [α-33P]GTP (Fig. 4C). This indicates cleavage at the β-γ phosphoanhydride bond of the triphosphate. Hydrolysis was Mg2+ dependent and copurified with p53 (results not shown). Negative control incubations included an extract from the Sf9 cells infected with wild-type baculovirus and subjected to the same purification scheme as the p53-producing cell extract. These controls showed no evidence of ATP or GTP hydrolysis (Fig. 4B and C, control lanes).
The efficiency of hydrolysis in the presence of p53 was low, and we were able to detect reaction products only under conditions when the enzyme concentration ([E]) was similar to or greater than the substrate concentration ([S]). Moreover, for both ATP or GTP, only part of the substrate was hydrolyzed by p53 within 1 h, even when the substrate was diluted up to 1:5 (Fig. 4B and C).
Additional experiments showed that when the amount of p53 protein was decreased relative to that of ATP or GTP, hydrolysis decreased in proportion to the amount of p53 present in the reaction (Fig. 4D and E). The starting protein to substrate ratio was 5:1 (picomoles of tetrameric p53 per picomole of nucleoside biphosphate), and quantitative analysis revealed that the efficiency of GTP hydrolysis was half that of ATP hydrolysis. Time course studies showed a slow steady-state process, reaching 75% of initial substrate hydrolyzed within 1 h for ATP and 40% for GTP (Fig. 4F). From our data, the ATP turnover number by p53 could be calculated as 0.012 molecule min−1. This low turnover is consistent with the activities of other molecular-switch proteins (see Discussion).
Hydrolysis of ATP is not required for p53-DNA dissociation.
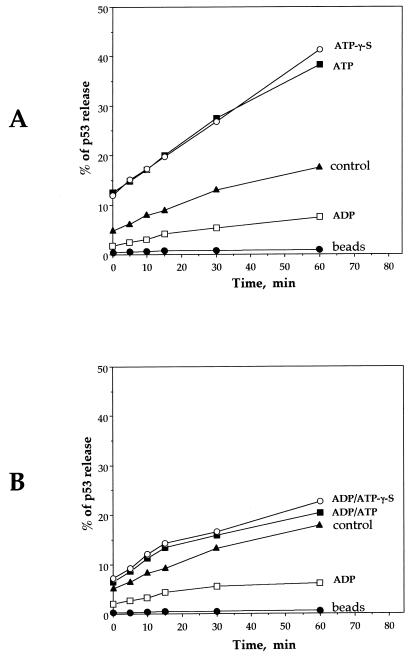
Having demonstrated that p53 has intrinsic ATPase and GTPase activities (Fig. 4), we asked whether this activity is coupled with the ability of ATP and GTP to dissociate p53-DNA complexes (Fig. 2). We reasoned that if hydrolysis is required for dissociation, the ATP analogue ATP-γ-S should not support dissociation, since this compound cannot be efficiently hydrolyzed at the β-γ phosphoanhydride bond. However, ATP-γ-S and ATP gave virtually identical results over a period of 60 min (Fig. 5A), indicating that ATP hydrolysis is not required for dissociation of p53-DNA complexes. This suggests that the ability of ATP to dissociate p53-DNA complexes may involve allosteric changes induced by ATP (and ATP-γ-S) in the p53 protein.
FIG. 5.
Hydrolysis of ATP is not required for dissociation of p53-DNA complexes. (A) Time course analysis of p53 release from sequence-specific DNA. Samples were incubated with ATP, ATP-γ-S, or ADP. Controls involve incubations without nucleotide addition. After the given times (5, 10, 20, 30, and 60 min), fractions of released and DNA-bound p53 were quantitated by scintillation counting. (B) p53-DNA complexes were initially stabilized with ADP and subsequently incubated with nucleotides as described above. For both experiments, the data represent an average of four independent experiments. “beads,” control incubation of beads, to which p53 was bound without DNA and subsequently washed (see Fig. 1A and Materials and Methods) to remove unbound protein before the incubation.
