ABSTRACT
Alphaviruses (family Togaviridae) include both human pathogens such as chikungunya virus (CHIKV) and Sindbis virus (SINV) and model viruses such as Semliki Forest virus (SFV). The alphavirus positive-strand RNA genome is translated into nonstructural (ns) polyprotein(s) that are precursors for four nonstructural proteins (nsPs). The three-dimensional structures of nsP2 and the N-terminal 2/3 of nsP3 reveal that these proteins consist of several domains. Cleavage of the ns-polyprotein is performed by the strictly regulated protease activity of the nsP2 region. Processing results in the formation of a replicase complex that can be considered a network of functional modules. These modules work cooperatively and should perform the same task for each alphavirus. To investigate functional interactions between replicase components, we generated chimeras using the SFV genome as a backbone. The functional modules corresponding to different parts of nsP2 and nsP3 were swapped with their counterparts from CHIKV and SINV. Although some chimeras were nonfunctional, viruses harboring the CHIKV N-terminal domain of nsP2 or any domain of nsP3 were viable. Viruses harboring the protease part of nsP2, the full-length nsP2 of CHIKV, or the nsP3 macrodomain of SINV required adaptive mutations for functionality. Seven mutations that considerably improved the infectivity of the corresponding chimeric genomes affected functionally important hot spots recurrently highlighted in previous alphavirus studies. These data indicate that alphaviruses utilize a rather limited set of strategies to survive and adapt. Furthermore, functional analysis revealed that the disturbance of processing was the main defect resulting from chimeric alterations within the ns-polyprotein.
IMPORTANCE Alphaviruses cause debilitating symptoms and have caused massive outbreaks. There are currently no approved antivirals or vaccines for treating these infections. Understanding the functions of alphavirus replicase proteins (nsPs) provides valuable information for both antiviral drug and vaccine development. The nsPs of all alphaviruses consist of similar functional modules; however, to what extent these are independent in functionality and thus interchangeable among homologous viruses is largely unknown. Homologous domain swapping was used to study the functioning of modules from nsP2 and nsP3 of other alphaviruses in the context of Semliki Forest virus. Most of the introduced substitutions resulted in defects in the processing of replicase precursors that were typically compensated by adaptive mutations that mapped to determinants of polyprotein processing. Understanding the principles of virus survival strategies and identifying hot spot mutations that permit virus adaptation highlight a route to the rapid development of attenuated viruses as potential live vaccine candidates.
KEYWORDS: RNA replication, adaptive mutations, alphavirus, promoters, proteases, replicase
INTRODUCTION
Alphaviruses (family Togaviridae) are positive-strand RNA viruses. Many mosquito-transmitted alphaviruses are human pathogens that cause debilitating symptoms. Chikungunya virus (CHIKV) has recently emerged worldwide, resulting in epidemics that require increasing medical attention (1). The need for effective antiviral compounds and vaccines is not currently met and is steadily increasing together with the spread of the virus (2).
The genome of alphaviruses contains two main open reading frames (ORFs) that encode nonstructural (ns) and structural polyproteins (3). The ns-polyprotein P1234 is autocatalytically cleaved into four individual proteins (nsP1 to 4). Processing occurs in a temporally and sequentially controlled manner and is synchronized with other essential viral functions (4, 5). nsPs have specific enzymatic activities and, together with host-derived factors, form a membrane-bound replicase complex (RC) that ensures viral genome replication and synthesis of subgenomic (SG) RNA that is translated into structural proteins (6).
Viral RC comprises several functional modules that correspond structurally to one or several protein domains. To date, the three-dimensional structures of three alphavirus nsPs have been published. nsP1 serves as a membrane anchor of RC. Twelve copies of nsP1 form a crown-shaped ring with a central 7- to 7.5-nm pore (7, 8). nsP2 consists of two functional regions, the N-terminal RNA helicase region (nsP2h) and the C-terminal protease region (nsP2p), which function in a coordinated manner (9, 10). nsP2h (for CHIKV, residues 1 to 464) consists of a uniquely folded N-terminal domain (NTD) followed by a superfamily 1 RNA helicase fold including two Rec-A-like domains, designated RecA1 and RecA2, and an extended α-helical connector domain (11). nsP2p (for CHIKV, residues 471 to 798) consists of papain-like protease and methyltransferase-like (MTL) domains (12, 13). The domains are connected via a flexible linker that has an important role in viral RNA synthesis (14). The N-terminal macrodomain of nsP3 has ADP-ribosyl-binding and hydrolase activities that are essential for virus infectivity and virulence (15–18). The central portion of nsP3 is referred to as the alphavirus unique domain (AUD); it binds zinc ions and RNA (19) and has multiple roles in RNA replication (20). The C-terminal hypervariable domain (HVD) of nsP3 is intrinsically disordered (21). The HVD is phosphorylated (22, 23), is involved in reprogramming of host cell metabolic activity (24), and serves as a binding platform for host proteins that are important for alphavirus replication (25–30).
In contrast to relatively well-characterized specific activities, the functional interactions of different domains of nsP2 and nsP3 with one another and with other viral proteins have not been sufficiently elucidated. Such interactions have traditionally been studied using point mutations, deletions, or insertions (4, 31, 32). This approach, however, is time- and labor-intensive because it relies on the meticulous introduction of specific mutations that have effects that are large enough to allow investigators to draw meaningful conclusions. Scanning mutagenesis has been used to map functionally important regions of nsPs (33). As a more general genetic shortcut, the homologous domain swapping approach, which can be viewed as the simultaneous introduction of multiple substitution mutations, has been used to map some of the determinants that are responsible for the specific phenotypes of alphavirus strains (34, 35). Additionally, the exchange of regions that encode homologous proteins or their domains has been used to study alphavirus RNA replication and virus-host interactions (36, 37; see companion article [38]), as well as in the analysis of determinants of the host range restrictions of an insect-specific alphavirus (39).
In this study, we used swapping of homologous domains to examine the functional interactions of different parts of nsP2 and nsP3. It was found that Semliki Forest virus (SFV) genomes harboring nsP2 or nsP2p of Sindbis virus (SINV) were not infectious, whereas those harboring nsP2 NTD or the nsP3 macro region of CHIKV had wild-type (WT)-like infectivity. Swapped genomes harboring nsP2 or nsP2p derived from the CHIKV or nsP3 macrodomain from SINV had low infectivity, but the corresponding viruses replicated to high titers after accumulating adaptive changes. In total, seven different adaptive substitutions that considerably improved the infectivity of the swapped genomes were identified. Interestingly, only three of these mutations also resulted in clearly increased RNA synthesis in the trans-replicase assay, and one acted by increasing the activity of the SG promoter. Our data highlight the importance of regions involved in the formation of alphavirus RC for virus infection, provide valuable information for understanding the interactions of nsPs, and describe an attractive approach that can be used in virus attenuation.
RESULTS
SFV tolerates swapping of its nsP2 and nsP3 domains with those of CHIKV.
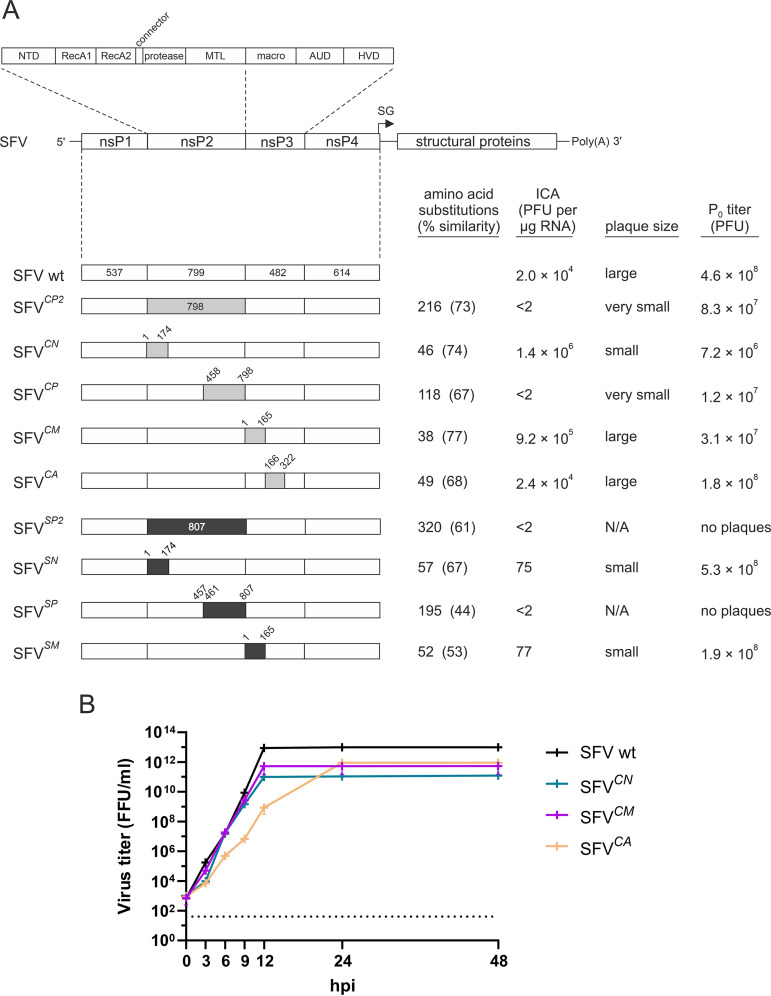
The domains of alphavirus polyproteins may function as a production line; in this case, the substitution of one section with an analogous working unit should have minimal effect on the overall architecture of the viral replicase. Alternatively, polyprotein modules may work closely together, forming a coevolved protein interaction network. Accordingly, replacement of one node would have profound effects on the functionality of the replication machinery, resulting in dysfunctional chimeras or in the accumulation of numerous compensatory changes aimed at restoration of protein interfaces. To discriminate between these possibilities, advantage was taken of an observation made in our previous studies in which, it was found that the processing of P1234 of SFV depends on the NTD and nsP2p regions of nsP2 and on the macro domain of nsP3 (31, 40). CHIKV belongs to the SFV serocomplex of alphaviruses (41). The nsP2-nsP3 macro regions of SFV and CHIKV have 74% amino acid sequence identity, and CHIKV P1234 is processed in a manner similar to that in which SFV P1234 is processed (32, 42), indicating that the regions involved in the processing of P1234 in these viruses may be interchangeable. To verify this hypothesis, infectious cDNAs (icDNA) of SFV in which nsP2 or the predicted NTD, nsP2p, or nsP3 macro regions were replaced with the corresponding regions of CHIKV were constructed. In addition, an SFV variant in which the nsP3 AUD domain was replaced with its CHIKV counterpart was constructed as a control (Fig. 1A). An infectious center assay (ICA) revealed that the rescue efficiencies of SFVCN, SFVCM, and SFVCA were at least as high as that of WT SFV (Fig. 1A; Table 1). To confirm the possible attenuation, multistep growth curves of these viruses were compared to the multistep growth curve of WT SFV. Notably, only minimal attenuation was observed for SFVCM. At 3 h postinfection (p.i.), the growth of SFVCN was slightly reduced, and an even greater reduction was observed for SFVCA from 3 to 12 h p.i. However, from 24 h p.i. onward, the titers of all of the mutant viruses were high and rather similar to that of WT SFV (Fig. 1B). In contrast, the infectivity of the SFVCP and SFVCP2 genomes was below the limit of detection. Nevertheless, viruses were rescued and grew to titers that were only ∼10-fold lower than that of WT SFV (Fig. 1A; Table 1). Thus, SFV tolerates swapping of its analyzed regions with their CHIKV counterparts, although larger swaps apparently create a need for subsequent adaptation(s).
FIG 1.
Swapping of fragments of SFV with homologous regions of CHIKV or SINV. (A) Schematic diagram of the SFV genome showing the domain organization of the nsP2-nsP3 region and the ns-regions of the chimeric genomes. NTD, N-terminal domain of nsP2; RecA, RecA-like domain; MTL, methyltransferase-like domain; AUD, alphavirus unique domain; HVD, hypervariable domain. The numbers indicate the amino acid residues flanking the swapped domains; sequences from CHIKV are shown in gray, and sequences from SINV are shown in black. The number of amino acid substitutions introduced by each of the swaps is shown, and the percent amino acid identity of the proteins encoded by the swapped regions is shown in parentheses. ICA titers are presented as PFU per 1 μg RNA and represent the averages of the data obtained in four independent experiments; <2 indicates values below the detection limit. Plaque sizes are indicated as follows: large, >3 mm; small, 1 to 2 mm; very small, <1 mm in diameter. P0 titers are presented as PFU per ml and represent the averages of the titers measured in three independent experiments. Please note that P0 stocks were harvested upon detection of CPE, i.e., at different times for different viruses. (B) Multistep growth curves of WT SFV, SFVCN, SFVCM, and SFVCA. BHK-21 cells were infected with P0 stocks of the indicated viruses at an MOI of 0.1. The culture medium was sampled immediately (0 h) and at 3 h, 6 h, 9 h, 12 h, 24 h, and 48 h p.i., and the amount of infectious virus present was measured using a focus-forming assay. Each data point represents the mean ± standard deviation (SD) from three independent experiments. The dotted line indicates the limit of detection.
TABLE 1.
