ABSTRACT
Interferon-induced transmembrane (IFITM) proteins inhibit a broad range of enveloped viruses by blocking entry into host cells. We used an inducible overexpression system to investigate if IFITM1, IFITM2, and IFITM3 could modulate early and/or late stages of influenza A virus (IAV) or parainfluenza virus 3 (PIV-3) infection in human A549 airway epithelial cells. IAV and PIV-3 represent respiratory viruses which utilize distinct cellular entry pathways. We verify entry by endocytosis for IAV, whereas PIV-3 infection was consistent with fusion at the plasma membrane. Following induction prior to infection, all three IFITM proteins restricted the percentage of IAV-infected cells at 8 hours postinfection. In contrast, prior induction of IFITM1 and IFITM2 did not inhibit PIV-3 infection, although a modest reduction was observed with IFITM3. Small interfering RNA (siRNA)-mediated knockdown of endogenous IFITM1, IFITM2, and IFITM3 expression, in the presence or absence of pretreatment with type I interferon, resulted in increased IAV, but not PIV-3, infection. This finding suggests that while all three IFITMs display antiviral activity against IAV, they do not restrict the early stages of PIV-3 infection. IAV and PIV-3 infection culminates in viral egress through budding at the plasma membrane. Inducible expression of IFITM1, IFITM2, or IFITM3 immediately after infection did not impact titers of infectious virus released from IAV- or PIV-3-infected cells. Our findings show that IFITM proteins differentially restrict the early stages of infection of two respiratory viruses with distinct cellular entry pathways but do not influence the late stages of replication for either virus.
IMPORTANCE Interferon-induced transmembrane (IFITM) proteins restrict the initial stages of infection for several respiratory viruses; however, their potential to modulate the later stages of virus replication has not been explored. In this study, we highlight the utility of an inducible overexpression system to assess the impact of IFITM proteins on either early- or late-stage replication of two respiratory viruses. We demonstrate antiviral activity by IFITM1, IFITM2, and IFITM3 against influenza A virus (IAV) but not parainfluenza virus 3 (PIV-3) during the early stages of cellular infection. Furthermore, IFITM induction following IAV or PIV-3 infection does not restrict the late stages of replication of either virus. Our findings show that IFITM proteins can differentially restrict the early stages of infection of two viruses with distinct cellular entry pathways and yet do not influence the late stages of replication for either virus.
KEYWORDS: IFITM, host restriction factors, influenza, parainfluenza virus
INTRODUCTION
Members of the interferon-induced transmembrane protein (IFITM) family are among the most widely studied interferon-stimulated genes (ISGs), with potent antiviral activity reported for a wide range of enveloped viruses, including several respiratory viruses (1–3). The mechanism of IFITM3-mediated restriction of influenza A virus (IAV) in epithelial cells is one of the most well-defined mechanisms for any virus to date (4–8). Specifically, localization of endogenous IFITM3 protein in the cellular endocytic compartment prevents IAV fusion with the endosomal membrane (6) and is thought to accelerate the trafficking of endosomal cargo to lysosomes for degradation (9). IFITM3 has also been identified as a major restriction factor controlling the early stages of IAV replication in immune cell subsets (5, 10–12). Hence, IFITM3 appears to be a potent restriction factor for viruses, including IAV, that utilizes the endocytic route for infectious entry (reviewed in reference 13).
In humans, there are a total of five IFITM family members, of which only IFITM1, IFITM2, and IFITM3 are induced by interferon (IFNs) (type I and II) (1). In addition to IFITM3, both IFITM1 and IFITM2 have also been reported to possess antiviral activity against a broad range of viruses, including IAV (reviewed in references 1 and 2). By virtue of its localization to the cell surface, IFITM1 is proposed play an important role in restricting viruses that enter via direct fusion with the plasma membrane, although the mechanism of antiviral action has not been defined (2, 14, 15). IFITM2 is also expressed within the cellular endocytic compartment like IFITM3, and both proteins display a high degree of similarity suggesting there could be some redundancy in antiviral action (1, 2, 16).
The orthomyxovirus IAV and paramyxovirus parainfluenza virus 3 (PIV-3) are two respiratory viruses that utilize distinct cellular entry pathways and replicate in different subcellular compartments during the early stages of infection. However, IAV and PIV-3 are likely to share common cellular processes relating to the egress of enveloped viruses at the late stages of replication (13, 17–19). Despite many studies exploring the impact of overexpression of IFITM1, IFITM2, and IFITM3 on restriction of the early stages of IAV infection by all three proteins (4), conflicting conclusions about the relative importance of IFITM1 and IFITM2 restriction have arisen from recent studies using the same IAV strain. Overexpression of IFITM1 and IFITM3, but not IFITM2, reduced IAV (A/PR/8/34, H1N1) infection as assessed by green fluorescent protein (GFP) reporter expression in Vero cells (14), whereas IFITM3 strongly inhibited, IFITM2 inhibited to a lesser extent, and IFITM1 had little effect on A/PR/8/34 infection when overexpressed in human HEK293T cells (20). Knockdown of endogenous IFITM2 and IFITM3 in HeLa cells enhanced IAV nucleoprotein (NP) expression following A/PR/8/34 infection, but there was no additional increase when IFITM1 was also knocked down, also suggesting IFITM1 does not play a significant antiviral role (9). Moreover, studies investigating the importance of IFITM proteins in the control of PIV-3 infection are limited to two reports using overexpression methods (14, 21). Specifically, overexpression of IFITM1 and IFITM3 but not IFITM2 in Vero cells was shown to limit the early stages of PIV-3 infection reflected in the reduced expression of a GFP reporter (14). Similarly, IFITM1 overexpression in HEK293 cells led to reduced PIV-3 hemagglutinin-neuraminidase (HN) protein expression, but IFITM3 displayed antiviral activity against PIV-3 only when the protein was localized to the plasma membrane through mutation of the tyrosine (Y20) in the YEML sorting signal (21). The antiviral role of IFITM2 was not investigated in this study. Therefore, the systematic comparison and conclusive validation of the role of IFITM1 and IFITM2, alongside IFITM3, in controlling the early stages of IAV and PIV-3 infection are warranted.
There is evidence to suggest that IFITM proteins are also able to exert antiviral activity on the late stages of virus infection. For example, IFITM1, IFITM2, and IFITM3 have been reported to antagonize the envelope glycoprotein of HIV-1 to reduce cell-to-cell infection (22). In addition, IFITM incorporation into the envelope of HIV-1, and a spectrum of other enveloped viruses, is proposed to reduce the infectivity of released viral particles (23, 24). Little is known about the influence of IFITM antiviral activity on the late stages of viral replication for respiratory viruses.
Here, we used a doxycycline (DOX)-inducible system to examine the impact of IFITM overexpression prior to or following infection of A549 cells with either IAV or PIV-3. Our findings clearly demonstrate differential antiviral activity by IFITM proteins against two respiratory viruses with distinct infectious entry pathways in A549 airway epithelial cells. IFITM1, IFITM2, and IFITM3 have the capacity to control the early stages of IAV infection following endocytic entry but do not appear to be important in the restriction of PIV-3 infection occurring via fusion at the plasma membrane. In contrast to other enveloped viruses, IFITM proteins do not influence the late stages of infection for either IAV or PIV-3.
RESULTS
IAV infection of A549 cells occurs via pH-dependent endocytosis, whereas PIV-3 infection occurs independently of endocytosis.
Following engagement with sialic acid-bearing receptors, IAV enters epithelial cells via clathrin-mediated endocytosis in a dynamin- and pH-dependent manner (25). These entry mechanisms are consistent with the potent restriction of IAV replication by host factors that localize to endocytic compartments, such as IFITM3 (5, 6). Conversely, it is widely accepted that most paramyxoviruses, including PIV-3, enter cells in a pH-independent manner through fusion at the plasma membrane following engagement with sialic acid-containing receptors (26–28). However, alternative entry routes, including dynamin-dependent endocytosis, have been shown for some members of the paramyxovirus family (29). We verified the infectious entry requirements for both IAV and PIV-3 in the A549 airway epithelial cell line before exploring the potential antiviral capacity of IFITM proteins in these cells.
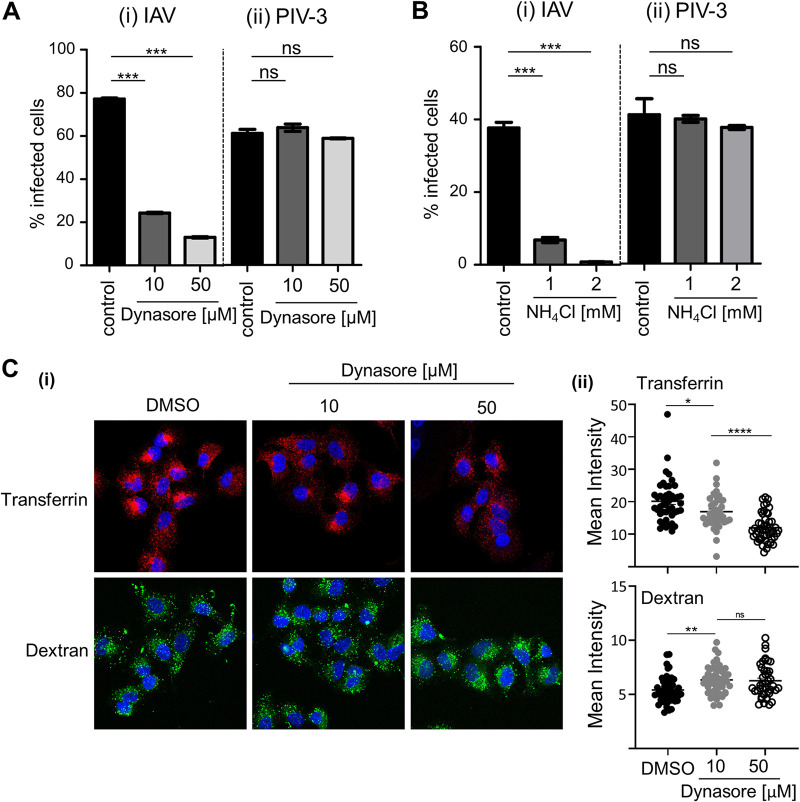
Using flow cytometry, IAV infection of A549 cells was assessed by identification of nucleoprotein (NP)-positive cells at 8 hours postinfection (hpi). As seen in Fig. 1Ai, infection was significantly blocked by pretreatment with 10 μM dynasore to prevent dynamin-dependent entry. Similarly, IAV infection of A549 cells was inhibited in the presence of 1 mM NH4Cl (Fig. 1Bi) to prevent endosome acidification, consistent with pH-dependent entry. In contrast, there was no significant reduction in the percentage of PIV-3 hemagglutinin-neuraminidase (HN)-positive cells at 18 hpi when A549 cells were pretreated with 10 μM or 50 μM dynasore (Fig. 1Aii) or infected with PIV-3 in the presence of 1 mM or 2 mM NH4Cl (Fig. 1Bii). These data are consistent with infectious entry of PIV-3 into A549 cells via fusion at the plasma membrane in a dynamin- and pH-independent manner. Internalization of transferrin (endocytic ligand) was significantly blocked using 10 μM or 50 μM dynasore (Fig. 1Ci and ii). Conversely, high-molecular-weight dextran (macropinocytic ligand) was not blocked by dynasore, and we observed a slight but significant increase in dextran uptake (Fig. 1Ci and ii). These data confirm specific inhibition of dynamin-dependent endocytosis at the concentrations of dynasore shown to prevent IAV infection.
