ABSTRACT
Some lytic proteins encoded by Epstein-Barr virus (EBV) suppress host interferon (IFN) signaling to facilitate viral replication. In this study, we sought to identify and characterize EBV proteins antagonizing IFN signaling. The induction of IFN-stimulated genes (ISGs) by IFN-β was effectively suppressed by EBV. A functional screen was therefore performed to identify IFN-antagonizing proteins encoded by EBV. EBV tegument protein BGLF2 was identified as a potent suppressor of JAK-STAT signaling. This activity was found to be independent of its stimulatory effect on p38 and JNK pathways. Association of BGLF2 with STAT2 resulted in more pronounced K48-linked polyubiquitination and proteasomal degradation of the latter. Mechanistically, BGLF2 promoted the recruitment of SHP1 phosphatase to STAT1 to inhibit its tyrosine phosphorylation. In addition, BGLF2 associated with cullin 1 E3 ubiquitin ligase to facilitate its recruitment to STAT2. Consequently, BGLF2 suppressed ISG induction by IFN-β. Furthermore, BGLF2 also suppressed type II and type III IFN signaling, although the suppressive effect on type II IFN response was milder. When pretreated with IFN-β, host cells became less susceptible to primary infection of EBV. This phenotype was reversed when expression of BGLF2 was enforced. Finally, genetic disruption of BGLF2 in EBV led to more pronounced induction of ISGs. Our study unveils the roles of BGLF2 not only in the subversion of innate IFN response but also in lytic infection and reactivation of EBV.
IMPORTANCE Epstein-Barr virus (EBV) is an oncogenic virus associated with the development of lymphoid and epithelial malignancies. EBV has to subvert interferon-mediated host antiviral response to replicate and cause diseases. It is therefore of great interest to identify and characterize interferon-antagonizing proteins produced by EBV. In this study, we perform a screen to search for EBV proteins that suppress the action of interferons. We further show that BGLF2 protein of EBV is particularly strong in this suppression. This is achieved by inhibiting two key proteins STAT1 and STAT2 that mediate the antiviral activity of interferons. BGLF2 recruits a host enzyme to remove the phosphate group from STAT1 thereby inactivating its activity. BGLF2 also redirects STAT2 for degradation. A recombinant virus in which BGLF2 gene has been disrupted can activate host interferon response more robustly. Our findings reveal a novel mechanism by which EBV BGLF2 protein suppresses interferon signaling.
KEYWORDS: Epstein-Barr virus, BGLF2, JAK-STAT signaling, SHP1 phosphatase
INTRODUCTION
Epstein-Barr virus is a gammaherpesvirus that infects more than 95% of adults all over the world. Infection with EBV causes epithelial and lymphoid malignancies such as Burkitt’s lymphoma and nasopharyngeal carcinoma in a small subset of individuals (1, 2). Although the primary infection is asymptomatic in early childhood, the infection during adolescence is accompanied by infectious mononucleosis. After initial phase of infection, EBV establishes latency in host B and epithelial cells throughout life span, which is characterized by specific latent gene expression, until it is reactivated to undergo productive phase of life cycle. During the lytic phase of EBV replication cycle, about 80 EBV lytic genes are expressed, along with active replication of the viral genome, resulting in virion production. These lytic proteins play essential roles in viral replication, transcription, virion assembly and packaging in the infected cells (3).
Upon recognition of pathogen-associated molecular patterns of EBV such as viral DNA and RNA, host innate immunity is activated to restrict viral infection, replication, and reactivation. To circumvent this host defense, EBV uses some of its lytic proteins to suppress the production and signaling of antiviral proteins such as interferons (IFNs). Particularly, EBV targets the signaling pathways of Toll-like receptors (TLRs), RIG-I, cGAS, NF-κB, and JAK-STAT to alter production and signaling of type I, type II, and type III IFNs (4, 5). For instance, EBV-encoded lytic protein BGLF5 targets TLR9 for degradation during productive cycle (6). BPLF1, a deubiquitinase and a late lytic tegument protein, interferes with the ubiquitination and activation of signal transducers in the TLR and RIG-I pathways, thereby inhibiting type I IFN production in infected cells (7–9). In addition, the master lytic inducer BZRF1/Zta and tegument protein LF2 interact with IRF7 transcription factor to prevent its dimerization and activation, leading to inhibition of type I IFN expression (10, 11). In addition to these viral proteins, EBV-encoded miRNAs can also target the translation of signal transducer and adaptor proteins in the RIG-I pathway to inhibit IFN induction (12). While it is generally believed that the interplay between EBV and innate immunity decides the fate of infection, maintenance, and reactivation of EBV inside cells, the molecular details of this interplay remain to be understood. In particular, EBV-encoded suppressors of IFN signaling have not been systematically identified and characterized.
The antiviral and immunomodulatory effects of all three types of IFNs are mediated through JAK-STAT signaling (13). Binding of IFNs to their respective receptors triggers the activation of JAK-STAT pathways. Whereas receptors IFNAR1 and IFNAR2 are specific to type I IFNs such as IFN-α, IFN-β, and IFN-ε, receptors IL-10-R2 and IFNLR1 are proprietary to type III IFNs, including IFN-λ. The binding with these receptors activates the associated tyrosine kinases TYK2 and JAK1, which in turn phosphorylate STAT1 and STAT2 transcription factors. Phosphorylated and activated STAT1 and STAT2 join with IRF9 to form a ternary complex, which binds to IFN-stimulated response elements (ISREs) in the promoter region to switch on the transcription of IFN-stimulated genes (ISGs) such as MxA, ISG15, ISG56, and OAS1 (14). Likewise, binding of IFN-γ with the receptors IFNGR1 and IFNGR2 activates JAK1 and JAK2 kinases that act on STAT1. Phospho-STAT1 forms a homodimer named γ-activated factor (GAF), which binds to a γ-activated sequence (GAS) in the promoter region of ISGs such as IP10, IRF1, and MCP1 (14, 15). JAK-STAT signaling is negatively regulated at different levels by protein tyrosine phosphatase such as SHP1 and by another protein family known as suppressor of cytokine signaling (SOCS) (16, 17). Plausibly, EBV also adopts multiple strategies to subvert JAK-STAT signaling. It will be of particularly great interest to see how EBV-encoded IFN antagonists might usurp cellular negative regulators or cripple cellular activators to perturb critical events in the JAK-STAT pathway.
In this study, we set out to perform a functional screen for novel EBV-encoded modulators of IFN signaling using a previously established expression library of 55 EBV proteins (18). Among multiple hits that were experimentally validated, we chose for further analysis a tegument protein, BGLF2, that suppresses the signaling of all three types of IFNs. BGLF2 is required for encapsulation of virions during late lytic phase of replication cycle (19). It plays an important role in EBV lytic reactivation by inducing BZLF1/Zta levels in EBV-infected cells (20). It interacts with BKRF4 and BBLF1 (18, 19, 21). It has also been shown to activate AP1 and p38 activity (20, 22), inhibit NF-κB signaling (23), induce cell cycle arrest (24), and trigger global SUMOylation (25). After completion of our study and initial report of our findings in international conferences (26), BGLF2 has recently been shown to suppress type I IFN signaling through association and inhibition of TYK2 tyrosine kinase (27). Although we came to the same conclusion on the suppression of IFN signaling by EBV BGLF2, our results from gain-of-function and infection experiments suggested additional intracellular targets of BGLF2, including SHP1 tyrosine phosphatase and STAT2. Furthermore, we created a BGLF2 mutant EBV to study its innate immunomodulatory function in infected cells.
RESULTS
EBV inhibits type I and type III IFN signaling.
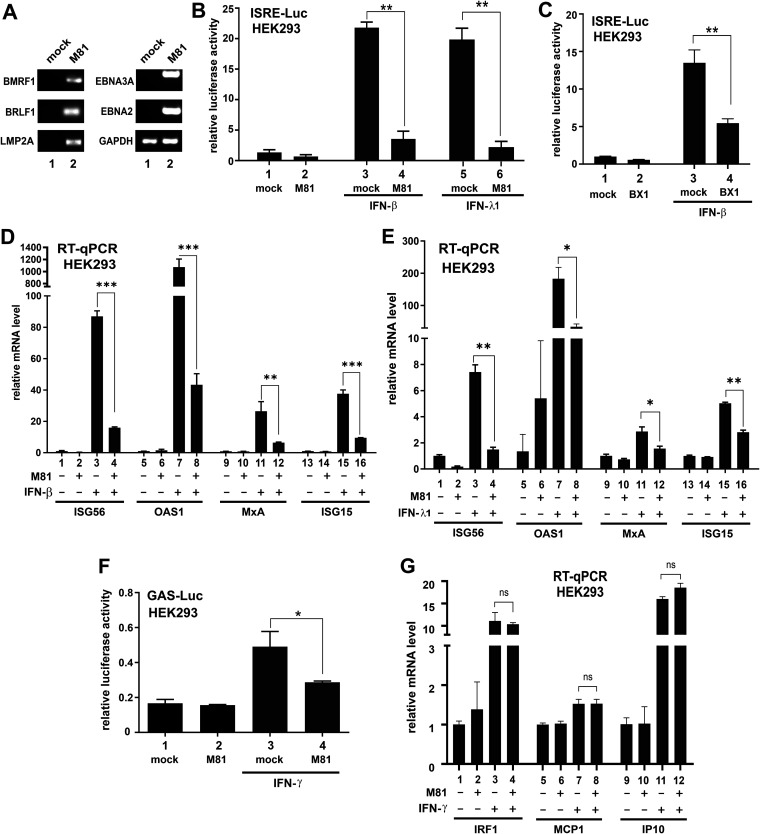
The interplay between IFNs and EBV is complex. Whereas IFN-α and IFN-γ exert inhibitory effects on EBV infection and transformation (28–31), they have no influence on EBV early antigen expression or lytic reactivation under some circumstances (32, 33). In some settings, EBV exhibits resistance to IFN signaling (27, 34, 35). Most studies on EBV were conducted with lymphotropic laboratory strains such as B95-8. The interplay of IFNs and epitheliotropic EBV such as M81, which is significantly different from B95-8 (36), has not been well documented. Thus, we set out to investigate the impact of infection with M81 strain of EBV on IFN signaling in HEK293 cells. This is not an ideal infection system. However, due to its high transfection and infection efficiency, it is widely used as the producer cell line for recombinant EBV (36) and as a model for mechanistic studies. At 48 h after infection, both lytic (BMRF1 and BRLF1/Rta) and latent (LMP2A, EBNA2, and EBNA3A) genes were expressed in these cells (Fig. 1A). Probably, some of the cells are still in lytic phase, but EBV has also started to express some latent genes. Little is known about how EBV might interact with type III IFNs. Since type I and type III IFN signaling regulates a common set of ISGs driven by ISREs (13, 14), a luciferase reporter plasmid under the control of ISREs (ISRE-Luc) served as the readout in our analysis of M81. Cells were infected with cell-free M81 virus. ISRE-dependent reporter expression was robustly induced by IFN-β and IFN-λ1, but this induction was blunted in M81-infected cells (Fig. 1B; bar 4 compared to 3, and bar 6 compared to 5). A similar observation was also made with BX1 strain of EBV (Fig. 1C, bar 4 compared to bar 3), indicating that the effect was unlikely strain specific. The inhibition of IFN-β and IFN-λ1 signaling by M81 was also confirmed by comparison of mRNA levels of four representative ISGs in mock-infected and M81-infected HEK293 cells by quantitative RT-PCR. The basal levels of ISG56, OAS1, MxA, and ISG15 transcripts were induced upon treatment with IFN-β and IFN-λ1. However, the transcription of all four ISGs that are specifically responsive to IFN-β and IFN-λ1 was inhibited in M81-infected cells (Fig. 1D and E, bars 4, 8, 12, and 16). Although M81 showed a modest suppressive effect on IFN-γ-induced activation of GAS-Luc activity (Fig. 1F), there was no significant difference in the mRNA levels of IP10, IRF1, and MCP1, which are type II IFN-responsive ISGs (Fig. 1G). Hence, expression of these three ISGs might be under complex control in EBV-infected cells. Taken together, EBV suppresses type I and type III IFN signaling potently. EBV might also exert a mild suppressive effect on IFN-γ-induced activation of GAF that binds to GAS, but it has no influence on the expression of key IFN-γ-responsive ISGs.
FIG 1.