ADP stabilized p53-DNA complexes during the above time course experiments (Fig. 5A), consistent with the results in Fig. 2. Given the opposing effects of ATP and ADP, we were interested in determining if ATP could overcome the stabilization of p53-DNA complexes induced by ADP. Preformed ADP-p53-DNA complexes were incubated with ATP or ATP-γ-S, and the release of p53 from the DNA over 60 min was assayed. The results show that both ATP and ATP-γ-S dissociate ADP-p53-DNA complexes but that dissociation was similar to the levels observed for control incubations (with no addition to the p53-DNA incubation [Fig. 5B]). Thus, in the presence of ADP, addition of ATP is unable to enhance the dissociation of p53-DNA complexes over control levels.
ADP-p53-DNA complexes can be dissociated by protein-protein interaction.
The results shown in Fig. 2, 3, and 5 indicate that ADP stabilizes p53-DNA complexes. This was investigated further by gel shift analysis with in vitro-translated p53 and radiolabelled CON-DNA. The anti-p53 monoclonal antibody PAb421 is routinely added to stabilize p53-DNA complexes for gel shift analysis (12). We now asked if ADP can substitute for PAb421 to give stable p53-DNA complexes detectable by a gel shift assay. Our results show that ADP stabilizes the p53-DNA complexes and that the effect is concentration dependent, with no further increase observed between 2.5 and 5.0 mM ADP (Fig. 6A, lanes 1 to 4).
Interestingly, the p53-CON complex stabilized by ADP was dissociated by anti-p53 monoclonal antibodies PAb246 and PAb1620 (Fig. 6A, lanes 5 and 6). These antibodies detect conformation-dependent epitopes in the core domain of p53, and PAb1620 (but not PAb246) dissociates DNA complexes of murine p53 in the presence of PAb421 (15). The fact that both PAb246 and PAb1620 dissociate ADP-p53-DNA complexes (Fig. 6A) indicates that the mechanism of stabilization by ADP may be distinct from that of stabilization by PAb421, since the latter is not sensitive to PAb246 dissociation (15). PAb421 or PAb248 formed additive complexes with ADP-p53-DNA and did not cause dissociation (Fig. 6A, lanes 7 and 8). When a twofold excess of ATP was added to the complex stabilized by ADP, a partial decrease in the amounts of ADP-p53-DNA complexes was observed (lane 9). This is consistent with the results presented in Fig. 5B.
The results shown in Fig. 3 indicate that dimer-dimer interaction is important for the stabilization of p53-DNA complexes by ADP. This was investigated further under gel shift conditions, and the results are shown in Fig. 6B. Dimeric p53-DNA complexes are clearly evident and can be stabilized by PAb421. However, in marked contrast to wild-type p53, the dimeric mutant-DNA complexes could not be stabilized in the presence of ADP alone (Fig. 6B, lane 10, compared with Fig. 6A, lane 4). These results confirm that an intact p53 oligomerization domain is required to provide the necessary quaternary environment for ADP-induced stabilization of p53-DNA complexes.
DISCUSSION
p53 functions as a molecular switch.
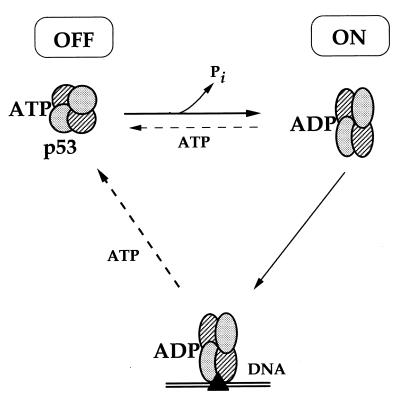
Our present results demonstrate that ATP and ADP have opposing effects on the interaction of p53 with DNA: ATP dissociates p53-DNA complexes, whereas ADP blocks dissociation (Fig. 2 and 6). We also show that p53 has ATPase activity and that this activity is not coupled to the dissociation of p53-DNA complexes (Fig. 4 and 5). Although ATP can dissociate p53-DNA complexes, it does not block the binding of p53 to ssDNA (38). We now show that ATP is able to induce only partial release of p53 from dsDNA (sequence-specific or nonspecific with mismatches). Considering the amounts of ATP in the reaction mixture (up to 5 mM) relative to DNA (100 pmol) and p53 (approximately 10 pmol for purified protein and in the femtomolar range for in vitro translated p53), we cannot explain the ATP-induced release of p53 by competition between ATP and DNA for the DNA-binding surface on p53. These overall observations lead us to suggest that the ATPase activity of p53 may serve to convert ATP-p53 to ADP-p53 and thus promote stable DNA binding (represented schematically in Fig. 7). The DNA-bound form of p53 can subsequently be dissociated by ATP, possibly by an allosteric mechanism (see below). In this model, the ATP- and ADP-bound p53 represent the off and on forms of p53 for DNA binding, respectively.