Summary of the properties of the swapped and mutated genomes analyzed in this studya
| Name | Region introduced by swap | Additional mutations | Infectivity (ICA) (PFU/μg) | P0 titer (BHK-21) (PFU/ml) | P1 titer (C6/36) (PFU/ml) | nsP2/P123 ratio (processing efficiency in in vitro translation) | Replication (trans-replicase) (avg increase in Fluc) | Transcription (trans-replicase) (avg increase in Gluc) |
|---|---|---|---|---|---|---|---|---|
| SFV WT | None | None | 2.0 × 104 | 4.6 × 108 | 4.7 × 109 | 100 | 1,040 | 235,000 |
| SFVCP2 | CHIKV nsP2 | None | <2 | 8.3 × 107 | 6.3 × 106 | >>100b | 52 | 168 |
| SFVCP2+1Y533D+1H534R | CHIKV nsP2 | Y533D (nsP1) H534R (nsP1) | 8 | 1.5 × 106 | ND | 720 ± 300 | ND | ND |
| SFVCP2+1G536V | CHIKV nsP2 | G536V (nsP1) | <2 | No plaques | ND | 51 ± 7c | ND | ND |
| SFVCP2+2E228K | CHIKV nsP2 | E228K (nsP2) | 1.7 × 103 | 3.8 × 108 | 5.9 × 107 | 100 ± 71 | 172 | 1,070 |
| SFVCP2+2E228K+2C798F | CHIKV nsP2 | E228K (nsP2)C798F (nsP2) | <2 | ND | ND | 0 | ND | ND |
| SFVCP2+2A796V | CHIKV nsP2 | A796V (nsP2) | 3.0 × 104 | 5.6 × 107 | 3.6 × 106 | 82 ± 84 | 554 | 1,690 |
| SFVCP2+4C483Y | CHIKV nsP2 | C483Y (nsP4) | 5.6 × 103 | 8.3 × 107 | 1.6 × 107 | >>100 | 67 | 286 |
| SFVCP2+4C483Y+3A1P | CHIKV nsP2 | A1P (nsP3)C483Y (nsP4) | <2 | ND | ND | 0 | ND | ND |
| SFVCP2+4R611KSame as SFVCP2-SG(−1)G/A | CHIKV nsP2 | R611K (nsP4)Same as SG(−1)G/A | 9.3 × 102 | 1.1 × 109 | 1.2 × 109 | >>100 | 53 | 177 |
| SFVCN | CHIKV nsP2 NTD | None | 1.4 × 106 | 7.2 × 106 | ND | >>100 | 39 | 10,100 |
| SFVCP | CHIKV nsP2 Pro | None | <2 | 1.2 × 107 | 1.7 × 107 | 21 ± 17 | 609 | 759 |
| SFVCP+1Y533D+1H534R | CHIKV nsP2 Pro | Y533D (nsP1) H534R (nsP1) | 600 | 3.8 × 107 | ND | 130 ± 24 | ND | ND |
| SFVCP+1G536V | CHIKV nsP2 Pro | G536V (nsP1) | <2 | No plaques | ND | 18 ± 8b | ND | ND |
| SFVCP+2V2F | CHIKV nsP2 Pro | V2F (nsP2) | 5.2 × 104 | 9.3 × 107 | 3.1 × 105 | 16 ± 15 | 497 | 1,200 |
| SFVCP+2G460S | CHIKV nsP2 Pro | G460S (nsP2) | 5.1 × 104 | 1.5 × 107 | 6.7 × 104 | 16 ± 13 | 595 | 808 |
| SFVCP+4R611KSame as SFVCP-SG(-1)G/A | CHIKV nsP2 Pro | R611K (nsP4)Same as SG(-1)G/A | 3.5 × 104 | 1.5 × 108 | 6.1 × 107 | 11 ± 7 | 590 | 434 |
| SFVCM | CHIKV nsP3 macro | None | 9.2 × 105 | 3.1 × 107 | ND | 110 ± 27 | 573 | 151,000 |
| SFVCA | CHIKV nsP3 AUD | None | 2.4 × 104 | 1.8 × 108 | ND | ND | 1,940 | 794,000 |
| SFVCH | CHIKV nsP3 HVD | None | ND | ND | ND | ND | 1,480 | 408,000 |
| SFVSP2 | SINVnsP2 | None | <2 | No plaques | ND | 19 ± 18b | 2 | 7 |
| SFVSN | SINVnsP2 NTD | None | 75 | 5.3 × 108 | 1.4 × 108 | 83 ± 22 | 83 | 1,610 |
| SFVSN+2A73V | SINVnsP2 NTD | A73V (nsP2) | 2–3 | 4.0 × 108 | 3.7 × 108 | 87 ± 46 | 145 | 3,400 |
| SFVSN+3K415M | SINVnsP2 NTD | K415M (nsP3) | 5–6 | 9.2 × 108 | 8.0 × 107 | 97 ± 41 | 50 | 1,480 |
| SFVSP | SINVnsP2 | None | <2 | No plaques | ND | 9 ± 8b | 2 | 4 |
| SFVSM | SINVnsP3 macro | None | 77 | 1.9 × 108 | 2.9 × 106 | 110 ± 65 | None | 168 |
| SFVSM+2V515E | SINVnsP3 macro | V515E (nsP2) | 2.2 × 104 | 1.9 × 109 | 1.9 × 106 | 34 ± 20 | 3 | 948 |
ND, not determined; >>, not possible to quantify. as P123 is not detectable.
Detected only in some replicas; in some, it was not possible to quantify, as P123 is not detectable.
No increase in the nsP2/P123 ratio over time; possibly no cleavage at all.
SFV tolerates swapping of the nsP2 NTD and nsP3 macrodomains with those of SINV.
From a genetic perspective, SFV and SINV belong to different serocomplexes of alphaviruses (41). The identity between the nsP2-nsP3 macro regions of SINV and those of SFV is 59%. Unlike SFV, the P34 polyprotein accumulates during the later stages of SINV infection (43), indicating that there are substantial differences in P1234 processing in the two viruses. Therefore, not surprisingly, replacement of nsP2 or nsP2p of SFV with their SINV counterparts resulted in nonviable chimeras. SFVSN and SFVSM were successfully rescued; however, the infectivity of the chimeric genomes was lower than that of WT SFV (Fig. 1A; Table 1). The observed effects may result from defects in P1234 processing, from mismatches within forming RC, and/or from disturbance of other functions that are crucial for successful infection.
Adaptation of chimeric viruses leads to the accumulation of mutations in functionally important regions of the genome.
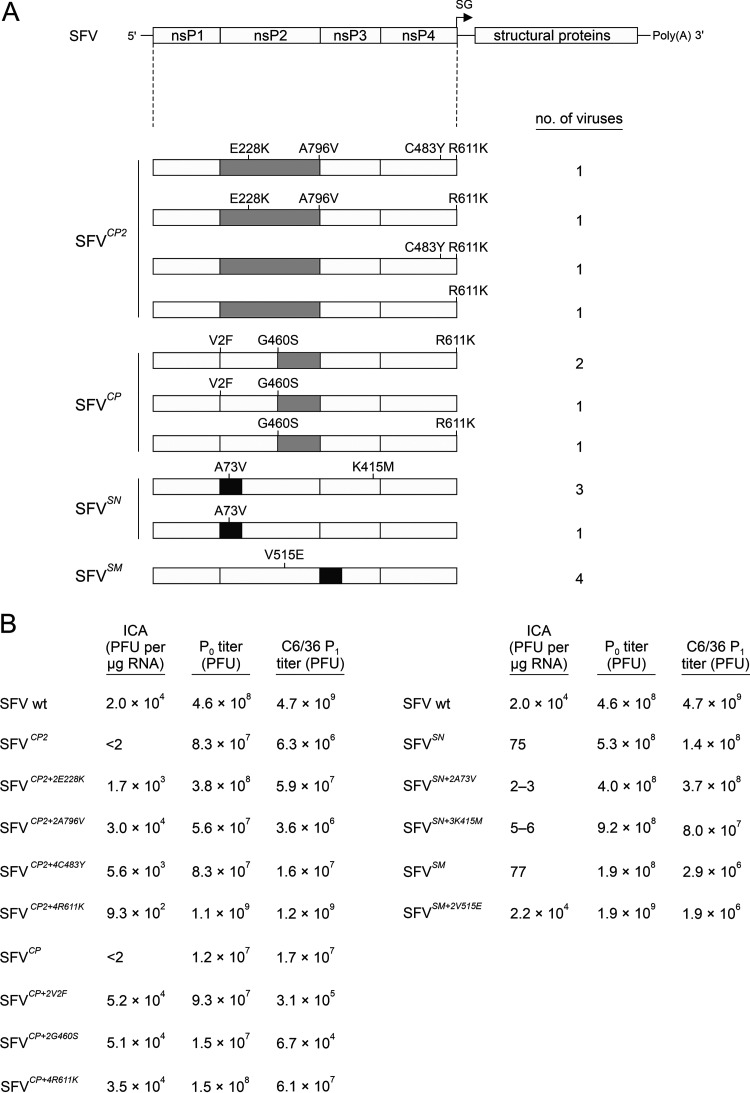
Mutant alphaviruses harboring one or a few substitutions often revert or pseudorevert (11, 44). Swapping of homologous domains, however, results in the simultaneous introduction of amino acid substitutions that are too numerous to be (pseudo)reverted (Fig. 1A). To identify the adaptive mutations that have the largest impact on the viability of rescued viruses, SFVCP2, SFVCP, SFVSN, and SFVSM were passaged five times. Four plaque-purified isolates were obtained for each of these in cellulo-evolved viruses, their ns-regions were sequenced, and the nonsynonymous substitutions present in these regions were identified (Fig. 2A; Table 1).
FIG 2.
Adaptation of chimeric viruses results in nonsynonymous mutations in the ns-region. (A) Locations of nonsynonymous mutations found in adapted stocks of SFVCP2, SFVCP, SFVSN, and SFVSM. The numbers on the right sides of the drawings represent the number of virus isolates in which the indicated combinations of mutations were found. (B) The identified nonsynonymous mutations increase the infectivity of the swapped genomes. P0 stocks were harvested, and their ICA titers and P0 titers were determined and are shown in Fig. 1A. Titers of P1 stocks collected from infected C6/36 cells are presented as PFU per ml and represent the averages of the values obtained in three independent experiments.
All of the isolates of SFVSM contained only a V515E substitution in nsP2. Residue 515 corresponds to the S4 subsite residue of nsP2p, and variation at this site is linked to the efficiency of processing of the cleavage site between nsP1 and nsP2 (1/2 site) (4, 45). All four isolates of SFVSN contained a V73A substitution in the NTD region, and three of these isolates also contained a K415M substitution in the HVD of nsP3 (Fig. 2A). Two isolates of SFVCP contained V2F and G460S substitutions in nsP2 and an R611K substitution in nsP4; these mutations also appeared in various combinations in the remaining SFVCP isolates (Fig. 2A). Interestingly, the guanine-to-adenine substitution (nucleotide [nt] G7371A) responsible for R611K occurs at position −1 of the SFV SG promoter and was also present in all isolates of SFVCP2. In addition, one isolate of SFVCP2 also contained the C483Y substitution in nsP4, a substitution that was previously shown to increase the fidelity of CHIKV replication (46, 47). Two isolates of SFVCP2 contained identical sets of three mutations (E228K and A796V in nsP2 and R611K in nsP4) and differed in the presence or absence of the C483Y substitution in nsP4 (Fig. 2A).
The mutations found in isolates of SFVSN decreased the infectivity of the recombinant genomes but did not prevent rescue and replication of infectious virus (Fig. 2B; Table 1). In contrast, E228K, A796V, C483Y, and R611K all markedly increased the infectivity of SFVCP2 (Fig. 2B; Table 1). A similar observation was made for all three substitutions identified in isolates of SFVCP and for the V515E substitution found in isolates of SFVSM (Fig. 2B; Table 1). Because mutations introduced into the ns-region can have different effects in vertebrate and mosquito cells (20, 48), Aedes albopictus C6/36 cells were infected with the obtained P0 stocks. Most of the rescued viruses replicated well in mosquito cells. The exceptions were SFVCP+2V2F and SFVCP+2G460S, which had titers >50-fold lower in mosquito cells than in BHK-21 cells. Because this feature was not observed for SFVCP+4R611K (Fig. 2B; Table 1), the defective replication of these viruses in mosquito cells (and/of at lower 28°C temperature) must originate from the V2F and G460S substitutions or from a combination of these substitutions with swapping of the nsP2p region.
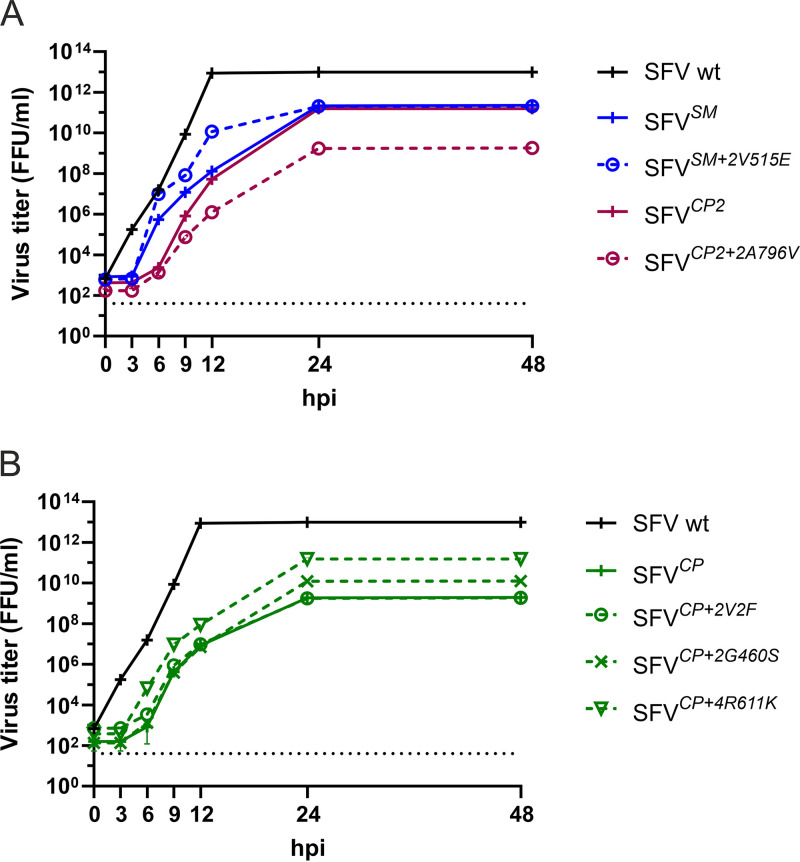
In cellulo-evolved viruses and viruses harboring adaptive mutations are attenuated.
The multistep growth curves of SFVSM+2V515E and SFVSM, obtained as a result of in cellulo evolution, were similar. Only a slight delay between 6 and 12 h p.i. was observed for SFVSM; thereafter, the titers of the two viruses were identical (Fig. 3A). This finding is consistent with the idea that V515E is the primary adaptive mutation found in the SFVSM stock (Fig. 2A). The growth dynamics of these viruses were similar to that of WT SFV; their titers were, however, consistently ∼10-fold lower, with the largest difference observed at 12 h p.i. (Fig. 3A). For SFVCP2 and SFVCP, the evolved stocks consist mostly of viruses that harbored combinations of the adaptive mutations (Fig. 2A). Analysis of the growth curve of evolved stock of SFVCP2 revealed that it grew more slowly than WT SFV; the difference was more prominent at early time points. The growth of SFVCP2+2A796V was slower than that of SFVCP2 (Fig. 3A), indicating that the adaptation that restored the infectivity of the RNA genome (Fig. 2B) did not fully restore replication of the virus. Compared to WT SFV, SFVCP grew more slowly and reached lower final titers (Fig. 3B). The growth rates of SFVCP+2V2F and of SFVCP+2G460S were almost identical to that of SFVCP (Fig. 3B). SFVCP+4R611K, on the other hand, grew faster than SFVCP and reached titers that were only ∼60-fold lower than those of WT SFV. Taken together, these findings indicate that the growth of all three derivatives of SFVCP that harbored single adaptive mutations was rather similar to that of SFVCP and suggest that the combinations of adaptive mutations found in the evolved stock (Fig. 2A) offered limited growth advantages.
