Abstract
Human enteroviruses (family Picornaviridae) are the major cause of aseptic meningitis and also cause a wide range of other acute illnesses, including neonatal sepsis-like disease, acute flaccid paralysis, and acute hemorrhagic conjunctivitis. The neutralization assay is usually used for enterovirus typing, but it is labor-intensive and time-consuming and standardized antisera are in limited supply. We have developed a molecular typing system based on reverse transcription-PCR and nucleotide sequencing of the 3′ half of the genomic region encoding VP1. The standard PCR primers amplify approximately 450 bp of VP1 for most known human enterovirus serotypes. The serotype of an “unknown” may be inferred by comparison of the partial VP1 sequence to those in a database containing VP1 sequences for the prototype strains of all 66 human enterovirus serotypes. Fifty-one clinical isolates of known serotypes from the years 1991 to 1998 were amplified and sequenced, and the antigenic and molecular typing results agreed for all isolates. With one exception, the nucleotide sequences of homologous strains were at least 75% identical to one another (>88% amino acid identity). Strains with homologous serotypes were easily discriminated from those with heterologous serotypes by using these criteria for identification. This method can greatly reduce the time required to type an enterovirus isolate and can be used to type isolates that are difficult or impossible to type with standard immunological reagents. The technique may also be useful for the rapid determination of whether viruses isolated during an outbreak are epidemiologically related.
Enteroviruses (EVs; family Picornaviridae) are responsible for 30,000 to 50,000 hospitalizations for aseptic meningitis per year in the United States. Other enteroviral diseases include mild illnesses such as common colds, hand-foot-and-mouth disease, and acute hemorrhagic conjunctivitis, as well as potentially life-threatening illnesses, including myocarditis, neonatal sepsis-like disease, and acute flaccid paralysis (18, 20). Sixty-six human EV serotypes have been identified antigenically by using an antibody neutralization test (3, 4, 18). The VP1 protein contains a number of important neutralization sites (for reviews, see references 15 and 19), but the specific epitopes responsible for serotype specificity have not been identified.
Although the neutralization test is generally reliable for EV typing, it is labor-intensive and time-consuming and may fail to identify the serotype of a clinical isolate because of antigenic drift, recombination, or the presence of virus mixtures in the specimen being tested. Several laboratories have used nucleotide sequencing of the 5′ nontranslated region (NTR) and the VP4-VP2 junction as diagnostic and epidemiologic tools, with some success (1, 6, 13), but the sequences in these regions do not always correlate with serotype (1, 14, 22). Since important neutralization sites reside in VP1, one would expect that the VP1 sequence or some portion thereof would correlate with serotype. We recently developed a database of complete VP1 sequences from all human EV serotypes and demonstrated that the VP1 sequence appears to correlate better with the serotype than does the sequence of either the 5′ NTR or the VP4-VP2 junction (22, 23). In the present study, we show that for clinical EV isolates of various serotypes, there is a 100% correlation between the nucleotide sequence of the 3′ half of VP1 and antigenic typing by the standard neutralization test.
MATERIALS AND METHODS
Viruses.
Fifty-one human EV isolates of 24 different serotypes were chosen from those processed in our laboratory during the period from 1991 to 1998 for routine nonpoliovirus EV (NPEV) reference testing. The viruses were from 19 different U.S. states and two other countries and were chosen to be representative of the serotypes in our collection for the period surveyed. To avoid the effects of sampling bias in the interpretation of sequence comparisons, no more than four isolates of any given serotype were chosen for sequencing. The isolates included examples of coxsackieviruses type A (CAs), coxsackieviruses type B (CBs), echoviruses, and numbered EVs.
Virus isolation and neutralization.
The virus strains were isolated from a wide range of clinical specimens, including blood (n = 1), cerebrospinal fluid (n = 7), conjunctival swab (n = 1), “lesion” (n = 1), postmortem lung (n = 1), nasopharyngeal swab (n = 2), sputum (n = 1), stool (n = 18), and throat swab (n = 8) specimens and nonspecified tissue specimens (n = 11). Forty-four of the 51 strains were isolated from original clinical material by the submitting laboratory, most of which were U.S. state public health laboratories. The remaining seven strains were isolated from original stool specimens at the Centers for Disease Control and Prevention. All isolates were typed antigenically at the Centers for Disease Control and Prevention with standard World Health Organization antiserum pools (17) supplemented with additional pooled and monospecific antisera such that all human EV serotypes, as well as antigenic variants of echovirus type 4 (E4), E6, E11, and E30, could be identified (6a).
RNA extraction and RT-PCR.
