ABSTRACT
Two separate introductions of human seasonal N2 neuraminidase genes were sustained in U.S. swine since 1998 (N2-98) and 2002 (N2-02). Herein, we characterized the antigenic evolution of the N2 of swine influenza A virus (IAV) across 2 decades following each introduction. The N2-98 and N2-02 expanded in genetic diversity, with two statistically supported monophyletic clades within each lineage. To assess antigenic drift in swine N2 following the human-to-swine spillover events, we generated a panel of swine N2 antisera against representative N2 and quantified the antigenic distance between wild-type viruses using enzyme-linked lectin assay and antigenic cartography. The antigenic distance between swine and human N2 was smallest between human N2 circulating at the time of each introduction and the archetypal swine N2. However, sustained circulation and evolution in swine of the two N2 lineages resulted in significant antigenic drift, and the N2-98 and N2-02 swine N2 lineages were antigenically distinct. Although intralineage antigenic diversity was observed, the magnitude of antigenic drift did not consistently correlate with the observed genetic differences. These data represent the first quantification of the antigenic diversity of neuraminidase of IAV in swine and demonstrated significant antigenic drift from contemporary human seasonal strains as well as antigenic variation among N2 detected in swine. These data suggest that antigenic mismatch may occur between circulating swine IAV and vaccine strains. Consequently, consideration of the diversity of N2 in swine IAV for vaccine selection may likely result in more effective control and aid public health initiatives for pandemic preparedness.
IMPORTANCE Antibodies inhibiting the neuraminidase (NA) of IAV reduce clinical disease, virus shedding, and transmission, particularly in the absence of neutralizing immunity against hemagglutinin. To understand antibody recognition of the genetically diverse NA in U.S. swine IAV, we characterized the antigenic diversity of N2 from swine and humans. N2 detected in swine IAV were derived from two distinct human-to-swine spillovers that persisted, are antigenically distinct, and underwent antigenic drift. These findings highlight the need for continued surveillance and vaccine development in swine with increased focus on the NA. Additionally, human seasonal N2 isolated after 2005 were poorly inhibited by representative swine N2 antisera, suggesting a lack of cross-reactive NA antibody-mediated immunity between contemporary swine and human N2. Bidirectional transmission between humans and swine represents a One Health challenge, and determining the correlates of immunity to emerging IAV strains is critical to mitigating zoonotic and reverse-zoonotic transmission.
KEYWORDS: influenza, swine, neuraminidase, vaccine, NA inhibition, antigenic cartography, antigenic drift, evolution, influenza vaccines
INTRODUCTION
The neuraminidase (NA) protein of influenza A virus (IAV) is a tetrameric, type II integral membrane protein that possesses sialidase activity (1–3). NA-mediated cleavage of sialic acid from glycosylated host cell proteins is required for the release of nascent virus particles from the surface of infected cells, completing the virus replication cycle. In addition, NA-mediated cleavage of mucins and disruption of viral aggregates promotes the initiation of infection (1, 4–6). Inhibition of the NA sialidase by antibodies or chemical compounds can be an effective antiviral therapeutic strategy. NA inhibitors (i.e., oseltamivir, peramivir, and zanamivir) are approved antiviral drugs proven to reduce disease severity and duration if administered soon after the onset of symptoms (7–9). The NA is also a target for the host adaptive immune response, and, similar to NA inhibitor drugs, NA-inhibiting (NAI) antibodies do not neutralize virus infection but can reduce the severity and duration of influenza infection (10–12). Monoclonal NAI antibodies were shown to protect mice from lethal influenza virus infection (13–15) in addition to displaying heterologous cross-reactivity within subtypes (16, 17). In humans, NAI antibodies are induced following infection and may provide broad immunity (18–20). Preexisting NAI antibodies can reduce the severity of influenza epidemics and pandemics and demonstrate a back boost against previously encountered historical antigens (21, 22). The NA, like the hemagglutinin (HA), undergoes antigenic drift over time, reducing the effectiveness of preexisting NA immunity (23, 24). These data have been used as rationale for increased attention to the NA component in human vaccines as a means to enhance vaccine efficacy (25).
Both N1 and N2 subtypes are endemic in U.S. swine populations, with N1 representing approximately 35% of recent NA detections and the remaining 65% of detections being N2. The classical N1 lineage is the predominant N1 isolated from U.S. swine (26, 27), along with consistent 2009 H1N1pdm N1 detection (28). Three N2 lineages currently cocirculate in U.S. swine and are the result of independent introductions of human seasonal N2 to swine in 1998 (N2-98), 2002 (N2-02), and 2016, and these lineages circulate with H1 and H3 HA in swine (29–33). The majority of N2 detections in passive surveillance are N2-02 lineage; the N2-98 lineage also has sustained detection (26, 34), but the 2016 lineage is infrequently detected (34). Similar to humans, pigs produce broadly cross-reactive NAI antibodies to the N2 protein following IAV infection or vaccination, though cross-reactivity did not extend between N2-98 and N2-02 lineages (35). Vaccine-associated enhanced respiratory disease (VAERD) is aggravated pneumonia in pigs manifested following a mismatch of HA in a whole-inactivated influenza vaccine (WIV) and the HA of a challenge virus (36, 37). However, matching the NA in the WIV, despite an HA mismatch, was shown to not only reduce IAV replication in the lung but also abrogate pathology related to VAERD (38). The antigenic diversity of contemporary swine N2 and the breadth of N2 NAI cross-reactivity is currently unknown. A more comprehensive understanding of the antigenic properties of NA in swine will improve vaccine efficacy while eliminating negative effects associated with a mismatched HA vaccine component.
The recent decade has shown an increase in the detections of zoonotic infections with IAV from swine, designated “variant” to differentiate from seasonal human IAV infections. From 2010 to 2018, 430 cases of variant H3N2, H3N2v, have been reported, most of which were associated with attendance of agricultural fairs or swine exhibits (39; https://www.cdc.gov/flu/weekly/fluviewinteractive.htm). Studies have shown some H3N2v to be capable of replication and transmission in ferrets, the gold-standard animal model for IAV replication and transmission in humans (40, 41). Following the introduction of H3N2 from humans in 1998, antigenic evolution of the H3 in swine was evident and demonstrated significant distance from human seasonal H3 as well (42–44). The relevance to human risk was demonstrated when ferrets immunized with human seasonal inactivated trivalent influenza vaccines (TIV) were shown to lack protection from infection with a swine-origin H3N2v (45, 46). Despite the documented antigenic evolution of the H3 from human and swine IAV, there were no reports characterizing the antigenic evolution of the swine N2 or the antigenic relationships between the N2 of human and swine IAV.
Here, we undertook the largest-scale antigenic characterization to date of the N2 lineages currently circulating in U.S. swine. Swine antisera were generated using recombinant, targeted N2 antigens that were tested in enzyme-linked lectin assays (ELLA) to measure NAI antibodies. Following the introduction of human seasonal strains, the N2 lineages in swine IAV diversified, resulting in the emergence of multiple clades of N2, increased antigenic distances between N2-98 and N2-02 lineages, and drift from contemporary human N2. Our data demonstrate a high degree of antigenic diversity in the N2 circulating in U.S. swine IAV and suggests vaccination efforts should include matching the NA component in addition to the HA to increase efficacy.
RESULTS
Genetic diversity of N2 in swine.