FIG 3.
In cellulo-evolved viruses with adaptive mutations display growth attenuation. (A and B) BHK-21 cells were infected at an MOI of 0.1 with WT SFV, with evolved stocks of (A) (SFVCP2, SFVSM) or (B) (SFVCP) or with P0 stocks of (A) (SFVCP2+2A796V, SFVSM+2V515E) or (B) (SFVCP+2V2F, SFVCP+2G460S, or SFVCP+4R611K). At 0 h, 3 h, 6 h, 9 h, 12 h, 24 h, and 48 h p.i., the growth medium was collected, and the virus in the medium was titrated using a focus-forming assay. Each data point represents the mean ± SD from three independent experiments. The dotted line indicates the limit of detection.
The 2/3 sites of SFVCP2+2E228K, SFVCP2+4C483Y, and SFVCP2+4R611K accumulate additional mutations.
Evolved SFVCP2 grew faster and had higher titers than SFVCP2+2A796V (Fig. 3A), indicating an additive effect of combinations of different mutations (Fig. 2A). It was therefore hypothesized that additional mutations with similar effects may also accumulate during rescue and passage of SFVCP2+2E228K, SFVCP2+2A796V, SFVCP2+4C483Y, and SFVCP2+4R611K. Sequence analysis of the region encoding the C-terminal fragment of nsP1, nsP2, and the N-terminal part of nsP3 revealed no additional mutations in the SFVCP2+2A796V stock. In contrast, genomes containing additional substitutions in the region corresponding to the 2/3 site of ns-polyprotein were found in the SFVCP2+2E228K, SFVCP2+4C483Y, and SFVCP2+4R611K stocks (Table 2), indicating that there was selection pressure for a decreased rate of processing at the 2/3 site. Unexpectedly, however, these changes did not provide any growth advantage; SFVCP2+2E228K+2C798F and SFVCP2+4C483Y+3A1P were both strongly attenuated; no plaques were detected in ICA, and only mild cytopathic effects (CPE) were observed (Table 1). This finding may indicate that the viruses in which the C798F and A1P mutations were found also contained additional mutation(s) in regions that were not analyzed in this experiment.
TABLE 2.
Amino acid sequences of the 2/3 sites of CHIKV, SFVCP2, and novel SFVCP2 variants found in the P1 stocka
| Amino acid no. | 793 | 794 | 795 | 796 | 797 | 798 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage site residue | P6 | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ |
| CHIKV WT, SFVCP2 | A | T | R | A | G | C | A | P | S | Y |
| SFVCP2+4R611K+2A796V | A | T | R | V | G | C | A | P | S | Y |
| SFVCP2+2E228K+2C798F | A | T | R | A | G | F | A | P | S | Y |
| SFVCP2+4C483Y+3A1P | A | T | R | A | G | C | P | P | S | Y |
Mutated residues found in novel SFVCP2 variants are shown in boldface and italics.
Swapping of nsP2, its regions, and the macrodomain of nsP3 results in defects in P1234 processing.
The known functions of swapped domains/proteins and the localization of the majority of adaptive mutations (Fig. 2A; Tables 1 and 2) strongly suggest that they may impact P1234 processing. Due to the lethal phenotype of SFVSP2 and SFVSP, the adaptations of SFVCP2, SFVCP, SFVSN, and SFVSM processing were studied using in vitro-translated polyproteins.
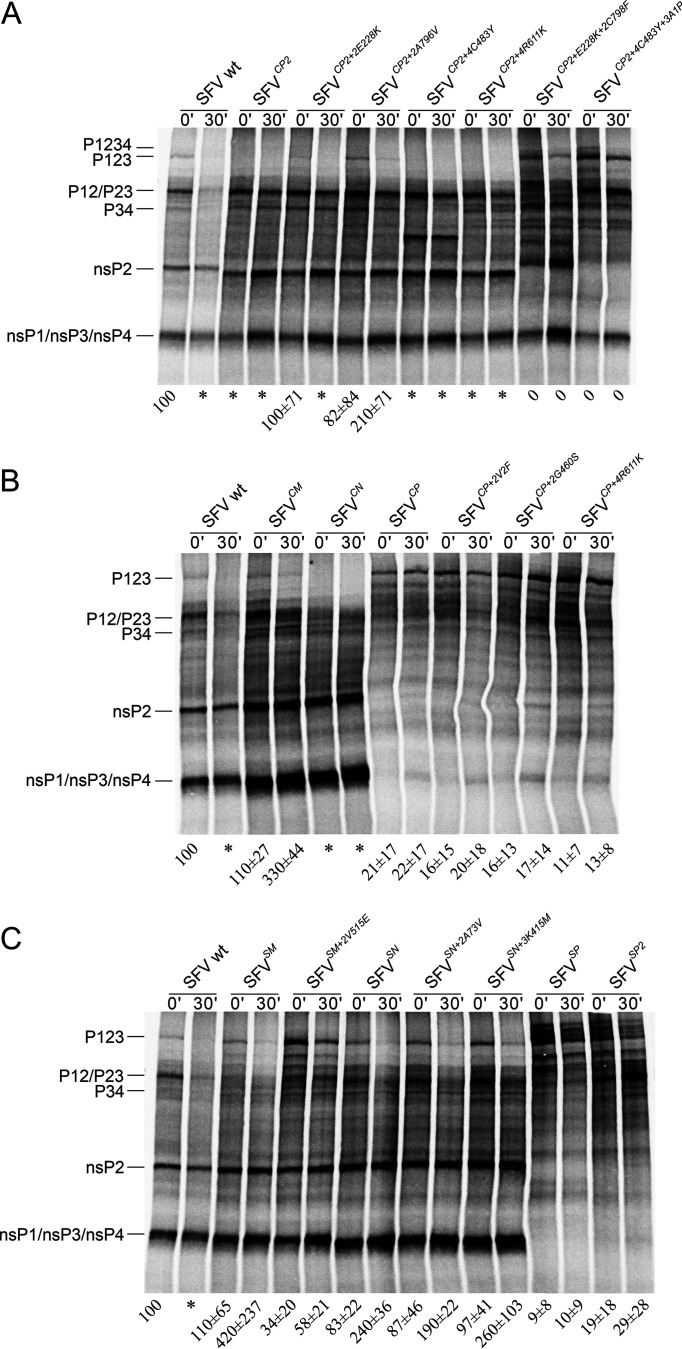
In WT SFV, the P123 and P12/P23/P34 polyproteins, as well as mature nsPs, were observed at the 0′ time point. During the 30-min chase, the amounts of P12/P23/P34 drastically decreased, and P123 became undetectable (Fig. 4). Processing of the 1/2 site of SFVCP2 was faster than that of WT SFV, most likely being the reason for the low infectivity of this construct and the need for adaptive mutations (4). Interestingly, the polyproteins with the mobilities of P12/P23 and P34 were more stable than their counterparts in WT SFV4 (Fig. 4A). As expected, A796V substitution reduced the speed of P123 processing. The E228K substitution, located in a RecA1 domain that is not known to affect the protease activity of nsP2, resulted in a similar, albeit less pronounced, effect. As expected, substitutions in nsP4 had no effect on P1234 processing (Fig. 4A). Adding secondary mutations affecting the 2/3 site to the ns-polyproteins of SFVCP2+2E228K or SFVCP2+4C483Y severely suppressed ns-polyprotein processing; P1234 became detectable, P123 was stabilized, and mature nsP2 could not be detected (Fig. 4A). Lack of release of mature nsP2, a major factor of cytotoxicity of Old World alphaviruses (49, 50), is therefore the most likely reason for the lack of CPE in cells infected with the corresponding viruses. The C798F substitution also resulted in the accumulation of unidentified processing/degradation products, while the A1P substitution resulted in the accumulation of a smaller polyprotein, most likely P23 (Fig. 4A).
FIG 4.
Swapping of the nsP2, nsP2 NTD, nsP2p, or nsP3 macrodomains often results in altered ns-polyprotein processing. In vitro translation was conducted using the TNT SP6 rabbit reticulocyte system, and plasmids containing icDNAs of the indicated viruses were used as templates. After 45 min, the reaction was stopped with cycloheximide, and samples were collected immediately (0′) or after incubation for an additional 30 min (30′). The produced ns-polyproteins and their processing products were separated using SDS-PAGE, and the densities of bands corresponding to nsP2 and P123 were quantified using ImageQuant software. (A) Translation of pSFVCP2 and its variants containing adaptive mutations. (B) Translation of other icDNAs harboring CHIKV-derived sequences. (C) Translation of icDNAs harboring SINV-derived sequences. The positions of P1234, P123, and P12/P23/P34 and those of these mature nsPs are shown. The ratio shows the amount of label that was incorporated into mature nsP2 relative to the amount that was incorporated into unprocessed P123; the nsP2/P123 ratio of WT SFV at 0′ was taken as 100. Each number represents the mean ± SD of the values obtained in three independent experiments. * indicates complete processing of P123 (corresponding density at background level). An additional band visible in samples of SFVCP2+4C483Y most likely represents C-terminally truncated P34. It was detected using only one batch of the TNT SP6 rabbit reticulocyte system and probably represents an artifact generated by premature termination of transcription in the region corresponding to the C483Y substitution.
SFVCM was found to have slightly reduced processing of P123 and somewhat increased stability of the processing intermediate (presumably P23). These effects are consistent with the previously described role of the nsP3 macrodomain in 2/3 site processing (31). We were unable to detect P123 in SFVCN (Fig. 4B). The acceleration of 1/2 site processing, which was probably not prominent enough to inhibit virus rescue (Fig. 1A; Table 1), may explain the attenuated growth of SFVCN (Fig. 1B). In contrast to SFVCP2, the ns-polyprotein of SFVCP was processed very inefficiently; P123 was stabilized, other processing intermediates were present in reduced amounts, mature nsPs were found only in trace amounts, and no significant increase in the nsP2 to P123 ratio was observed during the chase (Fig. 4B). Thus, chimeric nsP2 (Fig. 1A) cannot efficiently cleave the 1/2 site of SFV, and this in turn also inhibits processing of the 2/3 site (5). The V2F and G460S substitutions did not result in detectable changes (Fig. 4B), indicating that they increased SFVCP infectivity (Fig. 2B) by other mechanism(s) and/or that the effect of these mutations on P1234 processing is more prominent in infected cells.
The processing of P1234 of SFVSN was similar to that of WT SFV; the observed small reduction in processing efficiency is not likely to be the cause of the reduced infectivity of SFVSN genomes. Neither of the mutations found in the evolved SFVSN stock had a clear effect on processing (Fig. 4C). The processing of the ns-polyprotein of SFVSM was unique; the nsP2/P123 ratio was similar to that found in WT SFV, but the amount of P12/P23 was drastically reduced (Fig. 4C). As P23 is essential for SFV RC formation (51), the defect in this mutant probably originates from accelerated cleavage of the 2/3 site, an event in which the nsP3 macrodomain has an important role (31). Consistently, adding the V515E substitution to SFVSM not only resulted in a slowdown of P123 processing but also stabilized P12/P23 processing intermediates (Fig. 4C). Finally, swaps resulting in nonviable SFVSP2 and SFVSP (Fig. 1B) were found to inhibit the formation of individual nsPs. In contrast, P1234 was prominent in these mutants, indicating that nsP2 of SINV and a chimeric nsP2 harboring nsP2p of SINV were unable to process the 3/4 site of SFV ns-polyprotein. This property may also be the cause of the lethal phenotype, as cleavage of the 3/4 site is absolutely required for SFV infectivity (40).
Mutations introduced into the 1/2 cleavage site affect the rescue of SFVCP and SFVCP2 by changing ns-polyprotein processing.
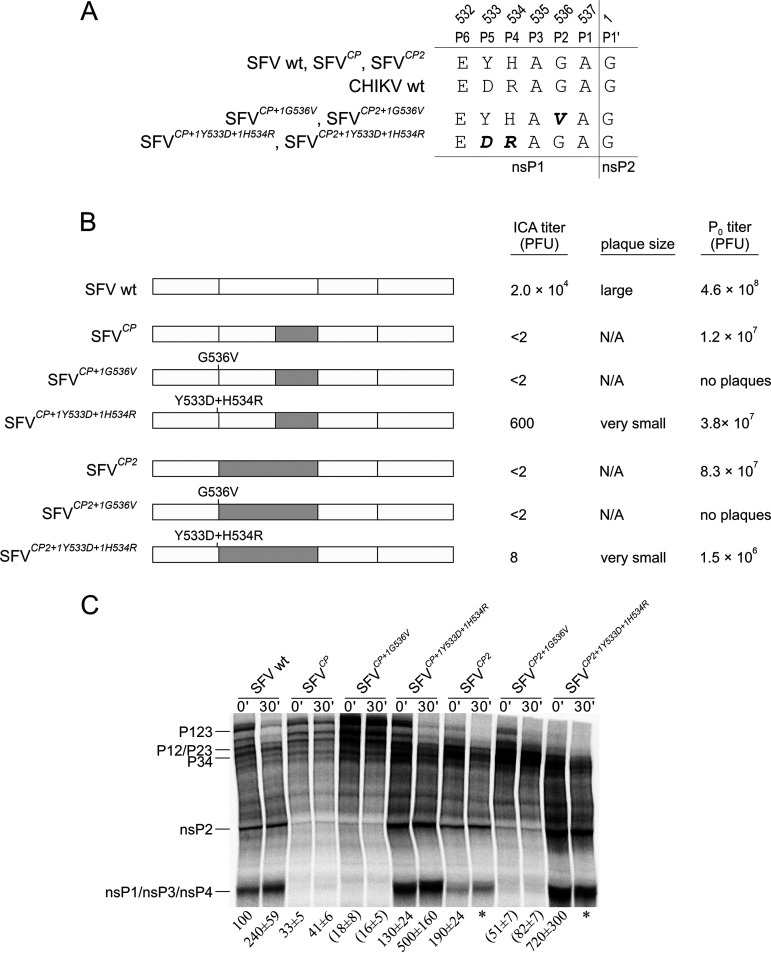
To confirm the link between ns-polyprotein processing and rescue of chimeric viruses, two types of mutations were introduced into the P-side of the 1/2 site in SFVCP and SFVCP2 (Fig. 5A and B; Table 1). In SFVCP+1G536V and SFVCP2+1G536V, the P2 Gly residue was substituted by Ala; this mutation abolishes cleavage of the corresponding site (52). In SFVCP+1Y533D+1H534R and SFVCP2+1Y533D+1H534R, the P5 Tyr and P4 His residues of the SFV 1/2 site were substituted by Asp and Arg, which are found in the 1/2 site of CHIKV. These two changes reintroduce a match between the sequence of the P-side of the 1/2 site and the nsP2p domain (Fig. 5B); they were also expected to accelerate the processing of the 1/2 site due to the introduction of a favorable P4 Arg residue (4, 53).
