ABSTRACT
Certain existing prebiotics meant to facilitate the growth of beneficial bacteria in the intestine also promote the growth of other prominent bacteria. Therefore, the growth-promoting effects of β-galactosides on intestinal bacteria were analyzed. Galactosyl-β1,4-l-rhamnose (Gal-β1,4-Rha) selectively promoted the growth of Bifidobacterium. Bifidobacterium longum subsp. longum 105-A (JCM 31944) has multiple solute-binding proteins belonging to ATP-binding cassette transporters for sugars. Each strain in the library of 11 B. longum subsp. longum mutants, in which each gene of the solute-binding protein was disrupted, was cultured in a medium containing Gal-β1,4-Rha as the sole carbon source, and only the BL105A_0502 gene-disruption mutant showed delayed and reduced growth compared to the wild-type strain. BL105A_0502 homolog is highly conserved in bifidobacteria. In a Gal-β1,4-Rha-containing medium, Bifidobacterium longum subsp. infantis JCM 1222T, which possesses BLIJ_2090, a homologous protein to BL105A_0502, suppressed the growth of enteric pathogen Clostridioides difficile, whereas the BLIJ_2090 gene-disrupted mutant did not. In vivo, administration of B. infantis and Gal-β1,4-Rha alleviated C. difficile infection-related weight loss in mice. We have successfully screened Gal-β1,4-Rha as a next-generation prebiotic candidate that specifically promotes the growth of beneficial bacteria without promoting the growth of prominent bacteria and pathogens.
KEYWORDS: Prebiotic, probiotic, bifidobacteria, microbiome, microbiota, Clostridioides difficile
Introduction
Humans maintain a complex gut microbiota, with the number of microorganisms estimated to be in the thousands based on a recent metagenomic study1. The number of bacterial cells is comparable to that of the host human cells.2 The gut microbiota is composed of beneficial bacteria with health-promoting effects, pathogenic bacteria that can cause disease, and opportunistic bacteria, which have not been shown to cause acute illness. However, neither the long-term health-promoting nor health-damaging effect of opportunistic bacteria is well known. In 2010, the most prominent 56 species in the human gut were reported using a non-cultivation approach, and most were found to be opportunistic bacteria.3
Probiotics are live bacteria that are ingested to improve the intestinal environment. Lactic acid bacteria and bifidobacteria are commonly used as probiotics. A prebiotic is defined as “a substrate that is selectively utilized by host microorganisms conferring a health benefit.”.4 Prebiotics are widely used as commercial ingredients to modify gut microbiota.
Previous research has reported that some prebiotics5 and prebiotic candidates6 have a growth-promoting effect on some of the opportunistic bacteria. Furthermore, it has been reported that some of opportunistic bacteria may cause autoimmune diseases7 and, in the presence of dietary fiber deficiency, may degrade the intestinal mucus barrier.8 Therefore, the development of next-generation prebiotics that specifically boost the growth of beneficial or probiotic bacteria is important to promote gut health and aid the treatment of gastrointestinal disease.
Clostridioides difficile colitis is commonly associated with antibiotic use. In the setting of a disrupted microbiota, C. difficile is able to colonize the intestine and produce toxins that ultimately cause inflammation. C. difficile is the most common pathogen in healthcare-associated infections in U.S. acute care hospitals.9 To cure C. difficile-related infection, the administration of metronidazole, fidaxomicin, or vancomycin has been used but often results in recurrent infections.10 Recently, fecal microbiota transplantation, the instillation of processed stool collected from a healthy donor into the intestinal tract of patients, has been recommended as an effective treatment for recurrent C. difficile colitis.10 However, there are cases in which new-onset obesity has been caused by the transplantation of an overweight donor’s stool,11 and one in which a patient died from infection by extended-spectrum β-lactamase-producing Escherichia coli after the transplantation.12 As an alternative treatment, probiotic administration has been studied. However, no controlled clinical trial has demonstrated significant and reproducible efficacy.10
In this study, we screened β-galactosides with various structures containing enzymatically synthesized oligosaccharides to develop next-generation prebiotics that specifically boost the growth of beneficial and probiotic bacteria without promoting the growth of opportunistic and pathogenic bacteria, by testing whether each bacterium can utilize each sugar. We then identified the gene required for the growth promotion specific to bifidobacteria. Furthermore, we conducted the co-culture test, fecal culture, and mouse test aimed at suppressing the growth of C. difficile by the obtained next-generation prebiotic candidate in combination with bifidobacteria.