Suppression of IFN signaling by EBV. The M81 or BX1 strain of EBV was used to infect HEK293 cells. Cells were treated with IFN after 24 h of infection and harvested after another 24 h. Error bars represent the SD (n = 3). (A) RT-PCR analysis of selected lytic and latent genes in M81-infected HEK293 cells. Cells were harvested 48 h after infection. (B and C) ISRE-Luc luciferase reporter assays. pISRE-Luc firefly luciferase reporter was transfected into HEK293 cells before infection, along with pSV-Rluc Renilla luciferase plasmid as an internal control. The ISRE promoter was induced with treatment of 1,000 U/ml IFN-β or 100 ng/ml IFN-λ1 for 24 h. Cells were lysed, and results are represented as the fold activation. The difference between mock- and M81- or BX1-infected samples was found to be statistically significant. (D and E) RT-qPCR analysis of ISG mRNAs responsive to IFN-β or IFN-λ1. Relative mRNA level was presented as the fold activation. The difference between mock- and EBV-infected samples was found to be statistically significant. (F) GAS-Luc reporter assay. Cells were treated with 50 ng/ml IFN-γ for 24 h. The results are represented as the fold activation. The difference between mock- and EBV-infected samples was found to be statistically significant. (G) RT-qPCR analysis of ISG mRNAs responsive to IFN-γ. The difference between mock- and EBV-infected samples was not significant (ns) statistically (P > 0.05).
Screening of EBV genes that influence IFN signaling.
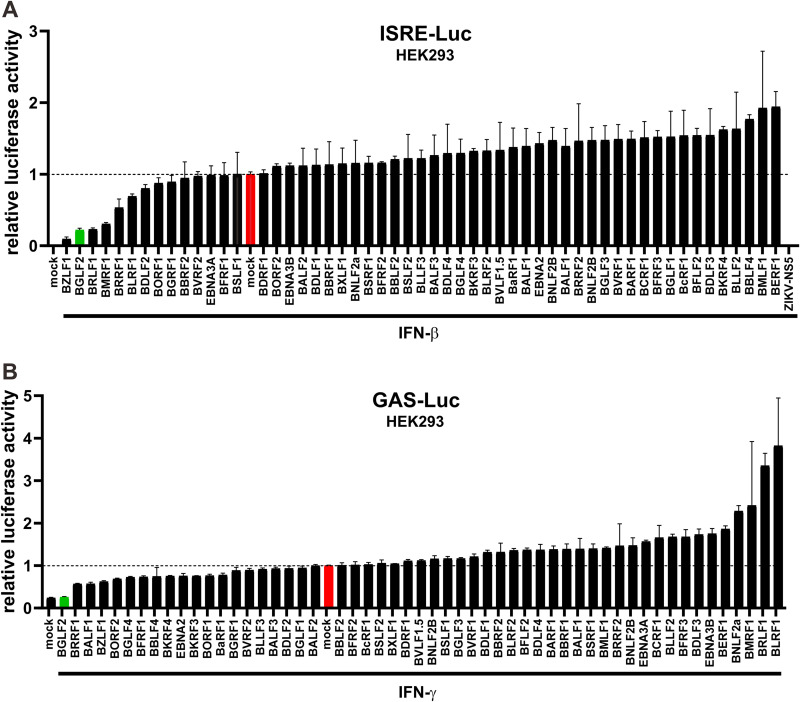
EBV expresses only a few viral proteins during latency, but the number of viral proteins expressed in the lytic phase of its replication cycle increases to ∼80 (3). Above, we demonstrated the suppression of IFN signaling by EBV. To identify the EBV proteins that are responsible for this suppression, an expression library consisting of 55 EBV latent and lytic genes was used in the functional screens for EBV-encoded IFN modulators. Each construct was individually transfected into HEK293 cells, along with ISRE-Luc and GAS-Luc reporter plasmids, respectively. Cells were then treated with IFN-β or IFN-γ. The mock-transfected and mock-treated cells were set as references (Fig. 2, red bars). Zika virus-encoded NS5 protein, which has previously been reported to inhibit type I IFN signaling (37), was used as a positive control in the IFN-β experiment (Fig. 2A). The effectiveness of the IFN-β screen was verified since BZLF1/Zta and BRLF1/Rta, which are known to suppress IFN-β signaling (10, 38), were identified in the screen (Fig. 2A). Likewise, BZLF1 that has been shown to suppress IFN-γ signaling (35) was also found in the IFN-γ screen, lending support to its validity. Interestingly, BGLF2 was singled out to be a more potent inhibitor of both type I and type II IFN signaling (Fig. 2, green bars).
FIG 2.
Screening of EBV proteins for modulators of type I and type II IFN signaling. Individual plasmid constructs expressing EBV proteins were transfected into HEK293 cells, along with pISRE-Luc (A) or pGAS-Luc (B) plus pSV-Rluc as an internal control. The ISRE and GAS promoters were induced by treatment with 1,000 U/ml of IFN-β and 50 ng/ml of IFN-γ, respectively, for 24 h. The relative luciferase activity is represented as the fold activation. The mock group transfected with an empty vector is highlighted in red, which was used as a reference to determine the activation or suppression of the two reporters. The BGLF2 group is indicated in green.
BGLF2 inhibits all three types of IFN signaling.
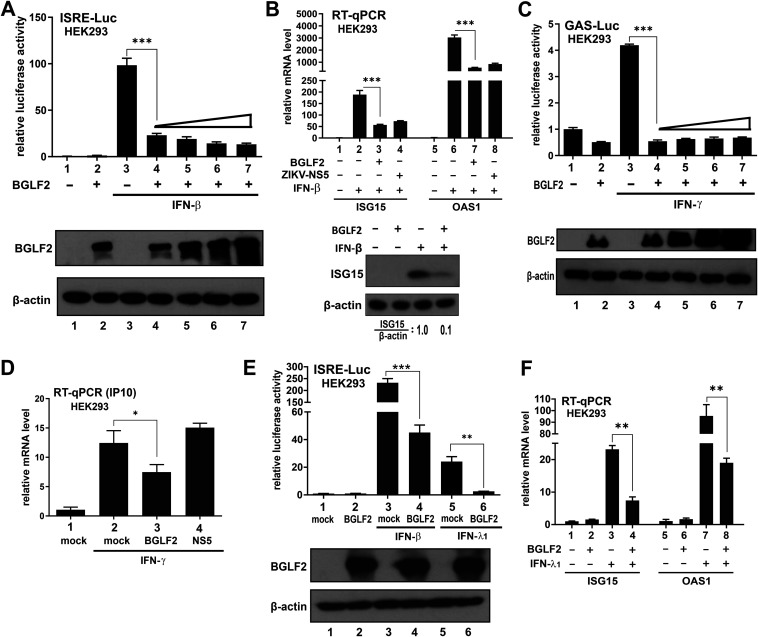
The inhibitory effect of BGLF2 on IFN signaling was further validated. HEK293 cells were chosen as a model cell line to study the effect of BGLF2 on IFN signaling because of its good transfection efficiency and integrity of the IFN signaling machinery. Although dose-dependent inhibition of ISRE-Luc activity by BGLF2 was observed, only a marginal difference was seen when the dose was escalated (Fig. 3A, bars 4 to 7). Quantification of the mRNA levels of ISG15 and OAS1 also confirmed the inhibitory effect of BGLF2 on IFN-β-induced transcription of these two IFN-β-responsive ISGs (Fig. 3B, bar 3 compared to bar 2, and bar 7 compared to bar 6). In addition, IFN-β-induced elevation of ISG15 protein expression was reversed by BGLF2 (Fig. 3B, lower panel). BGLF2 also inhibited IFN-γ-induced GAS-Luc activity when overexpressed in HEK293 cells; however, the inhibitory effect plateaued at the lowest dose of BGLF2 expression plasmid (Fig. 3C). Consistent with the result from reporter assay, IFN-γ-induced elevation of IP10 mRNA was dampened by BGLF2 (Fig. 3D, bar 3 compared to bar 2). Furthermore, expression of BGLF2 effectively suppressed IFN-λ1-induced activation of ISRE-Luc activity (Fig. 3E, bar 6 compared to bar 5), as well as ISG15 and OAS1 mRNA expression (Fig. 3F, bar 4 compared to bar 3, bar 8 compared to bar 7).
FIG 3.
Suppression of IFN signaling by BGLF2 in HEK293 cells. (A) Dose dependence of the suppressive effect of BGLF2. Increasing doses of BGLF2 plasmid were transfected into HEK293 cells. The indicated groups of cells were stimulated with IFN-β. Expression of BGLF2 was verified by Western blotting (bottom panel). (B) RT-qPCR analysis of ISG15 and OAS1 mRNA in response to IFN-β. Zika virus NS5 served as a positive control. The expression of ISG15 protein in IFN-β-treated cells was verified by Western blotting (bottom panel). The ratios of the ISG15 protein level normalized to that of β-actin were derived by densitometry. (C) GAS-Luc luciferase assay in IFN-γ-treated HEK293 cells. (D) RT-qPCR analysis of IP10 mRNA induced by IFN-γ. The mock group was transfected with an empty vector. (E) ISRE-Luc reporter assay in IFN-λ1-treated cells. The ISRE activity was induced by treatment with 1,000 U/ml of IFN-β or 100 ng/ml of IFN-λ1. An empty plasmid was used in the mock-transfected group. (F) RT-qPCR analysis of ISG15 and OAS1 mRNA in response to IFN-λ1.
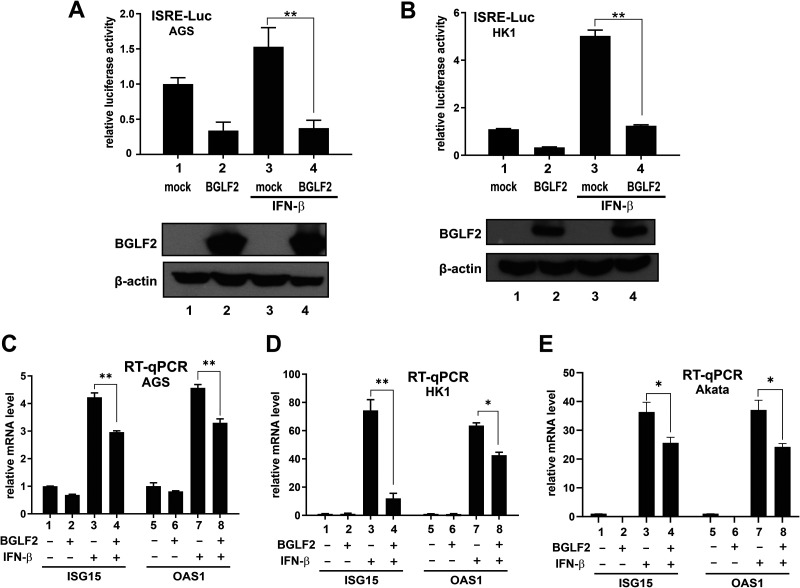
The inhibitory effect of BGLF2 on type I IFN signaling was further validated in epithelial and B cell lines that are susceptible to natural infection with EBV. EBV infection is etiologically associated with the development of gastric and nasopharyngeal carcinoma (1, 2). In this connection, an EBV-negative gastric carcinoma cell line AGS (39), and another nasopharyngeal carcinoma cell line, HK1, which has lost EBV (40), were chosen for further analysis. BGLF2 was found to inhibit the activation of ISRE-Luc reporter, as well as ISG15 and OAS1 mRNA expression, in response to IFN-β in both AGS (Fig. 4A and C) and HK1 (Fig. 4B and D) cells. Our analysis was also extended to EBV-negative Akata cells derived from Burkitt’s lymphoma (41). Expression of BGLF2 through a lentiviral vector was found to suppress IFN-β-induced expression of ISG15 and OAS1 mRNAs in Akata cells (Fig. 4E, bar 4 compared to bar 3, bar 8 compared to bar 7). The milder suppressive effect was plausibly due to suboptimal lentiviral transduction or BGLF2 expression. Collectively, BGLF2 acts as a potent suppressor of IFN signaling in both epithelial and B cells.
FIG 4.
Suppression of IFN signaling by BGLF2 in AGS, HK1 and Akata cells. (A to D) ISRE-Luc reporter assays and RT-qPCR analysis of ISG mRNAs in AGS and HK1 cell lines expressing BGLF2. An empty plasmid was used in the mock transfection. (E) RT-qPCR analysis of ISG mRNAs in Akata cells expressing BGLF2. EBV-negative Akata cells were transduced with a lentiviral vector expressing BGLF2. The negative control group was mock transduced with an empty lentiviral vector.