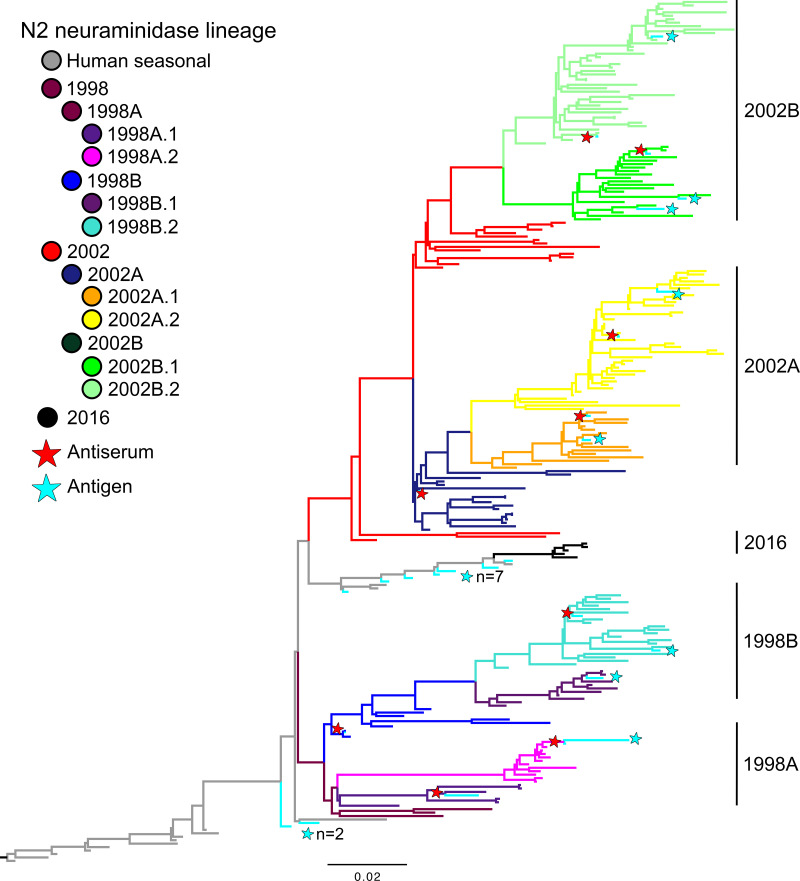
To understand the genetic evolution of N2 in swine, we inferred a maximum-likelihood phylogeny of 3,329 N2 sequences collected from 1998 to 2018 (Fig. 1). Major lineages in the phylogeny were derived from separate introductions of human seasonal N2 genes in 1998, 2002, and 2016. The 2016 N2 introduction (47) has persisted but is infrequently detected in passive surveillance (48). The 1998 and 2002 lineages had two distinct bifurcations, denoted as 1998A, 1998B, 2002A, and 2002B, that reflect strong statistical support and genetic divergence (33). Each clade began to diverge relatively close to the root of the tree, with short internal branches relative to external branches, indicating rapid expansion of both 1998 and 2002 lineages following the initial human-to-swine spillover. Additional statistically supported monophyletic clades were also identified within each lineage and clade, and these genetic divisions were denoted as “0.1” versus “0.2” (Fig. 1). The 2002 NA lineage had significantly more detections over the period sampled than the 1998 NA lineage, with 2,835 sequences compared to 494 sequences, respectively. The major clades of the 2002 N2 lineage, 2002A and 2002B, are similarly sized, though the minor clades 2002A.2 and 2002B.2 are the most numerous, with 1,056 and 1,161 sequences, respectively. The minor clade N2-1998B.2 was the predominant 1998 N2 clade within the lineage. We selected representative N2 genes from the two major N2 lineages with an amino acid sequence closest to clade consensus from each lineage and clade to generate recombinant influenza viruses with a heterosubtypic HA (H9). The reverse-engineered H9N2 was used to produce swine reference antisera for NAI assays and antigenic characterization (Fig. 1; Table 1).
FIG 1.
Representative maximum-likelihood phylogeny of 250 swine and human N2 NA genes. The two major N2 lineages in swine are the result of independent introductions of N2 from humans to swine and are grouped by 1998 and 2002. Each lineage was divided into multiple statistically supported clades (denoted A, B, 1, and 2) for which the branches have been colored. Selected reference and test antigens are annotated by stars, and branches are colored cyan; human seasonal N2 NA genes are colored in gray. The tree is rooted on the human seasonal N2 gene A/Port Chalmers/1/1973 (H3N2); all branch lengths are drawn to scale, and the scale bar indicates the number of nucleotide substitutions per site. Multiple antigens residing in closely located branches are denoted by text to the right of the star indicating the number (n) of antigens in this area of the tree. The phylogeny with tip labels included is presented in the supplemental materials and available at https://github.com/flu-crew/n2-characterization.
TABLE 1.
Antigens used for N2 antisera generation (reference antigen) and to test N2 antigenic relationships (contemporary and human antigens)
| Antigen | Abbreviation | Subtype | N2 clade | Virus propagation | Accession no. |
|---|---|---|---|---|---|
| Reference antigens | |||||
| A/swine/Texas/4199-2/1998 | TX/98 | H3N2 | 98B | MDCK | CY095677 |
| A/swine/Minnesota/02011/2008 | MN/08 | H1N2 | 98A.1 | MDCK | HM461804 |
| A/swine/South Dakota/A01349341/2013 | SD/13 | H1N2 | 98A.2 | MDCK | KC844211 |
| A/swine/Oklahoma/A01409770/2014 | OK/14 | H1N2 | 98B.2 | MDCK | KJ437590 |
| A/turkey/Ohio/313053/2004 | OH/04 | H3N2 | 2002 | MDCK | EU735820 |
| A/swine/Nebraska/A01492366/2014 | NE/14 | H1N2 | 02A.1 | MDCK | KJ549772 |
| A/swine/New York/A01104005/2011 | NY/11 | H3N2 | 02A.2 | MDCK | JN940424 |
| A/swine/Minnesota/A01678475/2016 | MN/16 | H3N2 | 02B.1 | MDCK | KY349114 |
| A/swine/Iowa/A01480656/2014 | IA/14 | H3N2 | 02B.2 | MDCK | KJ635930 |
| Contemporary N2 antigens | |||||
| A/swine/Illinois/A02216460/2017 | IL/17 | H1N2 | 1998A.2 | MDCK | MF375250 |
| A/swine/Indiana/A01894974/2016 | IN/16 | H1N2 | 1998B.1 | MDCK | KX255806 |
| A/swine/Iowa/A02217289/2017 | IA/17 | H1N2 | 1998B.2 | MDCK | MF455500 |
| A/swine/Iowa/A01778372/2016 | IA/8372 | H1N2 | 2002A.1 | MDCK | KX772386 |
| A/swine/Iowa/A01778106/2016 | IA/8106 | H3N2 | 2002A.2 | MDCK | KX761361 |
| A/swine/Illinois/A01671610/2016 | IL/16 | H1N2 | 2002B.2 | MDCK | KY486460 |
| A/swine/Oklahoma/A02157314/2018 | OK/18 | H1N2 | 2002B.1 | MDCK-L | MH259506 |
| A/swine/North Carolina/A02156991/2018 | NC/18 | H3N2 | 2002B.1 | MDCK | MH201009 |
| Human N2 antigens | |||||
| A/Wuhan/359/1995 | WU/95 | H3N2 | MDCK-L | CY112823 | |
| A/Sydney/5/1997 | SY/97 | H3N2 | MDCK-L | AJ291403 | |
| A/Moscow/10/1999 | MO/99 | H3N2 | MDCK-L | AJ458267 | |
| A/Fujian/411/2002 | FU/02 | H3N2 | MDCK-L | CY088485 | |
| A/Wisconsin/67/2005 | WI/05 | H3N2 | MDCK-L | CY034118 | |
| A/Brisbane/10/2007 | BR/07 | H3N2 | Egg | CY035024 | |
| A/Perth/16/2009 | PE/09 | H3N2 | Egg | CY081429 | |
| A/Victoria/361/2011 | VI/11 | H3N2 | MDCK | KJ942681 | |
| A/Hong Kong/4801/2014 | HK/14 | H3N2 | MDCK | EPI578429 |
Alignment of the amino acid sequences of reference N2 proteins showed frequent mutations within putative antigenic sites (Fig. S1 in the supplemental material). Mutations were identified in known NA epitopes, and generally, these were associated when comparing the 1998 to the 2002 lineage. However, there were mutations within epitopes associated with the finer-scale genetic clade divisions, i.e., F149 in N2-98B.2 versus I149 in N2-02A.1, T265 in N2-98A.2 versus V265 in N2-02B.1, V332 in N2-02A.2 versus L332 in N2-02B.1, and A435 in N2-98B.2 versus R435 in N2-02B.1, along with mutations only occurring in a single reference antigen (E367 in N2-98A.1, I370 in N2-98A.2, and S346 in N2-98A.2).
