ABSTRACT
Cedar virus (CedV) is a nonpathogenic member of the Henipavirus (HNV) genus of emerging viruses, which includes the deadly Nipah (NiV) and Hendra (HeV) viruses. CedV forms syncytia, a hallmark of henipaviral and paramyxoviral infections and pathogenicity. However, the intrinsic fusogenic capacity of CedV relative to NiV or HeV remains unquantified. HNV entry is mediated by concerted interactions between the attachment (G) and fusion (F) glycoproteins. Upon receptor binding by the HNV G head domain, a fusion-activating G stalk region is exposed and triggers F to undergo a conformational cascade that leads to viral entry or cell-cell fusion. Here, we demonstrate quantitatively that CedV is inherently significantly less fusogenic than NiV at equivalent G and F cell surface expression levels. We then generated and tested six headless CedV G mutants of distinct C-terminal stalk lengths, surprisingly revealing highly hyperfusogenic cell-cell fusion phenotypes 3- to 4-fold greater than wild-type CedV levels. Additionally, similarly to NiV, a headless HeV G mutant yielded a less pronounced hyperfusogenic phenotype compared to wild-type HeV. Further, coimmunoprecipitation and cell-cell fusion assays revealed heterotypic NiV/CedV functional G/F bidentate interactions, as well as evidence of HNV G head domain involvement beyond receptor binding or G stalk exposure. All evidence points to the G head/stalk junction being key to modulating HNV fusogenicity, supporting the notion that head domains play several distinct and central roles in modulating stalk domain fusion promotion. Further, this study exemplifies how CedV may help elucidate important mechanistic underpinnings of HNV entry and pathogenicity.
IMPORTANCE The Henipavirus genus in the Paramyxoviridae family includes the zoonotic Nipah (NiV) and Hendra (HeV) viruses. NiV and HeV infections often cause fatal encephalitis and pneumonia, but no vaccines or therapeutics are currently approved for human use. Upon viral entry, Henipavirus infections yield the formation of multinucleated cells (syncytia). Viral entry and cell-cell fusion are mediated by the attachment (G) and fusion (F) glycoproteins. Cedar virus (CedV), a nonpathogenic henipavirus, may be a useful tool to gain knowledge on henipaviral pathogenicity. Here, using homotypic and heterotypic full-length and headless CedV, NiV, and HeV G/F combinations, we discovered that CedV G/F are significantly less fusogenic than NiV or HeV G/F, and that the G head/stalk junction is key to modulating cell-cell fusion, refining the mechanism of henipaviral membrane fusion events. Our study exemplifies how CedV may be a useful tool to elucidate broader mechanistic understanding for the important henipaviruses.
KEYWORDS: attachment protein, receptor binding protein, head, stalk, syncytia, henipavirus, paramyxovirus, Hendra, Nipah, Cedar virus
INTRODUCTION
Henipavirus (HNV) is a genus of emerging viruses in the Paramyxoviridae family that includes the lethal Nipah (NiV) and Hendra (HeV) viruses. NiV infections cause severe encephalitis and pneumonia (1, 2), with an ∼61% average mortality rate in documented human outbreaks (3). NiV and HeV exhibit an unusually broad mammalian host range (4), having caused multiple zoonotic transmission events (3, 5). Currently, no vaccines or therapies are approved for human use and both viruses are designated as biosafety level 4 (BSL-4) select agents with potential to cause severe pandemic threats to human and animal health (6). Meanwhile, Cedar virus (CedV) is nonpathogenic in susceptible model organisms (7, 8) and is thus considered a model virus to further understand HNV pathogenesis (7–10). The nonpathogenic phenotype is primarily attributed to CedV’s inability to suppress host antiviral responses (7, 8) and to its lack of ephrin B3 recognition as an entry receptor (9, 10). However, the fusogenic properties of CedV glycoproteins remain unknown and may also contribute to the low pathogenicity.
HNV entry into cells and cell-cell fusion are orchestrated by concerted interactions between the viral attachment (G) and fusion (F) surface glycoproteins (11, 12). First, the receptor-binding head domain of G typically binds cell surface ephrin entry receptors. The broad host range of HNVs is attributed to ephrin recognition, as ephrins are highly conserved and ubiquitous across mammalian genera (13). For instance, NiV G and HeV G bind ephrins B2 and B3 (14–16), while CedV G binds ephrins A1 (murine, not human), A2, A5, B1, and B2 (9, 10). HNV G receptor binding then causes G to undergo a series of conformational changes that exposes its stalk domain, which triggers the metastable F to induce membrane fusion and thus viral entry or cell-cell fusion (syncytia formation) (17, 18). G and F immunogenicities, as well as their importance in mediating HNV entry and cell-cell fusion, make them major vaccine targets. For instance, a HeV vaccine for horses (Equivac) utilizes a soluble form of HeV G to elicit neutralizing protective antibodies (19).
The relative levels of syncytia formation across HNVs remain understudied. We recently reported that NiV and HeV induce similar cell-cell fusion and viral entry levels (20), likely attributed to their G/F high sequence similarities. However, NiV and CedV G/F have significantly lower amino acid sequence identities (Fig. 1A), which may cause different fusogenic capacities, which we explored in this study. We also investigated how CedV G modulates cell-cell fusion in comparison to other HNVs. We took advantage of a short G stalk C-terminal region that displays high conservation (71% identity) with NiV G and HeV G (Fig. 1B). This sequence includes 3 characteristic cysteine residues with critical roles in the oligomerization and stability of NiV G (21). Notably, several studies have shown that the stalk of paramyxoviral receptor binding proteins (RBPs: HN, H, or G) functionally link receptor binding to fusion activation. This is the case for NiV G (17, 18), measles virus (MeV H) (22), parainfluenza virus 5 (PIV5 HN) (23), Newcastle disease virus (NDV HN) (24), and mumps virus (MuV HN) (24). Headless HN/H/G constructs with exposed stalks spontaneously trigger F and induce robust fusion while bypassing receptor binding. For NiV, evidence points to a G stalk C-terminal region triggering NiV F, while the G head domain restricts fusion by blocking the stalk C terminus exposure, thus preventing premature F triggering (17, 18). Although NiV, MeV, PIV5, NDV, and MuV are representative members of their respective viral genera, the relative capacities of headless G proteins in fusion activation may vary not only among but also within genera. For instance, while headless MeV H yielded significant levels of fusion, headless canine distemper virus H (CDV H) yielded no fusion when coexpressed with wild-type CDV F, and fusion was only rescued when coexpressing a hyperfusogenic CDV F mutant, even though MeV and CDV are both morbilliviruses (25). Other than for NiV, the levels of fusogenicity of headless HNV Gs, as well as the mechanistic underpinnings of such findings, remain unknown.
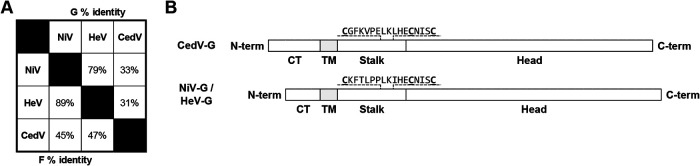
FIG 1.
Summary of amino acid sequence alignments between NiV and CedV receptor binding glycoproteins. (A) % aa identity between NiV, HeV, and CedV glycoproteins derived using BLAST. (B) Schematic representation of wild-type CedV G and NiV G/HeV G drawn to relative scale. The conserved stalk C-terminal residues have 71% identity (12/17) and are indicated above with the three distinct cysteines underlined and bolded. CT, cytoplasmic tail; TM, transmembrane domain; Stalk, stalk domain; Head, receptor-binding head domain.
Our study probed the fusogenic capacities of CedV G by testing full-length as well as several headless CedV G constructs. We revealed that the CedV G head “blocks” fusion more efficiently than the NiV G head. Further, addition of a linker-HA tag onto headless CedV G mutants enabled us to measure the effect head/stalk interfaces play in the fusogenic capacities of HNVs. This, in addition to uncovered heterotypic HNV functional bidentate interactions, provide evidence that the HNV head/stalk interface modulates membrane fusion steps apart from receptor binding and F triggering. Further, our study exemplifies the potential of utilizing CedV as a tool to elucidate HNV entry and pathogenesis mechanisms.