Results
Existing prebiotics promote the growth of multiple bacterial species of the human gut microbiota
To investigate the effect of existing prebiotics on the growth of human gut microbiota, we cultured beneficial bacteria, pathogenic bacteria, and prominent bacteria (Table 1) in a Gifu anaerobic medium (GAM) without sugar medium (GAM-wos) supplemented with prebiotics available on the market. The prominent bacteria used in this test were 32 dominant species that can be cultured in the GAM medium.15 However, five of these (Ruminococcus obeum, Eubacterium siraeum, Pseudoflavonifractor capillosus, Clostridium scindens, and Anaerotruncus colihominis) were excluded because their growth was unstable in GAM-wos. All five existing prebiotics and prebiotic candidates tested (namely, raffinose, 1-kestose, lactulose, galacto-oligosaccharides (GOS), and fructo-oligosaccharides (FOS)) promoted the growth of lactic acid bacteria and bifidobacteria (Figure 1, Supplemental Figure S1). However, raffinose, 1-kestose, lactulose, GOS, and FOS promoted the growth of four (Figure 1a), ten (Figure 1b), eight (Figure 1c), eight (Figure 1d), and seven species (Figure 1e), respectively, of other prominent bacterial species by more than 200%. Besides, 1-kestose, lactulose and GOS promoted the growth of the pathogen Clostridium perfringens (Figure 1b, 1c and 1d) that causes gas gangrene and food poisoning. Furthermore, 1-kestose also promoted the growth of pathogen C. difficile (Figure 1b).
Table 1.
List of bacterial strains used in this study
| SOURCE | IDENTIFIER | |
|---|---|---|
| Bacteroides thetaiotaomicron | JCM | JCM 5897 |
| Bacteroides uniformis | JCM | JCM 5828 |
| Bacteroides vulgatus | JCM | JCM 5826 |
| Bacteroides caccae | JCM | JCM9498 |
| Bacteroides ovatus | JCM | JCM 5824 |
| Bacteroides xylanisolvens | JCM | JCM15633 |
| Bacteroides dorei | JCM | JCM13471 |
| Bacteroides stercoris | JCM | JCM9496 |
| Bacteroides finegoldii | JCM | JCM13345 |
| Bacteroides intestinalis | JCM | JCM13265 |
| Bacteroides fragilis | JCM | JCM11019 |
| Parabacteroides distasonis | JCM | JCM 5825 |
| Parabacteroides merdae | JCM | JCM9497 |
| Dorea longicatena | DSMZ | DSM13814 |
| Dorea formicigenerans | ATCC | ATCC27755 |