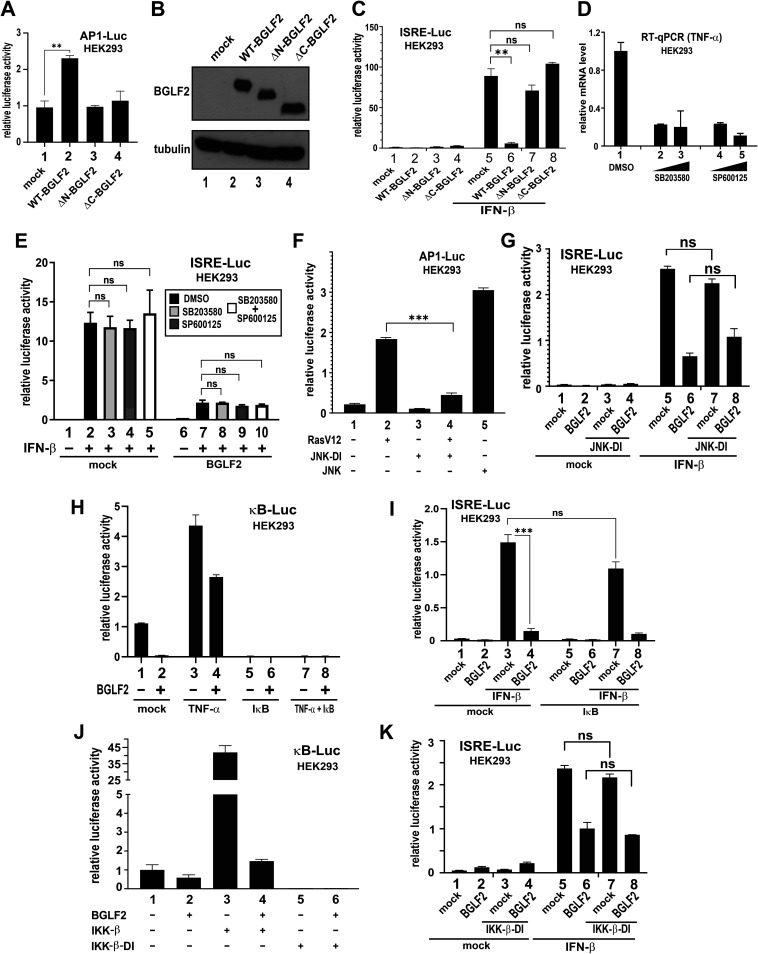
Inhibition of JAK-STAT signaling by BGLF2 is independent of JNK, p38, AP1, and NF-κB pathways.
BGLF2 has been reported to stimulate p38 and JNK activity leading to the activation of AP1 transcription factor (20, 22). It was therefore intriguing to investigate whether the inhibitory effect of BGLF2 on IFN signaling might be mediated through p38 or JNK. BGLF2 is known to lose its ability to activate AP1 upon removal of 66 amino acids from the C terminus of protein (22). Indeed, truncation of either the N or C terminus of BGLF2 resulted in almost complete abrogation of its AP1-activating property (Fig. 5A, bars 3 and 4 compared to bar 2). Both truncated forms of BGLF2, when expressed to comparable levels to that of wild-type (WT) BGLF2 in transfected cells (Fig. 5B), were also unable to suppress IFN-β-induced activation of ISRE-Luc activity (Fig. 5C, bars 7 and 8 compared to bar 6). These results suggested that an overlapping functional domain or conformation might be required for the ability of BGLF2 to modulate AP1 and ISRE activity. To further investigate this possibility, we employed SB203580 and SP600125, which are highly specific pharmaceutical inhibitors of p38 and JNK, respectively. In a control experiment, these inhibitors were fully capable of suppressing the expression of tumor necrosis factor α (TNF-α) mRNA (Fig. 5D), which requires p38 and JNK activity (42). The IFN-inhibitory effect of BGLF2 was unaffected upon treatment with SB203580 and SP600125, individually or combined (Fig. 5E, bars 8 to 10 compared to bar 7). Consistent with this, expression of a dominant-inactive (DI) form of JNK (43), which exhibited a pronounced suppressive effect on the activation of AP1-Luc activity by a dominant-active form of Ras named RasV12 (Fig. 5F, bar 4 compared to bar 2), a known activator of JNK (44, 45), had no influence on the suppression of IFN-β-induced activation of ISRE activity by BGLF2 (Fig. 5G, bar 8 compared to bar 6). Hence, the suppression of IFN signaling by BGLF2 was most likely independent of p38 and JNK activation, as well as the consequent activation of AP1.
FIG 5.
BGLF2 targets STATs in JAK/STAT signaling, independently of p38, AP1, or NF-κB. (A) AP1-Luc reporter assay with BGLF2 truncated mutants. ΔN-BGLF2 and ΔC-BGLF2 lack 66 amino acids at the N and C termini, respectively. (B) Western blot analysis of WT and truncated BGLF2 proteins. (C) ISRE-Luc reporter assay with BGLF2 truncated mutants in the absence or presence of IFN-β. (D) Effect of p38 and JNK inhibitors on TNF-α mRNA expression. HEK293 cells were mock treated with DMSO or treated with increasing concentrations of SB203580 and SP600125, respectively, for 2 h at 37°C. (E) Effect of p38 and JNK inhibitors on ISRE-Luc reporter activity activated by IFN-β. p38 inhibitor SB203580 (5 μM), JNK inhibitor SP600125 (20 μM), or both were added for 1 h before treatment with IFN-β for 24 h. (F) Effect of dominant-inactive JNK on RasV12-induced activation of AP1-Luc activity. A dominant-active form of Ras known as RasV12 (45) and a dominant-inactive form of JNK designated JNK-DI, which is the APF mutant of JNK1 (43), were used. The positive JNK control in this experiment is a dominant-active form of JNK1 known as MKK7B2Jnk1a1 (43). (G) Effect of dominant-inactive JNK on BGLF2-dependent activation of ISRE-Luc activity. (H) Effect of IκB super-repressor on BGLF2-induced suppression of NF-κB. κB-Luc reporter was stimulated by treatment with 30 ng/ml of TNF-α for 20 h. (I) Effect of IκB super-repressor on BGLF2-induced suppression of ISRE-Luc activity. IκB super-repressor was expressed in cells for 24 h. Cells were then treated with IFN-β for another 24 h. (J and K) Effect of dominant-inactive IKK-β (IKK-β-DI) on BGLF2 modulation of κB-Luc and ISRE-Luc activity. A previously described (44) dominant-inactive form of IKK-β (IKK-β-DI) was used. WT IKK-β served as a positive control. An empty plasmid was used in the mock transfection. ns, not significant statistically.
An inhibitory effect of BGLF2 on NF-κB signaling has also been suggested (23). Generally consistent with this, expression of BGLF2 was found to suppress TNF-α-induced activation of κB-Luc activity (Fig. 5H, bar 4 compared to bar 3). The NF-κB activity induced by TNF-α was completely erased when an IκB super-repressor, in which S32A S36A mutations had been introduced to inactivate the phosphorylation sites (42), was expressed to inhibit NF-κB signaling (Fig. 5H, bars 5 to 8). Since the expression of IκB super-repressor had no influence on the inhibitory effect of BGLF2 on IFN-β-induced activation of ISRE-Luc activity (Fig. 5I, bars 7 and 8 compared to bars 3 and 4), the potent suppression of IFN-β signaling by BGLF2 was unlikely due to its mild inhibitory effect on NF-κB signaling. Likewise, although the expression of a previously described (46) DI form of IKK-β (IKK-β-DI) effectively erased κB-Luc activity (Fig. 5J, bars 5 and 6 compared to bars 1 and 2), BGLF2-dependent suppression of IFN-β-induced activation of ISRE-Luc activity was not affected (Fig. 5K, bar 8 compared to bar 6). These results did not support the notion that the suppressive effect of BGLF2 on IFN-β signaling might be mediated through inhibition of NF-κB.
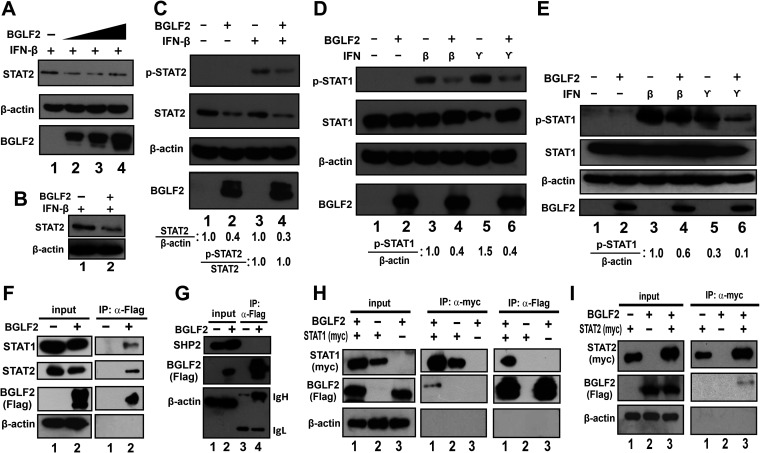
We next assessed the direct effect of BGLF2 on JAK-STAT signaling. As the first step, the impact of BGLF2 on STAT1 and STAT2 expression and phosphorylation was examined. When BGLF2 was expressed in IFN-β-treated HEK293 cells, the steady-state levels of STAT2 protein decreased (Fig. 6A, lanes 2 to 4). Likewise, less STAT2 protein was detected when BGLF2 was expressed in IFN-β-treated AGS cells (Fig. 6B). The inhibitory effect of BGLF2 on IFN-β-induced elevation of p-STAT2 level was also seen (Fig. 6C, lane 4 compared to lane 3). However, this was ascribed primarily to the reduction of total STAT2 protein since the relative amount of p-STAT2 versus total STAT2 remained unchanged (Fig. 6C, lane 4 compared to lane 3). In contrast, BGLF2 showed no effect on total STAT1 protein levels, instead the phosphorylation of STAT1 was reduced upon induction by IFN-β or IFN-γ (Fig. 6D). The same pattern was also observed in AGS cells (Fig. 6E). BGLF2 was found to interact with both STAT1 and STAT2 as analyzed by coimmunoprecipitation assay in HEK293 cells. Both STAT1 and STAT2 but not SHP2 as a negative control were detected in the immunoprecipitate that contains Flag-tagged BGLF2 (Fig. 5F, lane 2 on the right; Fig. 5G, lane 4; and Fig. 5H, right panel, lane 3). Reciprocally, BGLF2 was found in the immunoprecipitates containing myc-tagged STAT1 or STAT2 (Fig. 5H and I, left panel, lane 1 or 3). Thus, BGLF2 interacts with STAT1 and STAT2 to modulate their expression and activation.
FIG 6.
BGLF2 targets STATs in JAK/STAT signaling. (A to E) Influence of BGLF2 on STAT1 and STAT2 expression and phosphorylation. BGLF2 was overexpressed in HEK293 (A, C, and D) and AGS (B and E) cells; after 48 h, the cells were left untreated or treated with 1,000 U/ml of IFN-β or 50 ng/ml of IFN-γ for 30 min. Cell lysates were collected for analysis of p-STAT1, p-STAT2, total STAT1, and total STAT2 protein levels by Western blotting. β-Actin was used as an internal control. The BGLF2-negative control group was mock transfected with an empty vector. The relative STAT2 and p-STAT1 amounts normalized to β-actin, as well as the relative amount of p-STAT2 versus total STAT2, were assessed by densitometry. (F to I) Association of BGLF2 with STAT1 and STAT2 but not SHP2. BGLF2-Flag alone (F and G) or together with either STAT1-myc (B) or STAT2-myc (C) were overexpressed in HEK293 cells for 48 h before treatment with 10 μM MG132 for 4 h. Cell lysates were collected, and coimmunoprecipitation was performed using anti-Flag or anti-myc antibody. Endogenous or myc-tagged STAT1, STAT2, and SHP2 were probed by Western blotting.
BGLF2 reduces STAT1 phosphorylation by recruiting SHP1 phosphatase.