RESULTS
Cedar virus is significantly less fusogenic than Nipah virus.
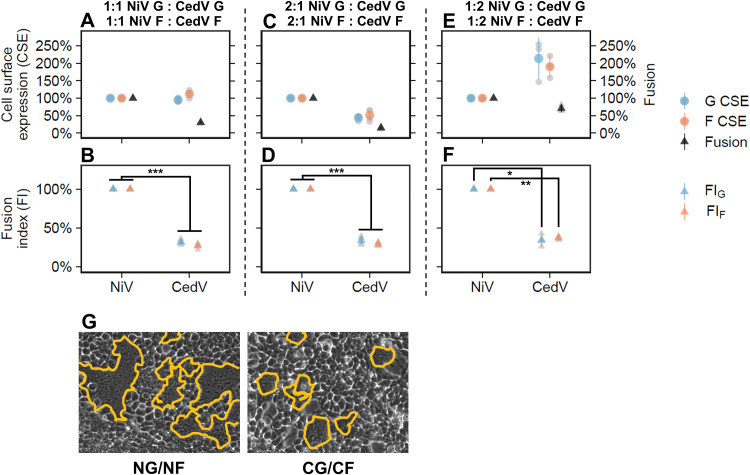
To accurately quantify and compare fusogenicity (cell-cell fusion) and cell surface expression (CSE) levels between NiV and CedV, we introduced extracellular hemagglutinin (HA) and FLAG tags into codon-optimized CedV G and F, respectively, to match the functional NiV G and F constructs (17, 18, 20). Then, since cell-cell fusion levels depend on CSE levels (26), we optimized transfections of NiV G/NiV F and CedV G/CedV F plasmid pairs into HEK 293T cells to obtain comparable CSE levels. We then quantified the number of nuclei inside syncytia per area 20 h posttransfection (hpt), as previously performed (17, 18, 20, 27). As the CSE levels of HNV glycoproteins affect their fusogenicity levels (20), to accurately compare NiV and CedV intrinsic fusogenic G/F capacities, we calculated fusion indices (FI), defined as the ratio of cell-cell fusion levels to G or F CSE levels. At ∼1:1 NiV G:CedV G and NiV F:CedV F CSE levels (Fig. 2A), the FIs of CedV were significantly lower than those of NiV by ∼3- to 4-fold relative to those of NiV set at 100% (FIG = 32.0% ± 3.3%, P < 0.001; FIF = 27% ± 3.4%, P < 0.001) (Fig. 2B). To assess the robustness of this observation, we further adjusted the amounts of transfected plasmids to obtain ∼2:1 NiV:CedV CSE levels (Fig. 2C) and ∼1:2 NiV:CedV CSE levels (Fig. 2E) and observed that CedV’s FIs remained consistently ∼3- to 4-fold lower than those of NiV (Fig. 2D: FIG = 33.9% ± 5.3%, P < 0.001; FIF = 29% ± 2.6, P < 0.001; Fig. 2F: FIG = 34.4% ± 8.3%, P < 0.05, FIF = 37.2% ± 2.5%, P < 0.01), confirming that CedV is inherently less fusogenic than NiV. Examples of syncytia differences between NiV and CedV at ∼1:1 glycoprotein CSE are shown in Fig. 2G.
FIG 2.
CedV is significantly less fusogenic than NiV. The left panels (A, B, and G) show data for a 1:1 NiV:CedV CSE ratio, the middle panels (C and D) show data for a 2:1 NiV:CedV CSE ratio, and the right panels (E and F) show data for a 1:2 NiV:CedV CSE ratio. (A) Relative levels of G CSE, F CSE, and fusion for NG/NF and CG/CF pairs in HEK 293T cells (18 to 22 hpt) normalized to the NG/NF pair set at 100% (mean ± standard deviation [SD], n = 4). HEK 293T cells were transfected with varying plasmid quantities (see Materials and Methods) to obtain 1:1 CSE levels for NG:CG and NF:CF. (B) Relative fusion indices (FI) of NG/NF and CG/CF normalized to the NG/NF pair set at 100% (mean ± SD, n = 4). To determine fusion indices (defined as fusion over CSE), we first normalized our fusion and CSE data to those obtained for NG/NF, then divided the values of normalized fusion over normalized CSE (fusion/G CSE, fusion/F CSE). Panels C/D and E/F are repeats of A/B except with 2:1 or 1:2 CSE levels for NG:CG and NF:CF obtained by adjusting the quantities of transfected plasmids (see Materials and Methods). Gray points in the plots represent individual data points. (G) Representative fusion images (outlined in yellow) for NG/NF and CG/CF in HEK 293T cells (21 hpt) at 1:1 CSE.
Headless CedV G mutants induce atypically hyperfusogenic phenotypes.
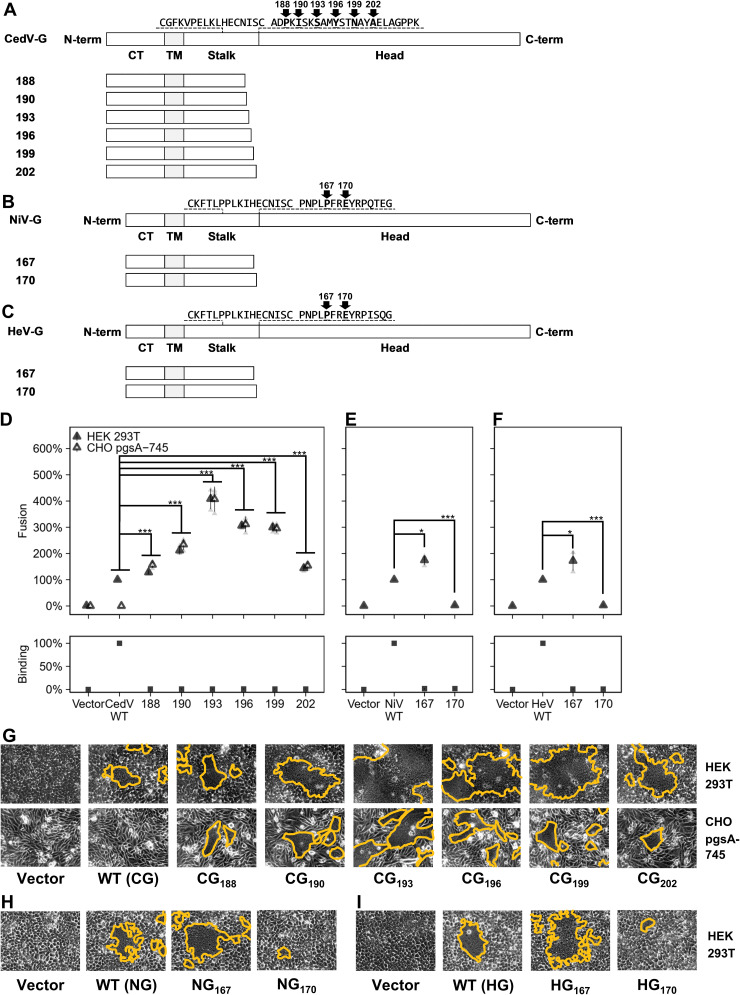
Our previous studies tested eight headless NiV G mutants (17, 18), but only one mutant, NG167, was fusogenic (fusion-promoting). Thus, we sought to determine if a headless CedV G mutant could induce cell-cell fusion. Since the stalk/head transition for CedV G is likely located between residues 186 to 209 (10), we constructed six head-truncated mutants in this region, named after their most C-terminal residue: CG188, CG190, CG193, CG196, CG199, and CG202 (Fig. 3A).
FIG 3.