Antigenic diversity of the N2 of IAV in swine.
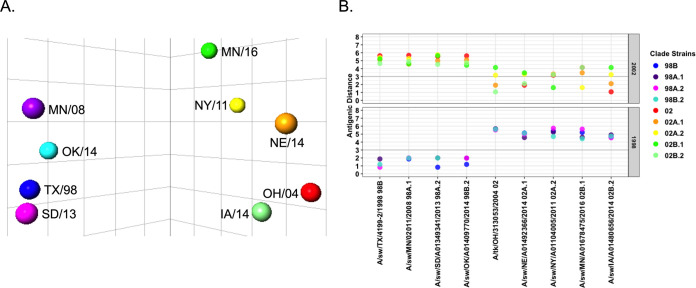
We hypothesized that the rapid expansion and cocirculation of multiple genetic clades of N2 were mirrored as a highly diverse antigenic landscape of N2 in swine. To assess whether genetic diversity impacted the antigenic diversity of swine N2, antisera with specific NAI titers were generated against a panel of recombinant influenza viruses carrying reference N2 genes and an irrelevant H9 HA to avoid interference by anti-HA antibodies. Two pigs per antigen were immunized twice with UV-inactivated recombinant H9N2 virus (256 HAU) combined with a commercial adjuvant. The resulting reference sera with an irrelevant H9 HA were then tested against wild-type H1N2 or H3N2 swine strains using an ELLA to detect NAI antibodies, with subsequent quantification with antigenic cartography. These ELLA data demonstrated there were minimal interlineage cross-reacting NAI antibodies between N2-98 and N2-02 antigens (Table S1). The limited cross-reactivity was reflected in the antigenic map as two linearly separable clusters of antigens grouped by N2-98 and N2-02 lineage (Fig. 2A). The calculated antigenic distance between N2-98 and N2-02 lineages was >4 antigenic units (AU) (Fig. 2B). Reference antigens from the N2-98 lineage formed an antigenic cluster, with all antigens being no more than 2.0 AU apart. The N2 antigens selected to represent the clades within the N2-02 lineage exhibited increased antigenic diversity compared to those within the N2-98 lineage. The N2-02B.2 (IA/14) reference antigen was in an antigenic cluster with the 2002 archetypal antigen from A/turkey/Ohio/313053/2004 (1.08 AU). The antigens of clades N2-02A.1 (NE/14) and N2-02A.2 (NY/11) were 2.11 and 3.24 AU from the N2-02B.2-N2-02 cluster, respectively. The reference antigens of the N2-02A clade, N2-02A.1 (NE/14) and N2-02A.2 (NY/11), were separated by an antigenic distance of 3.31 AU. The N2-02B.1 (MN/16) reference antigen was >4 AU from the N2-02 and N2-02B.2 (IA/14) cluster but was closest antigenically to the N2-02A.2 (NY/11, 1.61 AU) reference antigen.
FIG 2.
Antigenic distance of reference swine N2. The NAI of swine N2 antisera against homologous, reference antigens was determined using the ELLA. Antigenic cartography was used to chart the NAI data in three dimensions and calculate antigenic distances between antigens. (A) Antigenic map of N2 antigens shows a large antigenic distance exists between 1998 and 2002 lineages. (B) Graphical representation of antigenic distances between reference antigens. Each antigen is listed on the x axis, and the distances to reference antigens of the 1998 lineage (bottom) and 2002 lineage (top) are represented by colored dots. Bold line indicates an antigenic distance of 3 AU.
Antigenic drift of the N2 of IAV in swine.
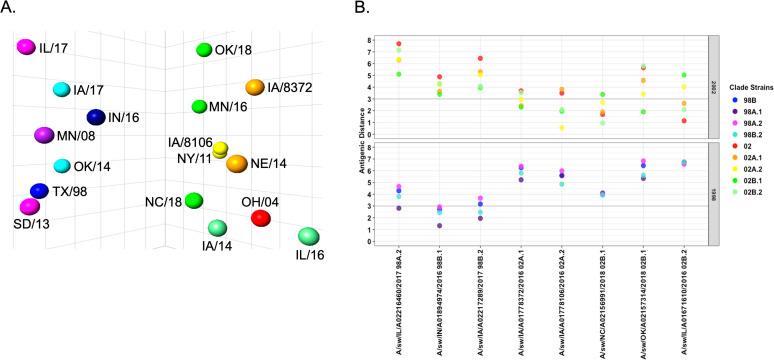
To assess the breadth of antigenic evolution within the N2-98 and N2-02 lineages, additional contemporary IAV antigens were selected (Table 1) and tested against our panel of swine H9N2 antisera. The antigenic map incorporating contemporary swine N2 antigens remained in two separate clusters, with each antigen placed within the cluster matching its genetic classification of N2-98 or N2-02 lineage (Fig. 3A). However, antigenic distances between these contemporary N2 antigens varied within genetic clade (Fig. 3B), with evidence for expanding antigenic diversity within each swine N2 lineage.
FIG 3.
Antigenic distance of reference swine N2 to contemporary swine N2. (A) Antigenic map of N2 antigens shows a large antigenic distance exists between N2-98 and N2-02 lineages. The NAI of swine N2 antisera against homologous, reference, and contemporary antigens was determined using the ELLA. Antigenic cartography was used to chart the NAI data in three dimensions and calculate antigenic distances between antigens. (B) Graphical representation of antigenic distances between reference and contemporary antigens. Contemporary antigens are listed on the x axis, and the distances to reference antigens of the N2-98 lineage (bottom) and N2-02 lineage (top) are represented by colored dots. Bold line indicates an antigenic distance of 3 AU.
Within the N2-98 lineage, the contemporary N2-98A.2 antigen, A/swine/Illinois/A02246460/2017 (IL/17), was 4.66 AU from the N2-98A.2 reference antigen, A/swine/South Dakota/A01349341/2013 (SD/13). This was the largest increase in antigenic distance between contemporary and reference antigens. The contemporary N2-98B.2 antigen, A/swine/Iowa/A02217289/2017 (IA/17), was 2.47 AU from the reference antigen, A/swine/Oklahoma/A01409770/2014 (OK/14). An antigen selected from the emerging minor clade N2-98B.1, A/swine/Indiana/A01894974/2016 (IN/16), was antigenically distant from other N2-98B antigens and showed greater antigenic similarity to antigens from the N2-98A.1 (A/swine/Minnesota/02011/2008, MN/08) and N2-98B.2 (A/swine/Oklahoma/A01409770/2014, OK/14 and A/swine/Iowa/A02217289/2017, IA/17) clades. Variation in putative antigenic sites in the N2-98 clades was mainly confined to residues 249, 254, 258, 263, 329, 331, 346, 369, 400, 401, and 402 (Fig. S2). Of these, the I254V, E258K, I263N, and K369T amino acid changes appear to be the major amino acid signatures associated with antigenic drift in the N2-98A.2 (SD/13 and IL/17) subclade.