FIG 1.
Pathways of infectious entry of IAV and PIV into A549 cells. Monolayers of A549 cells were pretreated with dynasore prior to infection with IAV (MOI, 1) or PIV-3 (MOI, 2) (A) or pretreated and infected in the presence of NH4Cl with IAV (MOI, 1) or PIV-3 (MOI, 2) (B). The percentage of infected cells was determined by quantitation of IAV NP-positive cells at 8 hpi or PIV-3 HN-positive cells at 18 hpi using flow cytometry. Data from 1 of 3 independent experiments performed in triplicate is shown. (C) Alexa Fluor-647-conjugated transferrin (red) or FITC-conjugated high-molecular-weight (MW) dextran (70,000) (green) was used to validate the specificity of dynasore in blocking endocytic entry of transferrin into A549 cells via fluorescence microscopy. (C) (i) Representative merged images, including DAPI staining for the nucleus (blue) are shown. ×63 magnification. (C) (ii) Mean intensity of fluorescent signal for transferrin or dextran was measured across the cell area (area mean) using ImageJ. Each data point represents the mean fluorescence intensity of one cell (n ≥ 40 for each treatment). (A, B, and C) Error bars = SEM. (****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05; significance by one-way analysis of variance [ANOVA] with Tukey’s multiple comparative analysis).
Generation and characterization of A549 cell lines with inducible overexpression of human IFITM1, IFITM2, and IFITM3 proteins.
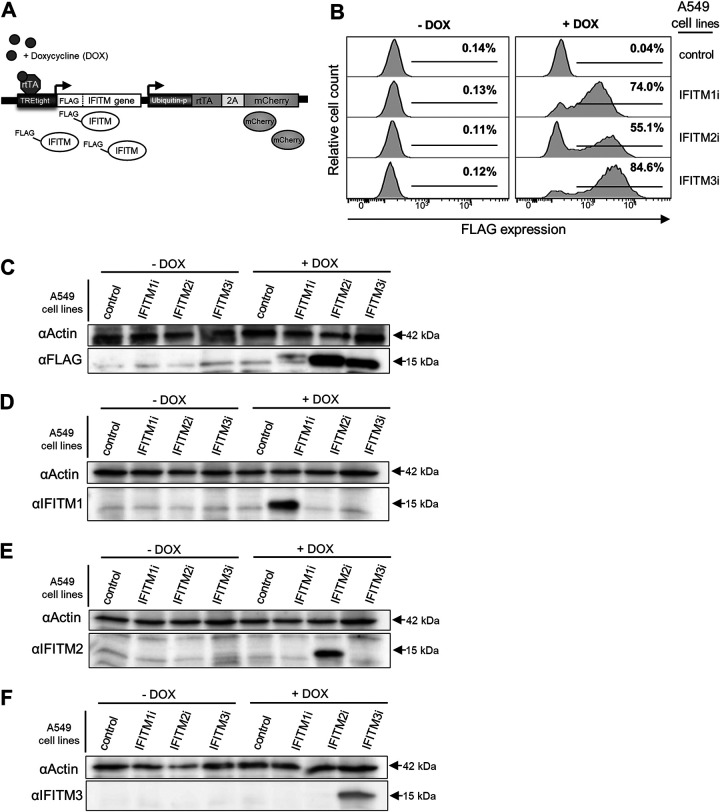
To investigate the role of human IFITM proteins as restriction factors during the early and late stages of respiratory virus infection, A549 cells were transduced using a lentiviral system to allow for inducible overexpression of IFITM1, IFITM2, and IFITM3 proteins, of which each were engineered to possess an N-terminal FLAG tag (Fig. 2A). Flow cytometry showed that in the absence of doxycycline (DOX), there was little evidence of IFITM protein expression (detected by staining for FLAG) in A549 cells relative to cells expressing a control protein that lacked the FLAG epitope (Fig. 2B). In contrast, 24 hours following the addition of DOX, expression of IFITM1, IFITM2, and IFITM3 was readily observed demonstrating robust expression. Quantitation of IFITM mRNA by reverse transcriptase PCR (RT-PCR; using primers designed to detect both endogenous and overexpressed IFITM1, IFITM2, and IFITM3) demonstrated that expression levels of IFITM1, IFITM2, and IFITM3 mRNA following DOX induction were comparable to the levels of endogenous IFITM1, IFITM2, and IFITM3 mRNA induced in parental A549 cells by IFN-α treatment (data not shown).
FIG 2.
Characterization of A549 cells with inducible IFITM1, IFITM2, or IFITM3 overexpression. (A) Lentiviral transduction was used to generate A549 cell lines with DOX-inducible expression of IFITM1, IFITM2, or IFITM3 proteins, each with an N-terminal FLAG tag (IFITM1i, IFITM2i, and IFITM3i cell lines). Constitutive expression of mCherry was used to determine transduction efficiency during the generation of each cell line. Control A549 cells express an irrelevant inducible protein without a FLAG-tag. (B) Flow cytometric analysis of FLAG expression in IFITM1i, IFITM2i, and IFITM3i A549 cell lines following culture for 24 hours in media supplemented with (+DOX) or without (−DOX) doxycycline. (C, D, E, and F) IFITMs were induced in A549 cells by DOX treatment for 24 hours, and expression of each specific IFITM (approximately 15 kDa in size) in cell lysates was confirmed by Western blotting. Proteins were detected using anti-FLAG antibody (C) or antibodies specific for IFITM1 (D), IFITM2 (E), or IFITM3 (F). Beta-actin (42 kDa) was used as a protein loading control.
Western blot analysis of IFITM-A549 cell lysates showed detectable FLAG-tagged proteins of the expected size for each respective IFITM protein (approximately 15 kDa) following induction via DOX treatment for 24 hours (Fig. 2C). IFITM1, IFITM2, and IFITM3 exhibit a high degree of similarity (1), with the highest amino acid sequence identity between IFITM2 and IFITM3 (30). The inducible expression of the IFITM proteins of the predicted molecular weight in the transduced A549 cells was then confirmed by Western blotting with antibodies specific for IFITM1, IFITM2, and IFITM3, respectively (Fig. 2D to F).
Distinct subcellular distribution patterns following induction of IFITM1, IFITM2, and IFITM3 protein expression in A549 cells.
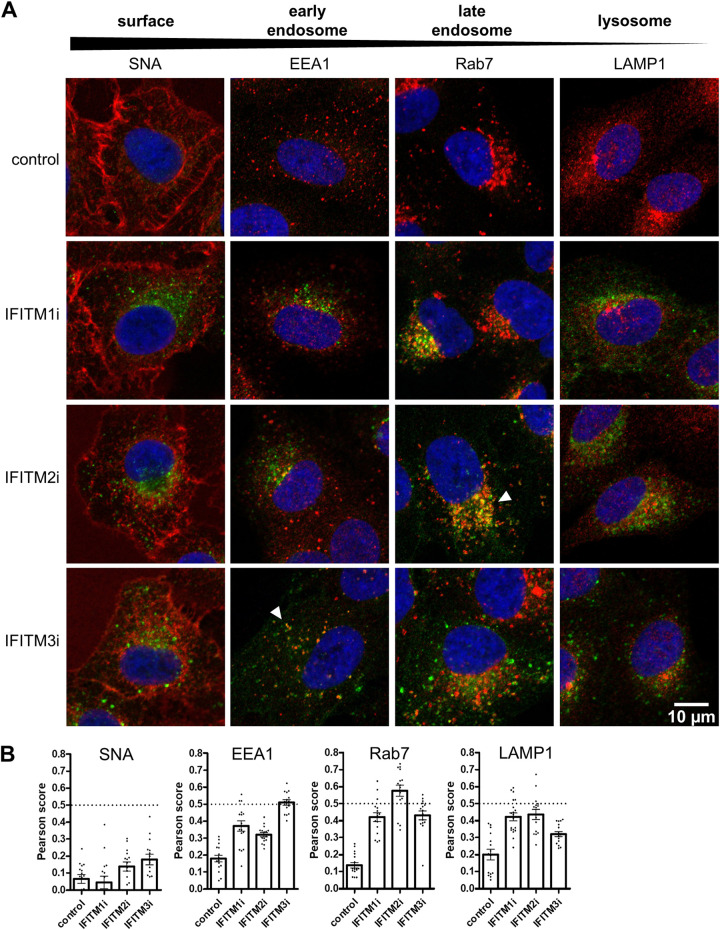
To extend our analyses of inducible IFITM protein expression in A549 cells, the subcellular distribution of each induced protein was assessed by immunofluorescence microscopy. The localization of each IFITM protein was visualized by anti-FLAG staining (Fig. 3A). Sambucus nigra agglutinin (SNA) lectin was used as a marker to delineate the cell surface, while intracellular endosomal compartments were detected using antibodies to EEA1 (early endosomes), Rab7 (late endosomes), and LAMP1 (lysosomes) (Fig. 3A). IFITM1, IFITM2, and IFITM3 showed distinct patterns of subcellular distribution. IFITM2 colocalized with Rab7-positive late endosomes (Fig. 3A, white arrows; Fig. 3B), whereas IFITM3 partially colocalized with EEA1-positive early endosomes (Fig. 3A, white arrows; Fig. 3B). These findings are consistent with previous reports describing the localization of endogenous (5, 9) and overexpressed (31–33) IFITM2 and IFITM3 to the endocytic compartment of epithelial cells. The localization of IFITM1 was largely distinct from that of IFITM2 and IFITM3, exhibiting reduced colocalization with either EEA1, Rab7, or LAMP1 (Fig. 3B).