FIG. 7.
Schematic representation of the ATP/ADP-dependent molecular switch for p53-DNA binding (see Discussion).
These results bear a number of striking similarities to the novel type of molecular-switch mechanism recently described for the human mismatch recognition complex hMSH2-hMSH6 (10, 14). First, both the hMSH2-hMSH6 heterodimer and the p53 protein bind mismatched DNA and possess low-level steady-state ATPase activity. Second, the effects of ATP on p53 and hMSH2-hMSH6 are similar in that when ATP is bound, both p53 and hMSH2-hMSH6 are in the “off” form, with low affinity for DNA. Moreover, ATP can release both p53 and hMSH2-hMSH6 from DNA without itself being hydrolyzed. However, when the ATP-protein complex is released from the DNA template, the intrinsic ATPase activity of the protein can regenerate the ADP-bound, “on” form thus completing the cycle of molecular switch regulation (14; see above). Third, for both p53 and hMSH2-hMSH6, the rate-limiting step in the ATP/ADP exchange appears to be the replacement of the adenosine nucleotide in the active centre. Thus, an excess of ATP did not dissociate all p53-DNA complexes (Fig. 2, 3, 5 and 6) and p53 protein hydrolyzes ATP with the same efficiency in the presence or absence of DNA (results not shown). Limiting the molecular switch at the step of nucleotide exchange will provide a situation where ADP-p53 (on) has a longer half-life than the opposing ATP-p53 (off) form of the molecular switch. It is interesting that neither hMSH2-hMSH6 nor p53 is released from DNA by AMP-PNP (14; see above) (Fig. 2). This suggests a similar spatial organization of the active-site centers between these two proteins and the importance of the O3β atom between Pβ and Pγ in a mechanism involved in the switch.
Despite the above similarities to hMSH2-hMSH6, additional observations serve to distinguish the p53 molecular switch and identify it as a novel mechanism for the regulation of the protein-DNA interaction. First, analysis of the p53 protein sequence does not reveal any homology to known ATP or GTP binding motifs (43, 46). Second, the ATP/ADP switch regulates the binding of p53 to two distinct types of DNA target, nonspecific DNA and sequence-specific DNA targets (binding to these targets involves different structural domains of the p53 protein [see Introduction]). Perhaps the positioning of the ATP/ADP regulatory switch within the oligomerization domain may provide a structural basis which allows a common regulatory mechanism to operate with different DNA targets (see Results, Fig. 3, and the following discussion). A third property thus far unique to p53 is that it can also use GTP as a release factor to dissociate p53-DNA complexes (Fig. 2C). This places p53 as a link between the two groups of other known molecular switch proteins, which are regulated exclusively either by ATP/ADP or by GTP/GDP mechanisms (reviewed in reference 10).