Within the N2-02 lineage, there was also considerable antigenic diversity within contemporary phylogenetic clades, but this was typically constrained to antigenic distances of less than 3 AU. The antigenic distances between reference and contemporary antigens within the minor clades N2-02A.1, N2-02A.2, and N2-02B.2 were 2.41 AU, 0.55 AU, and 2.09 AU, respectively (Fig. 3B). The N2-02B.1 reference antigen, A/swine/Minnesota/A01678475/2016 (MN/16), was antigenically closer to the N2-02A.2 reference NY/11 (1.61 AU), despite sharing a more recent common ancestor with the N2-02B.2 antigen IA/14 (4.14 AU) (Fig. 1). Since the MN/16 antigen from the clade N2-02B.1 was an antigenic outlier, we tested two additional contemporary antigens, A/swine/Oklahoma/A02157314/2018 (OK/18) and A/swine/North Carolina/A02156991/2018 (NC/18), in the ELLA against our N2 antisera panel. These two antigens displayed discordance in antigenic distances to N2-02 reference antigens (Fig. 3A). The OK/18 antigen was antigenically closest to the N2-02B.1 reference antigen (MN/16, 1.9 AU), while the NC/18 antigen was 3.4 AU from the MN/16 N2-02B.1 reference but was closer to the N2-02A.1 (NE/14, 1.9 AU), N2-02A.2 (NY/11, 2.7 AU), and N2-02B.2 (IA/14, 0.95 AU) reference antigens. Amino acid changes in putative antigenic sites potentially affecting antigenic distance in the N2-02 clades were located at 199, 263, 332, 344, 367, 369, 370, and 400 (Fig. S3). The E381K amino acid change was present only in N2-02B proteins, but this change has not been previously described as associated with any known antibody epitopes. When comparing contemporary N2-02B.1 clade antigens, there were amino acid changes at E199K, K344E, and T369K.
The N2 of IAV in swine were antigenically distinct from the N2 of human IAV.
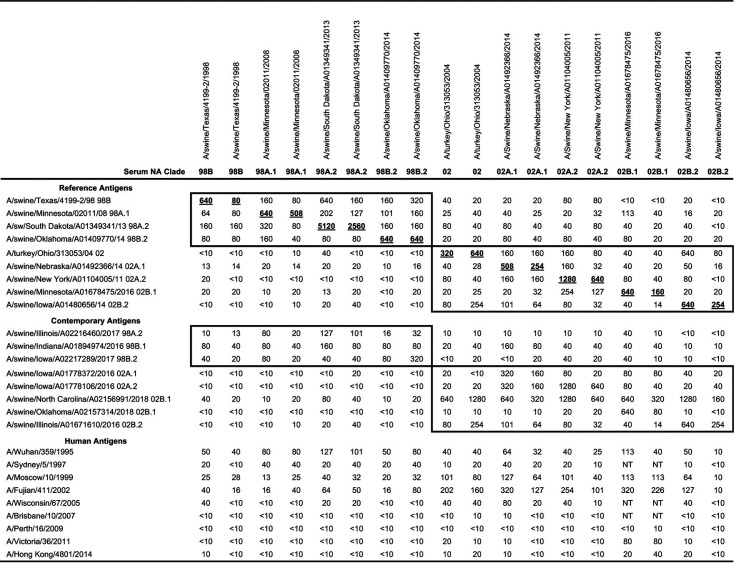
All N2 of IAV circulating in swine originated from human seasonal IAV. We tested representative human N2 against our swine N2 antiserum panel to determine whether there was cross-reactivity between our swine N2 against human N2 vaccine antigens. Low reactivity was observed in ELLA NAI assays between the swine N2 antisera tested against human vaccine N2 from strains isolated in the years between 1995 to 2014 (Table 2). Swine antisera against N2-98 lineage antigens retained some cross-reactivity against A/Wuhan/359/1995 but had limited cross-reactivity against A/Sydney/5/1997, A/Moscow/10/1999, and A/Fujian/411/2002. NAI titers of swine N2-02 lineage antisera were highest against A/Fujian/411/2002 but had minimal cross-reactivity against A/Wisconsin/67/2005 and N2 from human seasonal strains isolated prior to 2002 (Table S1). Both of the swine N2-98 and N2-02 lineage antisera showed little to no cross-reactive titers against human seasonal N2 from strains isolated after 2007.
TABLE 2.
Geometric mean of neuraminidase inhibition reciprocal titers of homologous, contemporary, and human N2 antigens against a swine reference antisera panela
H9N2 swine antisera (n = 2 pigs per antigen) were tested against wild-type H3N2 or H1N2 influenza A viruses in the enzyme-linked lectin assay. The geometric means of the reciprocal 50% inhibition titers were extracted from merged antigenic cartography data, and homologous titers are bold and underlined. Boxes indicate titers from same lineage (i.e., N2-98 or N2-02). NT, not tested.
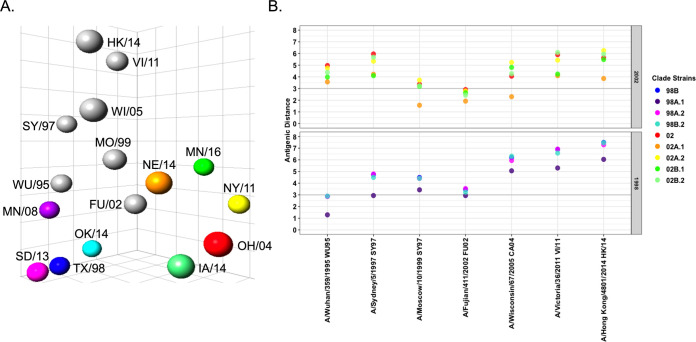
Antigenic cartography was used to visualize and quantify the antigenic distances between human and swine reference antigens. The human N2 vaccine antigens genetically and temporally closest to the human seasonal N2 introductions into swine were positioned nearest to the subsequent swine N2 lineage strains (Fig. 4A). A/Wuhan/359/1995 mapped closer to the N2-98 swine antigens, whereas A/Fujian/411/2002 was closest to N2-02 swine antigens. The most recent human N2 vaccine antigens demonstrated significant antigenic distance to all swine N2 antigens (Fig. 4B). The pairwise antigenic distances of N2-98 antigens were closest to A/Wuhan/359/1995, and N2-02 antigens were closest to A/Fujian/411/2002 (Fig. 4B) as predicted by the cross-NAI titers (Table S1). The N2-98A.1 reference antigen, A/swine/Minnesota/02011/2008 (MN/08), had the smallest antigenic distance to the progenitor human N2, 1.28 AU away from A/Wuhan/359/1995. The remaining N2-98 lineage antigens were approximately 3 AU from the human N2 A/Wuhan/359/1995. The distance from N2-02 lineage antigens to A/Wuhan/359/1995 was consistently greater than 3 AU. Instead, the majority of swine N2-02 antigens were more antigenically similar to A/Fujian/411/2002: the swine N2-02 antigen NE/14 was 1.9 AU from this human strain, and all other antigens tested were no more than 3.5 AU away. The greatest antigenic distances were between the swine N2 antigens and the most recent human seasonal N2, A/Victoria/361/2011 and A/Hong Kong/4801/2014.
FIG 4.
Antigenic distance of reference swine N2 to the N2 from human seasonal vaccines. (A) Antigenic distance between swine and human N2 increased over time. The NAI of swine N2 antisera against human seasonal N2 was determined using the ELLA. Antigenic cartography was used to chart the NAI data and calculate antigenic distances. The antigenic map reflects the introduction of human N2 into the U.S. swine population in 1998 and 2002 by A/Wuhan/359/1995-like and A/Fujian/411/2002-like viruses, respectively. (B) Graphical representation of antigenic distances between N2 of selected reference swine and human seasonal vaccine strains. Each contemporary antigen is listed on the x axis, and the distances to reference antigens of the N2-98 lineage (bottom) and N2-02 lineage (top) are represented by colored dots. Bold line indicates an antigenic distance of 3 AU.