BGLF2 was found to reduce the phosphorylation of STAT1 when induced with IFN-β or IFN-γ, while the total STAT1 remained unchanged in the absence and presence of BGLF2. SOCS proteins are not only expressed as negative-feedback regulators for IFN signaling but also induced by viral proteins to downregulate IFN signaling (17). Particularly, SOCS1 and SOCS3 have been reported to be mostly induced by viral proteins to inhibit JAK or STAT phosphorylation (47–49). Therefore, the possible involvement of SOCS1 or SOCS3 in BGLF2-mediated inhibition of STAT1 phosphorylation was explored. Although mRNA levels of SOCS1 and SOCS3 were induced up to 1.5-fold in the presence of BGLF2, as measured by RT-qPCR (Fig. 7A), no significant difference in their protein levels was found in Western blotting, in the absence or presence of BGLF2 (Fig. 7B).
FIG 7.
BGLF2 recruits SHP1 tyrosine phosphatase to result in STAT1 dephosphorylation. (A and B) Effect of BGLF2 on SOCS1 and SOCS3 expression. BGLF2 was overexpressed in HEK293 cells. The mock group was transfected with an empty vector. The levels of SOCS1 and SOCS3 mRNA and protein were analyzed by RT-qPCR (A) and Western blotting (B). (C) Effect of Na3VO4 on suppression of IFN-β-induced STAT1 phosphorylation by BGLF2. BGLF2 was overexpressed in HEK293 cells. Cells were then treated with 200 μM Na3VO4 for 1 h at 37°C, after 48 h of transfection. Cells were subsequently treated with IFN-β for 30 min. Cell lysates were collected for Western blotting. (D to F) Impact of SHP1 knockdown on suppression of IFN-β-induced elevation of p-STAT1 by BGLF2. siSHP1 was used to knock down the expression of SHP1 in HEK293 cells. The mRNA and protein levels of SHP1 were analyzed by RT-qPCR (D) and Western blotting (E), respectively. The effect of siSHP1 on p-STAT1 levels was analyzed by Western blotting (F). (G) Impact of BGLF2 on SHP1 protein expression. (H) BGLF2 recruits SHP1 to STAT1 complex. SHP1 and BGLF2-Flag were overexpressed in HEK293 cells for 48 h. Cell lysates were collected, and coimmunoprecipitation was performed using anti-STAT1 antibody to pull down endogenous STAT1. Then, the association with SHP1 was analyzed by Western blotting in the absence or presence of BGLF2. (I) Impact of BGLF2 on JAK1 and TYK2 phosphorylation. Cells were left untreated or treated with 1,000 U/ml of IFN-β for 5 and 10 min after 48 h of transfection. The cell lysate was collected and analyzed for p-JAK1, p-TYK2, total JAK1, and total TYK2 protein levels by Western blotting.
In addition to SOCS proteins, tyrosine phosphatases are also critically involved in the regulation of JAK-STAT signaling (16). To shed light on this, HEK293 cells overexpressing BGLF2 were treated or left untreated with sodium orthovanadate (Na3VO4), a broad-spectrum inhibitor of tyrosine phosphatases (50). Upon treatment with IFN-β, the level of p-STAT1 was restored when treated with Na3VO4, even in the presence of BGLF2 (Fig. 7C, lane 6 compared to lanes 4 and 5). To identify the tyrosine phosphatase involved, a series of RNA interference (RNAi) experiments were performed with small interfering RNAs (siRNAs) and SHP1 was singled out as a strong candidate. SHP1 is a protein tyrosine phosphatase which acts as a negative regulator of JAK-STAT signaling by targeting key components of the pathway (16, 50). Further experimental validation was therefore carried out. SHP1 was knocked down in HEK293 cells using standard siRNA. A knockdown efficiency of 60% to 80% at the level of SHP1 transcript or protein was achieved for siSHP1 (Fig. 7D and E, bar 2 compared to bar 1). The level of p-STAT1 was restored in the presence of BGLF2 and siSHP1, suggesting the involvement of SHP1 tyrosine phosphatase in BGLF2-mediated p-STAT1 reduction (Fig. 7F, lane 6 compared to lane 4). Although total protein levels of SHP1 remained unchanged in the presence and absence of BGLF2 (Fig. 6G), both SHP1 and BGLF2 were found to associate with STAT1 (Fig. 7H, lane 2 compared to lane 1 on the right). These results are consistent with the notion that BGLF2 might recruit SHP1 to the STAT1 signalosome.
Since SHP1 might also mediate dephosphorylation of other key kinases in JAK-STAT signaling such as JAK1 and TYK2 (16), which have been shown to be targeted by BGLF2 (27), we expanded our analysis to p-JAK1 and p-TYK2 in HEK293 cells overexpressing BGLF2. Interestingly, when JAK1 and TYK2 phosphorylation was induced by treatment with IFN-β for 5 or 10 min, the levels of p-JAK1 and p-TYK2 were diminished when BGLF2 was overexpressed (Fig. 7I, lane 4 compared to lane 3, lane 6 compared to lane 5). It will be of interest to see whether this suppressive effect might also be mediated through SHP1.
BGLF2 promotes STAT2 degradation through proteasomal degradation pathway.
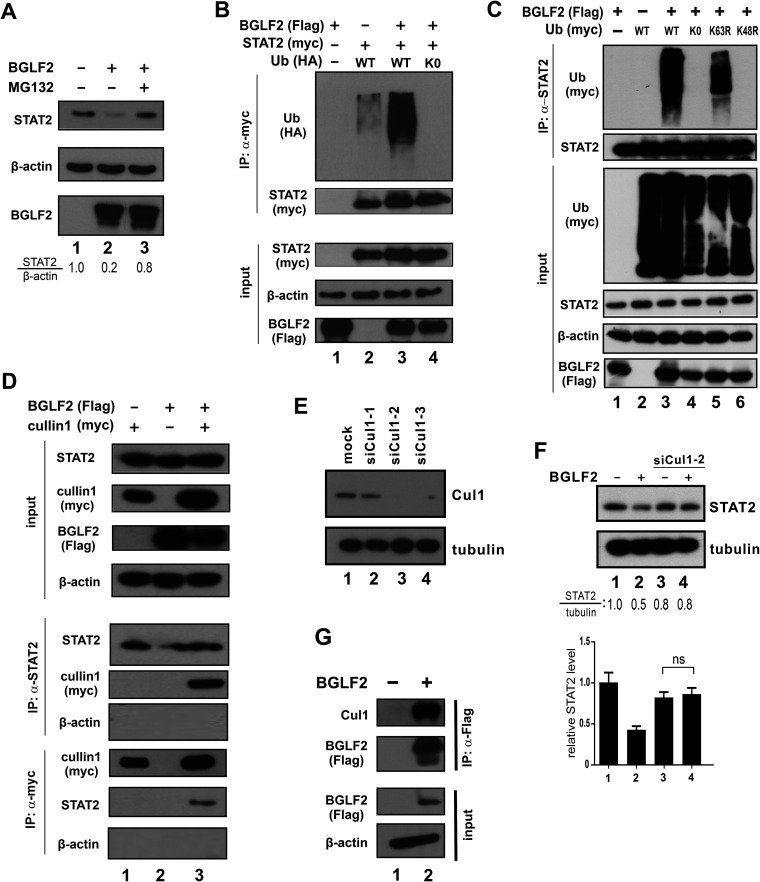
The possible destabilizing effect of BGLF2 on STAT2 protein prompted us to explore further whether proteasomal degradation of STAT2 might be affected. Treatment of HEK293 cells with proteasomal inhibitor MG132 restored STAT2 protein level in the presence of BGLF2 (Fig. 8A, lane 3 compared to lane 2), which suggested that BGLF2 might facilitate proteasomal degradation of STAT2.
FIG 8.
BGLF2 promotes K48-linked polyubiquitination and degradation of STAT2. (A) Effect of MG132. BGLF2 was overexpressed in HEK293 cells. Cells were treated with 10 μM MG132 for 6 h. The total STAT2 protein levels in the presence or absence of MG132 and BGLF2 were determined by Western blotting. β-Actin served as an internal control. (B) STAT2 ubiquitination assay. BGLF2-Flag and STAT2-myc were overexpressed in HEK293 cells individually or together with HA-tagged ubiquitin (Ub). After 40 h of transfection, cells were treated with MG132 for 6 h. Cell lysates were collected, and a coimmunoprecipitation assay was performed using anti-myc antibody. Ubiquitination of STAT2 was analyzed by Western blotting. (C) Analysis of ubiquitin mutants. BGLF2-Flag plus wild-type (WT) or mutant forms of ubiquitin-myc were overexpressed in HEK293 cells. Cells were treated with MG132 for 6 h after 40 h of transfection. Cell lysates were collected; next, a coimmunoprecipitation assay was performed using anti-STAT2 antibody, and ubiquitination was analyzed by Western blotting. (D) Recruitment of cullin 1 to STAT2. BGLF2-Flag and cullin 1-myc were overexpressed in HEK293 cells individually or together. Cells were treated with MG132 for 6 h after 40 h of transfection. A coimmunoprecipitation assay was performed reciprocally, using anti-STAT2 and anti-myc antibodies, followed by Western blotting. (E and F) Three siRNAs were used to knock down cullin 1 in HEK293 cells. (E) BGLF2 was overexpressed in HEK293 cells with control siRNA or siCul1-2. (F) Cell lysates were collected after 48 h of transfection and analyzed by Western blotting for STAT2 protein levels. Results from three independent experiments are shown in the lower panel. The difference between the indicated samples was not significant (ns) statistically (P > 0.05). (G) Association of BGLF2 with cullin 1. Coimmunoprecipitation was performed with anti-Flag antibody, and the precipitates were probed with anti-cullin 1.
To test whether BGLF2 can affect the ubiquitination state of STAT2, Flag-tagged BGLF2, myc-tagged STAT2, and hemagglutinin (HA)-tagged ubiquitin were expressed in combinations, as shown in Fig. 8B in HEK293 cells. STAT2 was immunoprecipitated using anti-myc antibody, and the precipitates were blotted for STAT2-myc and ubiquitin-HA. Notably, STAT2 ubiquitination was enhanced in the presence of BGLF2 compared to without BGLF2 (Fig. 8B, lane 3 compared to lane 2). Ubiquitin-K0 was used as another control since it is the lysine-free form of ubiquitin with all lysine residues replaced by arginine, thereby preventing ubiquitination (51). Indeed, no STAT2 ubiquitination ladder was observed when ubiquitin-K0 was expressed (Fig. 8B and C, lane 4). Furthermore, coimmunoprecipitation was also performed to verify K48-linked polyubiquitination of STAT2. Flag-tagged BGLF2 and myc-tagged ubiquitin were expressed in various combinations in HEK293 cells (Fig. 8C). Endogenous STAT2 was pulled down with anti-STAT2 antibody, and the samples were analyzed by Western blotting. A smear of polyubiquitin chains was observed upon expression of wild-type ubiquitin and its K63R mutant, in which lysine 63 had been replaced by arginine (Fig. 8C, lanes 3 and 5). This suggested that K63-linked ubiquitination is unlikely involved. In contrast, no ubiquitination smear was observed in the case of K48R mutant where lysine 48 had been replaced by arginine (Fig. 8C, lane 6). Hence, K48-linked ubiquitination of STAT2 was enhanced when BGLF2 was expressed.
Cullins that are key components of an E3 ubiquitin ligase complex have previously been implicated in viral induction of STAT2 degradation (52, 53). After multiple attempts to explore possible interaction between cullins and STAT2, cullin 1 was found to associate with STAT2 in the presence of BGLF2 (Fig. 8D, lane 3 compared to lane 2). Among three siRNAs that target cullin 1, siCul1-2 was found to be capable of achieving more than 90% knockdown efficiency constantly (Fig. 8E) and was therefore chosen for further knockdown experiments in HEK293 cells. When cullin 1 expression was depleted by siCul1-2, the level of STAT2 in BGLF2-expressing cells was restored (Fig. 8F, lane 4 compared to lanes 2 and 3). As inferred from coimmunoprecipitation experiment, BGLF2 interacts with cullin 1 in HEK293 cells (Fig. 8G, lane 2 compared to lane 1). Thus, BGLF2 might interact with cullin 1 to facilitate its recruitment to STAT2, leading to ubiquitination and degradation of the latter.
BGLF2 facilitates EBV infection and reactivation by perturbing type I IFN signaling.