| Parabacteroides johnsonii | JCM | JCM13406 |
| Coprococcus comes | ATCC | ATCC 27758 |
| Ruminococcus torques | ATCC | ATCC 27756 |
| Ruminococcus lactaris | ATCC | ATCC 29176 |
| Ruminococcus gnavus | ATCC | ATCC29149 |
| Collinsella aerofaciens | JCM | JCM 7790 |
| Blautia hansenii | JCM | JCM14655 |
| Eubacterium ventriosum | ATCC | ATCC 27560 |
| Clostridium nexile | ATCC | ATCC27757 |
| Enterococcus faecalis V583 | ATCC | ATCC700802 |
| Roseburia intestinalis | DSMZ | DSM 14610 |
| Clostridium asparagiforme | DSMZ | DSM15981 |
| Clostridium perfringens | JCM | JCM1290 |
| Lactobacillus casei subsp. casei | JCM | JCM1134 |
| Lactobacillus casei subsp. rhamnosus | ATCC | ATCC7469 |
| Lactobacillus paragasseri | JCM | JCM1130 |
| Lactobacillus johnsonii | JCM | JCM8794 |
| Lactobacillus plantarum | JCM | JCM1149 |
| Lactococcus lactis | JCM | JCM1158 |
| Lactobacillus reuteri | JCM | JCM1112 |
| Leuconostoc mesenteroides subsp. mesenteroides | JCM | JCM6124 |
| Bifidobacterium bifidum | JCM | JCM1254 |
| Bifidobacterium longum subsp. longum | JCM | JCM1217 |
| Bifidobacterium breve | JCM | JCM1192 |
| Bifidobacterium pseudocatenulatum | JCM | JCM1200 |
| Bifidobacterium adolescentis | JCM | JCM1275 |
| Bifidobacterium animalis subsp. lactis | JCM | JCM10602 |
| Bifidobacterium catenulatum | JCM | JCM1194 |
| Bifidobacterium pseudolongum | JCM | JCM1205 |
| Clostridioides difficile | JCM | JCM1296 |
| Bifidobacterium longum subsp. longum 105-A | JCM | JCM31944 |
| Bifidobacterium longum subsp. infantis | JCM | JCM1222 |
| B. longum subsp. longum strain 105-A bl105A_0501::pRH4 | 13 | N/A |
| B. longum subsp. longum strain 105-A bl105A _1817::pRH7 | This paper | N/A |
| B. longum subsp. longum strain 105-A bl105A _1867::pRH8 | This paper | N/A |
| B. longum subsp. longum strain 105-A bl105A _1888::pRH9 | This paper | N/A |
| B. longum subsp. longum strain 105-A bl105A _1890::pRH10 | This paper | N/A |
| B. longum subsp. longum strain 105-A bl105A _1896::pRH11 | This paper | N/A |
| B. longum subsp. longum strain 105-A bl105A _0201::pRH2 | This paper | N/A |
| B. longum subsp. longum strain 105-A bl105A _0500::pRH3 | 13 | N/A |
| B. longum subsp. longum strain 105-A bl105A _1223::pRH6 | This paper | N/A |