DISCUSSION
We present results detailing the existence of substantial antigenic diversity within the established N2 NA genes circulating in U.S. swine. Following two distinct human-to-swine spillovers, the N2-98 and N2-02 lineages have been established in swine and antigenically evolved across the past 20 years. Using a representative panel of swine antisera raised against H9N2 viruses, we showed that high homologous titer N2 antisera generated against the selected antigens of the N2-98 lineage did not inhibit the sialidase activity of N2-02 lineage antigens and vice versa (35). The antigenic diversity of the two circulating swine N2 lineages and statistically supported genetic clades within each lineage was consistent with the high degree of genetic diversity found in N2 of U.S. swine strains. Accordingly, we documented a number of amino acid mutations across each clade in putative antigenic sites that were previously associated with antibody escape mutants (24, 49–52) or antibody binding (53–55); specifically, substitutions at amino acid positions 147, 199, 249, 329, 344, 346, and 370 were identified in the swine N2 as previously reported in studies with N2 from human seasonal IAV. Further, amino acids changes in residues adjacent to previously identified NA epitopes were found in swine N2 at positions 263, 265, 331, 332, 367, 369, 400, 401, 402, and 435. In each case, these amino acid mutations were correlated with antigenic drift in each N2 clade, suggesting clades that are rapidly expanding in genetic diversity were also more antigenically diverse. Our data showed minimal inhibition of the N2 from human seasonal IAV vaccines from 1995 to 2014 by swine N2 antisera. This corresponded to antigenic distances of >3 AU between most of the contemporary N2 of swine and human seasonal vaccine strains. The antigenic distance between swine and human antigens increased over time: despite swine N2 NA being derived from human seasonal N2, evolution in the swine host has resulted in little antigenic similarity. These data suggest there to be little to no cross-reactive NA-mediated immunity in swine or human populations to circulating N2 genes, and this may significantly impact the occurrence of future spillovers in either pigs or humans. Together, these data represent the most comprehensive antigenic analysis of the NA from IAV in swine and highlight the necessity for increased focus on the NA in increasing vaccine efficacy and breadth of protection.
Previous studies have shown vaccination and infection of pigs induced the production of NAI antibodies (35, 38, 56). Serum NAI antibody titers from pigs vaccinated with whole-inactivated influenza vaccines (WIV) or live-attenuated influenza vaccines (LAIV) did not differ from those that were produced in response to natural infection, though mucosal NAI antibodies were only expressed following administration of LAIV or infection. Sandbulte et al. (35) found that serum NAI antibodies generated against N2-98 cross-reacted with a drifted N2-98, but NAI titers against an N2-02 were not detected. We expanded the swine N2-98 and N2-02 antigen and antisera panels to investigate this pattern and determine the extent of antigenic diversity among contemporary swine H1N2 and H3N2 strains. These data demonstrated that the antigenic distance between N2-98 and N2-02 is substantial (>4.5 AU), suggesting a lack of cross-reactivity between the 2 lineages. Mutations, including E199K/N, K221N, R/K331S/R, N336H/Y, S372L, Q432E, and E/A435K/R, may be the primary drivers of the observed antigenic distance between N2-98 and N2-02 lineages. Importantly, Rajao et al. (38) showed that matching the NA between vaccine and challenge strains can negate the manifestation of VAERD. Thus, in order to increase vaccine efficacy and to reduce the potential for negative impacts of mismatched vaccines, N2 lineage should be considered when selecting vaccine strains for swine.
Epitope mapping and identification of major antigenic residues are essential to understanding inhibition of NA activity by antibodies and how it pertains to antigenic differences. Previous studies found four major epitopes exist in the N2 NA of human IAV, though antibody binding to only two inhibits NA sialidase activity (57). A major epitope in the N2 of human IAV, the Mem5 epitope, was mapped to four loops of the NA, 146 to 157, 195 to 202, 216 to 231, and 243 to 251 (52, 53). Additional mapping studies have implicated residues at positions 328 to 370 and 381 to 403 in antibody binding (50, 58). Analysis of the amino acid sequence of reference and contemporary N2 of U.S. swine IAV revealed high levels of sequence variation located in the putative epitopes extrapolated from N2 of human IAV, suggesting the molecular determinants of N2 antibody binding in swine are located in these regions. Mutations potentially driving the observed antigenic drift of the N2-98 were identified at positions 249, 329, 331, 346, 369, 400, 401, and 402, though the contribution and impact of individual mutations to antigenic drift remain unknown. There were other potential lineage-defining mutations occurring at positions 432 and 436. Mutations in the N2-02 lineage potentially contributing to antigenic drift included 147, 199, 263, 265, 329, 332, 344, 367, 369, 370, 400, 402, and 435. Additional studies using antibody escape mutants will be crucial in identifying which of these amino acids are central to antibody binding and whether single or combinations of mutations are responsible for driving antigenic diversity.
Over the past decade, variant influenza cases have been detected as sporadic clusters of infections associated with agricultural fairs (39, 40, 59). Human H3N2v cases were more prevalent in young children with exposure to infected swine (60, 61), which correlated with a lack of preexisting neutralizing antibodies against variant virus (62, 63). Characterization of multiple lineages of variant IAV found no barriers to infection using in vitro and naive in vivo models of human infection or replication (40, 41, 64). Previous studies in ferrets with a mismatched seasonal IAV showed that prior infection, which was correlated with increased mucosal immunity but not vaccination, reduced transmission to contact ferrets (45). These studies did not assess NAI titers, although NAI titers have been shown to be a correlate of protection against emerging viruses (20). The data presented here showed minimal inhibition of human N2 with swine N2 antisera in the ELLA assay. Analysis by antigenic cartography found dramatic antigenic distances between swine and human N2, and this distance increased over time following the initial establishment of each N2 lineage in the swine population. It is plausible that a reduction in NA-mediated immunity due to the increase in antigenic distance between swine and human N2, particularly since the mid-2000s, may increase the risk of variant infections in humans. Additional analysis of NAI antibody activity against swine N2 antigens in human population sera, as well as antisera generated in ferrets, is necessary to fully understand the potential role of NA immunity for predicting and mitigating risk at the human and swine IAV interface. However, these data support future consideration of NA-mediated immunity in pandemic preparedness vaccine strain selection to protect against variant influenza viruses is warranted.
Vaccination is the most effective countermeasure against infection with influenza virus in both swine and humans. Vaccine formulation is predominantly based on the antigenic properties of currently circulating HA proteins, particularly HAI antibodies (65). Recently, increased attention on the NA content has been suggested as a means to improve vaccine efficacy (66, 67), but this requires a more comprehensive understanding of the antigenic properties of NA from swine IAV to improve swine vaccines. The scope of the antigenic diversity shown here in the contemporary N2 NA in swine presents an opportunity for improved vaccine strain selection for swine. Functional studies, aimed at elucidating the effects of N2 antigenic distance on protection from IAV infection, disease, and transmission, are necessary to better inform vaccine strain selection.
MATERIALS AND METHODS
Sequence and phylogenetic analysis.
All available swine IAV NA-N2 sequences from H1N2 and H3N2 subtypes were downloaded from the Influenza Research Database (68) on 24 October 2018. To restrict to wild-type field viruses, we excluded data with “lab” or “laboratory” in the host record. As our goal was to describe genetic diversity in circulating swine NA N2 genes, strains that reflected single-outbreak events collected as part of active surveillance at agricultural fairs were removed from analyses (57). These data were compiled with a temporally matched set of N2 genes from the World Health Organization (WHO)-recommended human seasonal vaccine sequences. The sequences were aligned with default settings in MAFFT v7.294 (69), and sequences with 100% identity, those that were missing more than 50% of the gene, or those with more than 5 nucleotide base ambiguities were removed. A maximum-likelihood phylogeny for the alignment was inferred, following automatic model selection, using IQ-TREE v2 (70, 71). Monophyletic clades were identified using statistical support (Shimodaira-Hasegawa approximate likelihood-ratio test [SH-aLRT] > 90; ultrafast bootstrap > 70) and detection frequency (a minimum of 10 N2 genes from multiple locations across years). From each monophyletic clade, we deduced amino acids for the N2 gene, aligned the data using MAFFT v7.294 (69), and generated a clade consensus sequence using python code in the flutile package (https://github.com/flu-crew/flutile). We subsequently identified the best-matched field strain to the clade consensus sequence, and this strain was used in the construction of reverse genetics (rg)-engineered H9N2 viruses (Table 1). For visualization purposes, we downsampled the inferred maximum-likelihood tree using smot (https://github.com/flu-crew/smot): we implemented paraphyletic sampling, where monophyletic trees were identified within each named genetic clade, and a random selection within each clade were identified with the scale factor set to 0.5 (excluding human seasonal N2 and swine antigen genes) and maintained for presentation in Fig. 1 with all data and trees archived at https://github.com/flu-crew/n2-characterization.