Viral RNA was extracted from infected cell culture supernatant by using the QIAamp Viral RNA kit (QIAGEN, Inc., Santa Clarita, Calif.). Reverse transcription-PCR (RT-PCR) was carried out as described previously (22). From each viral cDNA, an amplicon of approximately 450 bp encompassing the 3′ half of VP1 and the 5′ end of 2A was produced by PCR with the primer pairs 012 and 011 or 040 and 011 (see Table 1). Primer specificity was tested by PCR amplification of the prototype strain of each human EV serotype with both primer pairs (see Fig. 1). Amplification products were visualized by agarose gel electrophoresis and ethidium bromide staining. PCR products from clinical isolates were gel isolated and purified for sequencing with the QIAquick Gel Extraction kit (QIAGEN, Inc.) and sequenced on an automated DNA sequencer with fluorescent dideoxy-chain terminators (PE-Biosystems, Foster City, Calif.).
TABLE 1.
Oligonucleotide primers used to amplify and sequence unknown EVs
| Primer | Sequence (5′→3′) | Gene | Position |
|---|---|---|---|
| 012 | ATGTAYGTICCICCIGGIGG | VP1 | 2875–2894 (CB1 numbering)a |
| 040 | ATGTAYRTICCIMCIGGIGC | VP1 | 2905–2924 (CA16 numbering)a |
| 011 | GCICCIGAYTGITGICCRAA | 2A | 3311–3292 (CB1 numbering) |
Primers 012 and 040 recognize homologous sites, as described in the text.
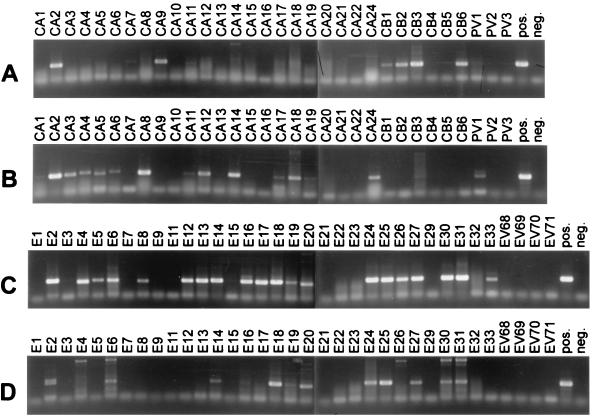
FIG. 1.
RT-PCR amplification of all prototype EV strains with primer pairs 012-011 and 040-011. PCR products were resolved by 1% agarose gel electrophoresis and were visualized by ethidium bromide staining and UV transillumination. (A) CAs, CBs, and PVs amplified with primer pair 012-011. (B) CAs, CBs, and PVs amplified with primer pair 040-011. (C) Echoviruses and numbered EVs amplified with primer pair 012-011. (D) Echoviruses and numbered EVs amplified with primer pair 040-011.
Sequence analysis.
The sequences were compared with the sequences in the EV VP1 sequence database (23) by sequential pairwise alignment of the query sequence with each sequence in the database by using the algorithm of Needleman and Wunsch (21) implemented in the program Gap (7). The results of the pairwise comparisons were compiled and sorted in descending order by percent identity with the query sequence. The frequency of pairwise identity scores for all comparisons (51 clinical isolates each compared with 64 prototype strains), rounded up to the nearest integer value, was plotted versus the identity score.
Nucleotide sequence accession numbers.
The sequences reported here were deposited in the GenBank sequence database under accession nos. AF081595 to AF081645.
RESULTS
Design of PCR primers.