| B. longum subsp. longum strain 105-A bl105A _0502::pRH5 | 13 | N/A |
| B. longum subsp. longum strain 105-A Δbl105A _1604 | 14 | N/A |
|
B. longum subsp. infantis strain ATCC15697T blij_2090::pRH12 |
This paper | N/A |
Figure 1.
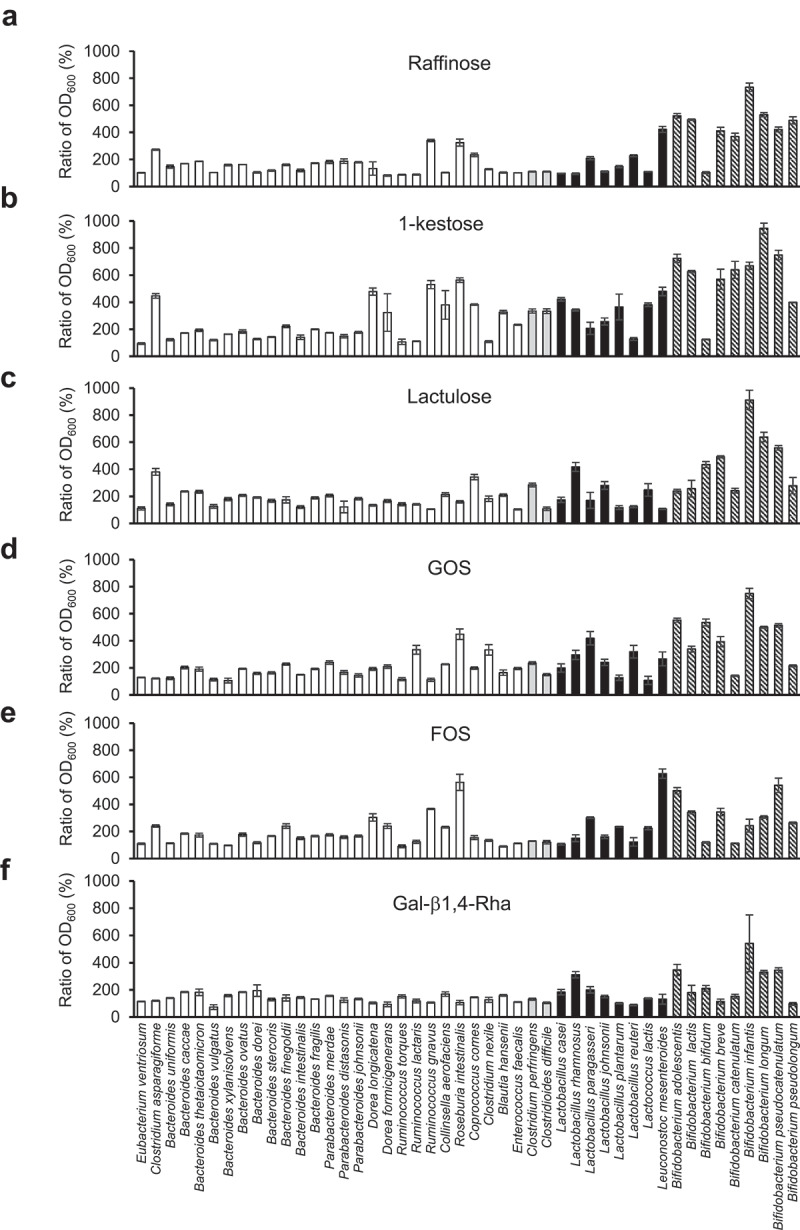
Growth-promoting effects of existing prebiotics and Gal-β1,4-Rha on beneficial bacteria and the most prominent 27 bacterial species in human gut microbiota
(a-f) Bacterial species were cultured in GAM-wos with or without 0.5% prebiotic supplements for 24 h. Bacterial growth was measured by determining the OD600. To compare the growth promotion ability by prebiotics for each bacterium, the growth-promoting value (Ratio of OD600) was obtained by dividing the average of growth (OD600) in the medium supplemented with prebiotics (n = 3) by that without prebiotics (n = 3). White bars, the most prominent 27 bacterial species in the human gut microbiota; gray bars, pathogenic bacterium; black bars, lactic acid bacteria; diagonal bars, bifidobacteria. GAM-wos was supplemented with raffinose (a), 1-kestose (b), lactulose (c), GOS (d), FOS (e), and Gal-β1,4-Rha (f). Data represent means ± standard deviation.
Gal-β1,4-Rha, a next-generation prebiotic candidate, has a bifidobacteria-specific growth-promoting effect
Next, we analyzed the growth-promoting effects of our enzymatically synthesized galactosyl-β1,4-l-rhamnose (Gal-β1,4-Rha),16,17 galactosyl-β1,3- N-acetylglucosamine (LNB),18 galactosyl-β1,3-glucose (Gal-β1,3-Glc),16 galactosyl-β1,3-galactose (Gal-β1,3-Gal)16 and galactosyl-β1,3 -N-acetylgalactosamine (GNB)19 in addition to lactose on gut bacteria (Supplemental Table S1, Supplemental Figures S2 and S3, and figure 1f). Gal-β1,4-Rha, a disaccharide previously synthesized enzymatically using the reverse reaction of d-galactosyl-β1→4-l-rhamnose phosphorylase,16,17 selectively promoted the growth of bifidobacteria, including Bifidobacterium longum subsp. infantis (B. infantis) and Bifidobacterium longum subsp. longum (B. longum) (figure 1f). Additionally, the growth of two lactic acid bacteria was promoted (figure 1f). The growth-promoting values by Gal-β1,4-Rha for C. perfringens, C. difficile, or prominent bacteria in the human gut microbiota3 were less than 200% (figure 1f).