FIG 3.
Localization of inducible IFITM1, IFITM2, and IFITM3 proteins expressed in A549 cells. Control, IFITM1i, IFITM2i, or IFITM3i A549 cells were cultured in the presence of DOX for 24 hours to induce IFITM expression before cells were fixed and stained with an anti-FLAG antibody in conjunction with Alexa Fluor-488-conjugated antibody (green). (A) Cellular compartments were visualized by staining with SNA (cell surface), EEA1 (early endosomes), Rab7 (late endosomes), or LAMP1 (lysosomes) antibodies in conjunction with Alexa Fluor-647-conjugated antibody (red). White arrows indicate areas of colocalization between FLAG staining and SNA, EEA1, Rab7, or LAMP1 staining. Nuclei were stained with DAPI (blue). Images were acquired at ×63 magnification. (B) The Pearson’s coefficient (R) value was calculated between signals in the green and red channels (from at least 15 cells for each sample). The dotted line at 0.5 represents the threshold for colocalization. Mean values are presented as a bar graph, and error bars represent SEM. Control A549 cells express an irrelevant inducible protein without a FLAG-tag.
Inducibility and stability of IFITM1, IFITM2, and IFITM3 protein expression in A549 cells.
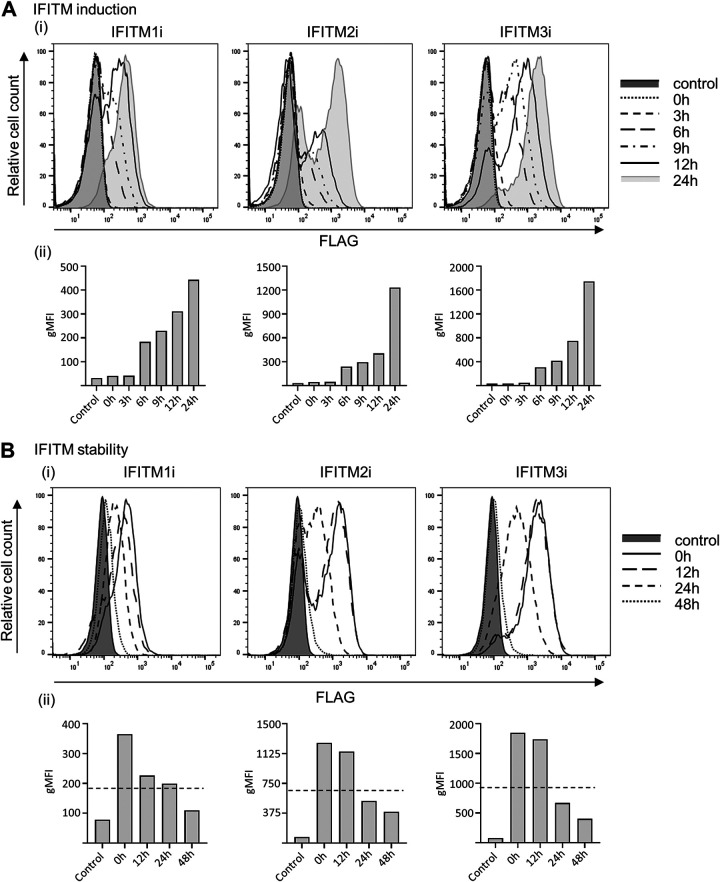
We characterized the kinetics of IFITM induction and the stability of IFITM expression following DOX induction in A549 cells to establish conditions that would enable antiviral activity to be assessed at early and late stages of IAV and PIV-3 infection. Following DOX treatment, IFITM protein expression was first detected in A549 cells at 3 to 6 hours, increased markedly by 8 to 12 hours, and reached peak levels at 24 hours postinduction (Fig. 4Ai and ii). The stability of IFITM protein expression was also assessed at various time points after cells were washed and then cultured in DOX-free media. Expression of IFITM2 and IFITM3 proteins was stable for 12 hours but had declined markedly (i.e., below half-maximal expression) by 24 hours. In contrast, IFITM1 protein levels had dropped markedly at 12 hours, but more gradually thereafter, reaching half-maximal expression after 24 hours (Fig. 4Bi and ii). All IFITM proteins reached levels similar to those of background controls at 48 h after removal of DOX-containing media (Fig. 4Bi and ii).
FIG 4.
Kinetics of induction and stability of IFITM1, IFITM2, and IFITM3 expression in A549 cell lines. (A and B) FLAG expression in IFITM1i, IFITM2i, and IFITM3i A549 cell lines at different times (0 to 24 hours) post-DOX induction (IFITM induction) (A) and different times after withdrawal of DOX, where cells were DOX induced for 24 hours and then washed and cultured in DOX-free media (0 to 48 hours) (IFITM stability) (B). For both (A) and (B), representative histograms from 2 independent experiments are displayed in (i) and the geometric mean fluorescent intensity (gMFI) of each histogram is graphed in (ii). The dotted-line in (Bii) represents half-maximal IFITM expression.
Induction of IFITM1, IFITM2, and IFITM3 protein expression confers potent restriction of IAV, but not PIV-3, during the early stages of infection.
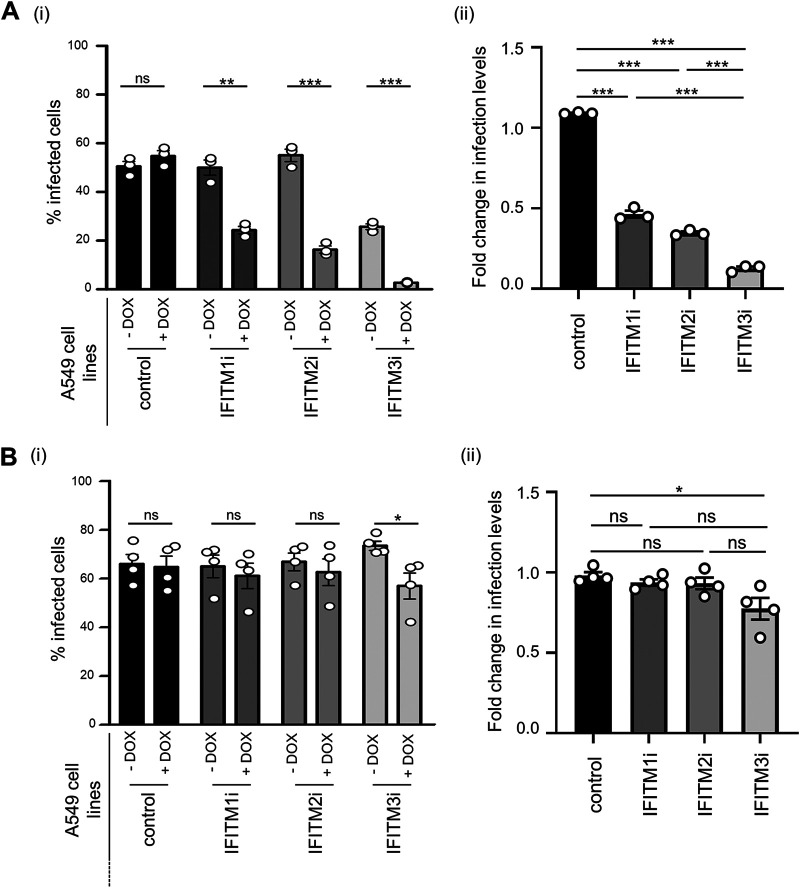
A549 cell lines with inducible IFITM1, IFITM2, and IFITM3 expression were used to investigate each IFITM protein for potential antiviral activity against IAV or PIV-3. Following DOX induction of IFITM expression, A549 cells were infected with IAV or PIV-3 and assessed for susceptibility to infection via flow cytometry using antibodies to detect newly synthesized IAV NP 8 hpi or PIV-3 HN at 18 hpi (Fig. 5). First, A549 control cells showed a similar percentage of IAV-infected cells when cultured in the presence or absence of DOX (Fig. 5Ai). As expected, DOX-inducible expression of IFITM3 resulted in a significant reduction in the percentage of IAV NP-positive cells (Fig. 5Ai). Early IAV infection was also restricted by the inducible expression of either IFITM1 or IFITM2 (Fig. 5Ai). When compared as a fold change in infection between the DOX-induced and uninduced condition within each cell type (Fig. 5Aii), IFITM3-mediated restriction of IAV was more potent in our system than restriction mediated by IFITM2 or IFITM1. This conclusion is supported by a 2.4-, 3.2-, and 8.6-fold reduction in infection following DOX-inducible expression of IFITM1, IFITM2, or IFITM3, respectively.
FIG 5.
Impact of inducible IFITM protein expression on IAV and PIV-3 infection of A549 cells. (A and B) A549 cells cultured for 24 hours in the absence (−DOX) or presence (+DOX) of doxycycline to induce IFITM expression were subsequently infected with IAV (MOI, 1) (A) or PIV-3 (MOI, 0.8) (B). (i) The percentage of virus-infected cells was determined by staining for IAV NP at 8 hpi or PIV-3 HN protein at 18 hpi, followed by flow cytometry. (***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05; unpaired, two-tailed t test). (ii) Fold change in the percentage of virus-infected cells in the presence or absence of each IFITM. (***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05; significance by one-way analysis of variance [ANOVA] with Tukey’s multiple comparative analysis). For IAV, data are representative of three independent experiments performed in triplicate. For PIV-3, data are pooled from four independent experiments performed in triplicate. Error bars represent the SEM. Control A549 cells express an irrelevant inducible protein.