A broad region of CedV G stalk induces robust cell-cell fusion. (A to C) Schematic representations of wild-type and headless CedV G, NiV G, and HeV G, respectively. Headless mutants were generated by mutating two downstream amino acids into stop codons and named after their C-terminal residues, as indicated. (D to F) Relative levels of fusion and ephrin B2 binding for wild-type or headless CedV G, NiV G, and HeV G, respectively, all normalized to their wild-type counterparts set at 100% (mean ± SD, n = 3). CedV G fusion was counted 18 to 22 hpt in HEK 293T cells (D, E, and F) and 22 to 26 hpt in CHO pgsA-745 cells (D). Fusion levels of CHO pgsA-745 cells were normalized based on the ratio of fusion for CG193 over wild-type CedV G in HEK 293T cells, to illustrate the fusogenicity of headless CedV G mutants. NiV G and HeV G fusion levels were measured at 12 to 14 hpt in HEK 293T cells. Different time points for CedV, NiV, and HeV fusion assays were chosen for optimal visualization of their respective fusion levels. (G to I) Representative fusion images (outlined in yellow) for CedV (G: 19 hpt for HEK 293T cells, 24 hpt for CHO pgsA-745 cells), or NiV and HeV (H and I: 13 hpt for HEK293T cells). Gray points in the plots represent individual data points.
Surprisingly, when normalized to wild-type CedV G, all six headless CedV G mutants induced substantial cell-cell fusion when coexpressed with wild-type CedV F (Fig. 3D and G). Further, while the CG193 fusion level was ∼4× higher than that of wild-type CedV G (in HEK 293T cells 408% ± 39.0%, P < 0.001; in CHO pgsA745 cells 407% ± 47.8%, P < 0.001) (Fig. 3D), the NG167 fusion level was only ∼1.7× higher than that of wild-type NiV G (in permissive HEK 293T cells 174% ±18.6%, P < 0.05) (Fig. 3E). As CG193 was the most hyperfusogenic mutant, we further generated one more NiV G mutant, NG170, to align with CG193 (Fig. 3B), in anticipation of potentially higher fusogenicity levels than that of NG167. However, coexpressing NG170 with NiV F produced minimal fusion (2.6% ± 0.5%, P < 0.001) (Fig. 3E). Since it is unknown whether headless HeV G is as fusogenic as headless NiV G, we also generated two headless HeV G mutants, HG167 and HG170, to correspond to NG167 and NG170 or CG190 and CG193, respectively (Fig. 3C). Coexpressing HG167 and HG170 with HeV F produced ∼1.5× hyperfusogenic (172% ± 37.7%, P < 0.05) and fusion-defective phenotypes (2.4% ± 0.8%, P < 0.001), respectively, compared to wild-type HeV G (Fig. 3F and I). These relative fusion levels are relatively more similar to those of NiV than CedV. Examples of syncytia elicited by wild-type and headless CedV G, NiV G, and HeV G mutants are shown in Fig. 3G to I. Note that we assayed cells transfected with CedV G/F at later time points (18 to 22 hpt) than those transfected with NiV or HeV G/F (12 to 14 hpt). This was because cells expressing CedV G/F took longer for observable fusion to form than cells expressing NiV G/F or HeV G/F; by the time CedV G/F cells had quantifiable amounts of fusion, NiV G/F and HeV G/F had basically fully fused cells to unquantifiable microscopic levels.
Since the receptor binding pockets of NiV G, HeV G, and CedV G are located on their head domains (9, 10, 28, 29), we confirmed the lack of binding of our headless mutants to soluble ephrin B2 via flow cytometry. As expected, only wild-type versions of G bound to ephrin B2 (Fig. 3D to F), which confirmed that headless NiV G, HeV G, and CedV G induced cell-cell fusion while bypassing receptor binding. Based on the proposed HNV stalk-exposure mechanism of fusion activation (17) in full-length G, receptor binding is required for G stalk exposure to initiate the fusion triggering cascade. To assess if CedV G similarly employs the stalk-exposure mechanism, we transfected wild-type or headless CedV G with CedV F into the ephrin ligand-negative cell line CHO pgsA-745 (9, 10). While headless CedV G mutants were also fusogenic in the receptor-negative CHO cells, wild-type CedV G was not (Fig. 3D and G), congruent with its receptor dependence (14, 15) and the stalk-exposure mechanism for CedV G.
Altogether, these results show that while the fusogenic capacities of the NiV G and HeV G stalks appear to be similar, the fusogenic capacity of CedV G stalk differs in two main aspects: (i) multiple different lengths of the CedV G stalk can induce robust levels of cell-cell fusion, and (ii) headless CedV G mutants are able to release unusually high levels of fusion relative to wild-type CedV G. Having multiple RBP stalk lengths capable of inducing hyperfusogenic phenotypes is unique among the paramyxoviruses thus far (12, 17, 18, 22–25). Why a broad region of the CedV G stalk is able to induce fusion remains to be determined. Nonetheless, wild-type CedV G similarly requires receptor binding to initiate the fusion cascade, which underlines a conserved induced stalk-exposure mechanistic model for viral entry across HNVs and paramyxoviruses.
Head domains of HNV Gs modulate fusogenicity to distinct extents.
Although CedV is significantly less fusogenic than NiV (Fig. 2), headless CedV G induced a higher fold increase in fusogenicity than headless NiV G when compared to their respective full-length counterparts (Fig. 3D and E). However, comparing fusogenicity is only quantitative when comparing the relative CSE levels of G and F, due to the dependence of cell-cell fusion on CSE levels. We chose CG193 as a representative headless CedV G mutant to proceed with our analysis because this was the most hyperfusogenic mutant (Fig. 3).
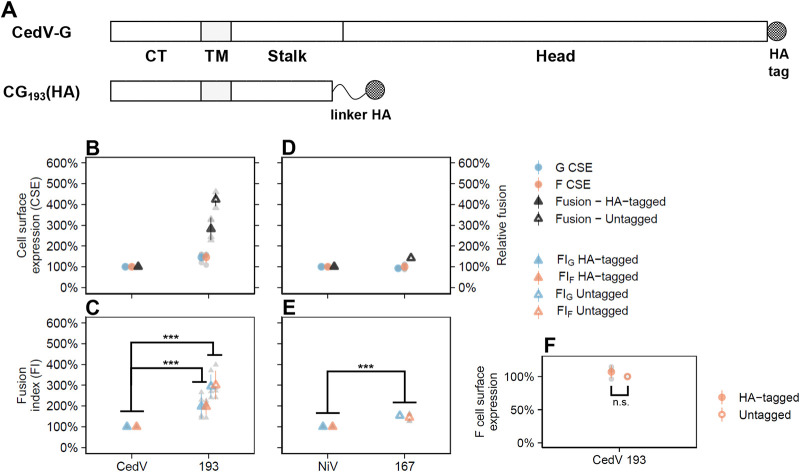
Since the wild-type G proteins have a C-terminal HA tag, we sought to attach an HA tag onto CG193. Our previous attempts to attach HA onto the C-terminal of NG167 resulted in a fusion-defective phenotype, indicating that direct attachment of tags onto functional sites can abrogate the G protein’s fusion-promotion function (17). Further, the use of polyclonal antibodies raised against NG167 presented challenges in comparing headless versus wild-type NiV G CSE, due to the differential binding of anti-NG167 antibodies to NG167 versus wild-type NiV G, in addition to potential variations in epitope affinities across different headless mutants (17). To avoid these problems, we attached an HA tag to the C terminus of CG193 via a flexible protein linker (GGGGS)3 designed to retain the function of the G stalk. We named this construct CG193(HA) (Fig. 4A). Linkers such as this are widely used to join protein segments and retain their functions (30).
FIG 4.
The CedV G head domain antagonizes fusion more than the NiV G head domain. (A) Schematic representation of HA-tagged CedV G and linker-HA tagged CG193(HA). The protein linker (GGGGS)3 motif retained the function of the CG193 stalk (30). (B and D) Relative levels of CSE and fusion for wild-type or headless CedV G coexpressed with CedV F and wild-type or headless NiV G coexpressed with NiV F, respectively, in HEK 293T cells (16 to 20 hpt) normalized to wild-type G/F set to 100% (mean ± SD, n = 4). (C and E) Relative fusion indices of wild-type or headless CedV G coexpressed with CedV F and wild-type or headless NiV G coexpressed with NiV F, respectively, normalized to wild-type G/F set at 100% (mean ± D, n = 4). To determine fusion indices (fusion/CSE), first fusion and CSE values were normalized to those of NG/NF, set to 100%, and then their ratios (fusion/G CSE or fusion/F CSE) were calculated. (F) CSE levels for CedV F when coexpressed with tagged CG193(HA) or untagged CG193, normalized to untagged CG193 set to 100% (mean ± SD, n = 4). Gray points in the plots represent individual data points.