Since the EV VP1 sequences appear to correlate with serotype (23), we targeted this region for development of sequence-based molecular diagnostics. We have previously shown that group- and serotype-specific PCR primers which target conserved amino acid epitopes within the VP1 gene of polioviruses (PVs) could be designed (11, 12). Using this approach, we designed degenerate deoxyinosine-containing PCR primers that specifically recognize regions within or near the VP1 gene of NPEVs. To choose NPEV PCR primers with the broadest specificity within the Enterovirus genus, we relied on the conservation of specific amino acid motifs within VP1 and immediately 3′ to VP1 in 2A. E22 and E23 were excluded, because it is likely that they will be reclassified as members of a new picornavirus genus, Parechovirus (16). The motif MYVPPG was present in the deduced VP1 amino acid sequences of 44 prototype EV strains (23). Thirteen prototypes had I substituted for V, and CA type 7 (CA7) contained A instead of V. CA12, CA14, and EV type 71 (EV71) contained the motif MFVPPG. In EV68 and EV70, a slightly different motif, MYVPTG, was present. For viruses in the CB-like phylogenetic group, the M(Y/F)(V/I)PPG motif is followed by G, whereas in all other EVs, the motif is followed by A (23). To account for differences between the virus groups and for codon degeneracy, two different inosine-containing primers were designed to anneal to this region (Table 1). Primer 012 is based on the amino acid sequence MYVPPGG. Primer 040 is based on the amino acid sequence MY(V/I)P(P/T)GA. The selectivities of these two primers are primarily due to the 3′ base of each primer (i.e., 012 has a G at the 3′ end and 040 has a C at the 3′ end) (Table 1). In addition, 040 contains increased degeneracy at positions 8 and 14 from the 3′ end of the primer in order to detect those viruses that have an isoleucine (position 8) or a threonine (position 14) at these positions. For PCR, 012 and 040 were each paired with primer 011, which corresponds to the amino acid sequence FG(Q/H)QSGA, which is present near the 5′ end of the 2A gene and which is conserved among most EVs for which the 2A sequence is available.
Specificities of PCR primers.
To assess the breadth of specificity and therefore the general applicability of the two primer pairs, both pairs were tested in RT-PCR assays with template RNA derived from each of the prototype human EV strains (Fig. 1). Primer pair 040-011 amplified 14 of 23 prototype CA strains (Fig. 1B), as well as PV type 1 (PV1), E2, E6, E14, E16, E18, E19, E20, E24, E25, E27, E30, and E31 (Fig. 1A). Primer pair 012-011 amplified 23 of 30 prototype echovirus strains (Fig. 1C), as well as CA2, CA7, CA9, CA11, CB type 1 (CB1), CB2, CB3, CB6, and PV1 (Fig. 1D). Twenty-two prototype strains were not amplified by either primer pair (CA10, CA13, CA15, CA16, CA20, CA21, CA22, CB4, CB5, E1, E7, E9, E21, E22, E23, E32, PV2, PV3, and EV68 to 71), despite the presence of RNA that was amplifiable with other primer pairs (data not shown). However, as shown in Table 2, recent isolates of CA16, CA21, CB5, E7, E9, E21, and EV71 were successfully amplified (see below).
TABLE 2.
Correspondence between typing by sequence comparison and by neutralization
| Straina | Neutralization type | Highest-scoring prototypeb
|
Second-highest-scoring prototype(s)b
|
|||||
|---|---|---|---|---|---|---|---|---|
| Type | % nt sequence identity | % aa sequence identity | Type | % nt sequence identity | Type | % aa sequence identity | ||
| WA91-0374 | E6 | E6 | 83.3 | 95.6 | E1 | 69.7 | E29 | 74.3 |