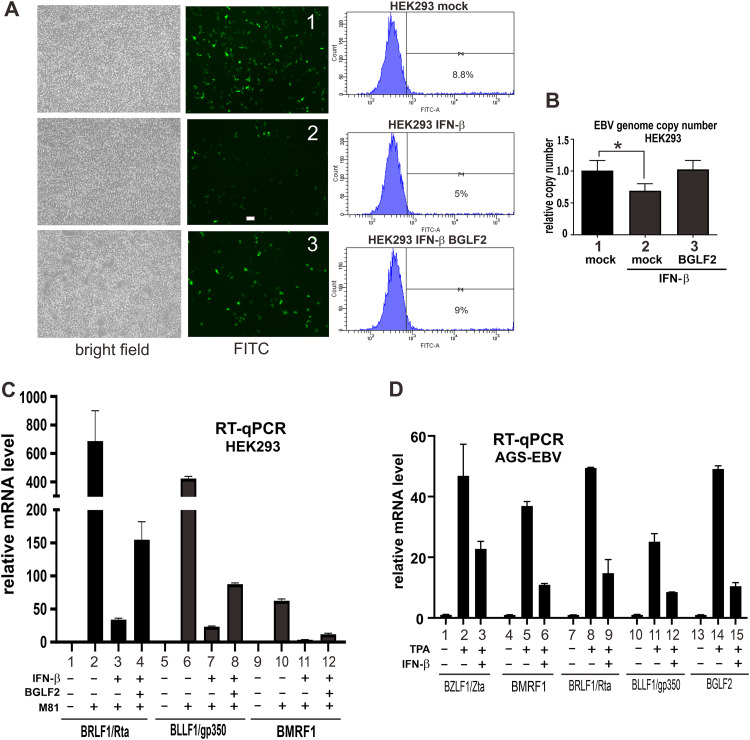
Since M81 strain of EBV is GFP marked, infected cells could be easily identified as GFP-positive cells. This was harnessed to study the impact of IFN-β and BGLF2 expression on EBV infection. In HEK293 cells pretreated with IFN-β, the infectivity of EBV was reduced compared to the mock-treated group (Fig. 9A, subpanel 2 compared to subpanel 1). This phenotype was reversed when BGLF2 was overexpressed in the cells before IFN treatment (Fig. 9A, subpanel 3 compared to subpanels 2 and 1). The trend was confirmed by flow cytometric analysis of GFP-positive cells (Fig. 9A, right panels). Consistently, the EBV genome copy number was also reduced when cells were treated with IFN-β (Fig. 9B, bar 2 compared to bar 1). The genome copy number was again restored when BGLF2 was expressed before IFN-β treatment. In addition, BGLF2 effectively reversed IFN-β-induced suppression of the expression of EBV lytic genes (BLLF1/gp350, BMRF1, and BRLF1/Rta) in M81-infected HEK293 cells (Fig. 9C, bars 4, 8, and 12). Therefore, BGLF2 prevents IFN-β-mediated inhibition of EBV infection.
FIG 9.
BGLF2 facilitates EBV primary infection in infected cells. (A and B) Impact of BGLF2 overexpression on EBV primary infection. HEK293 cells were treated with IFN-β for 12 h before EBV infection, with or without BGLF2 overexpression. (A) The infectivity of EBV was proportional to the number of GFP-positive cells observed under a microscope and quantified by flow cytometry. EBV-negative HEK293 cells were used as negative control, and HEK293-EBV-M81 cells served as a positive control for flow cytometric analysis. (B) For the same set of samples, the EBV genome copy number was measured by qPCR with TaqMan-LMP1 probe normalized to the glyceraldehyde 3-phosphate dehydrogenase probe. The mock group was transfected with an empty vector. (C) Impact of BGLF2 on IFN-β suppression of EBV infection in HEK293 cells. M81-infected cells were treated with 2,000 U/ml IFN-β for 12 h with or without BGLF2 overexpression. The expression of EBV immediate early (BRLF1), early (BMRF1), and late lytic (BLLF1) transcripts was analyzed by RT-qPCR. (D) Impact of IFN-β on EBV reactivation. EBV-positive AGS cells were treated with 20 ng/ml TPA for 24 h and then left untreated or treated with 2,000 U/ml IFN-β for 15 h. The expression of EBV immediate-early (BZLF1 and BRLF1), early (BMRF1), and late lytic (BLLF1) transcripts was analyzed by RT-qPCR.
Latently infected EBV-positive gastric carcinoma cell line AGS-EBV was next used to study the effect of type I IFN on EBV lytic reactivation. The cells were treated with 12-O-tetradecanoylphorbol-13-acetate (TPA) to induce lytic reactivation and showed a higher expression of EBV lytic genes, including immediate early genes BZLF1/Zta and BRLF1/Rta, early gene BMRF1, and late lytic gene BLLF1, compared to the control group which was only treated with DMSO (Fig. 9D). When cells were treated with IFN-β, expression of lytic genes induced by TPA was dampened (Fig. 9D, bars 3, 6, 9, and 12). Therefore, although treatment of EBV-positive cells with TPA induced the lytic cycle accompanied by higher expression of early and late lytic genes, their expression was inhibited by type I IFN.
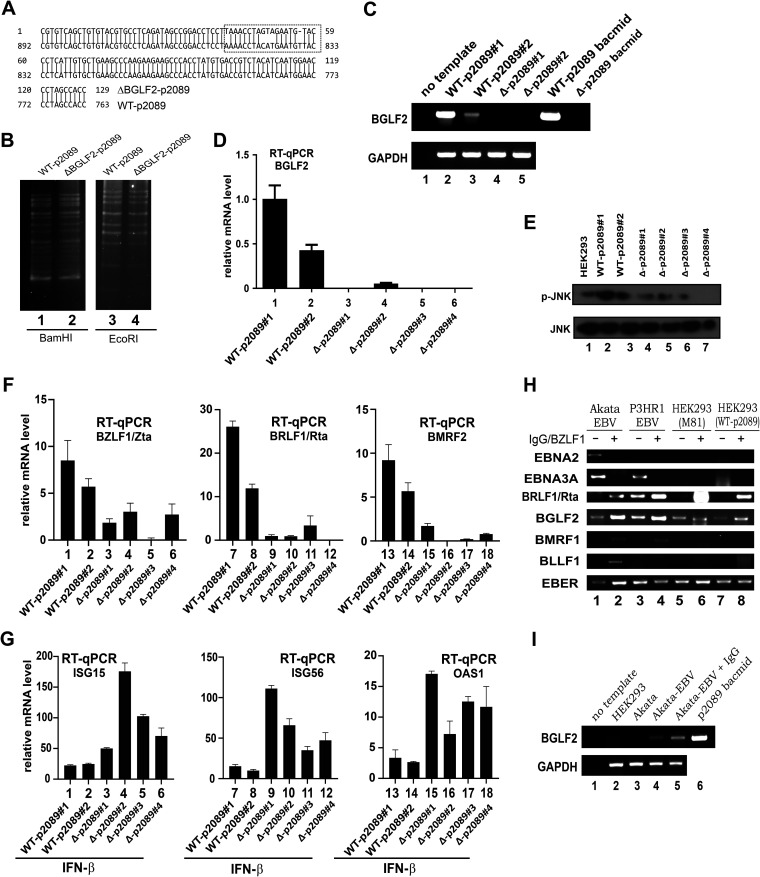
To shed light on the physiological significance of BGLF2 in the context of EBV replication, infection, and reactivation, recombinant EBV mutant defective of BGLF2 was constructed by bacmid recombineering technology on the p2089 bacmid of B95-8 strain of EBV (54, 55). Two codons in the BGLF2 region were replaced with stop codons, and one nucleotide was deleted to create a frameshift, leading to complete disruption of the BGLF2 open reading frame. The desired mutation was found in multiple bacmids as confirmed by sequencing (Fig. 10A). The genomic integrity of the mutant bacmids was determined by BamHI and EcoRI restriction patterning. The band patterns of WT-p2089 and its BGLF2-deficient mutant (ΔBGLF2-p2089) were identical (Fig. 10B), indicating that the mutant clones obtained were correct and intact. They were further used to establish stable HEK293 cell clones carrying the WT-p2089 or ΔBGLF2-p2089 bacmid. Loss of BGLF2 mRNA expression was verified by RT-PCR (Fig. 10C) and RT-qPCR with a TaqMan probe (Fig. 10D). p-JNK was used as another marker for BGLF2, as previously described (20). Cells with ΔBGLF2-p2089 were found to have comparatively lower levels of p-JNK than cells carrying WT-p2089 (Fig. 10E).
FIG 10.
BGLF2 suppresses IFN-β signaling in infected cells. (A) Verification of BGLF2-deficient mutant by sequencing. An alignment of mutant and wild-type sequence is shown. (B) Verification of genome integrity by BamHI and EcoRI patterning. WT-p2089 and ΔBGLF2-p2089 bacmids were digested with BamHI or EcoRI at 37°C for 24 h. The digested products were run on 2% agarose gel overnight at a low voltage. (C and D) Verification of BGLF2 mRNA expression by RT-PCR or RT-qPCR. The primers used can preferentially amplify WT-BGLF2 mRNA but not that of ΔBGLF2. A TaqMan probe was also used in the RT-qPCR. (E) Verification of BGLF2 deficiency through Western blot analysis of the p-JNK levels. (F) Expression of immediate-early and late lytic genes in ΔBGLF2-p2089 stable cells. Cell clones were treated with DMSO or 400 μM TPA plus 5 mM NaBt for 48 h. (G) ISG induction in ΔBGLF2-p2089 stable cells. Cell clones were treated with 1,000 U/ml of IFN-β. ISG15, ISG56 and OAS1 transcripts were analyzed by RT-qPCR. (H) Expression profiles of latent and lytic genes in HEK293 cells stably carrying WT-p2089 and other EBV-positive cells. Lytic reaction of Akata cells was induced by IgG cross-linking. A lytic reactivation of other cells was induced by transfection with a BZLF1/Zta expression plasmid. (I) Expression of BGLF2 gene in EBV-positive cell lines. EBV-negative and -positive Akata cells were induced for EBV lytic cycle using IgG and the expression of BGLF2 mRNA was analyzed by RT-PCR. HEK293 cells and p2089 bacmid were used as negative and positive controls, respectively.
The lytic cycle in the HEK293 cell clones carrying WT-p2089 and ΔBGLF2-p2089 was induced by treatment with TPA and sodium butyrate (NaBt). BZLF1/Zta expression was reduced in the ΔBGLF2-p2089 clones compared to those with WT-p2089 (Fig. 10F, bars 1 to 6). A similar expression pattern was also observed for two other lytic genes BRLF1/Rta and BMRF2, when lytic cycle was induced in these cell clones (Fig. 10F, bars 7 to 18). This result is generally consistent with the expression pattern of lytic genes in cells carrying ΔBGLF2-EBV constructed by others (20). When the cells were treated with IFN-β, the induction of ISG15, ISG56, and OAS1 mRNA expression was more robust in cells carrying ΔBGLF2-p2089 than in cells with WT-p2089 (Fig. 10G, bars 3 to 6, 9 to 12, and 15 to 18).
Both lytic and latent genes are expressed in HEK293 cells freshly infected with M81 (Fig. 1A). In contrast, EBV might enter latency in HEK293 cells stably carrying WT-p2089 and ΔBGLF2-p2089. In this regard, the expression of BGLF2 lytic gene in cells harboring WT-p2089 (Fig. 10C and D) was surprising. With this in mind, we sought to analyze the expression of BGLF2 and other lytic and latent genes by RT-PCR in three other lines of EBV-positive cells plus HEK293 cells carrying WT-p2089 (Fig. 10H). Akata and P3HR1 are B cell lines derived from Burkitt’s lymphoma (41, 56). HEK293 cells latently infected with M81 strain of EBV were also included. P3HR1 and HEK293-M81 cells were transfected with BZLF1/Zta expression plasmid to induce lytic reactivation. EBER was used a positive control for expression of EBV genes. EBNA2 and EBNA3A are latency III associated genes expressed in B cell line Akata. In contrast, only EBNA3A can be expressed in P3HR1 due to deletion of EBNA2 gene (57). The expression patterns of EBNA2 and EBNA3A mRNAs observed in Akata and P3HR1 cells (Fig. 10H, lanes 1 to 4) were generally as expected. Although BRLF1/Rta was detected at relatively low levels in all cells and particularly in P3HR1 cells without lytic induction, BMRF1 and BLLF1 were not detected without IgG cross-linking in Akata cells or enforced expression of BZLF1/Zta in P3HR1 cells (Fig. 10H, lanes 1 and 3). These cell lines served as a positive control for EBV lytic induction. EBNA2, EBNA3A, BMRF1, and BZLF1/Zta were undetectable in HEK293-M81 and HEK293-WT-p2089 cells, with only trace amount of BRLF1/Rta detected in both lines. BGLF2 was not detected in HEK293 or EBV– Akata cells (Fig. 10I, lanes 2 and 3 compared to lanes 4 and 5). However, BGLF2 was detected in all EBV+ cell lines irrespective of lytic induction (Fig. 10H and I). Thus, BGLF2 expression is not unique to HEK293 cells stably carrying WT-p2089. Our results are compatible with the interpretation that the difference in ISG induction by IFN-β should be ascribed to the expression of BGLF2.