Although CG193(HA) was relatively less fusogenic than untagged CG193 (66.3% ± 9.9%, P < 0.001), it was still significantly hyperfusogenic compared to wild-type CedV G (282% ± 53.1%, P < 0.001) (Fig. 4B). This showed that our linker-tag approach is viable for determining the effects of CSE levels. Importantly, the CSE levels of CedV F were almost identical when coexpressed with either CG193 or CG193(HA) (107% ± 9.5%, P > 0.05) (Fig. 4F). Under the assumption that CG193 and CG193(HA) also had similar CSE levels, CG193 was ∼3× more fusogenic than wild-type CedV G when coexpressed with CedV F (FIG = 295% ± 56.0%, P < 0.001; FIF = 300% ± 68.4%, P < 0.001) (Fig. 4B and C). In contrast, NG167 was only ∼1.5× more fusogenic than wild-type NiV G when co-expressed with NiV F (FIG = 153.4% ± 0.1%, P < 0.001; FIF = 144.6% ± 19.7%, P < 0.001) (Fig. 4D and E). These results indicate that the hyperfusogenic phenotypes observed for headless CedV G mutants in Fig. 3 were due to their inherent fusogenicities and not to an increase in G or F CSE levels. Considering that CedV is less fusogenic than NiV (Fig. 2), that CedV G and NiV G both follow a stalk-exposure fusion mechanism (Fig. 3), and that headless CedV G releases a higher fold increase in fusion than NiV G when compared to their respective wild-type Gs (Fig. 4C and E), our collective results suggest that the head domains of different HNV Gs are essential for modulating their fusogenicities, e.g., by being fusion repressive to different extents.
Heterotypic bidentate G/F interactions can exist across dissimilar HNVs.
Since previous studies have shown that some heterotypic combinations of HNV G and F could induce fusion (20, 31–33), we performed heterotypic fusion assays by coexpressing wild-type or headless NiV G with CedV F and coexpressing wild-type or headless CedV G with NiV F. Surprisingly, the NG/CF pair yielded cell-cell fusion whereas the NG167/CF pair did not (Fig. 5A). This was unexpected, as NG167 is a hyperfusogenic mutant when paired with NiV F (17, 18). To elucidate this finding, we next investigated the interactions of NiV and CedV glycoproteins, because such interactions are required to induce cell-cell fusion.
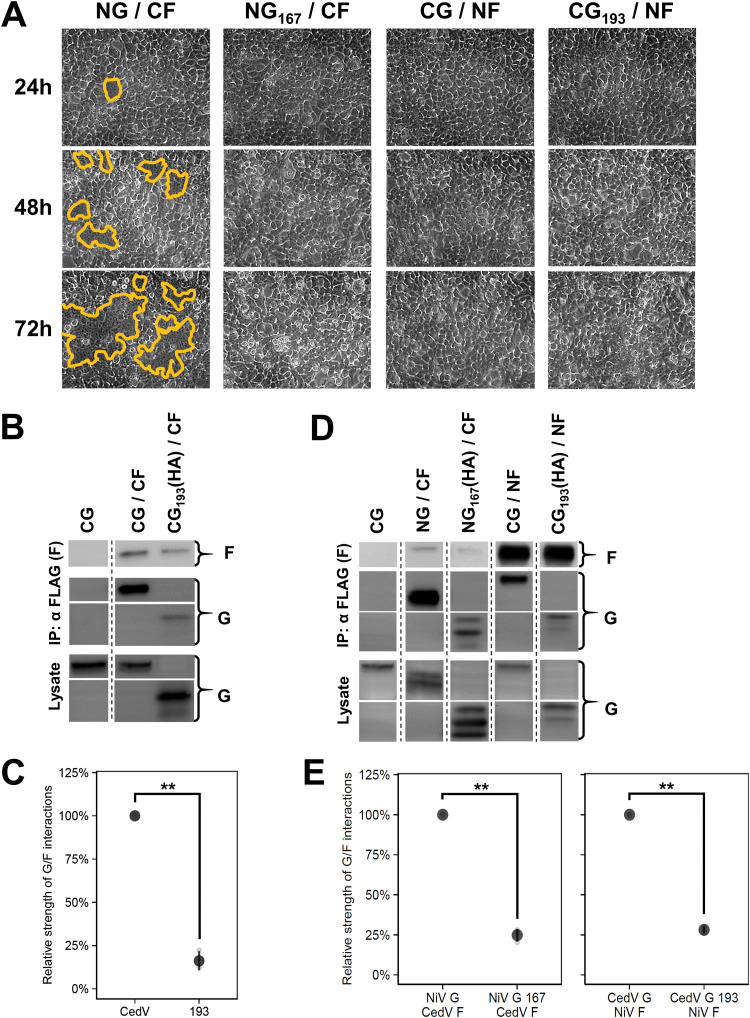
FIG 5.
Heterotypic assays between NiV and CedV glycoproteins reveal heterotypic bidentate interactions. (A) Representative fusion images (syncytia outlined in yellow) of heterotypic G/F combinations in HEK 293T cells. Cells were imaged every 24 hpt and fusion was only observed for NG/CF. (B) Representative blots of interactions between wild-type or headless CedV G and CedV F (24 hpt). The leftmost lanes represent the co-IP negative control. (C) Relative strength of G/F interactions between wild-type or headless CedV G with CedV F normalized to wild-type CedV G set to 100% (mean ± SD, n = 3). Relative G/F interaction strength was defined as ([GIP/GLysate] × FIP). Panels D and E are experimentally identical to panels B and C except for the use of heterotypic transfections as performed in A (48 hpt). All signals were obtained from the same blot, but the lanes were not necessarily adjacent. Gray points in the plots represent individual data points.
Our previous study showed that NiV glycoproteins engage in bidentate G/F interactions, where both the head and stalk of G interact with F (34). To assess if CedV glycoproteins also engage in bidentate G/F interactions, we coimmunoprecipitated wild-type or headless CedV G coexpressed with CedV F (Fig. 5B) and found that the relative strength of G/F interactions of headless CedV G193 and CedV F was significantly lower than that of wild-type CedV G and CedV F (16.2% ± 5.5%, P < 0.01) (Fig. 5C). The relative strength of G/F interactions was defined as ([G IP/G Lysate] × F IP), as previously established (20, 34). The decrease in relative G/F interaction strength between headless CG193/CF compared to wild-type CG/CF agrees with bidentate G/F interactions whereby both the head and stalk domains are important for such interactions (34). This underlies conserved bidentate G/F interactions across HNVs. Next, we coimmunoprecipitated heterotypic pairs of NiV and CedV glycoproteins and found that all heterotypic NiV/CedV combinations were capable of direct G/F interactions (Fig. 5D). Therefore, the nonfusogenic heterotypic combinations were not due to a lack of heterotypic G/F interactions. The relative strength of G/F interactions of headless Gs with Fs were also significantly lower than those of wild-type Gs, and thus consistent with heterotypic bidentate G/F interactions across HNVs (NG167/CF versus NG/CF = 24.8% ± 4.0%, P < 0.01; CG193/NF versus CG/NF = 28.1% ± 2.0%, P < 0.01) (Fig. 5E). Further, the requirement for the NiV G head domain for NG/CF fusion, but not for NG167/CF interactions, supports the notion that the HNV G head domains play further roles during membrane fusion beyond receptor binding (11, 12), stalk exposure (17, 18), and G/F interactions, presumably also contributing to F protein triggering.
CedV G forms a higher proportion of tetramers:dimers than NiV G.