FIG 5.
Mutations in the P-side of the 1/2 site affect the rescue of SFVCP and SFVCP2 and the processing of the corresponding ns-polyproteins. (A) Sequences of the P6-P1′ regions of the 1/2 sites of CHIKV, SFV, SFVCP, and SFVCP2. Mutations introduced to obtain SFVCP+1G536V, SFVCP+1Y533D+1H534R, SFVCP2+1G536V, and SFVCP2+1Y533D+1H534R are shown in boldface and italics. (B) Localization of swapped regions and introduced mutations in the ns-regions of the chimeric genomes. ICA titers are shown as PFU per 1 μg RNA and represent the averages of the values obtained in three independent experiments; <2 indicates values below the detectable limit. Plaque sizes are indicated as follows: large, >3 mm; very small, <1 mm in diameter. P0 titers are shown as PFU per ml and represent the averages of the values obtained in three independent experiments. The data for WT SFV, SFVCP, and SFVCP2 are replotted from Fig. 1A. (C) Results of in vitro translation and processing. The assay was conducted as described in the legend to Fig. 4. Each number represents the mean ± SD from three independent experiments. * indicates complete processing of P123 (corresponding density at background level); the numbers in parentheses indicate a lack of increase in the amount of product with the same mobility as that of nsP2 during the chase.
ICA revealed no plaque formation by SFVCP+1G536V or SFVCP2+1G536V. Furthermore, in contrast to SFVCP and SFVCP2, no infectious progeny was obtained (Fig. 5B; Table 1). An in vitro translation and processing assay revealed the presence of minor quantities of a protein with an electrophoretic mobility corresponding to that of nsP2. However, the amount of this product did not increase during the chase, and the increase in the nsP2/P123 ratio was due to a decrease in the amount of P123 present, presumably due to slow processing of the 2/3 site (Fig. 5C). These data indicate that processing at the 1/2 site is essential for replication of the virus and that the viability of SFVCP2, which is characterized by accelerated processing of P123 (Fig. 4A; 5C), cannot be restored by blocking the cleavage of the 1/2 site.
Substitution of the P5 and P4 residues increased the rescue of the corresponding viruses. For SFVCP+1Y533D+1H534R, the number of plaques in ICA was greatly increased, and the rescued virus grew to final titers ∼3-fold higher than those of SFVCP (Fig. 5B; Table 1). An in vitro translation and processing assay revealed that, in contrast to selected adaptive mutations, the substitution of P5 and P4 residues accelerated ns-polyprotein processing, which became similar to that in WT SFV (Fig. 5C). These data show that the defect caused by swapping of the nsP2p region can, like other defects resulting from swapping, be rescued by changes in ns-polyprotein processing, confirming processing as one of the key factors in the viability of viruses with swapped genomes. Interestingly, mutations at the P5 and P4 positions resulted in detectable rescue of SFVCP2, although that mutant suffers from accelerated processing of P123 (Fig. 4A and 5C). The effect on the rescue was, however, very minor, as SFVCP2+1Y533D+1H534R formed extremely small plaques that were observed only in some of the independent experiments. Furthermore, the rescued virus grew to titers that were ∼50-fold lower than those of SFVCP2 (Fig. 5B), indicating that further acceleration of 1/2 site processing was unfavorable for the virus. It is therefore plausible that the observed rescue in ICA was not due to the change in 1/2 site processing speed per se; it is more likely that the match between the P-side of the 1/2 site and the nsP2p region allowed better coordination of events leading to the formation of functional replicases that compensated to some degree for the unfavorable acceleration of ns-polyprotein processing.
Swaps and adaptive mutations have an impact on viral RNA synthesis.
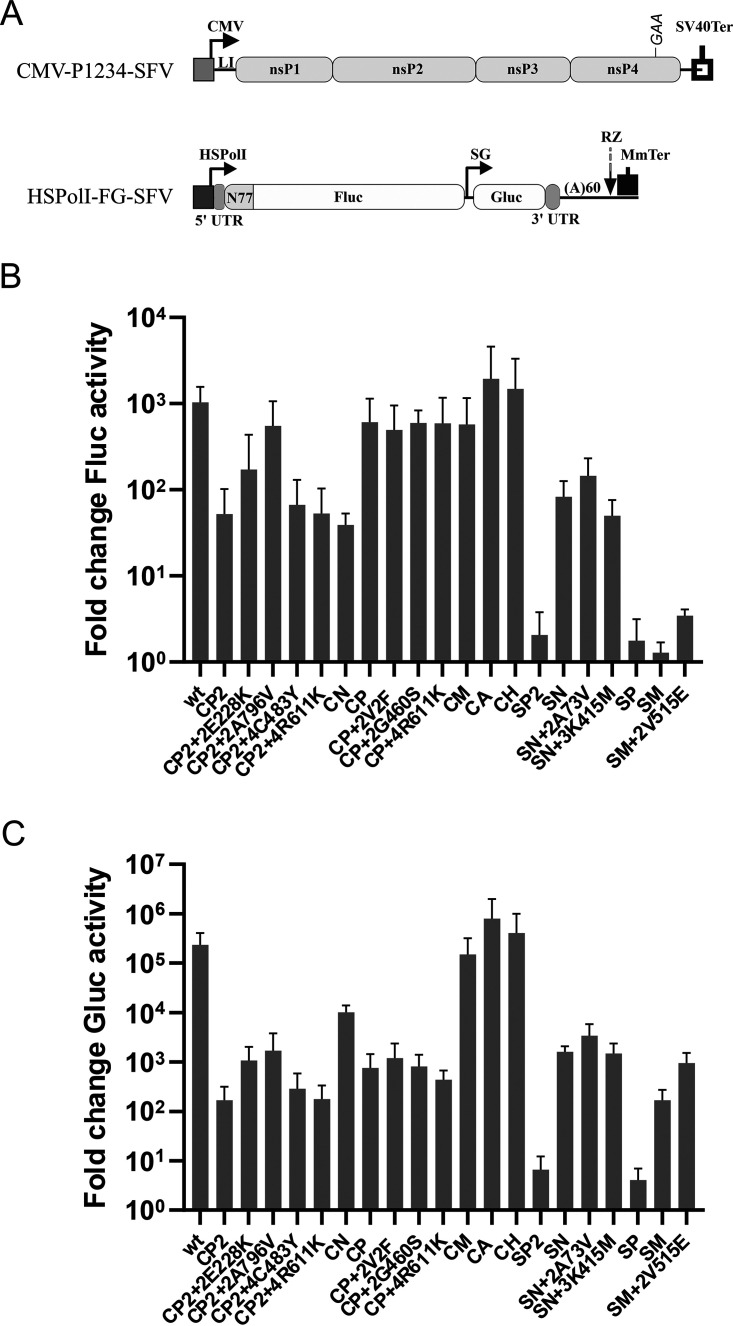
Mutations in the ns-region of the alphavirus genome can alter virus infection by affecting the activity of viral RNA replicase. Hence, a highly efficient SFV trans-replication assay (53) was used to evaluate the effects of swaps and adaptive mutations. In this system, ns-polyprotein is expressed from CMV-P1234-SFV, and template RNA is expressed from HSPolI-FG-SFV plasmids (Fig. 6A). Increases in firefly luciferase (Fluc) and Gaussia luciferase (Gluc) activities in the presence of active replicase serve as proxies for the synthesis of full-length “genomic” RNAs (here referred to as “replication”) and SG RNAs (here referred to as “transcription”), respectively (54).
FIG 6.
Swaps and adaptive mutations introduced into the ns-region of SFV have different impacts on the activity of viral replicase. (A) Organization of the plasmids used for the trans-replication assay. CMV, immediate early promoter of human cytomegalovirus; LI, leader sequence of the herpes simplex virus thymidine kinase gene; SV40Ter, simian virus 40 late polyadenylation region. The position of the inactivating mutation in the nsP4 catalytic site of CMV-P1234GAA-SFV is indicated. HSPolI-FG-SFV contains the following: HSPolI, a truncated (−211 to −1) promoter for human RNA polymerase I; SFV 5′ UTR and partial 3′ UTR; N77, a region encoding the 77 N-terminal amino acid residues of nsP1; SG, the SFV SG promoter; RZ, an antisense-strand ribozyme of the hepatitis delta virus; and MmTer, a terminator for RNA polymerase I in mice. The drawings are not to scale. (B and C) U2OS cells in 12-well plates were cotransfected with 1 μg of HSPolI-FG-SFV and 1 μg of CMV-P1234-SFV or with a plasmid carrying the swaps and point mutations indicated on the horizontal axis. As a negative control, CMV-P1234GAA-SFV, which lacks polymerase activity, was used. The cells were incubated at 37°C and lysed at 16 h p.t. The activities of (B) Fluc, a proxy of replication, and (C) Gluc, a proxy of transcription, in the presence of the analyzed replicases were normalized to those of the P1234GAA controls. The means ± SD of the values obtained in four independent experiments are shown.
The replicases of SFVCM and SFVCA had activities similar to those of WT SFV and SFVCH containing HVD of CHIKV nsP3 (37). Thus, the replacement of any domain of SFV nsP3 with its CHIKV counterpart had no significant negative effect (P > 0.1 in all cases) on viral RNA synthesis (Fig. 6B and C; Table 1). The replicase of SFVCN had significantly reduced replication activity (P < 0.01) and a less pronounced but still significant (P < 0.05) reduction in SG RNA synthesis. Consistent with the lethal phenotype (Fig. 1B) and lack of/strong reduction in 3/4 site processing (Fig. 4C), the activities of SFVSP2 and SFVSP replicases were close to the background level (Fig. 6B and C; Table 1). Taken together, the results of the trans-replication assay accurately reflected the properties of the corresponding viruses and their genomes.
The replication and transcription activities of the SFVSM replicase were notably low, but introduction of the V515M substitution into nsP2 significantly increased both of these activities (P < 0.001 and P < 0.05, respectively) (Fig. 6B and C; Table 1). Thus, adaptation of SFVSM does occur through an increase in the ability of its replicase to synthesize viral RNAs. The ability of SFVSN replicase to replicate template RNAs was similar to that of SFVCN replicase. At the same time, its capacity to synthesize SG RNAs was ⁓6-fold lower, and the difference was statistically significant (P < 0.01) (Fig. 6C). A73V substitution in nsP2 or K415M substitution in nsP3 failed to increase the activities of SFVSN replicase significantly, confirming that these changes are not associated with virus adaptation.
The ability of SFVCP replicase to replicate template RNA was similar to that of WT SFV replicase, and none of the adaptive mutations increased it significantly any further (Fig. 6B). In contrast, the ability of this replicase to produce SG RNAs was significantly reduced (P < 0.05). It was slightly increased by the V2F and G460S substitutions (Fig. 6C; Table 1); however, the differences associated with these mutations were not statistically significant. The most likely reason for these effects is highly inefficient P123 processing (Fig. 4B). Both the replication and the transcription activities of the SFVCP2 replicase were significantly lower (P < 0.01 and P < 0.05, respectively) than those of the WT SFV replicase (Fig. 6B and C). A796V substitution increased both of these activities ⁓10-fold. E228K substitution had a smaller activating effect on both replication and SG RNA synthesis (Fig. 6B and C; Table 1), but neither effect was statistically significant. Despite the lack of statistical significance, the observed trend suggests that both of these mutations activate RNA synthesis, likely by reducing the speed of P123 processing (Fig. 4A). C483Y substitution in nsP4 also resulted in an increase in the transcriptional activity of the SFVCP2 replicase, although the effect was minor (Fig. 6C) and statistically not significant. In contrast, R611K substitution in nsP4 failed to increase any of the activities of the SFVCP2 replicase (Fig. 6B and C; Table 1), indicating that the adaptation of the viruses with this substitution was not due to changes in their replicase activities.
Mutation resulting in the R611K substitution is cis-active and results in increased activity of the SFV SG promoter.
R611K was the only adaptive mutation found in two different swapped viruses (Fig. 2A). It was also the only mutation that rescued the infectivity of the swapped genomes but did not affect ns-polyprotein processing and/or increase the activity of RNA replicase (Fig. 4 and 6). In viruses with the R611K substitution, the nucleotide located at position −1 of the SFV SG promoter is changed from guanine to adenine. A corresponding variation has been found in natural SFV isolates (45), and it was demonstrated that in the context of recombinant SINV, an inserted SFV SG promoter with adenine at position −1 was ∼7-fold more efficient than its counterpart with guanine at that position (55). As nsP2 is a protein that recognizes the SG promoter (56), it was logical to assume that the −1 guanine-to-adenine substitution in the SG promoter found in viruses with swapped nsP2 or nsP2p region represents an adaptation that increases the activity of the SFV SG promoter.