B. longum requires BL105A_0502 to use Gal-β1,4-Rha as a carbon source
To identify the gene responsible for selective growth promotion of bifidobacteria (figure 1f), we used a genetically tractable strain B. longum 105-A (JCM31944).20,21 Sugar utilization systems of B. longum species mainly depend on different ABC transporters.22 We created a library of 11 B. longum mutant strains (Table 1) harboring a disruption of the gene encoding ABC transporter solute-binding protein. Wild-type B. longum and the mutant strains were grown in modified MRS without sugar medium (mMRS-wos) containing Gal-β1,4-Rha (Figure 2a, 2c) as a sole carbon source. Most mutant strains showed normal growth compared with the wild-type strain. However, the BL105A_0502 (GenBank protein ID: BAP83151.1) mutant showed delayed and decreased growth compared to the wild-type. On the other hand, when cultured in mMRS-wos containing galactose as a sole carbon source, BL105A_0502 mutant showed the same growth as the wild-type (Figure 2b and 2d). These results demonstrated that the solute-binding protein BL105A_0502 of an ABC transporter plays a vital role in the utilization of Gal-β1,4-Rha by B. longum 105-A and may contribute to Gal-β1,4-Rha transport.
Figure 2.
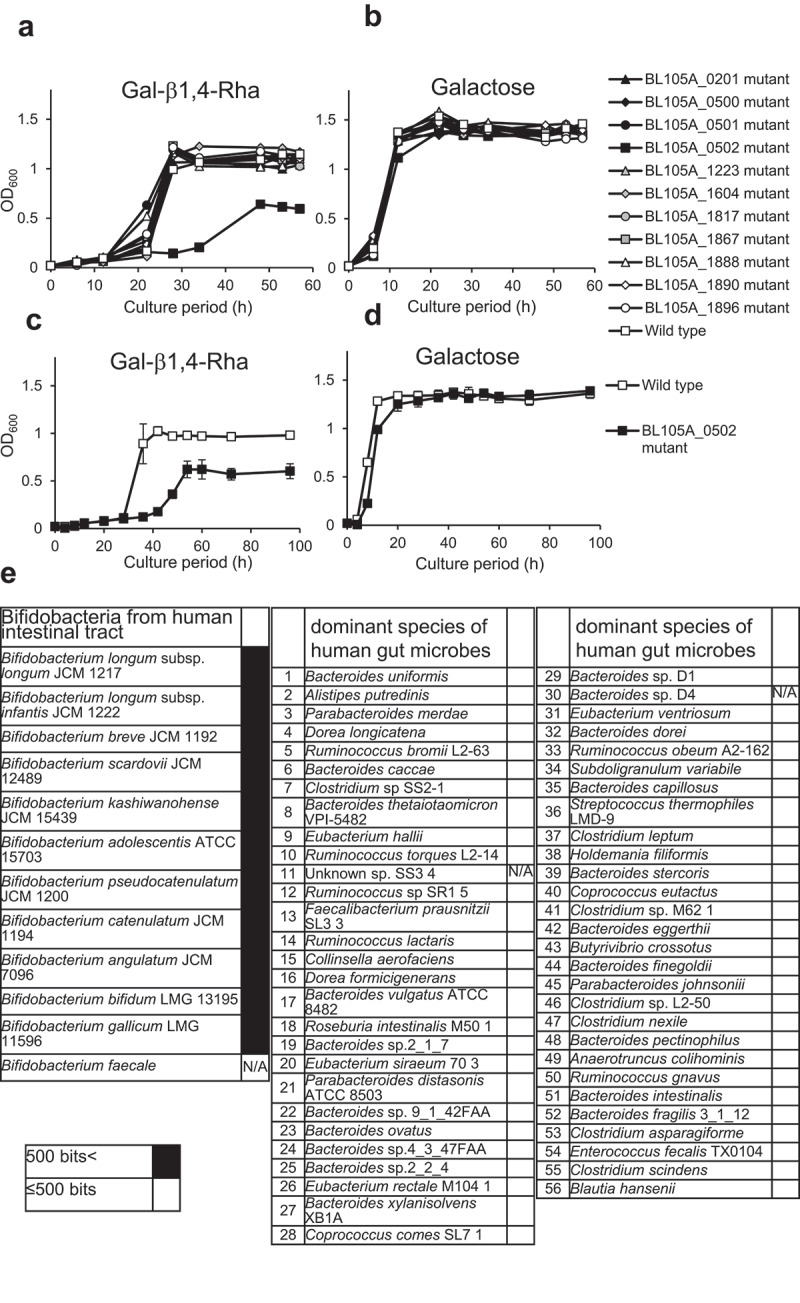
The importance of the BL105A_0502 gene for the utilization of Gal-β1,4-Rha in B. longum 105-A
(a-b) Growth curve of B. longum 105-A wild-type and mutants with a disruption of the gene encoding ABC transporter solute-binding protein in mMRS-wos containing 0.5% Gal-β1,4-Rha (a) or 0.5% galactose (b) as a carbon source (n = 1). The disrupted genes were indicated by the locus tag. Black triangle, BL105A_0201 mutant; black rhombus, BL105A_0500 mutant; black circle, BL105A_0501 mutant; black square, BL105A_0502 mutant; gray triangle, BL105A_1223 mutant; gray rhombus, BL105A_1604 mutant; gray circle, BL105A_1817 mutant; gray square, BL105A_1867 mutant; white triangle, BL105A_1888 mutant; white rhombus, BL105A_1890 mutant; white circle, BL105A_1896 mutant; white square, wild-type. (c-d) Growth of B. longum wild-type (white squares) and the BL105A_0502 mutant (black squares) in mMRS-wos containing 0.5% Gal-β1,4-Rha (c), or 0.5% galactose (d) as a carbon source (n = 3). Data represent means ± standard deviation. (e) Presence of BL105A_0502 homolog in bifidobacteria and 56 prominent bacteria in the human gut microbiota. The results shown are based on the Protein BLAST analysis of genomes in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The color of each column indicates the score from the Protein BLAST analysis: (black) >500 bits and (white) ≤500 bits. If no genome information was available in the NCBI database, it was indicated by N/A.