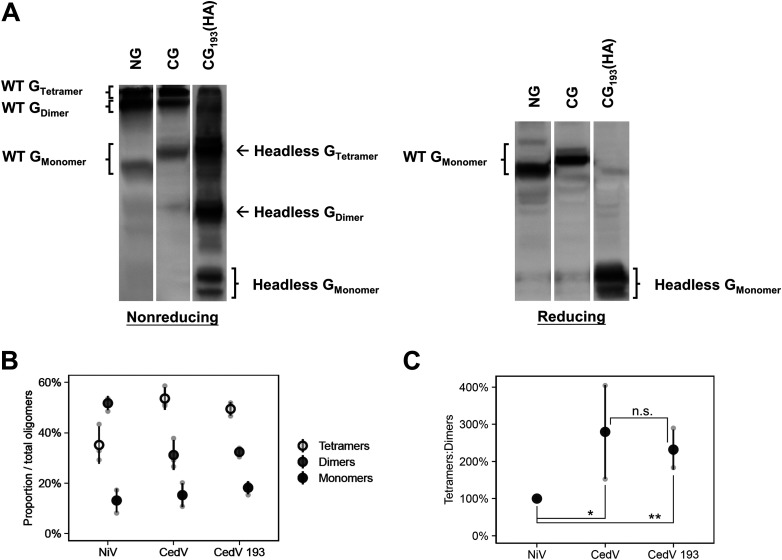
The stalk of HNV G oligomerizes into active tetramers (dimer of dimers); thus the proportion of tetramers to dimers and monomers gives a sense of their oligomeric stability, which in turn may affect fusogenicity by altering G’s oligomerization status. We compared nonreducing gels of NiV G and CedV G (Fig. 6A) and found that CedV G showed a nearly 3-fold higher proportion of tetramers:dimers (280% ± 126%, P < 0.05) compared to NiV G (Fig. 6B and C). Since CedV is less fusogenic than NiV (Fig. 2), these findings reinforce the inverse correlation previously noticed between HNV G’s relative oligomerization strength and HNV fusogenicity (31, 35). In addition, the nonreducing gels of wild-type versus headless CedV G (Fig. 6A) showed that they both formed similar proportions of tetramers, dimers, and monomers (Fig. 6B and C). This suggests that the head domain of CedV G plays minimal roles in modulating the oligomeric stability of the whole G protein and reinforces the stalk-mediated oligomerization model of HNV G (21).
FIG 6.
CedV G forms a higher proportion of tetramers:dimers than NiV G. (A) Representative nonreducing and reducing blots of wild-type NiV G, wild-type CedV G, and headless CedV G (24 hpt). (B) Oligomerization of the proteins (mean ± SD, n = 3); percent oligomers was defined as the percentage of (Goligomer/Gtetramer+dimer+monomer). (C) Ratios of tetramers:dimers of the G proteins normalized to that of NiV G set to 100%. All signals were obtained from the same blot, but the lanes were not necessarily adjacent. Gray points in the plots represent individual data points.
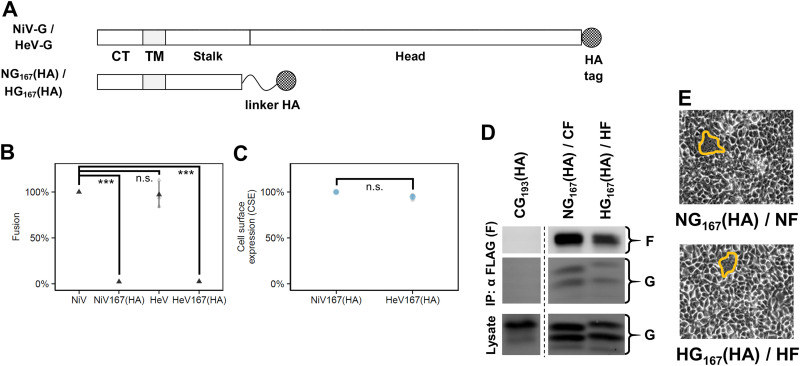
Linker tags abrogate fusogenicity of headless NiV G and HeV G.
Since linker-tagged headless CedV-G, CG193(HA), could induce fusion (Fig. 4), we also engrafted linker-HA tags on NG167 and HG167 and named them “NG167(HA)” and “HG167(HA),” respectively (Fig. 7A). However, in contrast to CG193(HA), NG167(HA) and HG167(HA) produced minimal levels of fusion (Fig. 7B). We found that both mutants had similar CSE and maintained interactions with a cognate F (Fig. 7C and D) (17, 18). These findings show that the fusion-defective phenotypes were primarily due to the linker tags and not to altered CSE levels, and further highlight the importance of the HNV stalk in promoting fusion (17, 18). Thus, although we were unable to adequately compare absolute fusion levels between headless NiV G, HeV G, and CedV G, the partial retention of fusogenicity for CG193(HA) showed that our linker tag strategy could situationally be a viable alternative approach for mutagenesis studies for some G proteins. More importantly, it supports our finding that the CedV G stalk is atypically hyperfusogenic (Fig. 3) and strengthens the notion that the head/stalk junctions of different HNVs are differentially sensitive to tags and are essential for modulating their respective fusogenicities.
FIG 7.
Linker-HA tags abrogate fusogenicity of headless NiV G and HeV G. (A) Schematic representation of HA-tagged NiV G/HeV G and linker-HA tagged NG167(HA)/HG167(HA). The protein linker is a (GGGGS)3 motif designed to retain the function of both linked proteins, headless G and HA tag (30). Note that all wild-type G used throughout the study had a C-terminal HA tag even if not shown in other schematics. (B) Relative levels of fusion for wild-type or headless linker-tagged NiV G and HeV G coexpressed with cognate F in HEK 293T cells at 16 to 20 hpt normalized to NG/NF set to 100% (mean ± SD, n = 3). (C) Relative levels of G CSE for NG167(HA)/NF and HG167(HA)/HF normalized to NG167(HA)/NF set to 100% (mean ± SD, n = 3). (D) Representative blots of coimmunoprecipitated supernatants and lysates of NG167(HA)/NF and HG167 (HA)/HF (24 hpt). The leftmost lanes represent the co-IP negative control. (E) Representative fusion images (outlined) for NG167(HA)/NF and HG167(HA)/HF in HEK 293T cells (24 hpt). All signals were obtained from the same blot, but the lanes were not necessarily adjacent. Gray points in the plots represent individual data points.
DISCUSSION
The current understanding of the lack of CedV pathogenicity is the lack of mRNA-editing capabilities required to express immunosuppressive V and W proteins encoded within the phosphoprotein (P) gene (7) and an inability to recognize ephrin B3 as a receptor (9, 10). This is because NiV mRNA-editing capabilities yield V and W proteins, drivers of NiV pathogenesis by antagonizing interferon responses (36, 37), and ephrin B3 is abundant in the central nervous system and associated with NiV acute encephalitis (38). Our data indicate that, in addition, CedV is inherently hypofusogenic compared to NiV (Fig. 2), perhaps further contributing to its lower pathogenicity. Syncytia formation is a hallmark of henipaviral and paramyxoviral infections, and syncytia prevalence appears to correlate with pathogenesis from postmortem histological examinations in NiV and HeV disease animal models (39, 40). Further studies are needed to reconcile the quantitative immune versus fusogenic impacts on CedV pathogenicity. Interestingly, indirect evidence suggests spillovers of other HNVs, such as Kumasi virus (KuV) and Mojiang virus (MojV), into human populations (5, 41), but whether these viruses are pathogenic in humans remains uncertain. In parallel, KuV and MojV also appear to be significantly less fusogenic than NiV or HeV (31), although their fusogenicities have not been compared quantitatively to consider glycoprotein CSE. Future studies may determine whether in vitro fusogenicity may serve as a proxy to assess the pathogenic potential of HNVs.