In contrast, we observed no significant reduction in the percentage of PIV-3 HN-positive cells following DOX-inducible expression of IFITM1 or IFITM2 (Fig. 5Bi); however, a modest but significant reduction in PIV-3-infected cells was noted following induction of IFITM3 (Fig. 5Bi). Similarly, a modest but significant difference was noted when examining the fold reduction in PIV-3 infection of IFITM3-expressing cells in the DOX-induced and uninduced conditions (Fig. 5Bii). Overall, our data indicate that prior induction of IFITM1, IFITM2, or IFITM3 resulted in potent inhibition of the early stages of IAV infection, but only inducible IFITM3 mediated a modest but significant inhibition of early PIV-3 infection.
siRNA knockdown of endogenous IFITM expression in the presence or absence of type I IFN treatment increases susceptibility of A549 cells to IAV, but not PIV-3, infection.
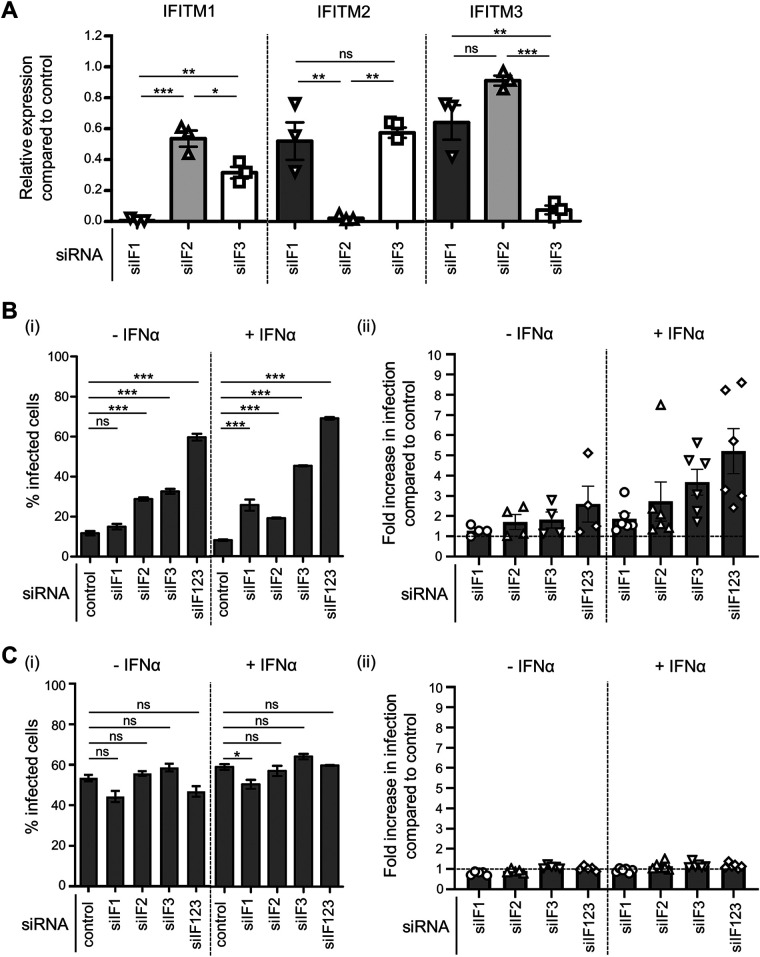
We extended our investigation to incorporate siRNA silencing of endogenous IFITM1, IFITM2, and IFITM3 expression in A549 cells, prior to infection with IAV or PIV-3. Our rationale for these experiments was if IFITM1, IFITM2, or IFITM3 were bona fide restriction factors for either virus, their silencing would lead to enhanced infection. Given the high sequence similarity between the IFITM1, IFITM2, and IFITM3 genes, it was essential to demonstrate effective and specific siRNA-mediated silencing of each target IFITM in A549 cells. To this end, we transfected A549 cells with siRNA targeting IFITM1, IFITM2, or IFITM3, as well as a nontargeting control siRNA. The potency and specificity of the knockdown were assessed by reverse transcriptase quantitative PCR (RT-qPCR) analyses of mRNA levels in A549 cells for each IFITM (Fig. 6A). All three IFITM-targeting siRNAs, namely, siIF1 (IFITM1 siRNA), siIF2 (IFITM2 siRNA), and siIF3 (IFITM3 siRNA) potently downregulated expression of their respective targets compared with the nontargeting siRNA-treated control (Fig. 6A). Moreover, siIF1 treatment significantly reduced IFITM1 mRNA expression compared with siIF2 treatment (P ≤ 0.001) and siIF3 treatment (P ≤ 0.05), siIF2 treatment significantly reduced IFITM2 mRNA expression compared with siIF1 treatment (P ≤ 0.01) and siIF3 treatment (P ≤ 0.01), and siIF3 treatment significantly reduced IFITM3 mRNA expression compared with siIF1 treatment (P ≤ 0.01) and siIF2 treatment (P ≤ 0.001) (Fig. 6A). Of note, siIF3 did lead to a modest but significant reduction in IFITM1 mRNA expression compared with siIF2 treatment (Fig. 6A).
FIG 6.
siRNA knockdown of endogenous IFITM1, IFITM2, and IFITM3 in A549 cells. (A) Quantitation of IFITM1, IFITM2, and IFITM3 mRNA in A549 cells treated with siRNA targeting IFITM1 (siIF1), IFITM2 (siIF2), or IFITM3 (siIF3) or nontargeting siRNA (control). A549 cells were transfected with 5 nM siRNA for 72 hours, including during overnight treatment with IFN-α prior to RNA isolation. Expression of each IFITM gene was normalized to the expression of GAPDH. The relative expression levels of IFITM1, IFTIM2, and IFITM3 mRNA in siIF-treated A549 cells compared with those of control-siRNA treated A549 cells are shown. Data are pooled from three independent experiments performed in triplicate. (Bi) A549 cells were treated with siIF1, siIF2, siIF3, pooled siRNA (siIF123), or control siRNA and 48 hours later were incubated in the presence (+IFN-α) or absence (−IFN-α) of IFN-α for an additional 18 to 24 hours. After being washed, cells were then infected with IAV at an MOI of 1 in the absence of IFN-α or at an MOI of 10 in the presence of IFN-α, such that similar baseline levels of infection were obtained in the presence or absence of IFN-α. The percentage of infected cells was determined by staining with anti-IAV NP at 8 hpi. Data are representative of four independent experiments performed in triplicate. Error bars depict SEM. (***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05; significance by one-way analysis of variance [ANOVA] with Tukey’s multiple comparative analysis). (Bii) Fold increase in IAV infection between A549 cells treated with IFITM-specific siRNA compared with infection levels in cells treated with control siRNA (dotted line). Data were pooled from four independent experiments performed in triplicate. Error bars represent SEM. (Ci) A549 cells were treated with siIF1, siIF2, siIF3, pooled siRNA (siIF123), or control siRNA and 48 hours later were incubated in the presence (+IFN-α) or absence (−IFN-α) of IFN-α for an additional 18 to 24 hours. After washing, cells were then infected with PIV-3 at an MOI of 0.8 in the absence of IFN-α or at an MOI of 8 in the presence of IFN-α, such that similar baseline levels of infection were obtained in the presence or absence of IFN-α. The percentage of infected cells was determined by staining with anti-PIV-3 HN antibody at 18 hpi. Data are representative of four independent experiments performed in triplicate. Error bars depict SEM. (***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05; significance by one-way analysis of variance [ANOVA] with Tukey’s multiple comparative analysis). (Cii) Fold increase in PIV-3 infection between A549 cells treated with IFITM-specific siRNA compared with infection levels in cells treated with control siRNA (dotted line). Data were pooled from four independent experiments performed in triplicate. Error bars represent SEM.
The role of each endogenous IFITM protein in modulating the susceptibility of A549 cells to the early stages of infection by IAV (Fig. 6B) or PIV-3 (Fig. 6C) was assessed following siRNA treatment. For IAV, A549 cells pretreated with pooled siRNA directed against all three IFITM genes (siIF123) were significantly more susceptible to IAV infection, resulting in an increase of IAV NP-positive cells from 12% in control siRNA-treated cells to 60% in siIF123-treated cells (Fig. 6Bi). While siIF1 treatment of A549 cells did not alter the percentage of IAV-infected cells, treatment with siIF2 or siIF3 increased the percentage of IAV-infected cells to 29% and 33%, respectively (Fig. 6Bi).
We also cultured siRNA-treated A549 cells with recombinant IFN-α, prior to infection with IAV, to increase endogenous IFITM expression at the time of infection. We hypothesized that this increase would further amplify the effect of IFITM silencing on IAV infection. Consistent with previous reports (2), preliminary experiments demonstrated that IFN-α pretreatment of A549 cells resulted in a 1,000-fold increase in IFITM1 mRNA, a 5-fold increase in IFITM2 mRNA, and a 44-fold increase in IFITM3 mRNA, compared with the basal mRNA levels for each IFITM (data not shown). Cells treated with pooled siRNA directed against all three IFITM genes prior to IFN-α treatment showed enhanced susceptibility to IAV infection, with the percentage of IAV NP-positive cells increasing from 8% in control siRNA-treated cells to 69% in siIF123-treated cells (Fig. 6Bi). In the presence of IFN-α, there was a significant increase in the number of IAV-infected cells not only following siIF2 (19%) and siIF3 (45%) treatment but also following siIF1 treatment (26%) (Fig. 6Bi). The effect of siRNA-mediated IFITM knockdown in A549 cells on IAV infection is summarized as an overall fold increase in infection relative to control nontargeting siRNA treatment (Fig. 6Bii). IAV infection increased by 1.9-fold for siIF1, 2.7-fold for siIF2, 3.7-fold for siIF3, and 5.2-fold for pooled siIF123-treated A549 cells in the presence of IFN-α, which is consistent with the ability of all three all three IFITM proteins to mediate antiviral activity against IAV.
A549 cells pretreated with siRNA targeting individual IFITMs or pooled siRNA directed to all three IFITMs showed no significant differences in the level of PIV-3 infection, as assessed by the number of PIV-3 HN-positive cells (Fig. 6Ci). Furthermore, there were no significant differences in PIV-3 infection when A549 cells were treated with IFN-α following siRNA treatment but prior to infection for any specific siIF treatment, including pooled siIF123 (Fig. 6Ci). In summary, there was no fold increase in the percentage of PIV-3-infected A549 cells following siRNA treatment, either in the presence or absence of IFN-α, even when pooled siRNA to all three IFITM family members were used (Fig. 6Cii). This finding indicates that endogenous IFITM1, IFITM2, and IFITM3 do not restrict the early stages of PIV-3 infection.