| OR91-1426 | E30 | E30 | 85.8 | 92.9 | E21 | 69.5 | E21 | 81.7 |
| CT92-1465 | E16 | E16 | 83.4 | 93.6 | E5 | 72.2 | E5 | 78.6 |
| FL92-1512 | CB2 | CB2 | 86.5 | 98.5 | CB4 | 68.3 | CB4 | 75.2 |
| WA92-1516 | E11′ | E11 | 77.1 | 90.1 | E19 | 72.9 | E19 | 83.0 |
| NC92-1612 | E9 | E9 | 77.8 | 94.6 | E17 | 70.2 | E16 | 72.9 |
| GA92-1616 | E11 | E11 | 77.6 | 89.4 | E19 | 72.2 | E19 | 82.3 |
| TX92-1647 | CA14 | CA14 | 86.8 | 97.1 | CA7 | 63.4 | CA7 | 67.9 |
| MD92-1649 | E25 | E25 | 77.1 | 91.5 | E1 | 68.5 | E21 | 77.6 |
| DOR93-1657 | CA24v | CA24 | 77.4 | 92.8 | CA20 | 67.9 | CA17 | 75.9 |
| FL93-1759 | E11′ | E11 | 78.5 | 90.1 | E19 | 72.6 | E19 | 83.0 |
| GA93-1763 | CA9 | CA9 | 83.8 | 95.3 | E4 | 68.6 | E4 | 70.8 |
| GA93-1765 | E7 | E7 | 79.7 | 95.7 | E32 | 68.8 | E32 | 77.1 |
| MO93-1808 | E25 | E25 | 77.6 | 91.5 | E33 | 67.5 | E21 | 76.9 |
| ME93-1814 | CB5 | CB5 | 95.2 | 98.5 | CB1 | 71.3 | CB1 | 77.7 |
| NM93-1816 | CB3 | CB3 | 80.3 | 97.7 | CB6 | 69.8 | CB1 | 81.5 |
| OR93-1817 | E25 | E25 | 77.8 | 91.5 | E1 | 68.5 | E21 | 76.9 |
| WA93-1821 | E4 | E4 | 81.1 | 96.1 | E1 | 73.1 | E1 | 80.9 |
| MN94-1828 | E25 | E25 | 76.9 | 92.2 | E29 | 67.9 | E21 | 77.6 |
| WA94-1849 | E3 | E3 | 79.6 | 93.0 | E7 | 68.2 | E12 | 80.0 |
| AR94-1884 | E30 | E30 | 86.0 | 93.6 | E21 | 70.0 | E21 | 82.4 |
| GA93-2460 | CB5 | CB5 | 95.8 | 98.5 | CB1 | 70.8 | CB1 | 77.7 |
| GA93-1892 | E30 | E30 | 85.5 | 93.6 | E21 | 69.5 | E21 | 83.4 |
| GA93-1894 | E7 | E7 | 79.7 | 95.7 | E32 | 69.1 | E32 | 77.1 |
| NM94-1919 | EV71 | EV71 | 80.6 | 93.4 | CA16 | 66.0 | CA16 | 76.6 |
| AZ94-1925 | CA14 | CA14 | 86.5 | 97.0 | CA7 | 63.8 | CA7 | 68.2 |
| RI94-1959 | E21 | E21 | 78.3 | 93.7 | E30 | 69.6 | E30 | 80.0 |
| CT94-2006 | EV71 | EV71 | 80.3 | 93.4 | CA16 | 66.0 | CA16 | 76.6 |
| MD95-2037 | EV71 | EV71 | 79.9 | 92.7 | CA16 | 67.0 | CA16 | 76.6 |
| AZ94-2060 | CA21 | CA21 | 90.9 | 98.6 | CA24 | 68.7 | CA24 | 75.5 |
| PA94-5753 | CA16 | CA16 | 77.9 | 94.2 | EV71 | 68.7 | EV71 | 78.1 |
| NM95-2070 | E6 | E6 | 76.8 | 94.1 | E29 | 68.1 | E29 | 75.5 |
| TX95-2089 | E13 | E13 | 72.4 | 88.7 | EV69 | 71.5 | EV69 | 83.0 |
| GA95-2093 | CA21 | CA21 | 91.4 | 98.6 | CA24 | 67.5 | CA24 | 75.5 |
| GA95-2095 | CA16 | CA16 | 77.9 | 94.9 | EV71 | 69.4 | EV71 | 77.4 |
| NC95-2135 | CB2 | CB2 | 83.2 | 99.2 | CB4 | 68.3 | CB4 | 76.2 |
| AR95-2139 | E9 | E9 | 75.7 | 92.8 | E17 | 70.0 | E1 | 71.8 |
| TX95-2147 | CA16 | CA16 | 76.5 | 94.9 | EV71 | 70.4 | EV71 | 77.4 |
| VA95-2154 | E11′ | E11 | 78.3 | 90.8 | E19 | 71.7 | E19 | 83.7 |
| WI95-2151 | E9 | E9 | 75.7 | 93.5 | E17 | 69.4 | E16 | 71.4 |
| VA95-2157 | E30 | E30 | 85.3 | 92.1 | E21 | 70.0 | E21 | 82.1 |
| GA96-2175 | CA9 | CA9 | 82.5 | 92.6 | E19 | 68.4 | E11 | 72.3 |
| CT96-2181 | E5 | E5 | 86.5 | 92.9 | E31 | 71.5 | E31 | 82.1 |
| CT96-2182 | E18 | E18 | 75.7 | 93.6 | E17 | 69.9 | E4 | 75.4 |
| TX96-2184 | CA21 | CA21 | 91.6 | 98.6 | CA24 | 68.2 | CA24 | 75.5 |
| TX97-2320 | E18 | E18 | 78.8 | 92.9 | E17 | 69.7 | E17 | 74.5 |
| NH97-2342 | CB3 | CB3 | 77.4 | 98.5 | CB5 | 67.9 | CB1 | 84.6 |
| PER98-2528 | E6 | E6 | 86.0 | 95.6 | CB1 | 71.6 | E29 | 74.3 |
| PER98-2533 | E7 | E7 | 80.4 | 95.7 | E32 | 68.1 | E12 | 78.6 |
| PER98-2537 | E11 | E11 | 78.5 | 94.3 | E19 | 71.9 | E19 | 82.3 |
| PER98-2558 | E33 | E33 | 79.3 | 96.9 | CB1 | 70.3 | E4 | 75.4 |
Strains are identified by the country (three-letter code [DOR, Dominican Republic; PER, Peru]) or U.S. state (two-letter code) of origin, year of isolation, and internal laboratory identifier.
nt, nucleotide; aa, amino acid.