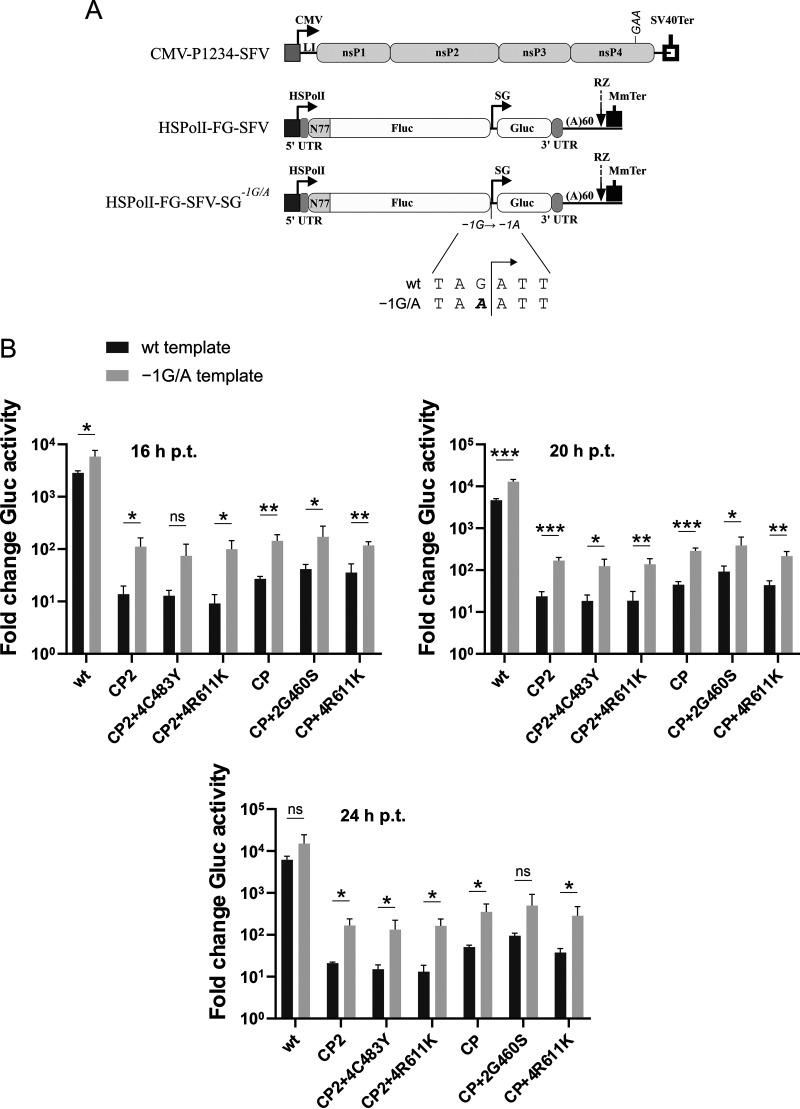
To test this hypothesis, position −1 of the SFV SG promoter in the template construct was changed from guanine to adenine (Fig. 7A). The abilities of WT SFV replicase and of the replicases of SFVCP2, SFVCP2+4C483Y, SFVCP2+4R611K, SFVCP, SFVCP+2G460S, and SFVCP+4R611K to transcribe templates encoded by HSPolI-FG-SFV and HSPolI-FG-SFV-SG−1G/A were compared at 16, 20, and 24 h posttransfection (p.t.). It was observed that the template with the −1 adenine residue always outperformed its counterpart with the −1 guanine residue; at 20 h p.t., these differences were 3- to 7-fold and were statistically significant (Fig. 7B). These results confirm that the R611K mutation or, from the functional point of view, the −1 guanine-to-adenine substitution is a cis-active mutation that increases the activity of the SG promoter of SFV. For this reason, here, the R611K mutation is referred to as the SG(-1)G/A mutation, SFVCP2-4R611K is designated SFVCP2-SG(-1)G/A, and SFVCP2-4R611K is designated SFVCP-SG(-1)G/A.
FIG 7.
The presence of an adenine residue at position −1 of the SG promoter of SFV increases template transcription. (A) Organization of the plasmids used in the trans-replication assay. The substitution of the −1 guanine residue in the SG promoter with adenine, which yielded the HSPolI-FG-SFV-SG−1G/A template-expression plasmid, is shown in the drawing. The other designations are the same as in Fig. 6A. (B) U2OS cells in 12-well plates were cotransfected with 500 ng of HSPolI-FG-SFV or HSPolI-FG-SFV-SG−1G/A and with 500 ng CMV-P1234-SFV (WT) or with a plasmid carrying the swaps and point mutations indicated on the horizontal axis. As a negative control, CMV-P1234GAA-SFV, which lacks polymerase activity, was used. The cells were incubated at 37°C, and aliquots of the culture supernatants were collected at 16, 20, and 24 h p.t. The activities of Gluc produced in the presence of the analyzed replicases were normalized to those of the P1234GAA controls. The means ± SD of three independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (Student’s unpaired t test).
Unlike SFV4, which was used as the backbone in the swapped genomes, CHIKV has a −1 adenine residue in its SG promoter. Therefore, it appeared plausible that the −1 guanine-to-adenine substitution may also act by increasing the affinity of the nsP2 region of CHIKV for the SG promoter in the template RNA. Consistent with this hypothesis, the smallest increase in Gluc activity (from 2-fold at 16 h p.t. to 3-fold at 20 h p.t.) was observed for WT SFV replicase, and the largest increases (from 3-fold at 16 h p.t. to 12-fold at 24 h p.t.) were observed for SFVCP, SFVCP2, and their variants (Fig. 7B). Thus, the transcription activities of replicases harboring nsP2 or nsP2p of CHIKV indeed benefited more from the SG(-1)G/A mutation than did the transcription activity of the WT SFV replicase. However, although this may indeed reflect improvement in the nsP2/SG promoter interaction, the observed differences were relatively small and could also have resulted from other factors such as large differences in the baseline values of Gluc activation (expression from HSPolI-FG-SFV).
Swapping of nsP2p of SFV with nsP2p of CHIKV affects SG promoter activity in SFV replicon vectors and inhibits the formation of virus replicon particles.
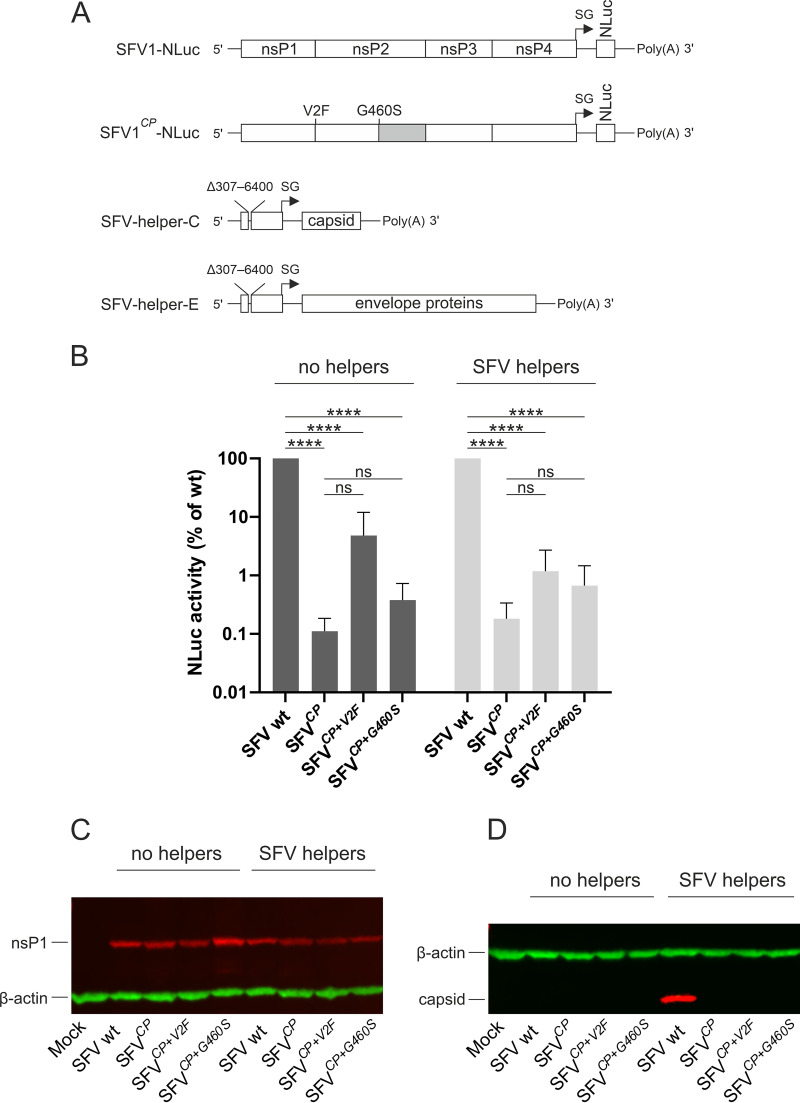
Swapping of nsP2p of SFV with that of CHIKV had minimal impact on the ability of trans-replicase to replicate the RNA template (Fig. 6B) but severely reduced its ability to transcribe the template (Fig. 6C; Table 1). The adaptive V2F and G460S substitutions found in nsP2 of SFVCP only caused small and statistically nonsignificant increases in transcription in the trans-replication assay (Fig. 6C; Table 1). At the same time, these mutations markedly increased the rescue efficiency of the swapped viruses (Fig. 2B), indicating that their impact on self-replicating RNA may be different from their effect on trans-replicase. Furthermore, as nsP2 is likely involved in the packaging of alphavirus genomes and is even included in virions (57), it was hypothesized that the V2F and G460S substitutions may increase the efficiency of virion formation.
To analyze these possibilities, advantage was taken of the availability of self-replicating (replicon) vectors and packaging systems of SFV making it possible to obtain virus replicon particles (VRPs). nsP2p swaps without or with V2F and G460S substitutions were introduced into an SFV replicon vector containing a sequence encoding nanoluciferase (NLuc) under the control of the SG promoter (Fig. 8A). Transfection of BHK-21 cells with the corresponding RNA transcripts revealed that the nsP2p swap drastically (∼900-fold) reduced NLuc expression. The presence of V2F and G460S substitutions resulted in ∼40-fold and ∼3-fold increases, respectively, in NLuc expression (Fig. 8B). Thus, SG promoter-mediated reporter expression was affected in the same manner as was seen in the trans-replicase system (compare Fig. 6C and 8B). Furthermore, similar to the expression of Fluc by the trans-replicase, the nsP2p swap and adaptive mutations did not have a prominent effect on the expression of nsP1, which is translated directly from replicon RNA (Fig. 8C).
FIG 8.
Effect of swapping nsP2p and of adaptive mutations on NLuc expression and replication of SFV replicon vectors. (A) Schematic presentation of the SFV1-NLuc replicon, the chimeric SFV1CP-NLuc replicon, and SFV helper RNAs. Δ307-6400 indicates a deletion in the ns-region of helper RNAs. The adaptive mutations present in SFV1CP+2V2F-NLuc and SFV1CP+2G460S-NLuc replicons are shown. (B) BHK-21 cells were transfected with RNA transcripts corresponding to SP6-SFV1-NLuc (WT), SP6-SFV1CP-NLuc, SP6-SFV1CP+2V2F-NLuc, or SP6-SFV1CP+2G460S-NLuc replicons with or without transcripts corresponding to SFV helper RNAs. At 24 h p.t., the cells were harvested and lysed, and the NLuc activity present in an amount of lysate corresponding to 12,000 cells was measured; the value obtained for cells transfected with SP6-SFV1-Nluc (WT) transcript was taken as 100. The means ± SD of the values obtained in three independent experiments performed in triplicate are shown. ****, P < 0.0001; ns, not significant (Student’s unpaired t test). (C) Lysates of BHK-21 cells transfected with RNA transcripts corresponding to SP6-SFV1-NLuc (WT), SP6-SFV1CP-NLuc, SP6-SFV1CP+2V2F-NLuc, and SP6-SFV1CP+2G460S-NLuc replicons with or without transcripts corresponding to SFV helper RNAs and lysates from mock-transfected control cells were subjected to SDS-PAGE and immunoblot analysis using antibodies against SFV nsP1 (left panel) and SFV capsid protein (right panel). β-actin was used as a loading control. The detection of capsid protein expression was performed three times with similar results; data from one experiment are shown.
To analyze whether nsP2p swaps and adaptive mutations affect VRP formation, BHK-21 cells were cotransfected with replicon and SFV helper RNAs (Fig. 8A). The presence of SFV helper RNAs did not significantly alter NLuc or nsP1 expression (Fig. 8B and C). Infection of naive BHK-21 cells with supernatant harvested from cells cotransfected with SFV helper RNAs and SFV1-NLuc RNA resulted in NLuc activities ∼20,000-fold greater than background levels, indicating effective formation of VRPs. In contrast, no expression of NLuc above the background level was observed for cells infected using supernatants harvested from cells cotransfected with SFV helper-RNAs and transcripts from pSP6-SFV1CP-NLuc, pSP6-SFV1CP+2V2F-NLuc, or pSP6-SFV1CP+2G460S-NLuc. The lack of VRP formation was likely due to the inability of the mutant replicons to activate SG promoter-mediated expression of capsid protein (Fig. 8C) and presumably also the expression of envelope proteins. Due to the lack of expression of structural proteins, it remains unknown whether there was also any specific effect of the mutations on VRP formation. To exclude the possibility that the observed inability to activate the SG promoter in helper RNAs was due to a mismatch between the CHIKV nsP2p region of the chimeric replicon and the SG promoter in SFV helper RNAs, the experiment was repeated using CHIKV helper RNAs. Again, the presence of these helper RNAs had no impact on NLuc reporter expression. No expression of NLuc above the background level was detected in BHK-21 cells infected with any of the harvested supernatants. The reason for this was a complete lack of CHIKV capsid protein expression (data not shown), indicating that none of the replicons used, including SFV1-NLuc, were able to replicate and transcribe CHIKV helper RNAs.
DISCUSSION
In this study, swapping of homologous domains and proteins was used to study the poorly understood functional interactions of alphavirus RC components. Complementary experiments, including analysis of the infectivity and adaptation of the obtained viruses and replicons, processing of ns-polyproteins, and the activities of RNA replicases and the SG promoter, enabled us to obtain unique insights into the process of alphavirus replication.
Sequences obtained from CHIKV were better tolerated by SFV than were their counterparts from SINV (Fig. 1A), indicating a better match between domains originating from more closely related viruses. Thus, replacing the entire nsP2 or nsP2p region with CHIKV sequences resulted in chimeric genomes with reduced infectivity (Fig. 1A) but did not prevent virus rescue and subsequent adaptation. In contrast, swapping of the nsP2 or nsP2p region with the corresponding regions of SINV resulted in noninfectious genomes and nonfunctional replicases (Fig. 1A and 6B and C). However, in a parallel study, an opposite trend was observed; swapping nsP4 of SFV with nsP4 of SINV (sequence identity 74%) increased the activity of the corresponding trans-replicase, whereas the use of nsP4 of CHIKV (sequence identity, 79%) reduced its activity, and replicase harboring P123 of SFV and nsP4 of Ross River virus (RRV; sequence identity, 81%) almost entirely lacked the ability to synthesize SG RNA. These phenotypes were also reflected in the properties of the corresponding recombinant viruses (see the companion article [38]). Thus, the similarity/identity of swapped sequences per se is not the main factor that determines the activity of chimeric RNA replicases of alphaviruses. It is likely that the most important factor is functional matching of the swapped regions; thus, inactivity of obtained chimeric replicases or genomes serves as an indication of the disturbance of crucial function(s). Here, for SFVSP and SFVSP2, the affected function was most likely processing of the 3/4 site (Fig. 4C). We previously observed that in trans, SINV nsP2 can cleave the 2/3 site of SFV but not the 1/2 or 3/4 sites (31). Current data indicate that cleavage of the SFV 3/4 site by SINV protease in cis is also not possible and/or that SINV protease requires the assistance of other SINV protein(s) to cleave at this site.