Viruses.
All swine influenza viruses were obtained from the USDA influenza A virus in swine surveillance system virus repository held at the National Veterinary Service Laboratories, USDA-APHIS. Viruses used as antigens or for generation of antiserum are listed in Table 1. All but three NA sequences from the clade consensus were generated by custom DNA synthesis (GenScript, Piscataway, NJ) and cloned into reverse genetics plasmids. NA from A/swine/Minnesota/02011/2008, A/swine/Nebraska/A01492366/2014, and A/swine/New York/A01104005/2011 were directly amplified from viral RNA and used for virus rescue using a PCR-based reverse-genetics strategy as previously described (72). To avoid interference from antibodies to H1 or H3 HA, all recombinant influenza viruses (rgH9N2) used for swine antiserum production were generated by pairing the respective NA gene with the H9 HA from A/guinea fowl/Hong Kong/WF10/99 (H9N2) on an attenuated internal gene cassette from A/turkey/Ohio/313053/2004 (H3N2) (OH/04 att) (73). All NA genes were sequenced before and after rescue and matched with the expected consensus sequence. Recombinant H9N2 and wild-type swine and human influenza viruses were propagated on MDCK or MDCK-London monolayers in Opti-MEM (Life Technologies, Waltham, MA, USA) supplemented with antibiotic/antimycotics and 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Worthington Biochemical Corp., Lakewood, NJ, USA). All virus work was performed in a biosafety level 2 laboratory in accordance with USDA NADC institutional biosafety protocols.
Swine antiserum production.
Three-week-old pigs were obtained from a herd free of IAV and porcine reproductive and respiratory syndrome virus and housed in a biosafety level 2 containment facility in compliance with the USDA NADC Institutional Animal Care and Use Committee. Two pigs per virus received an intramuscular immunization comprised of 2 ml of 128 HAU UV-inactivated, rgH9N2 virus in 20% Emulsigen-D adjuvant (MVP Laboratories, Omaha, NE, USA). Two weeks postvaccination, pigs were boosted with an identical dose of whole inactivated virus vaccine. Two weeks postboost, serum hemagglutination inhibition (HI) titers were assessed and confirmed to be greater than 160 HAU. Pigs were then humanely euthanized for blood collection. Whole blood was allowed to coagulate at 4°C overnight, and serum was collected via centrifugation at 2,000 rpm for 20 min.
Enzyme-linked lectin assay.
An enzyme-linked lectin assay (ELLA) was used to assess neuraminidase-inhibiting antibodies as previously described (74–76) with minor variations. As recombinant H9N2 viruses were used to generate antisera specific for swine N2 of interest, they could not be used as antigens in the ELLA, and thus, we used wild-type swine or human IAV (H1N2 or H3N2) as antigens for ELLA experiments. Briefly, 2-fold serial dilutions of H9N2 antisera were incubated with wild-type H1N2 or H3N2 IAV on fetuin-coated plates for 18 to 20 h at 37°C. Following incubation, plates were washed with PBS-Tween, incubated with horseradish peroxidase (HRP)-conjugated peanut agglutinin (Sigma-Aldrich, St. Louis, MO), and detected with TMB substrate (KPL Laboratories, Gaithersburg, MD). Plates were read at 650 nm and optical density at 650 nm (OD650) values were used to calculate the percent inhibition. The inverse of the highest serum dilution that showed a 50% inhibition of NA activity was considered the NA inhibition (NI) titer.
Antigenic cartography.
Three-dimensional antigenic maps were created from NI data (Table S1 in the supplemental material) using antigenic cartography as previously described (42, 77, 78). Spheres represent NA antigens and are color-coded by genetic lineage. The spheres representing each antigen were made to be equal size for ease of viewing, so antigens in the map may appear closer than the calculated distance due to the maps being in three dimensions, and a still image was taken from the rotation best displaying all antigens. Pairwise antigenic distances were extracted and graphed using the ggplot2 package within the R programming language (79).
Data availability.
All sequence data are publicly available in NCBI GenBank or GISAID with accession numbers provided in Table 1. Data and tree files used in analyses are archived at https://github.com/flu-crew/n2-characterization.
ACKNOWLEDGMENTS
We thank Michelle Harland and Jordyn Zoul for technical assistance, Jason Huegel, Alyssa Dannon, Randy Leon, Justin Miller, and Keiko Sampson for assistance with animal studies, and Zebulun Arendsee for a prerelease version of the smot tool.
We were supported by the U.S. Department of Agriculture (USDA) Agricultural Research Service (ARS project number 5030-32000-120-00-D), the USDA Animal and Plant Health Inspection Agency (ARS project numbers 5030-32000-120-28-A, 5030-32000-120-26-I, and 5030-32000-120-80-I), and by an NIH, National Institute of Allergy and Infectious Diseases (NIAID), interagency agreement associated with the Center of Research in Influenza Pathogenesis (CRIP), an NIAID-funded Center of Excellence in Influenza Research and Surveillance (CEIRS; HHSN272201400008C) to A.L.V. B.S.K. and J.C. were supported by an appointment to the USDA-ARS Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and USDA under contract number DE-AC05-06OR23100. This research used resources provided by the SCINet project of the USDA Agricultural Research Service (ARS project number 0500-00093-001-00-D). Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture, DOE, or ORISE. USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material is available online only.