PCR amplification of clinical isolates.
To determine the utility of using viral sequence analysis as an EV typing tool, we compared the method of typing by partial sequencing of VP1 with the conventional antigenic typing method using 52 clinical isolates typed in our laboratory from 1991 to 1998. Despite the failure of primer pair 012-011 to amplify the prototype E7, E9, E21, CB4, and CB5 strains, primer pair 012-011 successfully amplified recent clinical isolates of each of these serotypes (Table 2). Likewise, primer pair 040-011 amplified recent isolates of CA16, CA21, and EV71 but not the prototype strains of these serotypes (Table 2). These two primer pairs failed to amplify only one of the clinical isolates tested, a 1993 E6 isolate from Texas (isolate TX93-1673). The presence of amplifiable RNA in that specimen was confirmed by amplification of 5′-specific sequences with panenterovirus primers (data not shown). For the other 51 isolates, a VP1-specific fragment was amplified from purified RNA by RT-PCR with primer pair 012-011 or 040-011. In most cases, only one of the two primer pairs produced a product of the expected size (data not shown).
Molecular typing by sequence analysis.
Partial VP1 sequences of 51 recent clinical EV isolates, when compared with the VP1 sequences of all prototype human EV strains, completely correlated with the serotype determined by the conventional neutralization test (Table 2). The nucleotide sequences of recent clinical isolates were 72.4 to 95.2% identical to the sequences of their respective prototype strains and only 63.4 to 73.1% identical to the sequences of the highest-scoring heterologous prototype strains. The predicted partial VP1 amino acid sequences of the clinical isolates were 88.7 to 98.5% identical to that of the homologous prototype strain and 67.7 to 84.6% identical to that of the nearest heterologous prototype strain. With one exception, the difference between percent nucleotide sequence identity to the homologous prototype strain and percent identity to the highest-scoring heterologous prototype strain was at least 4.2%; this difference for TX95-2089, which was typed antigenically as E13, was only 0.9% (it was 72.4% identical to E13, whereas it was 71.5% identical to EV69), suggesting either that TX95-2089 has diverged significantly from the E13 prototype, that it may be a distinct new type which is closely related to both E13 and EV69, or that the prototype E13 strain (Del Carmen) is not representative of the serotype as a whole. The complete VP1 sequence of TX95-2089 was 72.6% identical to that of the prototype E13 strain, 70.1% identical to that of the prototype EV69 strain (second highest score), and 64.7% identical to that of the prototype E12 strain (third highest score) (data not shown). The predicted complete VP1 amino acid sequence of TX95-2089 was 88.2% identical to that of E13, 80.8% identical to that of EV69 (second highest score), and 70.0% identical to that of CB1 (third highest score), suggesting that TX95-2089 is probably a strain of E13 which has diverged in nucleotide sequence by accumulating mutations in the third codon position. TX95-2089 was neutralized by monospecific anti-E13 antisera but not by monospecific anti-EV69 antisera (data not shown).
Distribution of pairwise identity scores.