DISCUSSION
We performed a functional screen to identify EBV-encoded modulators of IFN response (Fig. 2) and identified BGLF2 as a multifunctional suppressor of JAK-STAT signaling (Fig. 3 and 4). This function of BGLF2 was independent of its modulation of JNK, p38, or NF-κB (Fig. 5). BGLF2 was found to interact with STAT1, STAT2, and cullin 1 (Fig. 6 and 8). It facilitates the recruitment of tyrosine phosphatase SHP1 to STAT1, leading to the decrease of p-STAT1 levels (Fig. 7). It also recruits cullin 1 to STAT2 and promotes STAT2 ubiquitination and proteasomal degradation (Fig. 8). BGLF2 suppresses JAK-STAT signaling and facilitates EBV primary infection (Fig. 9). BGLF2-deficient EBV was constructed and shown to express lytic genes less robustly upon stimulation but induce ISG expression more prominently (Fig. 10).
Our findings are generally consistent with a recent report on the suppression of type I IFN signaling and promotion of EBV reactivation by BGLF2 (27), although some mechanistic details differ. While the other study emphasizes on EBV reactivation, we showed that enforced expression of BGLF2 facilitates primary infection of EBV (Fig. 9). In addition, we constructed BGLF2-deficient EBV and demonstrated more robust IFN-β signaling in the absence of BGLF2 in infected cells (Fig. 10). The suppressive effect of BGLF2 on type I IFN signaling is thought to be mediated through association of BGLF2 with TYK2 kinase and consequent inhibition of STAT1 and STAT3 phosphorylation in the other study. In our work, the levels of p-TYK2 and p-JAK1 were also shown to decrease upon expression of BGLF2 (Fig. 7H). The two mechanisms suggested by the two studies might not be mutually exclusive. Hypophosphorylation of STAT1 could be ascribed to both inhibition of an upstream kinase such as TYK2 and the action of a tyrosine phosphatase such as SHP1. Because SHP1 is known to target not only STAT1 but also TYK2, JAK1, and other targets in JAK-STAT signaling (16, 50), it will be of particularly great interest to see whether BGLF2 might also recruit SHP1 to dephosphorylate and inactivate TYK2 and JAK1. We demonstrated the association of BGLF2 with STAT1 and STAT2 (Fig. 6), which is not seen in the other study. Since only a small fraction of STAT1 and STAT2 was found to be associated with BGLF2, it is not too surprising that the association is not observed in the earlier study (27). Plausibly, BGLF2 might interact with the JAK-STAT signalosome through one key component such as TYK2. In addition, BGLF2 could also recruit SHP1 tyrosine phosphatase to act on multiple components of the signalosome leading to suppression of JAK-STAT signaling. In our work, BGLF2 was capable of suppressing IFN-β- and IFN-λ1-induced expression of ISG15 and OAS1 (Fig. 3B and F). In contrast, BGLF2 was shown to suppress IFN-α-induced expression of IRF1, IRF7, and MxA, but not ISG15, in the earlier study (27). Whether the difference might be explained by the longer duration of IFN treatment in our study remains to be determined. Importantly, our results on BGLF2 (Fig. 3B and F) are consistent with the suppression of IFN-β- and IFN-λ1-induced expression of ISG15 by EBV infection (Fig. 1D and E). Whereas no suppressive effect of BGLF2 on IFN-γ signaling was seen in the earlier study (27), a suppressive effect of both M81 infection and BGLF2 expression on the sensitive GAS-Luc reporter activity was detected in our analysis (Fig. 1F and 3C). Further investigations are required to clarify the relevance of this suppressive activity during EBV life cycle.
IFN-λ1 displays potent antiviral activity against herpes simplex virus infection in vivo (58), but the roles of type III IFNs in EBV infection have not been well characterized. We demonstrated the suppressive effect of EBV and its BGLF2 protein on not only type I but also type III IFN signaling. Whether type III IFNs are induced by EBV and influential in EBV primary infection and reactivation remains to be determined. Our functional screen with an expression library of EBV proteins revealed that BGLF2 serves as the most potent inhibitor of IFN-γ signaling, as indicated by the highly sensitive GAS-Luc reporter (Fig. 2B). Its inhibitory effect was even stronger than BZLF1/Zta, which has previously been shown to be a major suppressor of IFN-γ signaling (35). The IFN-γ-antagonizing activity of BGLF2 was further verified by using GAS-Luc reporter and IP10 mRNA expression. However, BGLF2 does not suppress IFN-γ-induced phosphorylation of STAT1 in the published study (27). This discrepancy might be explained by the relatively mild suppressive effect of BGLF2 on IFN-γ signaling, which was best seen with the GAS-Luc reporter (Fig. 3C). We were unable to see a significant effect of EBV primary infection on IFN-γ signaling (Fig. 1G). Since the assay was performed after 48 h of infection, the role of IFN-γ remains to be better defined during the initial phase of infection. Interestingly, some other lytic proteins, including BRLF1/Rta, emerged as an activator of IFN-γ signaling in the same screen (Fig. 2B). Plausibly, the lack of suppression by EBV might be explained by the combined effect of different lytic proteins. It will be intriguing to see how EBV-encoded suppressors and activators of IFN-γ signaling work together in the infected cells to achieve differential regulation of IFN-γ activity in favor of EBV infection.
Our study identified STATs as the cellular targets of BGLF2. This effect is apparently independent of the previously demonstrated ability of BGLF2 to modulate p38, JNK, or NF-κB (20, 22, 23), suggesting that BGLF2 is a pleiotropic protein that targets multiple cell signaling pathways. Western blot analysis revealed that BGLF2 targets STAT1 and STAT2 for dephosphorylation and degradation, respectively, in HEK293 cells. Interestingly, BGLF2 is not influential on STAT1 degradation or STAT2 dephosphorylation. BGLF2 interacts with both STAT1 and STAT2 but exerts an inhibitory effect on them through distinct mechanisms. The differential effect of BGLF2 on STAT1 and STAT2 might be accounted for by different binding domains, different types of conformational changes or different partners recruited by BGLF2. All STATs have a conserved SH2 domain that recognizes and binds to specific phosphotyrosine residues of other STATs or JAKs (59). Unlike SH2 domain, other domains such as coiled-coil domain or transcriptional activation domain of STATs are less conserved. They might bind with BGLF2 to generate different effects. The binding of BGLF2 with STAT1 and STAT2 could also induce different conformation changes leading to different effects. Whereas SHP1 is the key mediator of the effect of BGLF2 on STAT1, cullin 1 might play a critical role in BGLF2-induced destabilization of STAT2. Although BGLF2 did not associate with SHP2 (Fig. 6G), whether SHP2 and additional protein tyrosine phosphatases (50, 60), as well as additional cullin-type E3 ubiquitin ligases (61), are involved in BGLF2-mediated suppression of STAT1 and STAT2 remains to be elucidated. Mapping of binding domain and characterization of STAT1 or STAT2 mutants and chimeric proteins might provide new mechanistic insight on the differential regulation of STAT1 and STAT2 by BGLF2.
In addition to cullins, several other E3 ubiquitin ligases such as UBR4 and PDLIM2 have also been shown to be recruited by different viral proteins to promote STAT2 ubiquitination and degradation (62–64). Other E3 enzymes involved in STAT2 destruction include FBW7 (65). It is not well studied as to exactly how different E3s might cooperate with each other to mediate STAT2 ubiquitination and degradation in different physiological and pathological contexts. Cullin 1 is a key component in the SCF complex representative of a large family of cullin-RING E3 ubiquitin ligases (61). Our finding lent further support to a possible role of SCF complex in the degradation of STAT2. To validate this, the role of SKP1 and cullin 1 in STAT2 ubiquitination and degradation should be further investigated.
Our work confirmed the inhibitory effect of IFN-β on not only EBV primary infection but also reactivation. Our results suggested a critical role of IFN-β in the expression of lytic genes and possibly also in lytic-latent switch. We further constructed ΔBGLF2-p2089 cell lines to determine the role of BGLF2 in the context of EBV infection. Consistent with a role of BGLF2 in the lytic phase (19, 21), the ΔBGLF2-p2089 cells were found to be less capable of expressing early (BZLF1 and BRLF1) and late lytic (BMRF2) genes compared to the WT-p2089 cells. The inhibition of lytic replication might be further investigated by quantifying EBV virions in the supernatant after induction of lytic cycle. Interestingly, the expression of Rta and EA-D/BMRF1 proteins were found to be unaffected in ΔBGLF2-p2089 cells transfected with a Zta plasmid in a recent study (19). One important difference between this study and ours is noteworthy. In our work, Zta induction by TPA plus NaBt was much less efficient in ΔBGLF2-p2089 cells (Fig. 10F). This is generally consistent with the notion that BGLF2 plays an important role in the induction of Zta expression (20). Whether enforced expression of Zta in the other study might be accounted for more pronounced expression of Rta and EA-D proteins in ΔBGLF2-p2089 cells remains to be investigated. The ΔBGLF2-p2089 cells were found to have a more robust induction of ISGs in response to IFN-β treatment (Fig. 10G). Plausibly, the reduced expression of lytic genes might also be ascribed to the enhanced expression of ISGs in ΔBGLF2-p2089 cells.
Interestingly, the difference in ISGs in WT-p2089 and ΔBGLF2-p2089 cells was observed in cell lines that were supposed to be in latent phase of EBV replication. Considering that the effect was solely due to loss of BGLF2, there could be two possibilities. First, the cells were undergoing a spontaneous lytic cycle, where BGLF2 was expressed in the WT-p2089 cells, causing the suppression of ISG expression. Second, BGLF2 was also expressed in latent phase similar to BZLF1 and BRLF1 (66). Further analysis of the latent and lytic gene expression in WT-p2089 and other cell lines provided some clues and hints. Weak detection of BRLF1 in cells that were not induced for lytic replication did lend some support to the notion that some cells might spontaneously undergo lytic reactivation or express some lytic genes, although BMRF1 and BLLF1 mRNAs were not very abundant in M81 and WT-p2089 cells. Intriguingly, BGLF2 was found to be expressed in all cell lines even without lytic induction, which could suggest that the cells might be expressing some levels of BGLF2 during latency as well. If this claim is true, the expression of BGLF2 should be assessed in EBV-associated nasopharyngeal carcinoma, gastric carcinoma and Burkitt’s lymphoma samples. Possible expression of BGLF2 during latency phase might have important implications in EBV biology. Further investigations are required to rectify this finding.
BGLF2 is highly conserved among all strains of EBV. It is also homologous structurally and functionally to UL16 of herpes simplex virus 1, ORF33 of murine gammaherpesvirus 68, UL94 of cytomegalovirus, ORF44 of varicella-zoster virus, and ORF33 of Kaposi’s sarcoma-associated herpesvirus. BGLF2 homologs in other herpesviruses can also induce cell cycle arrest (22, 24). In addition, they also inhibit type I IFN signaling through a similar mechanism (27). It would not be surprising if they can also recruit SHP1 and cullin 1 to suppress JAK-STAT signaling. They might also suppress IFN production and signaling through other mechanisms.
Other EBV-encoded tegument proteins such as BKRF4, BSRF1, BGLF4, and BBLF1 have previously been shown to play important roles in EBV infection (24, 67). A few EBV tegument proteins such as BPLF1, BLRF2, BNRF1, and BGLF4 have already been reported to exhibit innate immunomodulatory function (5). There is a common theme in herpesviruses that tegument proteins are used to antagonize IFN production and signaling. More examples will be revealed in the coming years. In this regard, BGLF2 is known to interact with other tegument proteins BKRF4 and BBLF1 (18, 19, 21). Whether and how BKRF4 and BBLF1 might influence the suppressive effect of BGLF2 on IFN signaling merit further investigations.
MATERIALS AND METHODS
Plasmid constructs, cell culture, transfection, and drug treatment.