The ability to utilize Gal-β1,4-Rha by bacteria in the human gut microbiota requires BL105A_0502 homologs
The distribution of BL105A_0502 homologs in other bifidobacterial species23,24 and the 56 prominent bacteria in the human gut microbiota3 were analyzed using BLAST.25 Proteins homologous to BL105A_0502 were not found in the 56 prominent bacteria in the human gut microbiota (Figure 2e). In contrast, all bifidobacteria examined were found to possess a BL105A_0502 homolog (Figure 2e). Therefore, the growth promotion by Gal-β1,4-Rha specific to bifidobacteria is likely to be caused by BL105A_0502 and its homologs.
B. infantis suppresses the growth of C. difficile in vitro
From the results obtained above, it is expected that Gal-β1,4-Rha promotes the growth of bifidobacteria selectively in competition against other intestinal bacteria that are unable to utilize Gal-β1,4-Rha. Therefore, we tried to suppress the enteric pathogen Clostridioides difficile, which cannot utilize Gal-β1,4-Rha (figure 1f, Supplemental Figure S4B), via the competition of B. infantis, which can utilize Gal-β1,4-Rha (figure 1f, Supplemental Figure S4A). After 48 h of incubation, in the co-culture of B. infantis and C. difficile in the presence of Gal-β1,4-Rha, the viable count of C. difficile dramatically decreased to 1/1,850 compared to the monoculture of C. difficile (Figure 3a). Contrastingly, no suppression of the viable count of C. difficile occurred in the co-culture when Gal-β1,4-Rha was absent at any time point (Figure 3c). These results (Figure 3a and 3c) suggest that the presence of both B. infantis and Gal-β1,4-Rha is necessary to significantly inhibit the growth of C. difficile. Weak suppression of the growth of C. difficile can be observed by co-culture in the presence of glucose after 48 h of incubation (Figure 3e), which is utilized by both B. infantis and C. difficile (Supplemental Figure S4A and S4B), but the viable count of C. difficile decreased only to 1/4 compared to the monoculture of C. difficile (Figure 3e). On the other hand, the viable count of B. infantis was not suppressed by co-culture in any medium (Figure 3b, 3d and 3f).
Figure 3.