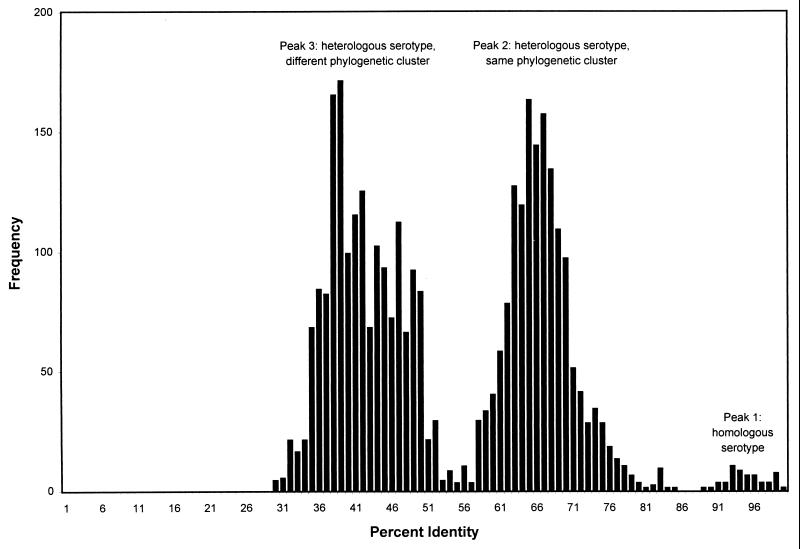
The pairwise identity scores, rounded up to the nearest integer value, were plotted as a histogram of score frequency to determine whether the serotype could be unambiguously assigned strictly on the basis of nucleotide or deduced amino acid sequence identity scores (Fig. 2). As shown previously for prototype strains (23), VP1 nucleotide sequence comparison scores were distributed in three major peaks: peak 1 consisted of scores for comparison of viruses of the same (homologous) serotype, peak 2 consisted of scores for comparison of viruses within the same major phylogenetic cluster, and peak 3 consisted of scores for comparison of viruses in different phylogenetic clusters. As described above, there was overlap between nucleotide sequence comparison peaks 1 (homologous serotype) and 2 (heterologous serotype, same cluster). Two heterologous-serotype pairwise comparison scores, WA92-1516 (E11) versus E19-Burke (72.9%) and WA93-1821 (E4) versus E1-Farouk (73.1%), were higher than the lowest homologous-serotype pairwise comparison score, TX95-2089 (E13) versus Del Carmen (E13) (72.4%) (Table 2). However, for a given clinical isolate, the homologous-serotype pairwise comparison score was always higher than the highest heterologous-serotype pairwise comparison score. The peak for homologous-serotype pairwise comparisons of deduced amino acid sequences was fully resolved from the peak for heterologous-serotype pairwise comparisons (Fig. 2). The minimum homologous-serotype amino acid identity score for homologous-serotype pairwise comparisons was 88.7% (TX95-2089), and the maximum score for heterologous-serotype pairwise comparisons was 84.6% (NH97-2342 [CB3] versus CB1).
FIG. 2.
Distribution of pairwise amino acid identity scores. Peak 1 corresponds to comparisons of homologous strains (same serotype), peak 2 corresponds to comparisons of heterologous strains (different serotype) of the same major phylogenetic cluster, and peak 3 corresponds to comparisons of heterologous strains of different major phylogenetic clusters.
DISCUSSION
Molecular methods allow the rapid and specific detection of human EVs, mainly by targeting the 5′ NTR (for a review, see reference 26). However, variability of the 5′ NTR sequence within a serotype (2, 5, 6, 14) has prevented the use of this region for identification of clinical isolates to the serotype level. Phylogenetic studies targeting the VP4-VP2 junction suggested that this region may be more suitable than the 5′ NTR for development of serotype-specific diagnostics (9, 24, 25), but this region also appears to correlate only partially with serotype (1, 22). The sequence at the VP1-2A junction has been used extensively to study PV transmission patterns, and PV VP1 sequences always cluster with sequences of isolates of the homologous serotype (10). EV serotypes were defined on the basis of neutralization, and VP1 contains a number of important neutralization epitopes (15, 19). Complete sequencing of the VP1 gene of all prototype human EV strains has also suggested that the VP1 sequence correlates well with serotype (23). Therefore, we have targeted VP1 for development of NPEV serotype-specific molecular diagnostics. Two generic PCR primer pairs, 012-011 and 040-011, were sufficient for amplification and sequencing of the 3′ half of VP1 from 44 of 66 prototype human EV strains (Fig. 1). Recent isolates of 7 of the remaining 22 serotypes were also amplified with primer pair 012-011 or 040-011. The ability of the primers to amplify some but not all isolates within a serotype probably reflects a high degree of intratypic genetic diversity. This hypothesis is supported by the sequence comparison scores presented in Table 2 and suggests that more extensive sequencing studies are required to define the limits of intratypic diversity and provide a basis for the development of improved primers.
These two primer pairs were used to amplify 51 EV strains isolated from clinical material between 1991 and 1998. Simple pairwise comparison of the sequence of the unknown to a database of human EV VP1 sequences showed that the partial VP1 sequence fully correlated with the serotype determined by the conventional neutralization test. The results of the nucleotide sequence comparisons reflect the high degree of genetic diversity among EVs and illuminate the challenge that such diversity represents in the systematic design of nucleic acid-based diagnostic reagents. Degenerate inosine-containing PCR primers were developed to overcome such nucleotide sequence diversity by specifically targeting regions of conserved amino acid sequences (11, 12). When the conserved sites flanked regions with high degrees of amino acid sequence diversity between serotypes, pairwise comparison of deduced amino acid sequences provided more resolution between homologous-serotype scores and those for heterologous serotypes than did nucleotide sequence comparisons, as previously shown for prototype EV strains (23). A similar method has recently been applied to the classification of plant viruses in the family Potyviridae (28).