All key plasmids and cell lines have been previously described (18, 37, 43–46, 51, 68–70). Expression construct for BGLF2 of the B95.8 strain of EBV was originally in the expression library of EBV genes (18, 71), kindly provided by Lori Frappier from University of Toronto (Toronto, Canada). Gene juice (Millipore) was used as a transfection reagent for HEK293 and HEK293-M81 cells according to the manufacturer’s protocol. Mirus, a keratinocyte transfecting reagent obtained from MirusBio, was used as a transfection reagent for AGS and HK1 cell lines. siRNA was transfected using Lipofectamine 2000 (Invitrogen). Proteasomal inhibitor MG132 was obtained from Sigma, and 10 μM MG132 was added to the cells for 6 h. Sodium orthovanadate was added to cells to obtain a final concentration of 200 μM, followed by incubation for 1 h at 37°C, followed in turn by IFN treatment for 30 min. p38 and JNK inhibitors SB203580 (InvivoGen) and SP600125 (Sigma) were added to obtain final concentrations of 5 and 20 μM, respectively, followed by incubation for 1 h at 37°C before IFN treatment. TPA was purchased from Cell Signaling. MG132 was from Sigma. Human IFN-β was from PBL Assay Science. Human IFN-λ1 and IFN-γ were from PeproTech.
EBV infection and lytic induction.
Infection of HEK293 cells with cell-free M81 or BX1 virus was performed as described previously (36). Lytic induction was conducted in HEK293-M81 cells by treatment with TPA or transfection with BZLF1/Zta construct. Cells in a 10-cm cell culture dish were treated with 20 ng/ml TPA or transfected with 4 μg each of BZLF1/Zta and gp110 constructs. The medium was changed after 24 h, and the supernatant was collected after 72 h of treatment or transfection. The medium was filtered through a 1.2-μm filter and then centrifuged in centrifugal filter unit at 3,000 rpm for 30 min. The concentrated virus was then used to infect the cells in a 6-well plate and incubated for 24 h at 37°C followed by replenishment with fresh RPMI 1640 medium. The virus-infected GFP-positive cells were visualized under microscope after 48 h of infection.
DNA, RNA, and protein analysis.
qPCR, RT-PCR, RT-qPCR, coimmunoprecipitation, and Western blotting were performed essentially as described previously (37, 68–70). Primer and probe sequences will be provided upon request. Antibodies to p-STAT1, p-JAK1, and p-TYK2 were from Cell Signaling. Anti-p-STAT2 was from R&D Systems. Antibodies to β-actin and Flag were from Sigma. Anti-V5 was from Invitrogen. Anti-SOCS1 was from Abcam. Mouse anti-myc for immunoprecipitation was from Roche. All other primary antibodies were from Santa-Cruz. ECL secondary antibodies were from GE Healthcare. Dynabeads M280 conjugated to anti-rabbit or anti-mouse secondary antibodies and recombinant protein G agarose beads were obtained from Invitrogen.
Construction of ΔBGLF2 mutant.
EBV bacmid p2089 derived from the B95.8 strain of EBV was kindly provided by Wolf Hammerschmidt from German Center for Infection Research (55). ΔBGLF2-p2089 was constructed by inserting two premature stop codons and one frameshift deletion into the region coding for the N terminus of BGLF2. Homologous recombination was carried out in Escherichia coli by using the galK selection method (72). In brief, the galK expression cassette for the first recombination was generated by PCR using pMOD4-galK-G, kindly provided by Soren Warming from National Cancer Institute (USA), as the template and primers 5′-TGTAGACGGT CACATAGGTG GGCTTCTTCT TGGGCTTCAG CACAATGAGG CCTGTTGACA ATTAATCATC GGCA-3′ (BGLF2-galK-forward) and 5′-TCTATACGGC CACAACTGCC GTGTCAGCTG TGTACGTGCC TCAGATAGCC TCAGCACTGT CCTGCTCCTT-3′ (BGLF2-galK-reverse). The replacement fragment with desired mutations for the second recombination was synthesized by Life Technology and annealed by PCR using the primers 5′-ACATAGGTGG GCTTCTTCTT GGGCTTCAGC ACAATGAGGG TACATTCTACT AGGTTTAAGG AGGTCCGGCT ATCTGAGGCA CGTACACAGC TGACACGG-3′ (BGLF2-mutation-sense) and 5′-CCGTGTCAGC TGTGTACGTG CCTCAGATAG CCGGACCTCC TTAAACCTAG TAGAATGTAC CCTCATTGTG CTGAAGCCCA AGAAGAAGCC CACCTATG-3′ (BGLF2-mutation-antisense). Recombination was confirmed by PCR, DNA sequencing, and BamHI and EcoRI restriction patterning. Primers for specific amplification of WT-BGLF2 but not Δ-BGLF2 by RT-PCR were 5′-GTTCCTCAAC AAGGAGTGCCT-3′ (forward) and 5′-CAATGAGGGT AACATTCATG-3′ (reverse). The primers and TaqMan probe for RT-qPCR analysis of WT-BGLF2 were 5′-TGAGAGTCAG GCTGTGTGG-3′ (forward), 5′-AATGGAACC CTAGCCACCG-3′ (reverse), and 5′-6FAM-GGACTGCCTT AGTGAAGAGA ACCTCG-MGB-3′ (probe). ΔBGLF2-p2089 bacmid was transfected into HEK293 cells to establish the virus-producing cell lines. Cells were selected by 70 μg/ml hygromycin B, and the GFP+ cell colonies were obtained after 2 weeks. Successful clones were verified by RT-PCR detection of EBER.
Statistical analysis.
All tests were performed with three biological replicates, and the statistical significance was assessed using two-tailed paired Student t tests. The results were expressed as means ± the standard deviations (SD). Significance is indicated in the figures by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0 0.001).
ACKNOWLEDGMENTS
We thank Henri-Jacques Delecluse from German Center for Infection Research (Heidelberg, Germany) for providing M81 bacmid, Wolfgang Hammerschmidt from German Center for Infection Research (Munich, Germany) for the gift of p2089 bacmid, Lori Frappier from University of Toronto (Toronto, Canada) for the gift of expression library of EBV proteins, and members of Jin laboratory for critical reading of the manuscript.
This study was supported by the Hong Kong Research Grants Council (C7027-16G) and the Hong Kong Health and Medical Research Fund (17160822 and 18170942).
Contributor Information
Michael George Botelho, Email: botelho@hku.hk.
Kit-San Yuen, Email: samyuen@hku.hk.
Dong-Yan Jin, Email: dyjin@hku.hk.
Joanna L. Shisler, University of Illinois at Urbana Champaign
REFERENCES
- 1.Jha HC, Pei Y, Robertson ES. 2016. Epstein-Barr virus: diseases linked to infection and transformation. Front Microbiol 7:1602. 10.3389/fmicb.2016.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrell PJ. 2019. Epstein-Barr virus and cancer. Annu Rev Pathol 14:29–53. 10.1146/annurev-pathmechdis-012418-013023. [DOI] [PubMed] [Google Scholar]
- 3.Kenney SC, Mertz JE. 2014. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin Cancer Biol 26:60–68. 10.1016/j.semcancer.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor GS, Long HM, Brooks JM, Rickinson AB, Hislop AD. 2015. The immunology of Epstein-Barr virus-induced disease. Annu Rev Immunol 33:787–821. 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 5.Jangra S, Yuen KS, Botelho MG, Jin DY. 2019. Epstein-Barr virus and innate immunity: friends or foes? Microorganisms 7:183. 10.3390/microorganisms7060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Gent M, Griffin BD, Berkhoff EG, van Leeuwen D, Boer IG, Buisson M, Hartgers FC, Burmeister WP, Wiertz EJ, Ressing ME. 2011. EBV lytic-phase protein BGLF5 contributes to TLR9 downregulation during productive infection. J Immunol 186:1694–1702. 10.4049/jimmunol.0903120. [DOI] [PubMed] [Google Scholar]
- 7.Saito S, Murata T, Kanda T, Isomura H, Narita Y, Sugimoto A, Kawashima D, Tsurumi T. 2013. Epstein-Barr virus deubiquitinase downregulates TRAF6-mediated NF-κB signaling during productive replication. J Virol 87:4060–4070. 10.1128/JVI.02020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Gent M, Braem SG, de Jong A, Delagic N, Peeters JG, Boer IG, Moynagh PN, Kremmer E, Wiertz EJ, Ovaa H, Griffin BD, Ressing ME. 2014. Epstein-Barr virus large tegument protein BPLF1 contributes to innate immune evasion through interference with Toll-like receptor signaling. PLoS Pathog 10:e1003960. 10.1371/journal.ppat.1003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Ylä-Anttila P, Callegari S, Tsai MH, Delecluse HJ, Masucci MG. 2018. Herpesvirus deconjugases inhibit the IFN response by promoting TRIM25 autoubiquitination and functional inactivation of the RIG-I signalosome. PLoS Pathog 14:e1006852. 10.1371/journal.ppat.1006852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn AM, Huye LE, Ning S, Webster-Cyriaque J, Pagano JS. 2005. Interferon regulatory factor 7 is negatively regulated by the Epstein-Barr virus immediate-early gene, BZLF-1. J Virol 79:10040–10052. 10.1128/JVI.79.15.10040-10052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, Fossum E, Joo CH, Inn KS, Shin YC, Johannsen E, Hutt-Fletcher LM, Hass J, Jung JU. 2009. Epstein-Barr virus LF2: an antagonist to type I interferon. J Virol 83:1140–1146. 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X, Wei L, Peng Q, Zheng Y, Ou C, Ye Q, Xiong W, Li G, Fu Y, Yan Q, Ma J. 2017. Epstein-Barr virus miR-BART6-3p inhibits the RIG-I pathway. J Innate Immun 9:574–586. 10.1159/000479749. [DOI] [PubMed] [Google Scholar]
- 13.Stark GR, DarnellJE, Jr.. 2012. The JAK-STAT pathway at twenty. Immunity 36:503–514. 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci USA 95:15623–15628. 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker T, Kovarik P, Meinke A. 1997. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res 17:121–134. 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Somani AK, Siminovitch KA. 2000. Roles of the SHP-1 tyrosine phosphatase in the negative regulation of cell signaling. Semin Immunol 12:361–378. 10.1006/smim.2000.0223. [DOI] [PubMed] [Google Scholar]
- 17.Durham GA, Williams JJL, Nasim MT, Palmer TM. 2019. Targeting SOCS proteins to control JAK-STAT signaling in disease. Trends Pharmacol Sci 40:298–308. 10.1016/j.tips.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Ho TH, Sitz J, Shen Q, Leblanc-Lacroix A, Campos EI, Borozan I, Marcon E, Greenblatt J, Fradet-Turcotte A, Jin DY, Frappier L. 2018. A screen for Epstein-Barr virus proteins that inhibit the DNA damage response reveals a novel histone binding protein. J Virol 24:e00262-18. 10.1128/JVI.00262-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung CH, Chiu YF, Wang WH, Chen LW, Chang PJ, Huang TY, Lin YJ, Tsai WJ, Yang CC. 2019. Interaction between BGLF2 and BBLF1 is required for the efficient production of infectious Epstein-Barr virus particles. Front Microbiol 10:3021. 10.3389/fmicb.2019.03021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konishi N, Narita Y, Hijioka F, Masud HMAA, Sato Y, Kimura H, Murata T. 2018. BGLF2 increases infectivity of Epstein-Barr virus by activating AP-1 upon de novo infection. mSphere 24:e00138-18. 10.1128/mSphere.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdullah Al Masud HM, Watanabe T, Yoshida M, Sato Y, Goshima F, Kimura H, Murata T. 2017. Epstein-Barr virus BKRF4 gene product is required for efficient progeny production. J Virol 24:e00975-17. 10.1128/JVI.00975-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Cohen JI. 2016. Epstein-Barr virus (EBV) tegument protein BGLF2 promotes EBV reactivation through activation of the p38 mitogen-activated protein kinase. J Virol 90:1129–1138. 10.1128/JVI.01410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T, Wang Y, Xu Z, Zou X, Wang P, Ou X, Li Y, Peng T, Chen D, Li M, Cai M. 2019. Epstein-Barr virus tegument protein BGLF2 inhibits NF-κB activity by preventing p65 Ser536 phosphorylation. FASEB J 33:10563–10576. 10.1096/fj.201901196RR. [DOI] [PubMed] [Google Scholar]