Chimeric viruses with highly infectious genomes, such as SFVCN, SFVCM, and SFVCA, can replicate without acquiring adaptive mutations. Surprisingly, however, no mutation with beneficial effects was found for any isolate of SFVSN. The P1234 processing of SFVSN was similar to that of WT SFV (Fig. 4C), and its replicase displayed activities higher than those of the replicases of some adapted viruses (Fig. 6B and C). Thus, compromised RNA synthesis was unlikely to be the reason for the low infectivity of SFVSN RNAs (Fig. 1A). Because evidence suggests that there is a functional connection between the nsP2 NTD and capsid protein (4, 44, 58), the capsid protein regions of SFVSN isolates were also sequenced. No mutation was found, leading to the hypothesis that the effect of defects in SFVSN is probably limited to reduced rescue efficiency.
Adaptive changes were found in the genomes of evolved SFVCP2, SFVCP, and SFVSM (Fig. 2A). Each of these substitutions was found in at least two virus isolates; the extreme case was SFVSM, for which all four isolates contained the V515E substitution (Fig. 2A). Other substitutions in nsP2 were located close to its termini, in the RecA1 domain (E228K), or in the connector domain (G460S) of nsP2h (11). The functional defect compensated for by the adaptive mutations was evident for the E228K, A796V, and V515E substitutions, all of which improved the RNA synthesis abilities of the corresponding replicases (Fig. 6B and C). In all three cases, this effect can be connected to the conversion of P1234 processing from abnormally fast to more like that of WT SFV (Fig. 4A and C). Interestingly, all these mutations appear to act by increasing the stability of both P123 and smaller polyprotein(s), presumably including P23. Indeed, in SFVCP, the 2/3 site has been made more favorable for cleavage by a change in the P4 residue of the site from unfavorable threonine (SFV) to favorable arginine (CHIKV). Single substitutions resulting in similar effects are well tolerated by SFV; however, when coupled with accelerated processing at the 1/2 site, they reduce the infectivity of the virus RNA and trigger the accumulation of adaptive mutations (4). It is likely that the SINV macro domain, which is present in SFVSM, also facilitates the processing of the 2/3 site. Thus, these adaptive mutations have likely been selected for to counteract the accelerated processing at these cleavage sites.
Substitution of nsP2p of SFV with that of CHIKV resulted in diminished synthesis of SG RNAs (Fig. 6B and C) and a major defect in P123 processing (Fig. 4B). Surprisingly, however, the adaptive mutations found in nsP2 of SFVCP had no effect on ns-polyprotein processing (Fig. 4A), and their beneficial effect on the synthesis of SG RNAs was minor and statistically not significant (Fig. 6C). It is possible that the ability of chimeric nsP2 to process the 1/2 site cannot be efficiently increased by a single adaptive substitution. However, when substitutions were introduced into both the P5 and P4 positions of the 1/2 site, the WT-like processing of P123 was restored, and the infectivity of the mutant genome was increased (Fig. 5). It is therefore possible that genomes such as that of SFVCP+1Y533D+1H534R remained attenuated and were eliminated during in cellulo adaptation. However, the observation that SFVCP+1Y533D+1H534R was capable of replicating to titers similar to those produced by in cellulo-adapted SFVCP, SFVCP+2V2F, SFVCP+2G460S, and SFVCP-SG(-1)G/A (compare Fig. 2B and 5B) argues against this possibility.
The mechanism of action of the V2F and G460S substitutions apparently includes activation of SG RNA synthesis. This trend was observed for trans-replicase (Fig. 6C) and was more prominent in the context of self-replicating RNA replicons (Fig. 8B). However, the impact of these mutations was relatively mild, and the observed increase in SG RNA-mediated expression did not reach statistical significance. Therefore, additional mechanism(s), such as more efficient use of a limited supply of structural proteins to increase virion production, remain possible. The V2 residue is located in the region of nsP2 that has been shown to be functionally linked to the capsid protein (4). The G460 residue is located in the connector region, and mutations in this region may affect the interaction of the nsP2 domains with each other or with other proteins. As nsP2 is likely involved in the packaging of the alphavirus genome and is even included in virions (57), it is possible that the V2F and G460S substitutions may increase the efficiency of virion formation. Our attempts to investigate this possibility using SFV replicon vectors and their packaging system were, however, unsuccessful, as none of the mutant replicons proved capable of triggering replication and transcription of the provided helper RNAs that are required for the expression of SFV structural proteins (Fig. 8C). Interestingly, SFVCP+2V2F and SFVCP+2G460S were the only mutant viruses that were specifically attenuated in mosquito cells (Fig. 2B). The defect is unlikely to be caused by inefficient processing, as increased P123 stability activates alphavirus RNA replication in mosquito cells (48). Therefore, it is possible that the V2F and G460S substitutions, which were selected in mammalian cells and increase the propagation of SFVCP in these cells, may have different effects in mosquito cells.
The C483Y substitution in nsP4 was first discovered upon passaging CHIKV in the presence of ribavirin or 5-fluorouracil; it increases the fidelity of CHIKV replication and reduces virus fitness in vivo (46). The same selection conditions also selected for the G641D substitution in nsP2, which, in combination with the C483Y substitution, resulted in complete resistance to the mutagenic activity of ribavirin. Interestingly, position 641 of nsP2 of SFV is occupied by a glutamate residue, which is similar to the aspartate residue found in the high-fidelity CHIKV mutant. Furthermore, it was found that RCs isolated from cells infected by CHIKV harboring the C483Y substitution displayed elevated SG RNA synthesis (47). C483Y substitution also resulted in a modest increase in SG RNA synthesis by trans-replicase (Fig. 5C), and this may be one of the mechanisms responsible for the adaptive effect of the C483Y substitution. Even if there are additional mechanism(s) through which the C483Y mutation increases the infectivity of SFVCP2 genomes, our data unequivocally confirm the functional connection between the nsP2 and nsP4 regions.
The majority of isolates of SFVCP2 and all isolates of SFVCP harbor combinations of adaptive mutations (Fig. 2A). Each of these substitutions alone restored the infectivity of the corresponding genome (Fig. 2B); furthermore, SG(-1)G/A was sufficient to increase the replication of SFVCP to a level exceeding that of the evolved stock (Fig. 3B). Why, then, were the combinations of mutations needed? Interestingly, most isolates harbored combinations of mutations that facilitate virus replication by different mechanisms; mutations affecting protease activity (E228K and A796V) were combined with cis-active SG(-1)G/A mutations in the SG promoter and/or with mutations that increase the fidelity and activity of RCs (C483Y). Similarly, a cis-active SG(-1)G/A mutation was also found in combination with mutation(s) that affect the ability of replicase to synthesize SG RNAs (Fig. 2A). Most likely, such combinations confer the greatest advantages for internal and external competition and were therefore selected. A simultaneous rise of several beneficial mutations is improbable. Sequential accumulation of mutations is also unlikely, because the advantages offered by additional mutations are relatively small. It is therefore possible that most of the combinations of adaptive mutations were created by homologous recombination. If this is the case, additional adaptive changes (Table 2) that had negative effects on individual viruses may represent a source for successive adaptation. On its own, SFVCP2+2E228K+2C798F has considerable growth disadvantages. However, viruses harboring such mutations typically lack superinfection exclusion (59, 60). Thus, cells infected by SFVCP2+2E228K+2C798F or similar viruses can be superinfected by other viruses, resulting in mixed infection and providing new possibilities for subsequent adaptation and recombination.
Although the initial aim of this study was to use swapping of functional modules in the SFV replicase to analyze interactions among protein surfaces by discovering compensatory mutations that occur at the interaction hot spots and adjust protein complementarity at the interfaces, we can now conclude that for the studied alphaviruses, such an experimental approach does not lead to this goal. Instead, taken together, the results of this study provide new insight into how alphavirus replicase proteins work together. Our data are consistent with a model in which functional modules in alphavirus replicase function relatively independently and thus are interchangeable. Such an organization of replicases is likely responsible for the robustness of alphavirus genomes. Therefore, even the replacement of the entire multifunctional nsP2, which accounts for ∼32% of the length of ns-polyprotein, was tolerated, and only a few amino acid changes were sufficient to adjust proteolytic homeostasis. Interestingly, we repeatedly detected the same adaptive mutations (e.g., nsP2 V515E, nsP4 C483Y) that were discovered previously through completely different experiments with alphaviruses, leading to the conclusion that these residues are true functionally crucial hot spots. We previously obtained a similar result for the P-side residues of the 1/2 site, which are involved in controlling replicase formation, the sensitivity of alphaviruses to G3BP depletion, and replication efficiency in mosquito cells as well as activation of innate immune responses and pathogenicity in mice (4, 45, 48, 53, 61, 62). Logically, such functional hot spots represent the best starting points for attenuation of alphaviruses (e.g., Mayaro virus) that have not yet been studied at the same level of detail as SFV and CHIKV. In addition, swapping of homologous domains of replicase proteins did result in viruses that remained attenuated, even in the presence of adaptive mutations. Thus, the approach can be used to obtain chimeric attenuated alphaviruses and possibly other positive-strand viruses that represent candidates for live attenuated vaccines.
It has also not escaped our notice that the chimeric virus genomes described in this study and in the companion article (38) may potentially find an application, e.g., in screening for inhibitors that target the replicase components of pathogenic alphaviruses under lower biosafety level (BSL) conditions. For example, instead of using CHIKV to test anti-CHIKV protease inhibitors, an experiment that would require BSL3 conditions, such experiments can be performed using newly developed SFVCP2 or SFVCP variants with viral fitness-improving mutations outside the region of interest [e.g., C483Y in nsP4 or SG(-1)G/A] because the swapped viruses are sufficiently stable and can undergo a full replication cycle; thus, they would faithfully present a target for inhibition and might even allow for the selection of resistance mutations that alter the binding pockets in response to selective pressure from inhibitors. We propose that similar chimeric viruses carrying foreign replication modules can be relatively easily created for other medically important alphaviruses such as RRV, Mayaro virus, and o’nyong’nyong virus, thus avoiding the need to establish a reverse genetics system for each alphavirus. In this case, because the genetic background of SFV is similar to that of these viruses, these chimeras can be used in parallel in screening for potential inhibitors.
Finally, our experiments revealed that alphaviruses use a rather limited set of strategies to survive and adapt; among these, the main strategy is likely downregulation of the efficiency of P1234 processing. In this case, when the corrupted replication process becomes evident, the processing rate decreases, and this likely promotes the production of an excessive number of copies of full-length genomes for prolonged replication instead of the usual switch to transcription of SG RNA culminating in virion production. It is not precisely known whether the fidelity of viral RNA polymerase in the context of early RCs is reduced compared to that of late replicase, but if that is the case, it may also increase the likelihood that mutations will be introduced into the viral RNA, and some of those mutations will subsequently facilitate virus adaptation. Importantly, since the threshold for creating viable chimeric alphaviruses appears to be surprisingly low, it can be expected that chimeric viruses with exchanged replication modules also emerge naturally by recombination, with consequences that are as yet difficult to predict.
MATERIALS AND METHODS
Cell culture.
BHK-21 cells (ATCC, CCL-10) were cultured in Glasgow’s minimal essential medium (GMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; GE Healthcare), 2% tryptose phosphate broth, and 20 mM HEPES (pH 7.2). U2OS cells (ATCC, HTB-96) were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Gibco) supplemented with 10% FBS and 2 mM l-glutamine. Aedes albopictus C6/36 cells were cultured in Leibowitz’s L-15 medium (Corning) with 10% FBS, 10% tryptose phosphate broth, and a 1/100 dilution of a stock of nonessential amino acids (Gibco). All growth media were supplemented with 100 U/ml penicillin and 0.1 mg/ml streptomycin (PAA). Mammalian cells were cultured at 37°C in a humidified incubator with 5% CO2, and C6/36 cells were maintained at 28°C without added CO2.
Construction of icDNA clones, replicons, and plasmids for the trans-replication assay.
All plasmids containing icDNAs of swapped viruses were constructed on the basis of the icDNA clone of pSP6-SFV4 (63); sequences of CHIKV were from the icDNA clone of the LR2006OPY1 strain (ECSA genotype) (64), and sequences of SINV were from the icDNA clone pToto1101 (65). The icDNAs with swaps shown in Fig. 1A were constructed using pSP6-SFV4, synthetic DNAs (GenScript), and standard subcloning procedures. The obtained constructs were designated pSFVCP2, pSFVCN, pSFVCP, pSFVCM, pSFVCA, pSFVSP2, pSFVSN, pSFVSP, and pSFVSM, and the corresponding viruses were SFVCP2, SFVCN, and so on. Variants of pSFVCP2, pSFVCP, pSFVSN, and pSFVSM harboring putative adaptive substitutions (Fig. 2A; Table 1) were obtained using PCR-based mutagenesis and standard cloning procedures; these clones were designated pSFVCP2+1Y533D+1H534R, pSFVCP2+1G536V, pSFVCP2+2E228K, pSFVCP2+2A796V, pSFVCP2+4C483Y, pSFVCP2+4R611K (pSFVCP2-SG(-1)G/A), pSFVCP+1Y533D+1H534R, pSFVCP+1G536V, pSFVCP+2V2F, pSFVCP+2G460S, pSFVCP+4R611K (pSFVCP-SG(-1)G/A), pSFVSN+2A73V, pSFVSN+3K415M, pSFVSM+2V515E, pSFVCP2+2E228K+2C798F, and pSFVCP2+C483Y+3A1P.
The replicons used in this study were based on the pSP6-SFV1 replicon (66). The sequence encoding the NLuc reporter was cloned under the control of the SG promoter (Fig. 8A), resulting in pSP6-SFV1-NLuc. Swaps and adaptive mutations present in pSFVCP, pSFVCP+2V2F, and pSFVCP+2G460S were introduced using standard subcloning procedures; the obtained plasmids were designated pSP6-SFV1CP-NLuc, pSP6-SFV1CP+2V2F-NLuc, and pSP6-SFV1CP+2G460S-NLuc.