The technique of viral protein fingerprinting has recently been used for the typing and characterization of clinical EV isolates (8). The method was specific and 97% accurate, but the radiolabeled proteins generate radioactive waste and the method requires the use of specialized instrumentation for data acquisition and analysis. In addition, the database of protein patterns available for comparison contains representatives of fewer than one-third of the 66 known human EV serotypes. PCR is well established in most research and many clinical laboratory settings; nucleotide sequencing can be performed manually without radiolabeled compounds at low cost and with minimal initial investment, and automated sequencing instruments are also widely available. Sequencing may also be outsourced at reasonable cost to one of the many commercial sequencing laboratories. The computer analysis of sequences may be performed with any of the free or inexpensive sequence alignment programs that are available for several popular computing platforms. Finally, the complete VP1 sequences of the prototype strains of all 66 known human EV serotypes are freely available through GenBank, and the sequences of additional strains of many serotypes are also available. Another advantage of typing by PCR and sequencing is that the virus need not be cultivable in cell culture if the VP1 fragment can be directly amplified from the original clinical material. While we have not attempted direct amplification in the studies described here, others have successfully amplified EV-specific fragments from cerebrospinal fluid and other original clinical material (27); therefore, it should be possible to adapt their methods to our VP1 RT-PCR.
The sequence of the 3′ half of VP1 has proved to be an excellent genetic correlate for the EV serotype. In our own laboratory, we have adopted sequencing as the primary typing method, with confirmation by neutralization with monospecific antisera. Application of PCR and partial VP1 sequencing to the routine laboratory diagnosis of EV infections and serotyping of virus isolates will greatly reduce the time needed to type EV isolates (2 to 3 days versus 1 to 2 weeks). Rapid serotyping may also provide the clinician with greater diagnostic information within a clinically relevant time frame, enabling, for example, the use of type-specific immune globulin in the immunodeficient patient or the use of an antiviral agent which may exhibit differential efficacy for different EV serotypes. The VP1 sequence will also be useful in EV taxonomy, in the identification of new EV types, and in molecular epidemiologic studies of enteroviral disease outbreaks.
REFERENCES
- 1.Arola A, Santti J, Ruuskanen O, Halonen P, Hyypiä T. Identification of enteroviruses in clinical specimens by competitive PCR followed by genetic typing using sequence analysis. J Clin Microbiol. 1996;34:313–318. doi: 10.1128/jcm.34.2.313-318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailly J-L, Borman A M, Peigue-Lafeuille H, Kean K M. Natural isolates of ECHO virus type 25 with extensive variations in IRES sequences and different translational efficiencies. Virology. 1996;215:83–96. doi: 10.1006/viro.1996.0009. [DOI] [PubMed] [Google Scholar]
- 3.Committee on Enteroviruses. The enteroviruses. Am J Pub Health. 1957;47:1556–1566. [PMC free article] [PubMed] [Google Scholar]
- 4.Committee on Enteroviruses. Classification of human enteroviruses. Virology. 1962;16:501–504. [Google Scholar]
- 5.Diedrich S, Driesel G, Schreier E. Sequence comparison of echovirus type 30 isolates to other enteroviruses in the 5′ noncoding region. J Med Virol. 1995;46:148–152. doi: 10.1002/jmv.1890460212. [DOI] [PubMed] [Google Scholar]
- 6.Drebot M A, Nguan C Y, Campbell J J, Lee S H S, Forward K R. Molecular epidemiology of enterovirus outbreaks in Canada during 1991–1992: identification of echovirus 30 and coxsackievirus B1 strains by amplicon sequencing. J Med Virol. 1994;44:340–347. doi: 10.1002/jmv.1890440406. [DOI] [PubMed] [Google Scholar]
- 6a.Feorino, P. Personal communication.