- 24.Paladino P, Marcon E, Greenblatt J, Frappier L. 2014. Identification of herpesvirus proteins that contribute to G1/S arrest. J Virol 88:4480–4492. 10.1128/JVI.00059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De La Cruz-Herrera CF, Shire K, Siddiqi UZ, Frappier L. 2018. A genome-wide screen of Epstein-Barr virus proteins that modulate host SUMOylation identifies a SUMO E3 ligase conserved in herpesviruses. PLoS Pathog 14:e1007176. 10.1371/journal.ppat.1007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lui WY, Jangra S, Yuen KS, Botelho MG, Jin DY. 2020. Interferon antagonism of Epstein-Barr virus tegument proteins. Proceedings 50:74. 10.3390/proceedings2020050074. [DOI] [Google Scholar]
- 27.Liu X, Sadaoka T, Krogmann T, Cohen JI. 2020. Epstein-Barr virus (EBV) tegument protein BGLF2 suppresses type I interferon signaling to promote EBV reactivation. J Virol 24:e00258-20. 10.1128/JVI.00258-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams A, Strander H, Cantell K. 1975. Sensitivity of the Epstein-Barr virus transformed human lymphoid cell lines to interferon. J Gen Virol 28:207–217. 10.1099/0022-1317-28-2-207. [DOI] [PubMed] [Google Scholar]
- 29.Kure S, Tada K, Wada J, Yoshie O. 1986. Inhibition of Epstein-Barr virus infection in vitro by recombinant human interferons alpha and gamma. Virus Res 5:377–390. 10.1016/0168-1702(86)90030-4. [DOI] [PubMed] [Google Scholar]
- 30.Lin JC, Zhang ZX, Chou TC, Sim I, Pagano JS. 1989. Synergistic inhibition of Epstein-Barr virus: transformation of B lymphocytes by alpha and gamma interferon and by 3′-azido-3′-deoxythymidine. J Infect Dis 159:248–254. 10.1093/infdis/159.2.248. [DOI] [PubMed] [Google Scholar]
- 31.Thorley-Lawson DA. 1981. The transformation of adult but not newborn human lymphocytes by Epstein Barr virus and phytohemagglutinin is inhibited by interferon: the early suppression by T cells of Epstein Barr infection is mediated by interferon. J Immunol 126:829–833. [PubMed] [Google Scholar]
- 32.Tovey MG, Dron M, Gresser I. 1982. Interferon enhances the expression of Epstein-Barr virus early antigen in Daudi cells. J Gen Virol 60:31–38. 10.1099/0022-1317-60-1-31. [DOI] [PubMed] [Google Scholar]
- 33.Gujer C, Murer A, Müller A, Vanoaica D, Sutter K, Jacque E, Fournier N, Kalchschmidt J, Zbinden A, Capaul R, Dzionek A, Mondon P, Dittmer U, Münz C. 2019. Plasmacytoid dendritic cells respond to Epstein-Barr virus infection with a distinct type I interferon subtype profile. Blood Adv 3:1129–1144. 10.1182/bloodadvances.2018025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke PA, Sharp NA, Arrand JR, Clemens MJ. 1990. Epstein-Barr virus gene expression in interferon-treated cells. Implications for the regulation of protein synthesis and the antiviral state. Biochim Biophys Acta 1050:167–173. 10.1016/0167-4781(90)90161-t. [DOI] [PubMed] [Google Scholar]
- 35.Morrison TE, Mauser A, Wong A, Ting JP, Kenney SC. 2001. Inhibition of IFN-γ signaling by an Epstein-Barr virus immediate-early protein. Immunity 15:787–799. 10.1016/s1074-7613(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 36.Tsai MH, Raykova A, Klinke O, Bernhardt K, Gärtner K, Leung CS, Geletneky K, Sertel S, Münz C, Feederle R, Delecluse HJ. 2013. Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell Rep 5:458–470. 10.1016/j.celrep.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhary V, Yuen KS, Chan JF, Chan CP, Wang PH, Cai JP, Zhang S, Liang M, Kok KH, Chan CP, Yuen KY, Jin DY. 2017. Selective activation of type II interferon signaling by Zika virus NS5 protein. J Virol 24:e00163-17. 10.1128/JVI.00163-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu LH, Gao S, Jin R, Zhuang LL, Jiang L, Qiu LZ, Xu HG, Zhou GP. 2014. Repression of interferon regulatory factor 3 by the Epstein-Barr virus immediate-early protein Rta is mediated through E2F1 in HeLa cells. Mol Med Rep 9:1453–1459. 10.3892/mmr.2014.1957. [DOI] [PubMed] [Google Scholar]
- 39.Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. 2014. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J Virol 88:1389–1393. 10.1128/JVI.02662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang DP, Ho JH, Poon YF, Chew EC, Saw D, Lui M, Li CL, Mak LS, Lai SH, Lau WH. 1980. Establishment of a cell line (NPC/HK1) from a differentiated squamous carcinoma of the nasopharynx. Int J Cancer 26:127–132. 10.1002/ijc.2910260202. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu N, Yoshiyama H, Takada K. 1996. Clonal propagation of Epstein-Barr virus (EBV) recombinants in EBV-negative Akata cells. J Virol 70:7260–7263. 10.1128/JVI.70.10.7260-7263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C-Y, Mayo MW, BaldwinAS, Jr.. 1996. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NFκB. Science 274:784–787. 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 43.Lei K, Nimnual A, Zong WX, Kennedy NJ, Flavell RA, Thompson CB, Bar-Sagi D, Davis RJ. 2002. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH2-terminal kinase. Mol Cell Biol 22:4929–4942. 10.1128/MCB.22.13.4929-4942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao L, Lang W. 2000. A dominant role for the c-Jun NH2-terminal kinase in oncogenic ras-induced morphologic transformation of human lung carcinoma cells. Cancer Res 60:400–408. [PubMed] [Google Scholar]
- 45.Ching YP, Wong CM, Chan SF, Leung TH, Ng DC, Jin DY, Ng IOL. 2003. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J Biol Chem 278:10824–10830. 10.1074/jbc.M208310200. [DOI] [PubMed] [Google Scholar]
- 46.Jin DY, Giordano V, Kibler KV, Nakano H, Jeang KT. 1999. Role of adapter function in oncoprotein-mediated activation of NF-κB: human T-cell leukemia virus type I Tax interacts directly with IκB kinase γ. J Biol Chem 274:17402–17405. 10.1074/jbc.274.25.17402. [DOI] [PubMed] [Google Scholar]
- 47.Song MM, Shuai K. 1998. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem 273:35056–35062. 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 48.Yu CR, Hayashi K, Lee YS, Mahdi RM, Shen DF, Chan CC, Egwuagu CE. 2015. Suppressor of cytokine signaling 1 (SOCS1) mitigates anterior uveitis and confers protection against ocular HSV-1 infection. Inflammation 38:555–565. 10.1007/s10753-014-9962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chien H, Alston CI, Dix RD. 2018. Suppressor of cytokine signaling 1 (SOCS1) and SOCS3 are stimulated within the eye during experimental murine cytomegalovirus retinitis in mice with retrovirus-induced immunosuppression. J Virol 24:e00526-18. 10.1128/JVI.00526-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bone H, Dechert U, Jirik F, Schrader JW, Welham MJ. 1997. SHP1 and SHP2 protein-tyrosine phosphatases associate with betac after interleukin-3-induced receptor tyrosine phosphorylation. Identification of potential binding sites and substrates. J Biol Chem 272:14470–14476. 10.1074/jbc.272.22.14470. [DOI] [PubMed] [Google Scholar]
- 51.Cheng Y, Gao WW, Tang HM, Deng JJ, Wong CM, Chan CP, Jin DY. 2016. β-TrCP-mediated ubiquitination and degradation of liver-enriched transcription factor CREB-H. Sci Rep 6:23938. 10.1038/srep23938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elliott J, Lynch OT, Suessmuth Y, Qian P, Boyd CR, Burrows JF, Buick R, Stevenson NJ, Touzelet O, Gadina M, Power UF, Johnston JA. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the elongin-cullin E3 ligase. J Virol 81:3428–3436. 10.1128/JVI.02303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le-Trilling VTK, Becker T, Nachshon A, Stern-Ginossar N, Schöler L, Voigt S, Hengel H, Trilling M. 2020. The human cytomegalovirus pUL145 isoforms act as viral DDB1-cullin-associated factors to instruct host protein degradation to impede innate immunity. Cell Rep 30:2248–2260. 10.1016/j.celrep.2020.01.070. [DOI] [PubMed] [Google Scholar]
- 54.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA 95:8245–8250. 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feederle R, Bartlett EJ, Delecluse HJ. 2010. Epstein-Barr virus genetics: talking about the BAC generation. Herpesviridae 1:6. 10.1186/2042-4280-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinuma Y, Konn M, Yamaguchi J, Wudarski DJ, BlakesleeJR, Jr, GraceJT, Jr.. 1967. Immunofluorescence and herpes-type virus particles in the P3HR-1 Burkitt lymphoma cell line. J Virol 1:1203–1051. 10.1128/jvi.1.5.1045-1051.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen JI, Wang F, Mannick J, Kieff E. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA 86:9558–9562. 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol 80:4501–4509. 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta S, Yan H, Wong LH, Ralph S, Krolewski J, Schindler C. 1996. The SH2 domains of STAT1 and STAT2 mediate multiple interactions in the transduction of IFN-α signals. EMBO J 15:1075–1084. 10.1002/j.1460-2075.1996.tb00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Böhmer FD, Friedrich K. 2014. Protein tyrosine phosphatases as wardens of STAT signaling. JAKSTAT 3:e28087. 10.4161/jkst.28087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarikas A, Hartmann T, Pan ZQ. 2011. The cullin protein family. Genome Biol 12:220. 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, García-Sastre A. 2013. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog 9:e1003265. 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joyce MA, Berry-Wynne KM, Dos Santos T, Addison WR, McFarlane N, Hobman T, Tyrrell DL. 2019. HCV and flaviviruses hijack cellular mechanisms for nuclear STAT2 degradation: up-regulation of PDLIM2 suppresses the innate immune response. PLoS Pathog 15:e1007949. 10.1371/journal.ppat.1007949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avia M, Rojas JM, Miorin L, Pascual E, Van Rijn PA, Martín V, García-Sastre A, Sevilla N. 2019. Virus-induced autophagic degradation of STAT2 as a mechanism for interferon signaling blockade. EMBO Rep 20:e48766. 10.15252/embr.201948766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee CJ, An HJ, Kim SM, Yoo SM, Park J, Lee GE, Kim WY, Kim DJ, Kang HC, Lee JY, Lee HS, Cho SJ, Cho YY. 2020. FBXW7-mediated stability regulation of signal transducer and activator of transcription 2 in melanoma formation. Proc Natl Acad Sci USA 117:584–594. 10.1073/pnas.1909879116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martel-Renoir D, Grunewald V, Touitou R, Schwaab G, Joab I. 1995. Qualitative analysis of the expression of Epstein-Barr virus lytic genes in nasopharyngeal carcinoma biopsies. J Gen Virol 76(Pt 6):1401–1408. 10.1099/0022-1317-76-6-1401. [DOI] [PubMed] [Google Scholar]
- 67.Guo H, Wang L, Peng L, Zhou ZH, Deng H. 2009. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J Virol 83:10582–10595. 10.1128/JVI.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuen KS, Chan CP, Wong NM, Ho CH, Ho TH, Lei T, Deng W, Tsao SW, Chen H, Kok KH, Jin DY. 2015. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J Gen Virol 96:626–636. 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 69.Chaudhary V, Zhang S, Yuen KS, Li C, Lui PY, Fung SY, Wang PH, Chan CP, Li D, Kok KH, Liang M, Jin DY. 2015. Suppression of type I and type III IFN signalling by NSs protein of severe fever with thrombocytopenia syndrome virus through inhibition of STAT1 phosphorylation and activation. J Gen Virol 96:3204–3211. 10.1099/jgv.0.000280. [DOI] [PubMed] [Google Scholar]
- 70.Cheung PHH, Lee TTW, Kew C, Chen H, Yuen KY, Chan CP, Jin DY. 2020. Virus subtype-specific suppression of MAVS aggregation and activation by PB1-F2 protein of influenza A (H7N9) virus. PLoS Pathog 16:e1008611. 10.1371/journal.ppat.1008611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salsman J, Zimmerman N, Chen T, Domagala M, Frappier L. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog 4:e1000100. 10.1371/journal.ppat.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]