The plasmids designated CMV-P1234-SFV, CMV-P1234GAA-SFV, and HSPolI-FG-SFV have been previously described (53); they were used to express active SFV replicase, inactive SFV replicase containing an Asp466–467 to Ala466–467 substitution in the catalytic center of nsP4, and template RNA for SFV replicase harboring Fluc and Gluc markers (Fig. 6A), respectively. Plasmid HSPolI-FG-SFV-SG−1G/A, in which the −1 residue of the SG promoter was changed from guanine to adenine (Fig. 7A), was obtained using site-directed mutagenesis. The swaps and adaptive mutations present in pSFVCP2, pSFVCN, pSFVCP, pSFVCM, pSFVCA, pSFVCH, pSFVSP2, pSFVSN, pSFVSP, pSFVSM, pSFVCP2+2E228K, pSFVCP2+2A796V, pSFVCP2+4C483Y, pSFVCP2+4R611K (pSFVCP2-SG(-1)G/A), pSFVCP+2V2F, pSFVCP+2G460S, pSFVCP+4R611K (pSFVCP-SG(-1)G/A), pSFVSN+2A73V, pSFVSN+3K415M, and pSFVSM+2V515E and in a previously described icDNA clone of SFV harboring HVD of CHIKV (37) were introduced into CMV-P1234-SFV using standard subcloning procedures. The sequences of all constructs were verified by Sanger sequencing.
Infectious center assay (ICA) and virus rescue.
Plasmids containing icDNAs of SFV, swapped genomes, or swapped genomes harboring additional substitutions were linearized by digestion with SpeI, purified and transcribed in vitro using the mMESSAGE mMACHINE SP6 transcription kit (Ambion). BHK-21 cells were then transfected via electroporation (850 V, 25 μF, two pulses in a cuvette with a 4-mm electrode gap) with 10 μg of RNA transcripts as previously described (42). For ICA, 10-fold dilutions of transfected cells were seeded on subconfluent BHK-21 monolayers grown in 6-well cell culture plates. The plates were incubated at 37°C for 3 h; the culture medium was then removed and replaced with GMEM containing 0.8% carboxymethylcellulose (CMC) and 2% FBS. The cells were incubated at 37°C for 72 to 120 h and then stained with crystal violet. The plaques that had formed were counted, and the infectivity was calculated and expressed as PFU per μg RNA. The remaining transfected cells were seeded in 6-well plates and incubated at 37°C. P0 virus stocks were collected when CPE were observed or at 72 h p.t.
Infection, titration, and growth curve analysis.
P1 stocks were obtained as previously described (67). In brief, confluent BHK-21 monolayers in 6-well plates were infected with viral P0 stocks at a multiplicity of infection (MOI) of 0.1; after 1 h, the inoculation medium was removed and replaced with GMEM containing 2% FBS. The cells were incubated at 37°C, and P1 stocks were collected when extensive CPE were observed or at 72 h p.i. C6/36 cells grown in 6-well plates were infected with P0 stocks at an MOI of 5 in L-15 medium without added FBS. After 2 h, the inoculum was replaced with growth medium, and P1 stocks were collected at 24 h p.i.
The titers of all virus stocks were determined in BHK-21 cells using a plaque assay. Briefly, confluent BHK-21 monolayers in 6-well plates were infected with serial 10-fold dilutions of collected virus stocks prepared using GMEM supplemented with 0.2% bovine serum albumin (BSA). The infection mixtures were then removed, and the cells were overlaid with GMEM containing 0.8% CMC and 2% FBS. The plates were incubated at 37°C for 72 h; the cells were then stained with crystal violet, and plaques were counted.
For growth curve analysis, confluent BHK-21 monolayers in 6-well plates were infected with P0 stocks at an MOI of 0.1; the cells were then washed with PBS and incubated in GMEM containing 2% FBS. Immediately after infection (0 h) and at 3 h, 6 h, 9 h, 12 h, 24 h, and 48 h p.i., the entire medium was collected from each well and replaced with fresh GMEM containing 2% FBS. The titers of the obtained samples were determined using a focus-forming assay. In this assay, BHK-21 cell monolayers in 96-well plates were infected with 10-fold dilutions of virus stocks. After 2 h, an overlay of GMEM containing 0.8% CMC and 2% FBS was added. The plates were incubated at 37°C for 24 h and then fixed with ice-cold 80% acetone in PBS at −20°C for 1 h. The liquid was then removed, and the plates were dried fully for 3 h, blocked using Pierce clear milk blocking buffer (Thermo Scientific) in PBS/Tween 20, and probed with primary (in-house) antibodies against the SFV capsid protein. The plates were washed with PBS/0.05% Tween 20, incubated with IRdye800-conjugated secondary antibody (LI-COR), and washed once more. After drying, the signal was visualized using a LI-COR Odyssey Fc imaging system, and foci were counted.
In cellulo adaptation and plaque purification.
Confluent BHK-21 monolayers in 6-well plates were infected with 10-fold dilutions of P0 stocks of SFVCP2, SFVCP, SFVSN, or SFVSM. At 24 h p.i., the cells were monitored for CPE, and medium was collected from cultures in which the cells showed cytopathology and had been infected with the highest dilutions of virus stock. The collected medium was used to infect naive BHK-21 cells as described above. This process was repeated four times, after which the evolved stocks were collected and titers were determined.
Plaque purification was performed as described previously (67). Briefly, collected P1 or in cellulo-evolved stocks were used to infect monolayers of BHK-21 cells grown in 6-well plates at an MOI sufficient to obtain 10 to 20 plaques per well. The infected cells were overlaid with GMEM containing 2% FBS and 0.9% agar (Sigma-Aldrich) and incubated at 37°C. At 120 h p.i., the cells were stained with neutral red, and plaques were picked and placed in a medium that was then used to infect 70% confluent BHK-21 monolayers. At 24 h p.i., the medium was collected and used in subsequent analysis.
RNA extraction, reverse transcription, PCR, and sequencing.
Total RNA was extracted from virus stocks using an RNeasy minikit (Qiagen) and reverse-transcribed with the First-Strand cDNA synthesis kit (Thermo Fisher Scientific). For the detection of mutations selected during in cellulo adaptation of SFVCP2, SFVCP, SFVSN, and SFVSM, a set of PCR fragments covering the complete nonstructural region of the viral genome was obtained and sequenced for four individual plaque-purified isolates for each swapped virus. To detect additional mutations present in P1 stocks of SFVCP2+2E228K, SFVCP2+2A796V, SFVCP2+4C483Y, and SFVCP2+4R611K, the region between nucleotides 1199 and 4312 was reverse transcriptase PCR (RT-PCR) amplified, and the obtained DNA fragments were cloned into the pJet1.2 vector (Thermo Fisher Scientific). For each virus, four of the obtained clones were analyzed using Sanger sequencing.
In vitro translation.
In vitro translation was performed using the TNT SP6 Quick coupled transcription/translation system (Promega); 10 μl of each reaction mixture was supplemented with 4 μCi of [35S] methionine (PerkinElmer), 0.5 μg of icDNA-containing plasmid, and 4 mM dithiothreitol. The reaction mixtures were incubated at 30°C for 45 min, and translation was then stopped by the addition of 1 mM cycloheximide. Each reaction mixture was divided into halves; 10 ng of RNase A was immediately added to the first half, while the second half of the reaction mixture was incubated at 30°C for another 30 min before the addition of RNase A. In each case, after RNase A addition, the mixtures were incubated at 30°C for 5 min, and the proteins in the mixtures were denatured by boiling in Laemmli sample buffer (final concentrations, 50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 100 mM dithiothreitol, and 0.1% bromophenol blue) for 2 min and separated by SDS-PAGE in an 8% gel. The gels were fixed and dried, and signals were visualized using an Amersham Typhoon IP biomolecular imager (GE Healthcare). The densities of bands corresponding to nsP2 and P123 were quantified using ImageQuant software (GE Healthcare), and the ratios of label incorporated into nsP2 to label incorporated into P123 were calculated.
Trans-replication assay.
Trans-replication assays were performed as previously described (54). Briefly, U2OS cells grown in 12-well plates were cotransfected with 1 μg of HSPolI-FG-SFV and 1 μg of CMV-P1234-SFV or its variant containing swaps and adaptive mutations using Lipofectamine LTX and the Plus reagent kit (Thermo Fisher Scientific). Control cells were transfected with 1 μg of HSPolI-FG-SFV and 1 μg of CMV-P1234GAA-SFV. At 16 h p.t., the cells were collected and lysed, and the activities of the Fluc and Gluc reporters were quantified using the dual-luciferase reporter assay system and a GloMax 20/20 luminometer (Promega).
In experiments designed to analyze the role of the −1 residue of the SG promoter in SG RNA synthesis, U2OS cells in 24-well plates were cotransfected with 500 ng of HSPolI-FG-SFV or HSPolI-FG-SFV-SG−1G/A and 500 ng of CMV-P1234-SFV or its variant containing swaps and adaptive mutations using Lipofectamine LTX and the Plus reagent kit (Thermo Fisher Scientific). Control cells were transfected with 500 ng of HSPolI-FG-SFV or HSPolI-FG-SFV-SG−1G/A and 500 ng of CMV-P1234GAA-SFV. At 16, 20, or 24 h p.t., aliquots of the growth medium were collected, and the activity of the Gluc reporter was quantified using the Renilla luciferase assay system and a GloMax 20/20 luminometer (Promega).
Expression of NLuc by SFV replicon vectors and analysis of VRP formation.
Two-helper systems for SFV and CHIKV have been described previously (68, 69). pSP6-SFV1-NLuc, pSP6-SFV1CP-NLuc, pSP6-SFV1CP+2V2F-NLuc, pSP6-SFV1CP+2G460S-NLuc, and plasmids for SFV helper RNAs were linearized with SpeI, and plasmids for CHIKV helper RNAs were linearized with NotI. DNA was purified and transcribed in vitro using an mMESSAGE mMACHINE SP6 transcription kit (Ambion). Then, 7.5 × 105 BHK-21 cells were transfected via electroporation (850 V, 25 μF, two pulses in a cuvette with a 4-mm electrode gap) with 50 μg of RNA transcripts corresponding to replicons with or without transcripts corresponding to helper RNAs (50 μg of each) of SFV or CHIKV. The transfected cells were seeded in 12-well plates; after incubation of the plates at 37°C for 24 h, the cells were harvested and used in Western blot analysis (see below) and NLuc activity measurement. For transfections performed using helper RNA, supernatants containing VRPs were harvested and clarified by centrifugation at 1,000 × g for 10 min. Aliquots (150 μl) of the obtained VRP stocks were used to infect BHK-21 cells grown in 12-well plates. The cells were washed with PBS, incubated in GMEM containing 2% FBS, and harvested at 24 h p.i. To measure NLuc activity, cells infected with VRPs or transfected with replicon RNAs were lysed. An amount of lysate corresponding to 12,000 cells was used to measure NLuc activity using the Renilla luciferase assay system and a GloMax 20/20 luminometer (Promega).
Western blotting.
Cells transfected using in vitro transcripts of pSP6-SFV1-NLuc, pSP6-SFV1CP-NLuc, pSP6-SFV1CP+2V2F-NLuc, or pSP6-SFV1CP+2G460S-NLuc with or without in vitro transcripts corresponding to SFV or CHIKV helper RNAs were harvested at the same time as the samples used for NLuc activity measurement. After washing with 1 ml of PBS, the cells were lysed and boiled, and proteins were separated by SDS-PAGE in 10% or 12% gels for detection of nsP1 and capsid protein, respectively. The separated proteins were transferred to polyvinylidene difluoride membranes and detected using antibodies against SFV nsP1, SFV capsid protein, CHIKV capsid protein (all in-house), and β-actin (sc-47778; Santa Cruz Biotechnology). After incubation with appropriate secondary antibodies conjugated to fluorescent labels (LI-COR), the proteins on the membrane were imaged using the LI-COR Odyssey Fc imaging system.
Statistical analysis.
Each experiment was repeated to obtain a minimum of 3 independent biological repeats. Statistical analysis was performed using GraphPad Prism 8.2.0 software. The data were analyzed using Student’s unpaired one-tailed t test. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
The authors are grateful to Mark A. Kay for providing the pMC.BESPX vector.
This work was supported by the European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering, Estonia (2014-2020.4.01.15-013), by basic funding from the Institute of Technology, by institutional research funding (IUT20-27), and by a program grant (PRG1154) from the Estonian Research Council to A.M. and an Estonian Science Foundation grant 9421 to V.L. The funders had no role in the study design, data collection, or interpretation or in the decision to submit the work for publication.
We declare no conflicts of interest.
Contributor Information
Aleksei Lulla, Email: al840@cam.ac.uk.