Induction of IFITM1, IFITM2, and IFITM3 in A549 cells following exposure to IAV or PIV-3 does not influence late stages of infection.
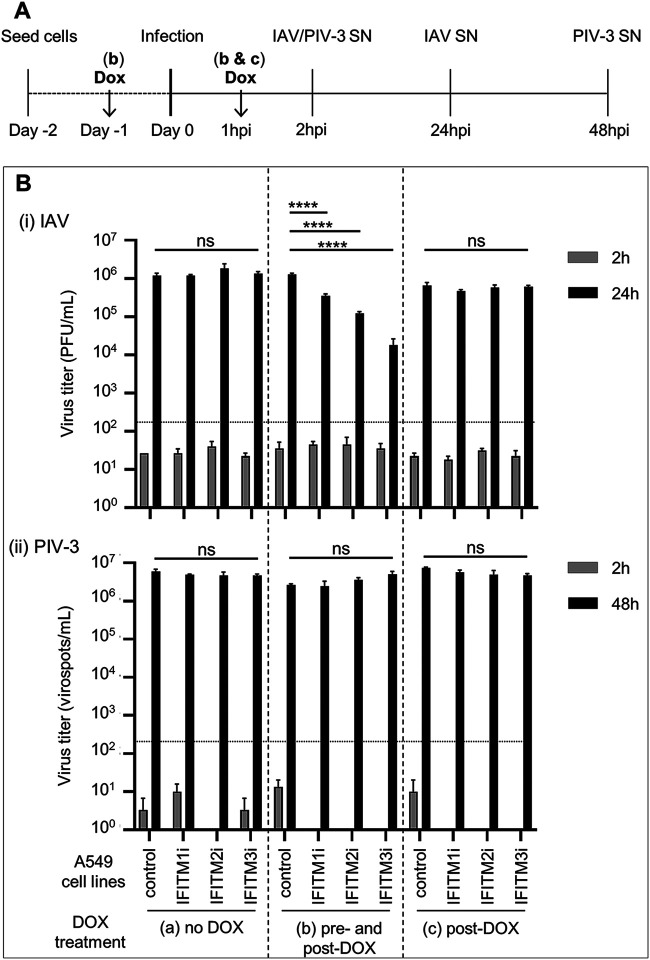
To uncouple the impact of IFITM expression on early versus late events in the virus replication cycle, cells were infected with IAV or PIV-3 prior to induction of IFITM proteins. This experiment allowed us to determine if induction of IFITMs following infectious entry of virus could modulate the infectivity and/or release of newly synthesized viral particles from infected cells. Therefore, A549 cells with inducible IFITM expression were either (i) cultured in DOX-free media before and after virus infection (no DOX, group A), (ii) pretreated with DOX and DOX was retained in the media throughout infection (pre- and post-DOX, group B), or (iii) infected with virus in DOX-free media before addition of DOX at 1 hpi (post-DOX, group C) (Fig. 7A). Titers of infectious virus released by infected cells were then determined in clarified cell supernatants harvested at 2 hpi (residual input virus) and at 24 (IAV) or 48 (PIV-3) hpi (Fig. 7B). Negligible input virus was detected in any sample at 2 hpi, with the titers of IAV below the limit of detection of the assay (Fig. 7Bi). No significant differences were noted in the titers of IAV recovered 24 hours postinfection of each cell line that was not exposed to DOX (group A). In contrast, if IFITM1, IFITM2, and IFITM3 expression was induced prior to infection and maintained throughout, IAV titers in the supernatant of infected cells as a measure of viral release were significantly reduced (Fig. 7Bi, group B). However, when IFITMs were induced only after IAV infection (post-DOX, group C) (Fig. 7Bi), no significant changes in titers of infectious virus were detected. Together, these data indicate that the inhibition of IAV release from infected cells by IFITMs is largely due to their ability to inhibit early events during IAV infection. Moreover, neither IFITM1, IFITM2, or IFITM3 showed a significant ability to inhibit IAV replication and release when induced following infectious entry.
FIG 7.
Effect of IFITM expression on the late stages of IAV and PIV-3 replication. Control A549 cells or A549 cells with inducible expression of IFITM1, IFITM2, or IFITM3 were cultured and infected with IAV (MOI, 1) (i) or PIV-3 (MOI, 0.5) (ii) in various experimental conditions, namely, in DOX-free media before and after virus infection (no DOX; group a), pretreated with DOX and with DOX retained in the media throughout the experiment (pre- and post-DOX; group b), or in DOX-free media during incubation with virus before the addition of DOX after washing at 1 hpi (post-DOX, group c). (A) Experimental design. (B) Cell-free supernatants (SNs) were harvested at 2 and 24 hpi for IAV or at 2 and 48 hpi for PIV-3. Titers of infectious virus were determined by plaque assay (IAV) or virospot assay (PIV-3) and expressed as PFU/ml or virospots/ml, respectively. In undiluted supernatant, a minimum of 10 plaques per well (133.4 PFUs/ml) or a minimum of 15 spots per well (150 virospots/ml) were required for an accurate determination of viral titers (limit of detection for the assay, represented as a dotted line). Data are representative of at least two independent experiments performed in triplicate. Error bars are SEM. (****, P ≤ 0.0001; ns, P > 0.05; one-way analysis of variance [ANOVA] with Tukey’s multiple comparative analysis).
In similar studies examining the release of infectious PIV-3 from infected cells, negligible input virus was detected in any sample at 2 hpi, with the titers of PIV-3 below the detection limit of the assay (Fig. 7Bii). Importantly, no significant differences in PIV-3 titers were recorded in any cell line cultured and infected under the no-DOX (group A), pre- and post-DOX (group B), or post-DOX (group C) culture conditions at 48 hpi (Fig. 7Bii). These data provide additional evidence that human IFITM proteins do not display potent antiviral activity against PIV-3.
DISCUSSION
Respiratory epithelial cells respond to virus infection through the production and sensing of type I and III IFN, which ultimately results in the induction of ISGs encoding a broad spectrum of host proteins that can antagonize and limit the infection (reviewed by 34). In this study, we systematically assessed and compared the ability of IFITM1, IFITM2, and IFITM3 to restrict the early and late stages of infection of an orthomyxovirus and a paramyxovirus. Each virus utilized a distinct cellular entry pathway during the early stages of infection in A549 cells, consistent with endocytosis of IAV and fusion with the plasma membrane for PIV-3 entry (Fig. 1). However, IAV and paramyxoviruses, such as PIV-3, are known to bud from the plasma membrane during release from infected cells (18, 19, 35) and are therefore likely to share at least some common cellular processes during the late stages of infection.
Our results using an inducible overexpression system in A549 cells corroborate previous findings regarding the well-established role for IFITM3 in restricting early stages of IAV infection, as well as confirm the anti-IAV activity of both IFITM1 and IFITM2. Importantly, these observations are validated by siRNA-mediated knockdown of endogenous IFITM expression in A549 cells (Fig. 6B), confirming that the anti-IAV activity observed in our overexpression system was not an artifact of IFITM protein overexpression per se.
While IFITM3 has been studied extensively and has been shown to prevent fusion of the IAV hemagglutinin with the endosomal membrane following endocytosis (6), relatively less is known about the antiviral activity and mechanism of action of IFITM1 and IFITM2 against IAV (4, 32, 36). Given the especially high sequence similarity between IFITM2 and IFITM3 (16) combined with the localization of IFITM2 within endocytic compartments (33), it is likely that IFITM2 also impairs the early stages of IAV infection during endocytosis. Indeed, Spence et al. (9) proposed a redundant yet synergistic mechanism of antiviral action between IFITM2 and IFITM3 against IAV infection in A549 and HeLa cells using IFITM knockdown approaches. In future studies, it will be interesting to explore if the subcellular localization of IFITM2 is critical for efficient antiviral function because localization of IFITM3 is known to impact antiviral activity. Mutation of a tyrosine residue at position 20 caused the accumulation of IFITM3 at the plasma membrane leading to a decrease of its anti-IAV capacity and an enhancement of HIV-1 inhibition (20). Furthermore, there is some evidence that the antiviral activity of IFITM1 against jaagsiekte sheep retrovirus (JSRV) and 10A1 murine leukemia virus (MLV) varies depending on its intracellular distribution. Endogenous IFITM1 localized to the lysosomes of HTX cells was associated with negligible antiviral activity against MLV, whereas a KR/AA mutant of IFTIM1 localized to CD63-positive multivesicular bodies and inhibited infection by both MLV and JSRV pseudoviruses (37). Of interest, the expression of IFITM2 and IFITM3 within endocytic compartments appears to vary between cell types, with some studies suggesting IFITM3 preferentially localizes to late endosomes, whereas other studies show IFITM3 localizes to early endosomes (5, 9, 32, 33). Therefore, it is conceivable that the relative efficiency of IFITM2 and IFITM3 to mediate potent antiviral activity could be linked to the variable expression of each protein within the endosomal compartment of different cells.
It is unclear how IFITM1 confers antiviral activity against IAV, but previous studies suggest that IFITM1 anti-IAV activity requires its cell surface expression (14). Targeting IFITM1 to the same endosomal compartments in which IFITM3 is normally present did not increase its anti-IAV activity (38). This finding suggests that IFITM1 has a distinct mechanism of action against IAV at the plasma membrane which is not dependent on expression within endocytic compartments. We have utilized overexpression and knockdown approaches to show that IFITM1 was less efficient at restricting the early stages of IAV infection than IFITM3 (Fig. 5A and 6B). The additive effect of using pooled siRNA against all three IFITM proteins also suggests each IFITM protein can restrict the early stages of IAV infection with differing efficiency (Fig. 6). Evidence from collective studies suggests that different IFITM family members also inhibit other viruses with various efficiency, although sometimes this is apparent only in certain cell types. For instance, early investigations using Vero E6, A549, and U2OS cells demonstrated that overexpression of each of the three IFITMs could mediate antiviral activity against West Nile virus (WNV) and dengue virus (DENV) (4), whereas overexpression studies in HEK293 cells revealed that IAV, vesicular stomatitis virus, WNV, and DENV infection were all inhibited by IFITM3, to a lesser extent by IFITM2, and not at all by IFITM1 (20, 39, 40). This discrepancy may reflect the distinct localization patterns of IFITM1, IFITM2, and IFITM3 to different cellular membranes in those overexpression systems, combined with the specific entry pathways utilized by these viruses to infect different cell types. The capacity of each IFITM to restrict IAV infection is also likely to be cell type dependent, which may relate to low basal expression levels within different cell types for some IFITM proteins. For example, IFITM1 depletion did not enhance IAV infectivity in HeLa cells (9), and the antiviral activity of endogenous IFITM1 was observed only following IFN-α stimulation to increase IFITM expression levels in A549 cells prior to treatment with IFITM1-specific siRNA and IAV infection in our study (Fig. 6).