This study expands our current knowledge on HNV entry mechanisms. NiV and HeV glycoproteins have relatively high protein sequence similarities and fusogenic capacities (20). Despite the low sequence identities of CedV glycoproteins (Fig. 1), we found that the fundamental steps of HNV entry, receptor binding leading to stalk-mediated fusion activation, are conserved (Fig. 3). In addition, we present evidence for differential HNV RBP head domain modulation of fusogenicity. Although we could not accurately compare absolute fusion levels between headless CedV G and NiV G due to the nonfusogenic phenotype of linker-tagged headless NiV G (Fig. 7), we found that headless CedV G revealed a higher fold increase in fusogenicity than did headless NiV G relative to their respective wild-type G fusion levels (Fig. 4). This suggests that different HNV G head domains modulate HNV fusion by antagonizing their G fusogenicities to different extents.
However, the absolute fusion capacity of headless CedV G is still lower than that of headless NiV G, thus the stalk and head, and not just the head alone, must both be important modulators of HNV fusogenicity. Interestingly, we also found that the NiV G head domain was required to promote CedV F-mediated fusion. While NiV G promoted CedV F fusion, we observed no fusion for the headless NiV G167/CedV F combination (Fig. 5A), despite their ability to interact (Fig. 5D). This suggests that the mere presence of a functional stalk domain is insufficient for promoting F triggering and that HNV G head domains are required beyond receptor binding and promoting G stalk exposure, thus indicating important mechanistic roles of HNV G head domains in the post-attachment step(s) of the fusion cascade.
We also found support for heterotypic bidentate G/F interactions across HNVs. For instance, both the head and stalk of NiV G were important for interacting with CedV F (Fig. 5). These results may help the future identification of key regions on HNV G and HNV F that enable such interactions. Targeting these regions to abrogate G/F associations may be an approach to arrest HNV entry because HNV G and F cannot mediate cell-cell fusion independently. Further, the relatively conserved stalk regions across HNV G, such as the conserved cluster of residues flanked by three cysteines at the C-terminal stalk (Fig. 1), may provide opportunities for developing broad-spectrum neutralizing antivirals against HNVs.
Although our study supports a conserved stalk-exposure fusion and entry mechanism across paramyxoviruses (23, 24), headless HNV Gs are particularly hyperfusogenic (Fig. 3) (17, 18), while other analyzed headless paramyxoviral Hs/HNs are hypofusogenic (22–25). Conceivably, the head domains of paramyxoviral RBPs may play an added central role in promoting fusion by regulating the flexibility of their stalks. This notion is supported by MeV H and PIV5 HN studies, which suggest that RBP flexible stalks (42–45) require stabilization by their head domains to regulate fusion (22, 46). Conversely, since headless HNV G mutants are hyperfusogenic (Fig. 3) (17, 18), their fusogenic potential may be in part due to their already lower flexibilities provided by the HNV G three stalk cysteine residues (18, 21, 35, 47), i.e., a fusogenicity “released” upon stalk exposure. Since multiple stalk lengths of headless CedV G mutants are capable of inducing substantial amounts of fusion (Fig. 3), another deviating feature from other headless paramyxovial RBPs studied thus far (17, 18, 22–25), we posit that stalk flexibility, rather than purely stalk length, is key to fusion activation. This is further supported by the nonfusogenic phenotypes of NG170 and HG170 despite sharing comparable stalk lengths with the highly fusogenic CG193 (Fig. 3). Further studies will be needed to refine the mechanism by which paramyxovial head/stalk interfaces modulate fusion activation.
Our oligomerization analysis of CedV G (Fig. 6) reinforces a notable trend whereby HNV G oligomerization strength and fusogenicity inversely correlate (31, 32), i.e., HNVs that preferably form more high-order G oligomers appear to be less fusogenic (20, 31, 35). For instance, NiV G and HeV G both oligomerize with lower tetramer:dimer ratios (20, 21, 31), and NiV and HeV are similarly fusogenic (20). On the contrary, CedV G forms relatively more stable tetramers (Fig. 6) and CedV is significantly less fusogenic than NiV (Fig. 2). In addition, KuV G and MojV G form stable tetramers and both viruses appear to be hypofusogenic compared to NiV (31, 35), although this still needs to be explored quantitatively, considering G/F CSE levels. These observations suggest underlying dynamics during the various steps of the membrane fusion process that may depend on G/F stoichiometric and G oligomeric interactions. In support of this notion, varying the stoichiometries of HeV glycoproteins significantly impacted cell-cell fusion levels (48).
Why CedV G acquired its unusual head/stalk junction remains to be determined. A possible explanation is that this is a result of natural selection that favors viruses with lower levels of virulence and higher transmissibility to facilitate adaptation within novel hosts. The pathogenicity of CedV is suppressed by its lack of CedV P mRNA-editing sites (7) and its inability to recognize ephrin B3 (9, 10), while acquiring recognition of ephrin B1, which is highly expressed in the oropharyngeal tract may enhance its transmissibility (9, 10). The stronger antagonism of CedV G’s head domain on its stalk’s fusogenicity may reduce viral pathogenicity. Thus, P/V/W gene expression, receptor usage, and G stalk flexibility differences may all be involved in the process of henipaviral evolution.
Functional differences between HNVs enhance our understanding of HNV pathogenicity and contribute to the repertoire of analyses that aim to evaluate zoonotic potential and health risks posed by HNVs, a rapidly expanding genus of significant concern (49). Our study elucidated novel functional roles of HNV G in mediating cell-cell fusion by studying CedV G mutants. A major advantage of studying CedV is that it is currently a BSL-3 virus, unlike the BSL-4 NiV and HeV, and there exists a recombinant CedV platform operable under BSL-2 conditions (50). As a result, further work on CedV could prove instrumental in accelerating the search for much-needed anti-HNV antivirals.
MATERIALS AND METHODS
Expression plasmids.
Codon-optimized NiV G, HeV G, and CedV G genes with HA tags at the C terminus, and NiV F, HeV F, and CedV F genes with internal FLAG tags, were constructed as previously described (17, 18, 20). Headless NiV G, HeV G, and CedV G mutants were constructed by mutating two downstream amino acids into stop codons at the corresponding sites (Fig. 2A to C) via site-directed mutagenesis. Headless NiV G, HeV G, and CedV G constructs tagged with hemagglutinin (HA) via a flexible (GGGGS)3 protein linker (30) at their C termini were ordered from Twist Bioscience. All genes are cloned between KpnI and XhoI restriction sites in the pcDNA3.1(+) vector.
Cell lines.
HEK 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and CHO pgsA-745 cells were cultured in minimal essential medium alpha (MEM-α). Cells were incubated at 37°C, and media were supplemented with 10% bovine calf serum (BCS) and 1% penicillin-streptomycin.
Transfection.
Cells were split into six-well plates and grown to ∼70% confluence prior to transfection. Each transfection (except parts of Fig. 2) comprised G and F plasmids (1 μg each), or empty pcDNA3.1(+) vector (2 μg) as negative control (designated “vector”). Polyethylenimine (PEI) was used as the transfection reagent at a 3:1 PEI:plasmid mass ratio. Cells were collected after various time periods (refer to figure captions) for further experimentation as detailed below. In Fig. 2, 1:1, 2:1, and 1:2 NiV:CedV G and F CSE ratios were obtained after transfecting a wide range of NiV G/F and CedV G/F plasmid quantities (and supplanted to a total of 3 μg plasmid using empty pcDNA3.1[+]), then selecting the data from those that produced NiV:CedV G and F CSE ratios of interest.
Syncytia quantification.
Transfected cells were observed under 200× magnification and syncytia were quantified by counting the number of nuclei inside multinucleated cells (≥4 nuclei) as previously established (17, 18, 20, 27). Each data point was derived from the average of five microscopic fields per well.
Cell surface expression and ephrin B2 binding quantification.