The regulation of molecular-switch proteins is determined by specific protein-protein or protein-DNA interactions. When alone, they exhibit marginal catalytic activity. ATP or GTP turnover rates for molecular-switch proteins span from 1 ATP molecule per min for MuB (51) to 0.03 per min for Ras (44) and as low as 0.003 per min for EF-Tu (18). Although the rate of p53 ATPase activity is low and is approximately 0.012 molecule min−1, it is within the range of turnover rates of known molecular-switch proteins (10, 43). However, interaction with specific protein partners can dramatically increase the turnover. For example, hydrolysis by Ras-type G proteins may be accelerated by 4 to 5 orders of magnitude by GTPase-activating proteins (GAPs) (43). For ras, it has been suggested that GAPs may provide a catalytic residue that ras itself lacks (43). Our future studies will investigate the possibility that ATPase and/or GTPase activity is similarly stimulated by p53 binding proteins present in the cell.
A possible mechanism of ATP/ADP-dependent modulation of p53.
Our results demonstrate that ATP/ADP operate a molecular switch to regulate DNA binding by p53. We also identify p53 dimer-dimer interaction as possibly important for this regulation. As yet, we do not know how ATP influences p53 at the structural level. One possibility is that ATP binding modifies the interhelical angles within the C-terminal tetramerization domain, with knock-on effects on the overall quaternary structure of the protein and its DNA binding properties. This would be consistent with the observed variation in the angles of subunit interaction at the dimer-dimer interface of the tetramerization domain (6, 7, 17, 24, 32) and with ATP interaction at a C-terminal DNA binding site (4). It would also accommodate the hypothesis that to form a stable complex with specific DNA, p53 must switch from dihedral symmetry with low affinity for DNA binding to an asymmetric state with high-affinity binding (50). Dihedral symmetry of the p53 tetramer is provided by the oligomerization domain. We now show evidence that the quaternary structure provided by this domain is required for ATP/ADP-dependent modulation of p53-DNA stability (Fig. 3). The switch from a dihedral to an asymmetric form may help to relieve possible steric clashes between protein subunits within the tetramer, allowing tetrameric p53 to accommodate itself to the structure of the DNA site (2, 35).
Interaction between p53 and DNA is essential for p53 function, and evidence from genetic studies indicates that p53 may play an important role in DNA repair as well as in the transcription of p53 target genes (11, 25, 29, 45, 47, 48). Any regulatory mechanism affecting the interaction of p53 and DNA is likely to be fundamentally important for maintenance of genetic integrity. We now present evidence that ATP/ADP can modulate p53-DNA interaction.
We also demonstrate that ADP-p53-DNA complexes retain accessible p53 binding sites for other proteins, namely, anti-p53 antibodies which bind epitopes at the N and C termini of p53 (PAb248 and PAb421) and within the core domain (PAb246 and PAb1620 [Fig. 6]). Antibodies binding at the N and C termini of p53 form additive complexes with ADP-p53-DNA, whereas the antibody-binding interaction within the core domain causes the dissociation of the ADP-p53-DNA complex (Fig. 6). These various characteristics raise the possibility that p53 functions as a “matchmaker” (41) for assembly of multiprotein complexes on DNA.
In summary, our experimental data show that p53 exhibits low ATPase activity and bears the hallmarks of an ATP/ADP-dependent molecular switch which regulates the stability of p53-DNA complexes. It is possible that stabilization by ADP is important to allow time for the ADP-p53-DNA complexes to mark DNA sites for correct assembly of macromolecular structures involved in repair and/or transcription in response to genotoxic stress.
ACKNOWLEDGMENTS
We thank Carlos Rubbi and Trevor Mee for many helpful discussions and critical reading of the manuscript, and we thank Meg Stark for photographic work.
This work was supported by Yorkshire Cancer Research.