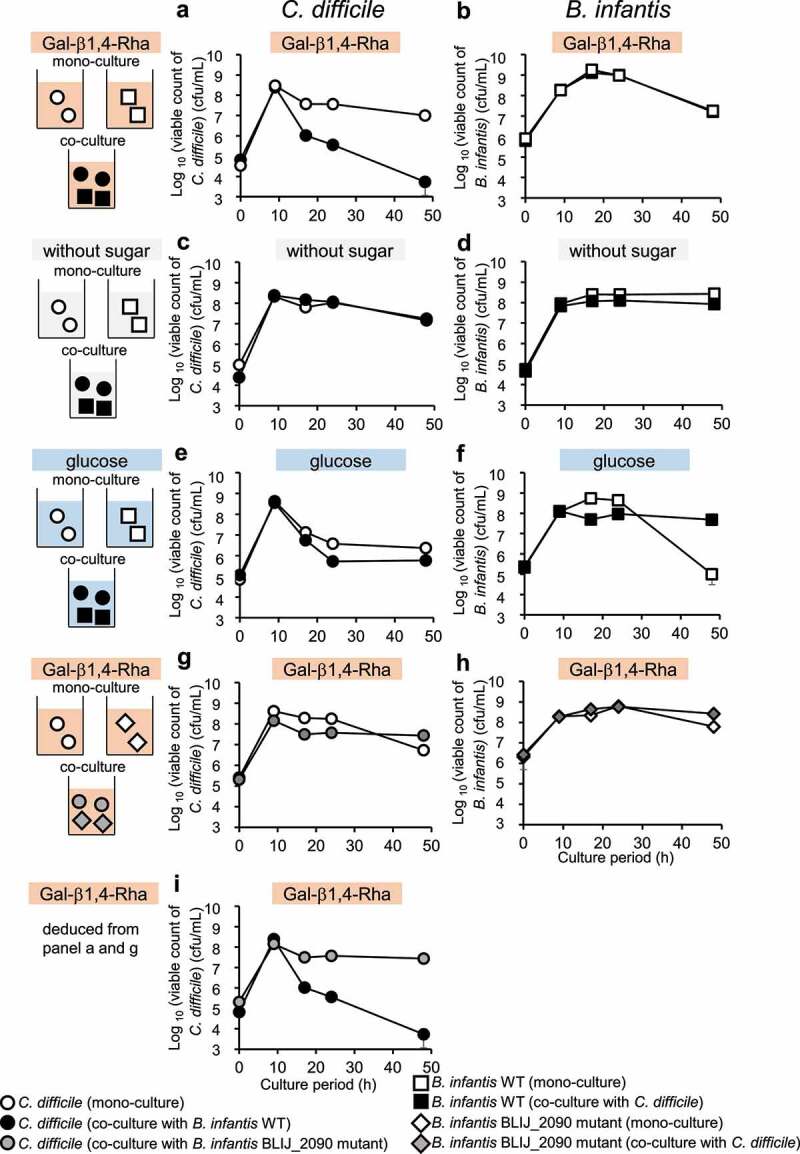
Growth inhibition of C. difficile by B. infantis in the presence of Gal-β1,4-Rha and the importance of BLIJ_2090 gene for the growth inhibition
(a, c, e, and g) Circles indicated the viable cell number of C. difficile. Monocultures, co-cultures with B. infantis wild-type, and co-culture with B. infantis BLIJ_2090 mutant are indicated by white, black, and gray symbols, respectively. (b, d, f, and h) The viable cell number of wild-type B. infantis and the BLIJ_2090 mutant is indicated by squares and rhombuses, respectively. Mono-cultures, co-cultures with B. infantis wild-type, and co-culture with the B. infantis BLIJ_2090 mutant are indicated by white, black, and gray symbols, respectively. GAM-wos was supplemented with 0.5% of Gal-β1,4-Rha (a-b, g-f and i), 0.5% of glucose (e-f), or without sugar (c-d). In i, viable cell numbers of C. difficile co-cultured with wild-type B. infantis (deduced from panel a) was compared to that with B. infantis BLIJ_2090 mutant (deduced from panel g). Data represent means ± standard deviation from biological replicates (n = 3).