Having confirmed the antiviral activity of IFITM1, IFITM2, and IFITM3 against the early stages of IAV infection, we extended our analysis to look at their potential to restrict PIV-3 infection. In contrast to two previous reports (14, 21), our results showed that IFITM1 overexpression did not restrict PIV-3 infection despite significant antiviral activity toward IAV within the same experiment (Fig. 5B). This disparity may reflect differences in the overexpression systems used. For example, Smith et al. (14) used a stable constitutive overexpression system in Vero cells to show that IFITM1 could mediate antiviral activity toward PIV-3. It is possible that constitutive overexpression leads to increased IFITM expression levels or different protein localization compared with our inducible system. Expression levels of IFITM proteins can certainly be an important determinant of antiviral activity. For example, clonal A549 cell lines expressing low or high levels of IFITM3 inhibited Zika virus infection to low or high levels, respectively (41). However, in previous studies, it was shown that Sendai virus (SeV), a paramyxovirus that infects mice, was not restricted by murine IFITM1 (42), supporting our conclusion that not all paramyxoviruses that fuse at the plasma membrane are necessarily restricted by IFITM1.
We did not expect IFITM2 or IFITM3 to mediate the potent inhibition of PIV-3 given that our data are consistent with PIV-3 entry into A549 cells via fusion at the plasma membrane (Fig. 1) and expression of DOX-inducible IFITM2/3 proteins in endocytic compartments (Fig. 3). While IFITM2-mediated restriction of PIV-3 was not observed, DOX-inducible expression of IFITM3 was associated with a modest but significant reduction in the percentage of PIV-3-infected cells. In previous studies, constitutive overexpression of IFITM3 also restricted the early stages of PIV-3 infection of Vero cells (14). It was therefore essential to validate the findings from our DOX-inducible overexpression system via complementary approaches which knocked down endogenous IFITM protein expression. There was no increase in the percentage of PIV-3-infected A549 cells following siRNA treatment to knockdown IFITM1, IFITM2, or IFITM3 alone or in combination, despite the significant enhancement of IAV infection in the same experiment (Fig. 6). This finding confirms that IFITM1, IFITM2, and IFITM3 are not major restriction factors controlling the early stages of PIV-3 infection in A549 cells. Furthermore, these findings emphasize the importance of complementary approaches to validate and substantiate conclusions regarding the antiviral activity of host proteins identified via overexpression studies (14, 21).
The antiviral activity of IFITM proteins at late stages in the viral life cycle has been demonstrated for some viruses. Studies from Compton et al. (23) and Tartour et al. (24) showed IFITM protein incorporation into nascent HIV-1 particles that was associated with a decrease in infectivity. In another study, expression of each of the three human IFITM proteins impaired HIV-1 Env protein processing and subsequent incorporation into virions, similarly resulting in the inhibition of viral infection (22). The inducible overexpression system used in our studies allowed us to uncouple the impact of IFITM expression on early versus late events in virus replication by inducing IFITM expression after infection with IAV and PIV-3. Our studies clearly showed that IFITM1, IFITM2, and IFITM3 do not impede the late stages of IAV or PIV-3 infection. A recent study by Lanz et al. used overexpression approaches to demonstrate that IFITM3 incorporation into IAV particles competes with the incorporation of the viral HA protein, but consistent with our findings relating to the late stages of infection, there was no reduction in the overall infectivity of viral particles produced in the presence or absence of exogenous IFITM3 (43). The authors propose that high HA density of IAV virions may be an antagonistic strategy used by the virus to evade this means of IFITM3-mediated restriction.
We conclude that IFITM1, IFITM2, and IFITM3 restrict IAV during the early stages of infection in A549 cells but that none of these host proteins display antiviral activity during the late stages of infection. We also provide evidence that IFITM1, IFITM2, and IFITM3 are not important for the restriction of PIV-3 infection at any stage of the viral life cycle. This study provides key insights into the differential antiviral activity of IFITM proteins against specific respiratory viruses with distinct cellular replicative requirements. Importantly, these findings highlight that the development of novel therapeutic strategies with broad-spectrum antiviral activity against a range of respiratory viruses will need to carefully consider the cellular replication strategies for each distinct virus family.
MATERIALS AND METHODS
Cells.
A549 human lung epithelial cells (American type culture collection [ATCC], Manassas, VA) were maintained and passaged in Kaighn’s modification of Ham’s F-12 medium (Gibco, Thermo Fisher Scientific, MA, USA) supplemented with 10% (vol/vol) fetal calf serum (FCS) (Sigma-Aldrich, MO, USA), 2 mM l-glutamine (Gibco), 1 mM sodium pyruvate (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco). 293T human kidney epithelial cells (ATCC) and Hep2 human laryngeal carcinoma epithelial cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented as above. Madin-Darby canine kidney (MDCK) cells (ATCC) were maintained and passaged in RPMI 1640 (Gibco) supplemented as above. LLC-MK2 monkey kidney epithelial cells (ATCC) were cultured in Opti-MEM I (Gibco) supplemented as above, but with 5% (vol/vol) FCS. All cell lines were mycoplasma free and tested on a regular basis with the Mycoalert mycoplasma detection kit (Lonza, Switzerland).
Viruses.
Human parainfluenza virus 3 (PIV-3) strain C243 was obtained from the ATCC (VR1540) and propagated in LLC-MK2 cells. Briefly, cells were inoculated at a multiplicity of infection (MOI) of 0.1 in serum-free media for 1 hour at 37°C and then maintained in 2% (vol/vol) FCS until a cytopathic effect (CPE) of approximately 80% was observed (5 to 6 days postinfection). Supernatants were collected and clarified by centrifugation, and virus titers were determined by titration on LLC-MK2 cells, followed by immunofluorescence staining using antibody directed to the PIV-3 hemagglutinin-neuraminidase (HN) protein (Abcam, UK) in conjunction with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Millipore, MA, USA). Virus titers were expressed as focus-forming units (FFU)/ml.
IAV strain RG-PR8-Br78HA (H1N1), generated by eight-plasmid reverse genetics (44), was a 7:1 reassortant comprising 7 genes from A/PR8/34 (PR8; H1N1) and the HA gene from A/Brazil/11/78 (Brazil 78; H1N1). IAV was propagated in the allantoic cavity of 10-day embryonated eggs, and titers of infectious virus were determined by plaque assay on MDCK cells (45). Embryonated eggs were provided by Seqirus, Parkville, Australia, and were infected with approval from the Biochemistry & Molecular Biology, Dental Science, Medicine, Microbiology & Immunology, and Surgery Animal Ethics Committee at The University of Melbourne in accordance with the NHMRC Australian code of practice for the care and use of animals for scientific purposes.
Generation of A549 cells with doxycycline (DOX)-inducible overexpression of human IFITM1, IFITM2, and IFITM3 proteins.
Coding sequences of human IFITM1, IFITM2, and IFITM3 proteins (NCBI consensus coding sequences of IFITM1, CCDS41584.1; IFITM2, CCDS41583.1; IFITM3, CCDS41585.1) containing an N-terminal FLAG tag (peptide sequence DYKDDDK) were synthesized as geneblocks (GeneArt Gene Synthesis; Thermo Fisher Scientific). Lentiviruses were produced for the generation of stable cell lines with doxycycline (DOX)-inducible expression of IFITM proteins (tetracycline on system) as previously described (46). Briefly, a two-step cloning strategy was used to generate a lentiviral transfer plasmid for the generation of lentiviral particles. Each IFITM gene was cloned from geneblocks into a plasmid carrying a modified tetracycline response element (TRE) promoter with seven tet operator sequences. Fragments containing the TRE promoter and gene of interest were then subcloned into the pFUVm1Cherry lentivirus transfer plasmid, expressing (i) the reverse tetracycline-controlled transactivator (rtTA) regulatory protein to confer DOX inducibility and (ii) mCherry under the control of a constitutive ubiquitin promoter to monitor successful lentiviral transduction. All lentiviral constructs were kindly provided by Marco Herold (Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia). Replication-defective lentiviral particles were packaged in 293T cells by cotransfection of the pFUVm1Cherry lentiviral transfer plasmid expressing human IFITM proteins with lentiviral packaging plasmids (pMDL and pRSV-REV) and the envelope plasmid (pMD2G.VSVg) using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific). At 48 hours posttransfection, supernatants (containing lentiviral particles) were harvested and used to transduce A549 cells. Transduced cells were sorted based on mCherry expression, using a FACSAria III cell sorter (BD Biosciences, CA, USA), before expansion in culture.
To induce the expression of IFITM proteins, A549 cells were incubated in media containing 1 μg/ml of DOX (Sigma-Aldrich) for 24 hours at 37°C. At 24 hours post-DOX induction, intracellular FLAG expression (as a measure of IFITM protein induction) was assessed by flow cytometry. A549 control cells were also engineered to express an irrelevant protein without a FLAG tag. The coding sequence of cytoplasmic hen egg ovalbumin (OVA) (lacking the sequence for cell-surface trafficking [47], obtained from a plasmid generously donated by A. M. Lew and J. L. Brady, The Walter and Eliza Hall Institute, Parkville, Australia) was used as an irrelevant control protein and cloned and packaged into lentiviral particles as described above.
Flow cytometric detection of IFITM proteins.
A549 cells expressing IFITM FLAG-tagged proteins were detached and stained with fixable viability dye eFlour780 or eFluor450 (eBioscience, CA, USA) before fixation with 4% (vol/vol) paraformaldehyde (PFA) in phosphate-buffered saline (PBS). After fixation, cells were permeabilized in PBS containing 0.5% (vol/vol) Triton X-100 (Sigma-Aldrich) and stained with anti-FLAG-allophycocyanin (APC) monoclonal antibody (MAb; Clone L5, Biolegend, CA, USA). Samples were analyzed on a FACSCanto II (BD Biosciences, CA, USA) or a LSRFortessa flow cytometer (BD Bioscience), before performing data analysis using FlowJo software version 10.6.2.