REFERENCES
- 1.Bakalkin G, Selivanova G, Yakovleva T, Kiseleva E, Kashuba E, Magnusson K, Szekely L, Klein G, Terenius L, Wiman K. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balagurumoorthy P, Sakamoto H, Lewis M S, Zambrano N, Clore G M, Gronenborn A M, Appella E, Harrington R E. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc Natl Acad Sci USA. 1995;92:8591–8595. doi: 10.1073/pnas.92.19.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 4.Brain R, Jenkins J R. Human p53 directs DNA strand reassociation and is photolabelled by 8-azido ATP. Oncogene. 1994;9:1775–1780. [PubMed] [Google Scholar]
- 5.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenicmutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 6.Clore G M, Ernst J, Clubb R, Omichinski J G, Kennedy W M, Sakaguchi K, Appella E, Gronenborn A M. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat Struct Biol. 1995;2:321–333. doi: 10.1038/nsb0495-321. [DOI] [PubMed] [Google Scholar]
- 7.Clore G M, Omichinski J G, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn A M. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science. 1994;265:386–391. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- 8.Davison T S, Yin P, Nie E, Arrowsmith C H. Abstracts of the 9th p53 Workshop 1998. 1998. Oligomerisation and p53 function; p. 40. [Google Scholar]
- 9.Davison T S, Yin P, Nie E, Kay C, Arrowsmith C H. Characterisation of the oligomerisation defects of two p53 mutants found in families with Li-Fraumeni and Li-Fraumeni-like syndrome. Oncogene. 1998;17:651–656. doi: 10.1038/sj.onc.1202062. [DOI] [PubMed] [Google Scholar]
- 10.Fishel R. Mismatch repair, molecular switches, and signal transduction. Genes Dev. 1998;12:2096–2101. doi: 10.1101/gad.12.14.2096. [DOI] [PubMed] [Google Scholar]
- 11.Ford J M, Hanawalt P C. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA-repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funk W D, Pak D J, Karas R H, Wright W E, Shay J W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb T, Oren M. p53 in growth-control and neoplasia. Biochim Biophys Acta Rev Cancer. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 14.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 15.Hall A R, Milner J. Specific p53-DNA complexes contain an mdm2-related protein. Oncogene. 1997;14:1371–1376. doi: 10.1038/sj.onc.1200962. [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman L, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 17.Jeffry P D, Gorina S, Pavletich N P. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 18.Kalbitzer H R, Goody R S, Wittinghofer A. Electron-paramagnetic-resonance studies of manganese(II) complexes with elongation factor Tu from Bacillus stearothermophilus. Observation of a GTP hydrolysis intermediate state complex. Eur J Biochem. 1984;141:591–597. doi: 10.1111/j.1432-1033.1984.tb08234.x. [DOI] [PubMed] [Google Scholar]
- 19.Kastan M. Signalling to p53—where does it all start. Bioessays. 1996;18:617–619. doi: 10.1002/bies.950180804. [DOI] [PubMed] [Google Scholar]
- 20.Ko L J, Prives C. p53—puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 21.Kondratov R V, Pugacheva E N, Kuznetsov N V, Prasolov V S, Kopnin B P, Chumakov P M. Human adenosine-deaminase gene contains p53-responsive element. Proc Russian Acad Sci. 1996;346:260–262. [PubMed] [Google Scholar]
- 22.Lee S, Elenbaas B, Levine A, Griffith J. p53 and its 14 kDa C-terminal domain recognize primary DNA-damage in the form of insertion/deletion mismatches. Cell. 1995;81:1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 23.Lee S M, Cavallo L, Griffith J. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J Biol Chem. 1997;272:7532–7539. doi: 10.1074/jbc.272.11.7532. [DOI] [PubMed] [Google Scholar]
- 24.Lee W, Harvey T S, Yin Y, Yau P, Litchfield D, Arrowsmith C H. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 25.Leveillard T, Andera L, Bissonnette N, Schaeffer L, Bracco L, Egly J M, Wasylyk B. Functional interactions between p53 and the TFIIH complex are affected by tumor-associated mutations. EMBO J. 1996;15:1615–1624. [PMC free article] [PubMed] [Google Scholar]
- 26.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 27.Linke S, Clarkin K, Dileonardo A, Tsou A, Wahl G. A reversible, p53-dependent G(0)/G(1) cell-cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 28.Lomax M E, Barnes D M, Hupp T R, Picksley S M, Camplejohn R S. Characterisation of p53 oligomerisation domain mutations isolated from Li-Fraumeni and Li-Fraumeni like family members. Oncogene. 1998;17:643–649. doi: 10.1038/sj.onc.1201974. [DOI] [PubMed] [Google Scholar]
- 29.McKay B C, Francis M A, Rainbow A J. Wildtype p53 is required for heat shock and ultraviolet light enhanced repair of a UV-damaged reporter gene. Carcinogenesis. 1997;18:245–249. doi: 10.1093/carcin/18.2.245. [DOI] [PubMed] [Google Scholar]
- 30.McLure K G, Lee P W K. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mee, T., and J. Milner. 1998. Unpublished data.