Transfected cells were transferred to 96-well plates and antibody staining was performed for 45 min on ice. To assess cell surface expression (CSE), primary rabbit anti-HA and mouse anti-FLAG antibodies (1:1,000) and secondary fluorescently labeled anti-rabbit Alexa Fluor 488 and anti-mouse Alexa Fluor 647 antibodies (1:1,000) were used. To determine ephrin B2 binding levels, primary soluble recombinant mouse ephrin B2/human Fc chimeric protein (10 nM) and secondary fluorescently-labeled anti-human Alexa Fluor 647 antibody (1:1,000) were used. After each stain, cells were washed twice with fluorescence-associated cell sorting (FACS) buffer (phosphate-buffered saline [PBS] with 1% BCS). Lastly, cells were resuspended in 0.5% paraformaldehyde (200 μl) and analyzed by flow cytometry (Guava easyCyte8 HT).
Coimmunoprecipitation.
Transfected HEK 293T cells were spun down at 350 × g for 5 min and the cell pellet was resuspended in 200 μl of 1× Triton X-100 radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors. Cells were further lysed with occasional vortexing over 20 min. Cells were then centrifuged at 15,000 × g for 20 min to remove cell debris. The supernatants containing cell lysates (100 μl) were incubated with anti-FLAG beads (40 μl) while rotating at 4°C for 1 h. Columns were equilibrated with 200 μl of RIPA buffer, followed by running the lysates over the columns. Each column was washed five times with RIPA buffer (400 μl per wash) followed by equilibration with 25 μl of preheated (95°C) elution buffer. Bound proteins were finally eluted with 70 μl of preheated elution buffer and analyzed by Western blotting.
Western blotting analysis.
Cell lysates (30 μl) were loaded with 1× loading dye containing 5% β-mercaptoethanol (BME) and heated to 95°C. BME was excluded for nonreducing gels. Lysates and coimmunoprecipitation (co-IP) eluates were then separated by 10% SDS-PAGE under 100V for ∼100 min, transferred to nitrocellulose membranes under 0.25A for ∼200 min, blocked overnight, and stained with primary and secondary antibodies as described above. Protein bands were detected using ChemiDoc MP Imager system and densitometry analyses were performed with Image Lab software (Bio-Rad).
Statistical analyses.
In general, statistical analyses were performed using mixed models. Such models enable accounting for the hierarchical structures due to the clustering of data collected within the same experimental session (51). Indeed, each experiment had at least 3 biological replicates and some experiments included technical replicates within biological replicates. Mixed models further enable post hoc pairwise comparisons between ≥3 groups using a Tukey test, including a correction for multiple testing (51). Mixed models were constructed using fusion, fusion index, ephrin B2 binding, or F G avidity as a response variable, and glycoprotein genotypes (wild-type or mutant CedV, NiV, or HeV) as an explicative variable, and experimental session was included as a random effect. As for a given response variable, the variance was visibly different among protein genotypes; for instance, the genotypes leading to higher levels of fusion also lead to higher variance of fusion (see Fig. 3). For each analysis, we considered a model allowing a genotype-dependent variance in addition to a more classical model considering a constant variance across all conditions. These two models were compared based on their Akaike information criterion (AIC) and the model with the lowest AIC was selected (51), which was in all cases the model with genotype-dependent variance. Data formatting and analyses were performed in R 3.6.2 using the tidyverse, nlme, and lsmeans packages (52–55). Results of pairwise comparisons are reported as mean estimated difference ± standard error of the mean (SEM) and associated P value, with a significance cutoff set at P < 0.05. Data, codes, and detailed results of the statistical analyses are available at: https://github.com/AmandineGamble/CedV_fusion_public.
Data visualization.
For visualization purposes, data were transformed into values relative to a reference condition. Within each biological replicate, technical replicates were averaged and then divided by the reference condition value to obtain relative values. However, statistical analyses were conducted on raw data. Only pairwise comparisons mentioned in the text were indicated as statistically significant (*) or not (n.s): *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical results reported in the text represent mean difference ± standard error between the two compared groups.
ACKNOWLEDGMENTS
This research was developed with funding from the Defense Advanced Research Projects Agency (DARPA) administered through Cooperative Agreement D18AC00031-PREEMPT, as well as NIH/NIAID R01 grant AI109022, to H.C.A.
We thank Qian Liu (McGill University) for providing us with data from our prior publication (17) that was incorporated for comparison reasons into Fig. 4D and E. We also thank Victoria Ortega and Eun Jin Choi (Cornell University) for technical assistance.
Conceptualization: Y.Y.Y. and H.C.A. Methodology: Y.Y.Y., D.W.B., A.G., M.J., and H.C.A. Investigation and data curation: Y.Y.Y. Formal analysis: Y.Y.Y., D.W.B., A.G., and H.C.A. Writing —original draft preparation: Y.Y.Y. Writing—review and editing: Y.Y.Y., D.W.B., A.G., M.J., and H.C.A. Visualization: Y.Y.Y., D.W.B., A.G., and H.C.A. Supervision and funding acquisition: H.C.A.
Contributor Information
Hector C. Aguilar, Email: ha363@cornell.edu.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Goh KJ, Tan CT, Chew NK, Tan PSK, Kamarulzaman A, Sarji SA, Wong KT, Abdullah BJJ, Chua KB, Lam SK. 2000. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med 342:1229–1235. 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- 2.Hossain MJ, Gurley ES, Montgomery JM, Bell M, Carroll DS, Hsu VP, Formenty P, Croisier A, Bertherat E, Faiz MA, Azad AK, Islam R, Molla MAR, Ksiazek TG, Rota PA, Comer JA, Rollin PE, Luby SP, Breiman RF. 2008. Clinical presentation of Nipah virus infection in Bangladesh. Clin Infect Dis 46:977–984. 10.1086/529147. [DOI] [PubMed] [Google Scholar]
- 3.Kenmoe S, Demanou M, Bigna JJ, Nde Kengne C, Fatawou Modiyinji A, Simo FBN, Eyangoh S, Sadeuh-Mba SA, Njouom R. 2019. Case fatality rate and risk factors for Nipah virus encephalitis: a systematic review and meta-analysis. J Clin Virol 117:19–26. 10.1016/j.jcv.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Pernet O, Wang YE, Lee B. 2012. Henipavirus receptor usage and tropism, p 59–78. In Lee B, Rota PA (ed), Henipavirus: ecology, molecular virology, and pathogenesis. Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- 5.Pernet O, Schneider BS, Beaty SM, LeBreton M, Yun TE, Park A, Zachariah TT, Bowden TA, Hitchens P, Ramirez CM, Daszak P, Mazet J, Freiberg AN, Wolfe ND, Lee B. 2014. Evidence for henipavirus spillover into human populations in Africa. Nat Commun 5:5342. 10.1038/ncomms6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. 2020. List of Blueprint priority diseases. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 7.Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, Payne J, Robinson R, Broz I, Crameri G, Field HE, Wang L-F. 2012. Cedar virus: a novel henipavirus isolated from Australian bats. PLoS Pathog 8:e1002836. 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schountz T, Campbell C, Wagner K, Rovnak J, Martellaro C, DeBuysscher BL, Feldmann H, Prescott J. 2019. Differential innate immune responses elicited by Nipah virus and Cedar virus correlate with disparate in vivo pathogenesis in hamsters. Viruses 11:291. 10.3390/v11030291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laing ED, Navaratnarajah CK, Silva SCD, Petzing SR, Xu Y, Sterling SL, Marsh GA, Wang L-F, Amaya M, Nikolov DB, Cattaneo R, Broder CC, Xu K. 2019. Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus. Proc Natl Acad Sci USA 116:20707–20715. 10.1073/pnas.1911773116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pryce R, Azarm K, Rissanen I, Harlos K, Bowden TA, Lee B. 2019. A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus. Life Sci Alliance 3:e201900578. 10.26508/lsa.201900578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguilar HC, Iorio RM. 2012. Henipavirus membrane fusion and viral entry, p 79–94. In Lee B, Rota PA (ed), Henipavirus: ecology, molecular virology, and pathogenesis. Springer Berlin Heidelberg, Berlin, Heidelberg. [DOI] [PubMed] [Google Scholar]