Contributor Information
Amy L. Vincent, Email: amy.vincent@usda.gov.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Liu C, Eichelberger MC, Compans RW, Air GM. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol 69:1099–1106. 10.1128/JVI.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palese P, Compans RW. 1976. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J Gen Virol 33:159–163. 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- 3.Palese P, Tobita K, Ueda M, Compans RW. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397–410. 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 4.Zanin M, Marathe B, Wong SS, Yoon SW, Collin E, Oshansky C, Jones J, Hause B, Webby R. 2015. Pandemic swine H1N1 influenza viruses with almost undetectable neuraminidase activity are not transmitted via aerosols in ferrets and are inhibited by human mucus but not swine mucus. J Virol 89:5935–5948. 10.1128/JVI.02537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, Gagneux P. 2013. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 10:321. 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Steukers L, Forier K, Xiong R, Braeckmans K, Van Reeth K, Nauwynck H. 2014. A beneficiary role for neuraminidase in influenza virus penetration through the respiratory mucus. PLoS One 9:e110026. 10.1371/journal.pone.0110026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yen HL, Herlocher LM, Hoffmann E, Matrosovich MN, Monto AS, Webster RG, Govorkova EA. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob Agents Chemother 49:4075–4084. 10.1128/AAC.49.10.4075-4084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leneva IA, Roberts N, Govorkova EA, Goloubeva OG, Webster RG. 2000. The neuraminidase inhibitor GS4104 (oseltamivir phosphate) is efficacious against A/Hong Kong/156/97 (H5N1) and A/Hong Kong/1074/99 (H9N2) influenza viruses. Antiviral Res 48:101–115. 10.1016/S0166-3542(00)00123-6. [DOI] [PubMed] [Google Scholar]
- 9.Baranovich T, Webster RG, Govorkova EA. 2011. Fitness of neuraminidase inhibitor-resistant influenza A viruses. Curr Opin Virol 1:574–581. 10.1016/j.coviro.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. 1974. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129:411–420. 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- 11.Webster RG, Hinshaw VS, Laver WG. 1982. Selection and analysis of antigenic variants of the neuraminidase of N2 influenza viruses with monoclonal antibodies. Virology 117:93–104. 10.1016/0042-6822(82)90510-4. [DOI] [PubMed] [Google Scholar]
- 12.Webster RG, Reay PA, Laver WG. 1988. Protection against lethal influenza with neuraminidase. Virology 164:230–237. 10.1016/0042-6822(88)90640-x. [DOI] [PubMed] [Google Scholar]
- 13.Wohlbold TJ, Chromikova V, Tan GS, Meade P, Amanat F, Comella P, Hirsh A, Krammer F. 2016. Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against lethal H10N8 influenza virus infection in mice. J Virol 90:851–861. 10.1128/JVI.02275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, Subramaniam S, Palese P, Krammer F. 2017. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol 2:1415–1424. 10.1038/s41564-017-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan H, Qi L, Gao J, Couzens LK, Jiang L, Gao Y, Sheng ZM, Fong S, Hahn M, Khurana S, Taubenberger JK, Eichelberger MC. 2018. Comparison of the Efficacy of N9 neuraminidase-specific monoclonal antibodies against influenza A(H7N9) virus infection. J Virol 92:e01588-17. 10.1128/JVI.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87:9290–9300. 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556. 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, DaveyRT, Jr., Taubenberger JK. 2016. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio 7:e00417-16. 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm AE, Stamper CT, Lan LY, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173:417–429.e10. 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE. 2015. Antibody to influenza virus neuraminidase: an independent correlate of protection. J Infect Dis 212:1191–1199. 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- 21.Monto AS, Kendal AP. 1973. Effect of neuraminidase antibody on Hong Kong influenza. Lancet 1:623–625. 10.1016/S0140-6736(73)92196-X. [DOI] [PubMed] [Google Scholar]
- 22.Rajendran M, Nachbagauer R, Ermler ME, Bunduc P, Amanat F, Izikson R, Cox M, Palese P, Eichelberger M, Krammer F. 2017. Analysis of anti-influenza virus neuraminidase antibodies in children, adults, and the elderly by ELISA and enzyme inhibition: evidence for original antigenic sin. mBio 8:e02281-16. 10.1128/mBio.02281-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westgeest KB, de Graaf M, Fourment M, Bestebroer TM, van Beek R, Spronken MI, de Jong JC, Rimmelzwaan GF, Russell CA, Osterhaus AD, Smith GJ, Smith DJ, Fouchier RA. 2012. Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution. J Gen Virol 93:1996–2007. 10.1099/vir.0.043059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. 2011. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci USA 108:20748–20753. 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS. 2018. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 9:e02332-17. 10.1128/mBio.02332-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walia RR, Anderson TK, Vincent AL. 2019. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 13:262–273. 10.1111/irv.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. 2008. Swine influenza viruses: a North American perspective. Adv Virus Res 72:127–154. 10.1016/S0065-3527(08)00403-X. [DOI] [PubMed] [Google Scholar]
- 28.Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. 2015. Continual reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol 89:6218–6226. 10.1128/JVI.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–8856. 10.1128/JVI.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. 2000. Evolution of swine H3N2 influenza viruses in the United States. J Virol 74:8243–8251. 10.1128/jvi.74.18.8243-8251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. 2004. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res 103:67–73. 10.1016/j.virusres.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Karasin AI, Carman S, Olsen CW. 2006. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005). J Clin Microbiol 44:1123–1126. 10.1128/JCM.44.3.1123-1126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeller MA, Chang J, Vincent AL, Gauger PC, Anderson TK. 2020. Coordinated evolution between N2 neuraminidase and H1 and H3 hemagglutinin genes increase influenza A virus genetic diversity in swine. BioRxiv 10.1101/2020.05.29.123828. [DOI] [PMC free article] [PubMed]
- 34.Zeller MA, Anderson TK, Walia RW, Vincent AL, Gauger PC. 2018. ISU FLUture: a veterinary diagnostic laboratory web-based platform to monitor the temporal genetic patterns of influenza A virus in swine. BMC Bioinformatics 19:397. 10.1186/s12859-018-2408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandbulte MR, Gauger PC, Kitikoon P, Chen H, Perez DR, Roth JA, Vincent AL. 2016. Neuraminidase inhibiting antibody responses in pigs differ between influenza A virus N2 lineages and by vaccine type. Vaccine 34:3773–3779. 10.1016/j.vaccine.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Gauger PC, Vincent AL, Loving CL, Lager KM, Janke BH, KehrliME, Jr., Roth JA. 2011. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (delta-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine 29:2712–2719. 10.1016/j.vaccine.2011.01.082. [DOI] [PubMed] [Google Scholar]
- 37.Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, KehrliME, Jr., Roth JA. 2012. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol 49:900–912. 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- 38.Rajao DS, Chen H, Perez DR, Sandbulte MR, Gauger PC, Loving CL, Shanks GD, Vincent A. 2016. Vaccine-associated enhanced respiratory disease is influenced by haemagglutinin and neuraminidase in whole inactivated influenza virus vaccines. J Gen Virol 97:1489–1499. 10.1099/jgv.0.000468. [DOI] [PubMed] [Google Scholar]