Western blot detection of IFITM proteins.
Whole-cell lysates were prepared using a buffer comprising 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% (vol/vol) Triton X-100, 1 mM CaCl2, 1 mM MgCl2, and a broad-spectrum protease inhibitor cocktail (Roche, Manheim, Germany). Samples were heated to 90°C for 5 min before separation by SDS-PAGE under reducing conditions using 4% to 15% gradient gels (Bio-Rad), followed by transfer to polyvinylidene difluoride (PVDF) membrane (Millipore) in Tris-glycine transfer buffer (25 mM Tris containing 192 mM glycine and 10% [vol/vol] methanol; pH 8.3). Membranes were blocked in PBS containing 5% (wt/vol) bovine serum albumin (BSA) (Sigma-Aldrich) and 0.1% (vol/vol) Tween 20 (Sigma-Aldrich). All subsequent wash and antibody binding steps were performed in PBS containing 0.05% (vol/vol) Tween 20. Cellular β-actin (approximately 43 kDa) was monitored to ensure equivalent protein loading of all samples using a mouse monoclonal antibody to β-actin (clone sc-47778; Santa Cruz, CA, USA) in conjunction with rabbit anti-mouse horseradish peroxidase (HRP; Dako). IFITM-specific antibodies for IFITM1 (Proteintech, IL, USA; number 60074-1-Ig), IFITM2 (number 66137-1-Ig; Proteintech), and IFITM3 (number 59212; Cell Signaling Technology [CST], MA, USA) or an anti-FLAG antibody (anti-FLAG [M2]; Sigma-Aldrich) were used to detect IFITM proteins in conjunction with HRP-conjugated secondary antibodies against mouse or rabbit immunoglobulins (Dako) and enhanced chemiluminescence (ECL) (Amersham) according to the manufacturer’s instructions and visualized on an Amersham imager 600 series system (GE Healthcare).
IFITM protein localization by fluorescence microscopy.
A549 cells were seeded into 8-well chamber slides (Nunc Lab-Tek, Millipore-Sigma, USA) and incubated overnight. Biotinylated Sambucus nigra agglutinin (SNA) lectin (Vector Laboratories, CA, USA) diluted in tris-buffered saline (TBS) with 10 mM CaCl2 and 1% (wt/vol) BSA, in conjunction with streptavidin-Alexa Fluor-647 (Life Technologies, Thermo Fisher Scientific), were used to stain the cell surface. For intracellular staining, cell monolayers were fixed in 4% (wt/vol) PFA, permeabilized in PBS with 0.1% (vol/vol) Triton X-100, and blocked in PBS with 5% (wt/vol) BSA and 5% (vol/vol) FCS. Intracellular IFITMs were detected via the FLAG tag using a mouse anti-FLAG antibody conjugated to FITC (M2 clone; Sigma-Aldrich), in conjunction with anti-mouse Alexa Fluor-488 antibody (Life Technologies) to enhance the FLAG signal. Early endosomes, late endosomes, and lysosomes were stained with an anti-EEA1 antibody (number 2900; Abcam), anti-Rab7 antibody (137029; Abcam), or anti-LAMP1 antibody (24170; Abcam), respectively, in conjunction with secondary donkey anti-rabbit Alexa Fluor-647 (Life Technologies). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) before mounting with ProLong Gold (Life Technologies). Images were acquired on a Zeiss LSM 780 laser scanning microscope and analyzed with ImageJ software (Fiji distribution [48]). A correlation analysis between IFITM protein expression (FLAG signal) and cellular membranes (surface or endosomal marker expression) was performed. The Pearson’s coefficient (R) was calculated with the JACoP plugin (49) as an indicator of colocalization.
Virus infection assays.
Cell monolayers in 24-well tissue culture plates (Corning) were infected with IAV or PIV-3 (at MOIs indicated) in serum-free media for 1 hour at 37°C in the presence of 5% CO2. The viral inoculum was removed, and cell monolayers were washed thoroughly to remove residual input virus and then maintained in serum-free media at 37°C in 5% CO2. Viral infection was quantitated by intracellular staining of newly synthesized viral proteins in conjunction with flow cytometric analysis. The newly synthesized IAV nucleoprotein (NP) was detected 8 h postinfection (hpi) using anti-IAV NP antibody (MAb MP3.10g2.1C7, from the WHO Collaborating Centre for Reference and Research on Influenza, Melbourne, Australia) in conjunction with FITC-conjugated goat anti-mouse IgG (Millipore) (50). The newly synthesized PIV-3 HN protein was detected 18 hpi as described previously (51) using anti-PIV-3 HN antibody (Abcam) in conjunction with FITC-conjugated goat anti-mouse IgG (Millipore).
Viral entry inhibition assays.
For viral entry inhibition assays, a variation of the infection experiments described above were performed. Cell monolayers were pretreated for 30 min at 37°C with various concentrations of dynasore (Sigma-Aldrich) or ammonium chloride (NH4Cl) (Chem-Supply, Gillman, Australia) and then inoculated with virus (noting that dynasore and NH4Cl were included in the inoculum). After incubation and a wash, cells were cultured in serum-free media containing NH4Cl. To avoid unwanted cytotoxic effects, cells inoculated in the presence of dynasore were treated with 100 mU/ml V. cholerae neuraminidase (Sigma-Aldrich) for 30 min at 37°C to remove residual virus bound to the cell surface and then cultured in serum-free media. Efficient inhibition of endocytosis by dynasore was confirmed by incubating cell monolayers in 8-well glass chamber slides with transferrin conjugated to Alexa Fluor-647 (Life Technologies), in the presence or absence of dynasore, for 15 min at 37°C. FITC-conjugated high-molecular-weight (70,000) dextran (Sigma-Aldrich) was used as a specificity control. Cells were fixed in 2% (wt/vol) PFA, counterstained with DAPI, and mounted before imaging as described above. Images were acquired on a Zeiss LSM 780 confocal microscope with a 63× oil objective. Transferrin or dextran uptake was quantified by determining average fluorescence intensity across the cell area using ImageJ software. At least 40 cells per treatment were measured and analyzed.
siRNA-mediated knockdown of endogenous IFITMs.
Endogenous IFITM proteins were knocked down using Dharmacon (CO, USA) siRNA smart pools targeting IFITM1 (M-019543-01-0005) and IFITM3 (M-014116-01-0005) or IFITM2-specific single RNA oligonucleotide (D-020103-02-002). A nontargeting (n.t.) siRNA (D-001206-14-05) (Dharmacon) was used as a control. A549 cell monolayers in 24-well cell culture plates (Corning) were transfected with 5 nM siRNA per well using the RNAiMAX reagent (Invitrogen) according to the manufacturer’s instructions, followed by incubation for 72 hours. Specific IFITM knockdown was validated by RT-qPCR (described below), and cells were used in viral infection assays. siRNA-treated cells were stimulated overnight with 1,000 U/ml of human IFN-α (Lonza) to induce endogenous IFITM expression prior to RNA extraction or before virus infection.
RT-qPCR to detect IFITM mRNA.
Total RNA was isolated from A549 cells using the RNeasy plus minikit (Qiagen, MD, USA) according to manufacturer’s instructions. cDNA was prepared using the SensiFAST cDNA synthesis kit (Bioline, TN, USA). qPCR was performed using the SensiFast 2× SYBR Lo-Rox kit (Bioline) with primer pairs specific for human IFITM1 (forward, AGCATTCGCCTACTCCGTGAAG; reverse, CACAGAGCCGAATACCAGTAACAG), human IFITM2 (forward, GAACCACATTGTGCAAACCTTCTCTC; reverse, TTCCTGCTCCTCCTTGAGCATC), or human IFITM3 (forward, ATCGTCATCCCAGTGCTGAT; reverse, ACGTGGGATACAGGTCATGG). Human GAPDH (forward, TGAAGGTCGGAGTCAACGG; reverse, GGCAACAATATCCACTTTACCAGAG) expression was used for normalization. Data acquisition was performed using the QuantStudio 7 Flex real-time PCR system and Analysis Software (Applied Biosystems). Statistical analysis was performed using Prism (GraphPad Software, San Diego, USA).
Viral exit assays.
The impact of IFITM expression on the late stages of viral replication was examined using IAV and PIV-3 exit assays, where IFITM overexpression was induced postinfection followed by quantification of virus released from infected cells. Briefly, cell monolayers were infected with IAV or PIV-3 as described above with 1 μg/ml of DOX added to the media after washing at 1 hour postinfection. Additional controls included cells without DOX induction and cells pretreated with DOX to induce IFITM expression before infection. Cell-free supernatants containing infectious virus were harvested at 2 and 24 hpi (IAV) or 2 and 48 hpi (PIV-3). IAV titers were quantitated by plaque assay on MDCK cells (52). PIV-3 titers were quantitated by a virospot assay. Briefly, Hep2 cells were seeded into 96-well tissue culture plates (Costar Cell Culture, Corning, NY) and cultured overnight. Confluent monolayers were inoculated with 10-fold serial dilutions of virus for 1 hour and then overlaid with serum-free DMEM containing 1.6% (wt/vol) carboxymethyl cellulose (CMC; Sigma-Aldrich) and incubated for 48 hours at 37°C and 5% CO2. PIV-3-positive cells were stained and visualized with a mouse anti-PIV-3 HN antibody (Abcam), followed by rabbit anti-mouse IgG conjugated to HRP (Dako) and TrueBlue peroxidase substrate (SeraCare Life Sciences). Images were taken on a ImmunoSpot analyzer (CTL, OH, USA), virus spots were counted using ImageJ software, and titers of infectious virus were expressed as virospots/ml.
Statistical analysis.
Graphing and statistical analysis of data were performed using Prism (GraphPad Software, San Diego, USA). P values were calculated by one-way analysis of variance (ANOVA) followed by post hoc analysis using Tukey’s multiple comparative test or unpaired, two-tailed t test as indicated.
ACKNOWLEDGMENTS
This study was supported by project grants APP1143154, APP1083307, and APP1184532 from the National Health and Medical Research Council (NHMRC) of Australia. P.C.R., S.L.L., and A.G.B. are all recipients of funding from the NHMRC. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health.