- 32.Miller M, Lubkowski J, Rao J K M, Danishefsky A T, Omichinski J G, Sakaguchi K, Sakamoto H, Appella E, Gronenborn A M, Clore G M. The oligomerization domain of p53: crystal structure of the trigonal form. FEBS Lett. 1996;99:166–170. doi: 10.1016/s0014-5793(96)01231-8. [DOI] [PubMed] [Google Scholar]
- 33.Milner J, Medcalf E A, Cook A. The tumor suppressor p53: analysis of wild-type and mutant complexes. Mol Cell Biol. 1991;11:12–19. doi: 10.1128/mcb.11.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molinari M, Okorokov A L, Milner J. Interaction with damaged DNA induces selective proteolytic cleavage of p53 to yield 40 kDa and 35 kDa fragments competent for sequence-specific DNA binding. Oncogene. 1996;13:2077–2086. [PubMed] [Google Scholar]
- 35.Nagaich A K, Zhurkin V B, Sakamoto H, Gorin A A, Clore G M, Gronenborn A M, Appella E, Harrington R E. Architectural accommodation in the complex of four p53 DNA binding domain peptides with the p21/waf1/cip1 DNA response element. J Biol Chem. 1997;272:14830–14841. doi: 10.1074/jbc.272.23.14830. [DOI] [PubMed] [Google Scholar]
- 36.Nelson W G, Kastan M B. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okorokov A L, Ponchel F, Milner J. Induced N- and C-terminal cleavage of p53: a core fragment of p53, generated by interaction with damaged DNA, promotes cleavage of the N-terminus of full-length p53, whereas ssDNA induces C-terminal cleavage of p53. EMBO J. 1997;16:6008–6017. doi: 10.1093/emboj/16.19.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavletich N P, Chambers K A, Pabo C O. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 40.Reed M, Woelker B, Wang P, Wang Y, Anderson M, Tegtmeyer P. The C-terminal domain of p53 recognizes DNA damaged by ionizing radiation. Proc Natl Acad Sci USA. 1995;92:9455–9459. doi: 10.1073/pnas.92.21.9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sancar A, Hearst J E. Molecular matchmakers. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 42.Sherley J L. Guanine nucleotide biosynthesis is regulated by the cellular p53 concentration. J Biol Chem. 1991;266:24815–24828. [PubMed] [Google Scholar]
- 43.Sprang S R. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 44.Temeles G L, Gibs J B, D’Alonzo J S, Sigal I S, Scolnick E M. Yeast and mammalian ras proteins have conserved biochemical properties. Nature. 1985;313:700–703. doi: 10.1038/313700a0. [DOI] [PubMed] [Google Scholar]
- 45.Venkatachalam S, Yu-Ping S, Jones S N, Vogel H, Bradley A, Pinkel D, Donehower L A. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker J E, Saraste M, Runswick M J, Gay N G. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Vermeulen W, Coursen J D, Gibson M, Lupold S E, Forrester K, Xu G W, Elmore L, Yeh H, Hoeijmakers J H J, Harris C C. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis pathway. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 48.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Friedberg E C, Evans M K, Taffe B G, Bohr V A, Weeda G, Hoeijmakers J H J, Forrester K, Harris C C. p53 modulation of TFIIH-associated nucleotide excision-repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 50.Waterman J, Shenk J, Halazonetis T. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA-binding. EMBO J. 1995;14:512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamauchi M, Baker T A. An ATP-ADP switch in MuB controls progression of the Mu transposition pathway. EMBO J. 1998;15:5509–5518. doi: 10.1093/emboj/17.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]