- 7.Genetics Computer Group. Wisconsin package, version 9.1. Madison, Wis: Genetics Computer Group; 1997. [Google Scholar]
- 8.Holland D T, Senne J, Peter C R, Urmeneta C, Connor J D. Differentiation and characterization of enteroviruses by computer-assisted viral protein fingerprinting. J Clin Microbiol. 1998;36:1588–1594. doi: 10.1128/jcm.36.6.1588-1594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huttunen P, Santii J, Pulli T, Hyypiä T. The major echovirus group is genetically coherent and related to coxsackie B viruses. J Gen Virol. 1996;77:715–725. doi: 10.1099/0022-1317-77-4-715. [DOI] [PubMed] [Google Scholar]
- 10.Kew O M, Mulders M N, Lipskaya G Yu, da Silva E E, Pallansch M A. Molecular epidemiology of polioviruses. Semin Virol. 1995;6:401–414. [Google Scholar]
- 11.Kilpatrick D R, Nottay B, Yang C-F, Yang S-J, Mulders M N, Holloway B P, Pallansch M, Kew O M. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol. 1996;34:2990–2996. doi: 10.1128/jcm.34.12.2990-2996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilpatrick D R, Nottay B, Yang C-F, Yang S-J, da Silva E, Peñaranda S, Pallansch M, Kew O M. Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol. 1998;36:352–357. doi: 10.1128/jcm.36.2.352-357.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim G R, Lee J S, Jung Y T, Chung Y J, Rhyu M G. Nucleotide sequencing of a part of the 5′-noncoding region of echovirus type 9 and rapid detection during the acute phase of aseptic meningitis. Arch Virol. 1997;142:853–860. doi: 10.1007/s007050050124. [DOI] [PubMed] [Google Scholar]
- 14.Kopecka H, Brown B, Pallansch M. Genotypic variation in coxsackie B5 isolates from three different outbreaks in the United States. Virus Res. 1995;38:125–136. doi: 10.1016/0168-1702(95)00055-u. [DOI] [PubMed] [Google Scholar]
- 15.Mateu M G. Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res. 1995;38:1–24. doi: 10.1016/0168-1702(95)00048-u. [DOI] [PubMed] [Google Scholar]
- 16.Mayo M A, Pringle C R. Virus taxonomy—1997. J Gen Virol. 1998;79:649–657. doi: 10.1099/0022-1317-79-4-649. [DOI] [PubMed] [Google Scholar]
- 17.Melnick J L, Rennick V, Hampil B, Schmidt N J, Ho H H. Lyophilized combination pools of enterovirus equine antisera: preparation and test procedures for the identification of field strains of 42 enteroviruses. Bull W H O. 1973;48:263–268. [PMC free article] [PubMed] [Google Scholar]
- 18.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Fields B N, Knipe D M, Howley P M, Channock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 655–712. [Google Scholar]
- 19.Minor P D. Antigenic structure of picornaviruses. Curr Top Microbiol Immunol. 1990;161:121–154. doi: 10.1007/978-3-642-75602-3_5. [DOI] [PubMed] [Google Scholar]
- 20.Morens D M, Pallansch M A. Epidemiology. In: Rotbart H A, editor. Human enterovirus infections. Washington, D.C: ASM Press; 1995. pp. 3–23. [Google Scholar]
- 21.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequences of two proteins. J Mol Biol. 1970;48:444–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 22.Oberste M S, Maher K, Pallansch M A. Molecular phylogeny of all human enterovirus serotypes based on comparison of sequences at the 5′ end of the region encoding VP2. Virus Res. 1998;58:35–43. doi: 10.1016/s0168-1702(98)00101-4. [DOI] [PubMed] [Google Scholar]
- 23.Oberste M S, Maher K, Kilpatrick D R, Pallansch M A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pöyry T, Kinnunen L, Hyypiä T, Brown B, Horsnell C, Hovi T, Stanway G. Genetic and phylogenetic clustering of enteroviruses. J Gen Virol. 1996;77:1699–1717. doi: 10.1099/0022-1317-77-8-1699. [DOI] [PubMed] [Google Scholar]
- 25.Pulli T, Koskimies P, Hyypiä T. Molecular comparison of coxsackie A virus serotypes. Virology. 1995;211:30–38. doi: 10.1006/viro.1995.1450. [DOI] [PubMed] [Google Scholar]
- 26.Rotbart H A, Romero J R. Laboratory diagnosis of enteroviral infections. In: Rotbart H A, editor. Human enterovirus infections. Washington, D.C: ASM Press; 1995. pp. 401–418. [Google Scholar]
- 27.Rotbart H A, Ahmed A, Hickey S, Dagan R, McCracken G H, Jr, Whitley R J, Modlin J F, Cascino M, O’Connell J F, Menegus M A, Blum D. Diagnosis of enterovirus infection by polymerase chain reaction of multiple specimen types. Pediatr Infect Dis J. 1997;16:409–411. doi: 10.1097/00006454-199704000-00014. [DOI] [PubMed] [Google Scholar]
- 28.Van Regenmortel M H V, Bishop D H L, Fauquet C M, Mayo M A, Maniloff J, Calisher C H. Guidelines to the demarcation of virus species. Arch Virol. 1997;142:1505–1518. [PubMed] [Google Scholar]