- 12.Jardetzky TS, Lamb RA. 2014. Activation of paramyxovirus membrane fusion and virus entry. Curr Opin Virol 5:24–33. 10.1016/j.coviro.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeltina A, Bowden TA, Lee B. 2016. Emerging paramyxoviruses: receptor tropism and zoonotic potential. PLoS Pathog 12:e1005390. 10.1371/journal.ppat.1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401–405. 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 15.Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang L-F, Eaton BT, Broder CC. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci USA 102:10652–10657. 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negrete OA, Wolf MC, Aguilar HC, Enterlein S, Wang W, Mühlberger E, Su SV, Bertolotti-Ciarlet A, Flick R, Lee B. 2006. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog 2:e7. 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Montano JAB, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. 2013. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog 9:e1003770. 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Bradel-Tretheway B, Monreal AI, Saludes JP, Lu X, Nicola AV, Aguilar HC. 2015. Nipah virus attachment glycoprotein stalk C-terminal region links receptor binding to fusion triggering. J Virol 89:1838–1850. 10.1128/JVI.02277-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middleton D, Pallister J, Klein R, Feng Y-R, Haining J, Arkinstall R, Frazer L, Huang J-A, Edwards N, Wareing M, Elhay M, Hashmi Z, Bingham J, Yamada M, Johnson D, White J, Foord A, Heine HG, Marsh GA, Broder CC, Wang L-F. 2014. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg Infect Dis 20:372–379. 10.3201/eid2003.131159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradel-Tretheway BG, Zamora JLR, Stone JA, Liu Q, Li J, Aguilar HC. 2019. Nipah and Hendra virus glycoproteins induce comparable homologous but distinct heterologous fusion phenotypes. J Virol 93:e00577-19. 10.1128/JVI.00577-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maar D, Harmon B, Chu D, Schulz B, Aguilar HC, Lee B, Negrete OA. 2012. Cysteines in the stalk of the nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J Virol 86:6632–6642. 10.1128/JVI.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. 2013. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol 87:11693–11703. 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci USA 109:E2625–E2634. 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose S, Song AS, Jardetzky TS, Lamb RA. 2014. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol 88:3925–3941. 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ader-Ebert N, Khosravi M, Herren M, Avila M, Alves L, Bringolf F, Örvell C, Langedijk JP, Zurbriggen A, Plemper RK, Plattet P. 2015. Sequential conformational changes in the morbillivirus attachment protein initiate the membrane fusion process. PLoS Pathog 11:e1004880. 10.1371/journal.ppat.1004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Bertolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, Su SV, Wolf MC, Kohatsu L, Baum LG, Lee B. 2006. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80:4878–4889. 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes Zamora JL, Aguilar HC. 2018. Flow virometry as a tool to study viruses. Methods 134–135:87–97. 10.1016/j.ymeth.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu K, Rajashankar KR, Chan Y-P, Himanen JP, Broder CC, Nikolov DB. 2008. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA 105:9953–9958. 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowden TA, Aricescu AR, Gilbert RJC, Grimes JM, Jones EY, Stuart DI. 2008. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol 15:567–572. 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Zaro J, Shen W-C. 2013. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65:1357–1369. 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rissanen I, Ahmed AA, Azarm K, Beaty S, Hong P, Nambulli S, Duprex WP, Lee B, Bowden TA. 2017. Idiosyncratic Mòjiāng virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses. Nat Commun 8:16060. 10.1038/ncomms16060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weis M, Behner L, Hoffmann M, Krüger N, Herrler G, Drosten C, Drexler JF, Dietzel E, Maisner A. 2014. Characterization of African bat henipavirus GH-M74a glycoproteins. J Gen Virol 95:539–548. 10.1099/vir.0.060632-0. [DOI] [PubMed] [Google Scholar]
- 33.Pernet O, Beaty S, Lee B. 2014. Functional rectification of the newly described African henipavirus fusion glycoprotein (Gh-M74a). J Virol 88:5171–5176. 10.1128/JVI.03655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone JA, Vemulapati BM, Bradel-Tretheway B, Aguilar HC. 2016. Multiple strategies reveal a bidentate interaction between the Nipah virus attachment and fusion glycoproteins. J Virol 90:10762–10773. 10.1128/JVI.01469-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behner L, Zimmermann L, Ringel M, Weis M, Maisner A. 2018. Formation of high-order oligomers is required for functional bioactivity of an African bat henipavirus surface glycoprotein. Vet Microbiol 218:90–97. 10.1016/j.vetmic.2018.03.031. [DOI] [PubMed] [Google Scholar]
- 36.Satterfield BA, Cross RW, Fenton KA, Agans KN, Basler CF, Geisbert TW, Mire CE. 2015. The immunomodulating V and W proteins of Nipah virus determine disease course. Nat Commun 6:7483. 10.1038/ncomms8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez JJ, Parisien J-P, Horvath CM. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J Virol 76:11476–11483. 10.1128/jvi.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Negrete OA, Chu D, Aguilar HC, Lee B. 2007. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J Virol 81:10804–10814. 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres-Velez FJ, Shieh W-J, Rollin PE, Morken T, Brown C, Ksiazek TG, Zaki SR. 2008. Histopathologic and immunohistochemical characterization of Nipah virus infection in the guinea pig. Vet Pathol 45:576–585. 10.1354/vp.45-4-576. [DOI] [PubMed] [Google Scholar]
- 40.Li M, Embury-Hyatt C, Weingartl HM. 2010. Experimental inoculation study indicates swine as a potential host for Hendra virus. Vet Res 41:33. 10.1051/vetres/2010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, Sun L, Zhu Y, Zhou H, Jin Q. 2014. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerging Infect Dis 20:1064–1066. 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navaratnarajah CK, Rosemarie Q, Cattaneo R. 2016. A structurally unresolved head segment of defined length favors proper measles virus hemagglutinin tetramerization and efficient membrane fusion triggering. J Virol 90:68–75. 10.1128/JVI.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JPM, Örvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. 2012. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger f protein refolding for membrane fusion. J Biol Chem 287:16324–16334. 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adu-Gyamfi E, Kim LS, Jardetzky TS, Lamb RA. 2016. Flexibility of the head-stalk linker domain of paramyxovirus HN glycoprotein is essential for triggering virus fusion. J Virol 90:9172–9181. 10.1128/JVI.01187-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol 85:12855–12866. 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adu-Gyamfi E, Kim LS, Jardetzky TS, Lamb RA. 2016. Mutagenesis of paramyxovirus hemagglutinin-neuraminidase membrane-proximal stalk region influences stability, receptor binding, and neuraminidase activity. J Virol 90:7778–7788. 10.1128/JVI.00896-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Q, Biering SB, Mirza AM, Grasseschi BA, Mahon PJ, Lee B, Aguilar HC, Iorio RM. 2013. Individual N-glycans added at intervals along the stalk of the Nipah virus G protein prevent fusion but do not block the interaction with the homologous F protein. J Virol 87:3119–3129. 10.1128/JVI.03084-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitman SD, Dutch RE. 2007. Surface density of the Hendra G protein modulates Hendra F protein-promoted membrane fusion: role for Hendra G protein trafficking and degradation. Virology 363:419–429. 10.1016/j.virol.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, Seebens A, Knörnschild M, Oppong S, Sarkodie YA, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stöcker A, Carneiro AJB, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EKV, Kruppa T, Franke CR, Kallies R, Yandoko ERN, Herrler G, Reusken C, Hassanin A, Krüger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. 2012. Bats host major mammalian paramyxoviruses. Nat Commun 3:796–713. 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laing ED, Amaya M, Navaratnarajah CK, Feng Y-R, Cattaneo R, Wang L-F, Broder CC. 2018. Rescue and characterization of recombinant cedar virus, a non-pathogenic Henipavirus species. Virol J 15:56–56. 10.1186/s12985-018-0964-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Queen JP, Quinn GP, Keough MJ. 2002. Experimental design and data analysis for biologists. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 52.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 53.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. 2019. Welcome to the Tidyverse. Joss 4:1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- 54.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2013. nlme: linear and nonlinear mixed effects models. R package version 31 111.
- 55.Lenth RV. 2016. Least-squares means: the R package lsmeans. J Stat Soft. 69. 10.18637/jss.v069.i01. [DOI] [Google Scholar]