We thank the Melbourne Flow Cytometry Core Platform for assistance with flow cytometric analysis. We also thank the staff at the Biological Optical Microscopy Platform for support with confocal microscopy analysis and Marco Herold at the Walter and Eliza Hall Institute for provision of the lentiviral constructs and useful discussions.
Contributor Information
Sarah L. Londrigan, Email: sarahll@unimelb.edu.au.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Bailey CC, Zhong G, Huang IC, Farzan M. 2014. IFITM-family proteins: the cell’s first line of antiviral defense. Annu Rev Virol 1:261–283. 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perreira JM, Chin CR, Feeley EM, Brass AL. 2013. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol 425:4937–4955. 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMichael TM, Zhang Y, Kenney AD, Zhang L, Zani A, Lu M, Chemudupati M, Li J, Yount JS. 2018. IFITM3 restricts human metapneumovirus infection. J Infect Dis 218:1582–1591. 10.1093/infdis/jiy361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, Brass AL. 2011. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7:e1002337. 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. 2014. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 10:e1004048. 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesarino NM, Compton AA, McMichael TM, Kenney AD, Zhang L, Soewarna V, Davis M, Schwartz O, Yount JS. 2017. IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep 18:1740–1751. 10.15252/embr.201744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesarino NM, McMichael TM, Yount JS. 2015. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog 11:e1005095. 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spence JS, He R, Hoffmann HH, Das T, Thinon E, Rice CM, Peng T, Chandran K, Hang HC. 2019. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol 15:259–268. 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Londrigan SL, Wakim LM, Smith J, Haverkate AJ, Brooks AG, Reading PC. 2020. IFITM3 and type I interferons are important for the control of influenza A virus replication in murine macrophages. Virology 540:17–22. 10.1016/j.virol.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Bedford JG, O'Keeffe M, Reading PC, Wakim LM. 2019. Rapid interferon independent expression of IFITM3 following T cell activation protects cells from influenza virus infection. PLoS One 14:e0210132. 10.1371/journal.pone.0210132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Infusini G, Smith JM, Yuan H, Pizzolla A, Ng WC, Londrigan SL, Haque A, Reading PC, Villadangos JA, Wakim LM. 2015. Respiratory DC use IFITM3 to avoid direct viral infection and safeguard virus-specific CD8+ T cell priming. PLoS One 10:e0143539. 10.1371/journal.pone.0143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edinger TO, Pohl MO, Stertz S. 2014. Entry of influenza A virus: host factors and antiviral targets. J Gen Virol 95:263–277. 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 14.Smith SE, Busse DC, Binter S, Weston S, Diaz Soria C, Laksono BM, Clare S, Van Nieuwkoop S, Van den Hoogen BG, Clement M, Marsden M, Humphreys IR, Marsh M, de Swart RL, Wash RS, Tregoning JS, Kellam P. 2019. Interferon-induced transmembrane protein 1 restricts replication of viruses that enter cells via the plasma membrane. J Virol 93:e02003-18. 10.1128/JVI.02003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkins C, Woodward J, Lau DT, Barnes A, Joyce M, McFarlane N, McKeating JA, Tyrrell DL, GaleM, Jr.. 2013. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57:461–469. 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X, Li J, Winkler CA, An P, Guo JT. 2018. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front Microbiol 9:3228. 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4:613–636. 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chlanda P, Zimmerberg J. 2016. Protein-lipid interactions critical to replication of the influenza A virus. FEBS Lett 590:1940–1954. 10.1002/1873-3468.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Najjar F, Schmitt AP, Dutch RE. 2014. Paramyxovirus glycoprotein incorporation, assembly and budding: a three way dance for infectious particle production. Viruses 6:3019–3054. 10.3390/v6083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Compton AA, Roy N, Porrot F, Billet A, Casartelli N, Yount JS, Liang C, Schwartz O. 2016. Natural mutations in IFITM3 modulate post-translational regulation and toggle antiviral specificity. EMBO Rep 17:1657–1671. 10.15252/embr.201642771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabbani MA, Ribaudo M, Guo JT, Barik S. 2016. Identification of interferon-stimulated gene proteins that inhibit human parainfluenza virus type 3. J Virol 90:11145–11156. 10.1128/JVI.01551-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Li M, Wilkins J, Ding S, Swartz TH, Esposito AM, Zheng YM, Freed EO, Liang C, Chen BK, Liu SL. 2015. IFITM proteins restrict HIV-1 infection by antagonizing the envelope glycoprotein. Cell Rep 13:145–156. 10.1016/j.celrep.2015.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, Liang C, Casartelli N, Schwartz O. 2014. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 16:736–747. 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tartour K, Nguyen XN, Appourchaux R, Assil S, Barateau V, Bloyet LM, Burlaud Gaillard J, Confort MP, Escudero-Perez B, Gruffat H, Hong SS, Moroso M, Reynard O, Reynard S, Decembre E, Ftaich N, Rossi A, Wu N, Arnaud F, Baize S, Dreux M, Gerlier D, Paranhos-Baccala G, Volchkov V, Roingeard P, Cimarelli A. 2017. Interference with the production of infectious viral particles and bimodal inhibition of replication are broadly conserved antiviral properties of IFITMs. PLoS Pathog 13:e1006610. 10.1371/journal.ppat.1006610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dou D, Revol R, Ostbye H, Wang H, Daniels R. 2018. Influenza A virus cell entry, replication, virion assembly and movement. Front Immunol 9:1581. 10.3389/fimmu.2018.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb RA. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1–11. 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 27.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43:189–219. 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porotto M, Palmer SG, Palermo LM, Moscona A. 2012. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J Biol Chem 287:778–793. 10.1074/jbc.M111.298059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith EC, Popa A, Chang A, Masante C, Dutch RE. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J 276:7217–7227. 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Liu J, Li M, Yang H, Zhang C. 2012. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 7:e49265. 10.1371/journal.pone.0049265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weston S, Czieso S, White IJ, Smith SE, Kellam P, Marsh M. 2014. A membrane topology model for human interferon inducible transmembrane protein 1. PLoS One 9:e104341. 10.1371/journal.pone.0104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, Elledge SJ, Bavari S, Denison MR, Choe H, Farzan M. 2011. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7:e1001258. 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, Lloyd AR, Beard MR. 2015. The Interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J Biol Chem 290:25946–25959. 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanifer ML, Guo C, Doldan P, Boulant S. 2020. Importance of type I and III interferons at respiratory and intestinal barrier surfaces. Front Immunol 11:608645. 10.3389/fimmu.2020.608645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bose S, Malur A, Banerjee AK. 2001. Polarity of human parainfluenza virus type 3 infection in polarized human lung epithelial A549 cells: role of microfilament and microtubule. J Virol 75:1984–1989. 10.1128/JVI.75.4.1984-1989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin TY, Chin CR, Everitt AR, Clare S, Perreira JM, Savidis G, Aker AM, John SP, Sarlah D, Carreira EM, Elledge SJ, Kellam P, Brass AL. 2013. Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep 5:895–908. 10.1016/j.celrep.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li K, Jia R, Li M, Zheng YM, Miao C, Yao Y, Ji HL, Geng Y, Qiao W, Albritton LM, Liang C, Liu SL. 2015. A sorting signal suppresses IFITM1 restriction of viral entry. J Biol Chem 290:4248–4259. 10.1074/jbc.M114.630780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.John SP, Chin CR, Perreira JM, Feeley EM, Aker AM, Savidis G, Smith SE, Elia AE, Everitt AR, Vora M, Pertel T, Elledge SJ, Kellam P, Brass AL. 2013. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J Virol 87:7837–7852. 10.1128/JVI.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weidner JM, Jiang D, Pan XB, Chang J, Block TM, Guo JT. 2010. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol 84:12646–12657. 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, Guo JT. 2010. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol 84:8332–8341. 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gobillot TA, Humes D, Sharma A, Kikawa C, Overbaugh J. 2020. The robust restriction of Zika virus by Type-I interferon in A549 cells varies by viral lineage and is not determined by IFITM3. Viruses 12:503. 10.3390/v12050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hach JC, McMichael T, Chesarino NM, Yount JS. 2013. Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J Virol 87:9923–9927. 10.1128/JVI.00621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanz C, Schotsaert M, Magnus C, Karakus U, Hunziker A, Sempere Borau M, Martinez-Romero C, Spieler EE, Gunther SC, Moritz E, Hale BG, Trkola A, Garcia-Sastre A, Stertz S. 2021. IFITM3 incorporation sensitizes influenza A virus to antibody-mediated neutralization. J Exp Med 218:e20200303. 10.1084/jem.20200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA 96:9345–9350. 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders EM, Hartley CA, Jackson DC. 1990. Bovine and mouse serum beta inhibitors of influenza A viruses are mannose-binding lectins. Proc Natl Acad Sci USA 87:4485–4489. 10.1073/pnas.87.12.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herold MJ, van den Brandt J, Seibler J, Reichardt HM. 2008. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci USA 105:18507–18512. 10.1073/pnas.0806213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyle JS, Koniaras C, Lew AM. 1997. Influence of cellular location of expressed antigen on the efficacy of DNA vaccination: cytotoxic T lymphocyte and antibody responses are suboptimal when antigen is cytoplasmic after intramuscular DNA immunization. Int Immunol 9:1897–1906. 10.1093/intimm/9.12.1897. [DOI] [PubMed] [Google Scholar]
- 48.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 50.Londrigan SL, Short KR, Ma J, Gillespie L, Rockman SP, Brooks AG, Reading PC. 2015. Infection of mouse macrophages by seasonal influenza viruses can be restricted at the level of virus entry and at a late stage in the virus life cycle. J Virol 89:12319–12329. 10.1128/JVI.01455-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillespie L, Roosendahl P, Ng WC, Brooks AG, Reading PC, Londrigan SL. 2016. Endocytic function is critical for influenza A virus infection via DC-SIGN and L-SIGN. Sci Rep 6:19428. 10.1038/srep19428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders EM, Hartley CA, Reading PC, Ezekowitz RA. 1994. Complement-dependent neutralization of influenza virus by a serum mannose-binding lectin. J Gen Virol 75:615–622. 10.1099/0022-1317-75-3-615. [DOI] [PubMed] [Google Scholar]