- 39.Duwell MM, Blythe D, Radebaugh MW, Kough EM, Bachaus B, Crum DA, PerkinsKA, Jr., Blanton L, Davis CT, Jang Y, Vincent A, Chang J, Abney DE, Gudmundson L, Brewster MG, Polsky L, Rose DC, Feldman KA. 2018. Influenza A(H3N2) variant virus outbreak at three fairs - Maryland, 2017. MMWR Morb Mortal Wkly Rep 67:1169–1173. 10.15585/mmwr.mm6742a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun X, Pulit-Penaloza JA, Belser JA, Pappas C, Pearce MB, Brock N, Zeng H, Creager HM, Zanders N, Jang Y, Tumpey TM, Davis CT, Maines TR. 2018. Pathogenesis and transmission of genetically diverse swine-origin H3N2 variant influenza A viruses from multiple lineages isolated in the United States, 2011-2016. J Virol 92:e00665-18. 10.1128/JVI.00665-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearce MB, Jayaraman A, Pappas C, Belser JA, Zeng H, Gustin KM, Maines TR, Sun X, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2012. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc Natl Acad Sci USA 109:3944–3949. 10.1073/pnas.1119945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. 2014. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol 88:4752–4763. 10.1128/JVI.03805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, Anderson TK, Rajao DS, Perez DR, Vincent AL. 2016. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza A virus. J Virol 90:8266–8280. 10.1128/JVI.01002-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolton MJ, Abente EJ, Venkatesh D, Stratton JA, Zeller M, Anderson TK, Lewis NS, Vincent AL. 2019. Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016. Influenza Other Respir Viruses 13:83–90. 10.1111/irv.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houser KV, Pearce MB, Katz JM, Tumpey TM. 2013. Impact of prior seasonal H3N2 influenza vaccination or infection on protection and transmission of emerging variants of influenza A(H3N2)v virus in ferrets. J Virol 87:13480–13489. 10.1128/JVI.02434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houser KV, Katz JM, Tumpey TM. 2013. Seasonal trivalent inactivated influenza vaccine does not protect against newly emerging variants of influenza A (H3N2v) virus in ferrets. J Virol 87:1261–1263. 10.1128/JVI.02625-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeller MA, Li G, Harmon KM, Zhang J, Vincent AL, Anderson TK, Gauger PC. 2018. Complete genome sequences of two novel human-like H3N2 influenza A viruses, A/swine/Oklahoma/65980/2017 (H3N2) and A/Swine/Oklahoma/65260/2017 (H3N2), detected in swine in the United States. Microbiol Resour Announc 7:e01203-18. 10.1128/MRA.01203-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, Lewis NS, Davis CT, Vincent AL. 2021. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med 11:a038737. 10.1101/cshperspect.a038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lentz MR, Webster RG, Air GM. 1987. Site-directed mutation of the active site of influenza neuraminidase and implications for the catalytic mechanism. Biochemistry 26:5351–5358. 10.1021/bi00391a020. [DOI] [PubMed] [Google Scholar]
- 50.Air GM, Els MC, Brown LE, Laver WG, Webster RG. 1985. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology 145:237–248. 10.1016/0042-6822(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 51.Gulati U, Hwang CC, Venkatramani L, Gulati S, Stray SJ, Lee JT, Laver WG, Bochkarev A, Zlotnick A, Air GM. 2002. Antibody epitopes on the neuraminidase of a recent H3N2 influenza virus (A/Memphis/31/98). J Virol 76:12274–12280. 10.1128/jvi.76.23.12274-12280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JT, Air GM. 2006. Interaction between a 1998 human influenza virus N2 neuraminidase and monoclonal antibody Mem5. Virology 345:424–433. 10.1016/j.virol.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 53.Venkatramani L, Bochkareva E, Lee JT, Gulati U, Graeme Laver W, Bochkarev A, Air GM. 2006. An epidemiologically significant epitope of a 1998 human influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. J Mol Biol 356:651–663. 10.1016/j.jmb.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 54.Tulip WR, Varghese JN, Webster RG, Air GM, Laver WG, Colman PM. 1989. Crystal structures of neuraminidase-antibody complexes. Cold Spring Harbor Symp Quant Biol 54 Pt 1:257–263. 10.1101/sqb.1989.054.01.032. [DOI] [PubMed] [Google Scholar]
- 55.Zhu X, Turner HL, Lang S, McBride R, Bangaru S, Gilchuk IM, Yu W, Paulson JC, CroweJE, Jr., Ward AB, Wilson IA. 2019. Structural basis of protection against H7N9 influenza virus by human anti-N9 neuraminidase antibodies. Cell Host Microbe 26:729–738.e4. 10.1016/j.chom.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Souza CK, Rajao DS, Sandbulte MR, Lopes S, Lewis NS, Loving CL, Gauger PC, Vincent AL. 2018. The type of adjuvant in whole inactivated influenza a virus vaccines impacts vaccine-associated enhanced respiratory disease. Vaccine 36:6103–6110. 10.1016/j.vaccine.2018.08.072. [DOI] [PubMed] [Google Scholar]
- 57.Webster RG, Brown LE, Laver WG. 1984. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology 135:30–42. 10.1016/0042-6822(84)90114-4. [DOI] [PubMed] [Google Scholar]
- 58.Laver WG, Air GM, Webster RG, Markoff LJ. 1982. Amino acid sequence changes in antigenic variants of type A influenza virus N2 neuraminidase. Virology 122:450–460. 10.1016/0042-6822(82)90244-6. [DOI] [PubMed] [Google Scholar]
- 59.Bowman AS, Sreevatsan S, Killian ML, Page SL, Nelson SW, Nolting JM, Cardona C, Slemons RD. 2012. Molecular evidence for interspecies transmission of H3N2pM/H3N2v influenza A viruses at an Ohio agricultural fair, July 2012. Emerg Microbes Infect 1:e33. 10.1038/emi.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, Boulton R, Haddy L, Biggerstaff M, Brammer L, Trock S, Burns E, Gomez T, Wong KK, Katz J, Lindstrom S, Klimov A, Bresee JS, Jernigan DB, Cox N, Finelli L, Influenza A (H3N2)v Virus Investigation Team. 2013. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis 57(Suppl 1):S4–S11. 10.1093/cid/cit272. [DOI] [PubMed] [Google Scholar]
- 61.Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, Beaudoin A, Berman L, Bidol S, Blanton L, Blythe D, Brammer L, D'Mello T, Danila R, Davis W, de Fijter S, Diorio M, Durand LO, Emery S, Fowler B, Garten R, Grant Y, Greenbaum A, Gubareva L, Havers F, Haupt T, House J, Ibrahim S, Jiang V, Jain S, Jernigan D, Kazmierczak J, Klimov A, Lindstrom S, Longenberger A, Lucas P, Lynfield R, McMorrow M, Moll M, Morin C, Ostroff S, Page SL, Park SY, Peters S, Quinn C, Reed C, Richards S, Scheftel J, Simwale O, Shu B, et al. 2013. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis 57:1703–1712. 10.1093/cid/cit649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skowronski DM, Janjua NZ, De Serres G, Purych D, Gilca V, Scheifele DW, Dionne M, Sabaiduc S, Gardy JL, Li G, Bastien N, Petric M, Boivin G, Li Y. 2012. Cross-reactive and vaccine-induced antibody to an emerging swine-origin variant of influenza A virus subtype H3N2 (H3N2v). J Infect Dis 206:1852–1861. 10.1093/infdis/jis500. [DOI] [PubMed] [Google Scholar]
- 63.Skowronski DM, De Serres G, Janjua NZ, Gardy JL, Gilca V, Dionne M, Hamelin ME, Rheaume C, Boivin G. 2012. Cross-reactive antibody to swine influenza A(H3N2) subtype virus in children and adults before and after immunisation with 2010/11 trivalent inactivated influenza vaccine in Canada, August to November 2010. Euro Surveill 17:20066. 10.2807/ese.17.04.20066-en. [DOI] [PubMed] [Google Scholar]
- 64.Pulit-Penaloza JA, Pappas C, Belser JA, Sun X, Brock N, Zeng H, Tumpey TM, Maines TR. 2018. Comparative in vitro and in vivo analysis of H1N1 and H1N2 variant influenza viruses isolated from humans between 2011 and 2016. J Virol 92:e01444-18. 10.1128/JVI.01444-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincent AL, Perez DR, Rajao D, Anderson TK, Abente EJ, Walia RR, Lewis NS. 2017. Influenza A virus vaccines for swine. Vet Microbiol 206:35–44. 10.1016/j.vetmic.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eichelberger MC, Monto AS. 2019. Neuraminidase, the forgotten surface antigen, emerges as an influenza vaccine target for broadened protection. J Infect Dis 219:S75–S80. 10.1093/infdis/jiz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eichelberger MC, Morens DM, Taubenberger JK. 2018. Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol 53:38–44. 10.1016/j.coi.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. 2017. Influenza Research Database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45:D466–D474. 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H, Ye J, Xu K, Angel M, Shao H, Ferrero A, Sutton T, Perez DR. 2012. Partial and full PCR-based reverse genetics strategy for influenza viruses. PLoS One 7:e46378. 10.1371/journal.pone.0046378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pena L, Vincent AL, Ye J, Ciacci-Zanella JR, Angel M, Lorusso A, Gauger PC, Janke BH, Loving CL, Perez DR. 2011. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol 85:456–469. 10.1128/JVI.01503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, Eichelberger M. 2014. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods 210:7–14. 10.1016/j.jviromet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 75.Sandbulte MR, Gao J, Straight TM, Eichelberger MC. 2009. A miniaturized assay for influenza neuraminidase-inhibiting antibodies utilizing reverse genetics-derived antigens. Influenza Other Respir Viruses 3:233–240. 10.1111/j.1750-2659.2009.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaplan BS, Vincent AL. 2020. Detection and titration of influenza A virus neuraminidase inhibiting (NAI) antibodies using an enzyme-linked lectin assay (ELLA). Methods Mol Biol 2123:335–344. 10.1007/978-1-0716-0346-8_24. [DOI] [PubMed] [Google Scholar]
- 77.Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, Gramer MR, Russell CA, Smith DJ, Lager KM, Lewis NS. 2011. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol 92:919–930. 10.1099/vir.0.027557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 79.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer Verlag, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4<br>. Download JVI.00632-21-s0001.pdf, PDF file, 8.7 MB (8.7MB, pdf)
Data Availability Statement
All sequence data are publicly available in NCBI GenBank or GISAID with accession numbers provided in Table 1. Data and tree files used in analyses are archived at https://github.com/flu-crew/n2-characterization.