Andres Merits, Email: andres.merits@ut.ee.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Waggoner JJ, Pinsky BA. 2015. How great is the threat of chikungunya virus? Expert Rev Anti Infect Ther 13:291–293. 10.1586/14787210.2015.995634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola T, Couderc T, Courderc T, Ng LFP, Hallengärd D, Powers A, Lecuit M, Esteban M, Merits A, Roques P, Liljeström P. 2015. Therapeutics and vaccines against chikungunya virus. Vector Borne Zoonotic Dis 15:250–257. 10.1089/vbz.2014.1681. [DOI] [PubMed] [Google Scholar]
- 3.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lulla V, Karo-Astover L, Rausalu K, Saul S, Merits A, Lulla A. 2018. Timeliness of proteolytic events is prerequisite for efficient functioning of the alphaviral replicase. J Virol 92:e00151-18. 10.1128/JVI.00151-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasiljeva L, Merits A, Golubtsov A, Sizemskaja V, Kääriäinen L, Ahola T. 2003. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem 278:41636–41645. 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 6.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96:2483–2500. 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones R, Bragagnolo G, Arranz R, Reguera J. 2021. Capping pores of alphavirus nsP1 gate membranous viral replication factories. Nature 589:615–619. 10.1038/s41586-020-3036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K, Law Y-S, Law MCY, Tan YB, Wirawan M, Luo D. 2021. Structural insights into viral RNA capping and plasma membrane targeting by Chikungunya virus nonstructural protein 1. Cell Host Microbe 29:757–764.e3. 10.1016/j.chom.2021.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Das PK, Merits A, Lulla A. 2014. Functional cross-talk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity. J Biol Chem 289:5635–5653. 10.1074/jbc.M113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasiljeva L, Valmu L, Kääriäinen L, Merits A. 2001. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J Biol Chem 276:30786–30793. 10.1074/jbc.M104786200. [DOI] [PubMed] [Google Scholar]
- 11.Law Y-S, Utt A, Tan YB, Zheng J, Wang S, Chen MW, Griffin PR, Merits A, Luo D. 2019. Structural insights into RNA recognition by the Chikungunya virus nsP2 helicase. Proc Natl Acad Sci USA 116:9558–9567. 10.1073/pnas.1900656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narwal M, Singh H, Pratap S, Malik A, Kuhn RJ, Kumar P, Tomar S. 2018. Crystal structure of chikungunya virus nsP2 cysteine protease reveals a putative flexible loop blocking its active site. Int J Biol Macromol 116:451–462. 10.1016/j.ijbiomac.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Russo AT, White MA, Watowich SJ. 2006. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 14:1449–1458. 10.1016/j.str.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Law Y-S, Wang S, Tan YB, Shih O, Utt A, Goh WY, Lian B-J, Chen MW, Jeng U-S, Merits A, Luo D. 2020. Interdomain flexibility of Chikungunya virus nsP2 helicase-protease differentially influences viral RNA replication and infectivity. J Virol 95:e01470-20. 10.1128/JVI.01470-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham R, Hauer D, McPherson RL, Utt A, Kirby IT, Cohen MS, Merits A, Leung AKL, Griffin DE. 2018. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc Natl Acad Sci USA 115:E10457–E10466. 10.1073/pnas.1812130115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPherson RL, Abraham R, Sreekumar E, Ong S-E, Cheng S-J, Baxter VK, Kistemaker HAV, Filippov DV, Griffin DE, Leung AKL. 2017. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc Natl Acad Sci USA 114:1666–1671. 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham R, McPherson RL, Dasovich M, Badiee M, Leung AKL, Griffin DE. 2020. Both ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain affect neurovirulence in mice. mBio 11:e03253-19. 10.1128/mBio.03253-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayabalan AK, Adivarahan S, Koppula A, Abraham R, Batish M, Zenklusen D, Griffin DE, Leung AKL. 2021. Stress granule formation, disassembly, and composition are regulated by alphavirus ADP-ribosylhydrolase activity. Proc Natl Acad Sci USA 118:e2021719118. 10.1073/pnas.2021719118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin G, Yost SA, Miller MT, Elrod EJ, Grakoui A, Marcotrigiano J. 2012. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci USA 109:16534–16539. 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, Goonawardane N, Ward J, Tuplin A, Harris M. 2019. Multiple roles of the non-structural protein 3 (nsP3) alphavirus unique domain (AUD) during Chikungunya virus genome replication and transcription. PLoS Pathog 15:e1007239. 10.1371/journal.ppat.1007239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meshram CD, Agback P, Shiliaev N, Urakova N, Mobley JA, Agback T, Frolova EI, Frolov I. 2018. Multiple host factors interact with the hypervariable domain of Chikungunya virus nsP3 and determine viral replication in cell-specific mode. J Virol 92:e00838-18. 10.1128/JVI.00838-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vihinen H, Saarinen J. 2000. Phosphorylation site analysis of Semliki forest virus nonstructural protein 3. J Biol Chem 275:27775–27783. 10.1074/jbc.M002195200. [DOI] [PubMed] [Google Scholar]
- 23.Teppor M, Žusinaite E, Merits A. 2021. Phosphorylation sites in the hypervariable domain in Chikungunya virus nsP3 are crucial for viral replication. J Virol 95:e02276-20. 10.1128/JVI.02276-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzon M, Castro C, Thaa B, Liu L, Mutso M, Liu X, Mahalingam S, Griffin JL, Marsh M, McInerney GM. 2018. Alphavirus-induced hyperactivation of PI3K/AKT directs pro-viral metabolic changes. PLoS Pathog 14:e1006835. 10.1371/journal.ppat.1006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, Frolova EI. 2016. New World and Old World alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog 12:e1005810. 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meertens L, Hafirassou ML, Couderc T, Bonnet-Madin L, Kril V, Kümmerer BM, Labeau A, Brugier A, Simon-Loriere E, Burlaud-Gaillard J, Doyen C, Pezzi L, Goupil T, Rafasse S, Vidalain P-O, Bertrand-Legout A, Gueneau L, Juntas-Morales R, Ben Yaou R, Bonne G, de Lamballerie X, Benkirane M, Roingeard P, Delaugerre C, Lecuit M, Amara A. 2019. FHL1 is a major host factor for chikungunya virus infection. Nature 574:259–263. 10.1038/s41586-019-1578-4. [DOI] [PubMed] [Google Scholar]
- 27.Meshram CD, Phillips AT, Lukash T, Shiliaev N, Frolova EI, Frolov I. 2020. Mutations in hypervariable domain of Venezuelan equine encephalitis virus nsP3 protein differentially affect viral replication. J Virol 94:e01841-19. 10.1128/JVI.01841-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mutso M, Morro A, Smedberg C, Kasvandik S, Aquilimeba M, Teppor M, Tarve L, Lulla A, Lulla V, Saul S, Thaa B, McInerney G, Merits A, Varjak M. 2018. Mutation of CD2AP and SH3KBP1 binding motif in alphavirus nsP3 hypervariable domain results in attenuated virus. Viruses 10:226. 10.3390/v10050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuvonen M, Kazlauskas A, Martikainen M, Hinkkanen A, Ahola T, Saksela K. 2011. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog 7:e1002383. 10.1371/journal.ppat.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukash T, Agback T, Dominguez F, Shiliaev N, Meshram C, Frolova EI, Agback P, Frolov I. 2020. Structural and functional characterization of host FHL1 protein interaction with hypervariable domain of Chikungunya virus nsP3 protein. J Virol 95:e01672-20. 10.1128/JVI.01672-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lulla A, Lulla V, Merits A. 2012. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J Virol 86:553–565. 10.1128/JVI.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rausalu K, Utt A, Quirin T, Varghese FS, Žusinaite E, Das PK, Ahola T, Merits A. 2016. Chikungunya virus infectivity, RNA replication and non-structural polyprotein processing depend on the nsP2 protease’s active site cysteine residue. Sci Rep 6:37124. 10.1038/srep37124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J Virol 81:5046–5057. 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martikainen M, Niittykoski M, von und zu Fraunberg M, Immonen A, Koponen S, van Geenen M, Vähä-Koskela M, Ylösmäki E, Jääskeläinen JE, Saksela K, Hinkkanen A. 2015. MicroRNA-attenuated clone of virulent Semliki Forest virus overcomes antiviral type I interferon in resistant mouse CT-2A glioma. J Virol 89:10637–10647. 10.1128/JVI.01868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuittila MT, Santagati MG, Roytta M, Maatta JA, Hinkkanen AE. 2000. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J Virol 74:4579–4589. 10.1128/jvi.74.10.4579-4589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxton-Shaw KD, Ledermann JP, Borland EM, Stovall JL, Mossel EC, Singh AJ, Wilusz J, Powers AM. 2013. O’nyong nyong virus molecular determinants of unique vector specificity reside in non-structural protein 3. PLoS Negl Trop Dis 7:e1931. 10.1371/journal.pntd.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thaa B, Biasiotto R, Eng K, Neuvonen M, Götte B, Rheinemann L, Mutso M, Utt A, Varghese F, Balistreri G, Merits A, Ahola T, McInerney GM. 2015. Differential phosphatidylinositol-3-kinase-Akt-mTOR activation by Semliki Forest and Chikungunya viruses is dependent on nsP3 and connected to replication complex internalization. J Virol 89:11420–11437. 10.1128/JVI.01579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lello LS, Bartholomeeusen K, Wang S, Coppens S, Fragkoudis R, Ariën KK, Utt A. 2021. nsP4 is a major determinant of alphavirus replicase activity and template selectivity. J Virol 95:e00355-21. doi: 10.1128/JVI.00355-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nasar F, Gorchakov RV, Tesh RB, Weaver SC. 2015. Eilat virus host range restriction is present at multiple levels of the virus life cycle. J Virol 89:1404–1418. 10.1128/JVI.01856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lulla A, Lulla V, Tints K, Ahola T, Merits A. 2006. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J Virol 80:5413–5422. 10.1128/JVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen R, Mukhopadhyay S, Merits A, Bolling B, Nasar F, Coffey LL, Powers A, Weaver SC, ICTV Report Consortium . 2018. ICTV virus taxonomy profile: Togaviridae. J Gen Virol 99:761–762. 10.1099/jgv.0.001072. [DOI] [PubMed] [Google Scholar]
- 42.Utt A, Das PK, Varjak M, Lulla V, Lulla A, Merits A. 2015. Mutations conferring a noncytotoxic phenotype on chikungunya virus replicons compromise enzymatic properties of nonstructural protein 2. J Virol 89:3145–3162. 10.1128/JVI.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Groot RJ, Hardy WR, Shirako Y, Strauss JH. 1990. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J 9:2631–2638. 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lulla V, Kim DY, Frolova EI, Frolov I. 2013. The amino-terminal domain of alphavirus capsid protein is dispensable for viral particle assembly but regulates RNA encapsidation through cooperative functions of its subdomains. J Virol 87:12003–12019. 10.1128/JVI.01960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saul S, Ferguson M, Cordonin C, Fragkoudis R, Ool M, Tamberg N, Sherwood K, Fazakerley JK, Merits A. 2015. Differences in processing determinants of nonstructural polyprotein and in the sequence of nonstructural protein 3 affect neurovirulence of Semliki Forest virus. J Virol 89:11030–11045. 10.1128/JVI.01186-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coffey LL, Beeharry Y, Borderia AV, Blanc H, Vignuzzi M. 2011. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci USA 108:16038–16043. 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stapleford KA, Rozen-Gagnon K, Das PK, Saul S, Poirier EZ, Blanc H, Vidalain P-O, Merits A, Vignuzzi M. 2015. Viral polymerase-helicase complexes regulate replication fidelity to overcome intracellular nucleotide depletion. J Virol 89:11233–11244. 10.1128/JVI.01553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartholomeeusen K, Utt A, Coppens S, Rausalu K, Vereecken K, Ariën KK, Merits A. 2018. A Chikungunya virus trans-replicase system reveals the importance of delayed nonstructural polyprotein processing for efficient replication complex formation in mosquito cells. J Virol 92:e00152-18. 10.1128/JVI.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akhrymuk I, Kulemzin SV, Frolova EI. 2012. Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol 86:7180–7191. 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hellström K, Kallio K, Utt A, Quirin T, Jokitalo E, Merits A, Ahola T. 2017. Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. J Virol 91:e00787-17. 10.1128/JVI.00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golubtsov A, Kääriäinen L, Caldentey J. 2006. Characterization of the cysteine protease domain of Semliki Forest virus replicase protein nsP2 by in vitro mutagenesis. FEBS Lett 580:1502–1508. 10.1016/j.febslet.2006.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Götte B, Utt A, Fragkoudis R, Merits A, McInerney GM. 2020. Sensitivity of alphaviruses to G3BP deletion correlates with efficiency of replicase polyprotein processing. J Virol 94:e01681-19. 10.1128/JVI.01681-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Utt A, Rausalu K, Jakobson M, Männik A, Alphey L, Fragkoudis R, Merits A. 2019. Design and use of Chikungunya virus replication templates utilizing mammalian and mosquito RNA polymerase I-mediated transcription. J Virol 93:e00794-19. 10.1128/JVI.00794-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hertz JM, Huang HV. 1992. Utilization of heterologous alphavirus junction sequences as promoters by Sindbis virus. J Virol 66:857–864. 10.1128/JVI.66.2.857-864.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suopanki J, Sawicki DL, Sawicki SG, Kääriäinen L. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J Gen Virol 79:309–319. 10.1099/0022-1317-79-2-309. [DOI] [PubMed] [Google Scholar]
- 57.Schuchman R, Kilianski A, Piper A, Vancini R, Ribeiro JMC, Sprague TR, Nasar F, Boyd G, Hernandez R, Glaros T. 2018. Comparative characterization of the Sindbis virus proteome from mammalian and invertebrate hosts identifies nsP2 as a component of the virion and sorting Nexin 5 as a significant host factor for alphavirus replication. J Virol 92:e00694-18. 10.1128/JVI.00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim DY, Atasheva S, Frolova EI, Frolov I. 2013. Venezuelan equine encephalitis virus nsP2 protein regulates packaging of the viral genome into infectious virions. J Virol 87:4202–4213. 10.1128/JVI.03142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehrengruber MU, Goldin AL. 2007. Semliki Forest virus vectors with mutations in the nonstructural protein 2 gene permit extended superinfection of neuronal and non-neuronal cells. J Neurovirol 13:353–363. 10.1080/13550280701393204. [DOI] [PubMed] [Google Scholar]
- 60.Sawicki DL, Perri S, Polo JM, Sawicki SG. 2006. Role for nsP2 proteins in the cessation of alphavirus minus-strand synthesis by host cells. J Virol 80:360–371. 10.1128/JVI.80.1.360-371.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X, Mutso M, Utt A, Lepland A, Herrero LJ, Taylor A, Bettadapura J, Rudd PA, Merits A, Mahalingam S. 2018. Decreased virulence of Ross River virus harboring a mutation in the first cleavage site of nonstructural polyprotein is caused by a novel mechanism leading to increased production of interferon-inducing RNAs. mBio 9:e00044-18. 10.1128/mBio.00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan Y, Teo T, Utt A, Tan JJ, Amrun SN, Abu Bakar F, Yee W, Becht E, Lee CY, Lee B, Rajarethinam R, Newell E, Merits A, Carissimo G, Lum F, Ng LF. 2019. Mutating chikungunya virus non‐structural protein produces potent live‐attenuated vaccine candidate. EMBO Mol Med 11:e10092. 10.15252/emmm.201810092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liljeström P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol 65:4107–4113. 10.1128/JVI.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pohjala L, Utt A, Varjak M, Lulla A, Merits A, Ahola T, Tammela P. 2011. Inhibitors of alphavirus entry and replication identified with a stable Chikungunya replicon cell line and virus-based assays. PLoS One 6:e28923. 10.1371/journal.pone.0028923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polo JM, Davis NL, Rice CM, Huang HV, Johnston RE. 1988. Molecular analysis of Sindbis virus pathogenesis in neonatal mice by using virus recombinants constructed in vitro. J Virol 62:2124–2133. 10.1128/JVI.62.6.2124-2133.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liljeström P, Garoff H. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (N Y) 9:1356–1361. 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 67.Lulla V, Karo-Astover L, Rausalu K, Merits A, Lulla A. 2013. Presentation overrides specificity: probing the plasticity of alphaviral proteolytic activity through mutational analysis. J Virol 87:10207–10220. 10.1128/JVI.01485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gläsker S, Lulla A, Lulla V, Couderc T, Drexler J, Liljeström P, Lecuit M, Drosten C, Merits A, Kümmerer B. 2013. Virus replicon particle based Chikungunya virus neutralization assay using Gaussia luciferase as readout. Virol J 10:235. 10.1186/1743-422X-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smerdou C, Liljeström P. 1999. Two-helper RNA system for production of recombinant Semliki Forest virus particles. J Virol 73:1092–1098. 10.1128/JVI.73.2.1092-1098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]