The suppression of growth of C. difficile by B. infantis in medium containing Gal-β1,4-Rha requires BLIJ_2090, a homolog of BL105A_0502
B. infantis JCM1222T has BLIJ_2090, a protein with a 96% amino acid sequence identity to BL105A_0502 in B. longum. To investigate whether BLIJ_2090 is involved in growth inhibition of C. difficile by B. infantis in the presence of Gal-β1,4-Rha, a B. infantis BLIJ_2090 insertional mutant and C. difficile were co-cultured, and the viable cell count of C. difficile was compared to the co-culture of wild-type B. infantis and C. difficile. As a result, no strong growth inhibition of C. difficile by the B. infantis BLIJ_2090 mutant occurred in the co-culture even in the presence of Gal-β1,4-Rha (Figure 3g and 3i). The viable count of B. infantis was not affected by co-culture (Figure 3h). These results suggest that BLIJ_2090, the putative solute-binding protein of Gal-β1,4-Rha of B. infantis, is essential for the inhibition of growth of C. difficile in the presence of Gal-β1,4-Rha.
Analysis of the growth inhibition mechanism of C. difficile by co-culturing with B. infantis
To understand the mechanism of growth inhibition when C. difficile was co-cultured with B. infantis in the presence of Gal-β1,4-Rha, analyses of the culture supernatant were performed. When B. infantis was mono-cultured in GAM-wos supplemented with Gal-β1,4-Rha or glucose, the pH of the culture medium decreased considerably from 7.1 to 4.8 (when Gal-β1,4-Rha was supplemented) or 4.7 (when glucose was supplemented) (Supplemental Figure S4D). In contrast, the decrease in pH caused by monoculture of B. infantis was weaker in GAM-wos (Supplemental Figure S4D).
When C. difficile was mono-cultured in GAM-wos supplemented with glucose, the pH decreased from 7.1 to 6.2 (Supplemental Figure S4C). However, when C. difficile was mono-cultured in GAM-wos supplemented with Gal-β1,4-Rha, which was not available to C. difficile, the pH did not decrease (Supplemental Figure S4C). In the co-culture of C. difficile and B. infantis, the pH decreased from 7.1 to approximately 6.0 when cultured in GAM-wos supplemented with Gal-β1,4-Rha or glucose (Supplemental Figure S4E and S4F). Although these two culture supernatants had similarly low pH values, the viable count of C. difficile when co-cultured with wild-type B. infantis in GAM-wos supplemented with Gal-1,4-Rha was 110 times lower than that when co-cultured in GAM-wos supplemented with glucose (Supplemental Figure S4E). Furthermore, when C. difficile was co-cultured with wild-type B. infantis or the BLIJ_2090 mutant in GAM-wos supplemented with Gal-β1,4-Rha, the pH decreased from 7.1 to approximately 6.0 in both the cases. However, when co-cultured with wild-type B. infantis, the viable cell count of C. difficile was approximately 5,000 times lower than that when co-cultured with the BLIJ_2090 mutant (Supplemental Figure S4E).
When C. difficile was cultured in a medium supplemented with the pH-unadjusted cell-free supernatant of B. infantis (Supplemental Figure S4G), the growth of C. difficile was slightly delayed when the cell-free supernatant of B. infantis cultured in GAM-wos supplemented with Gal-β1,4-Rha (pH 5.0) was used compared to when the cell-free supernatant of B. infantis cultured in GAM-wos was used (pH 6.4, Supplemental Figure S4G). The extent of the growth inhibition of C. difficile by the cell-free supernatant of wild-type B. infantis (pH 5.0) and the BLIJ_2090 mutant was equivalent (pH 5.1, Supplemental Figure S4G).
When C. difficile was cultured in a medium supplemented with the pH-adjusted (to 7.1) cell-free supernatant of B. infantis (Supplemental Figure S4H), there was no delay in the growth of C. difficile regardless of the type of cell-free supernatant used (Supplemental Figure S4H).
C. difficile toxin production is inhibited by Gal-β1,4-Rha and B. infantis
We examined whether the growth inhibition of C. difficile by B. infantis in medium supplemented with Gal-β1,4-Rha occurs under conditions of coexistence of various gut bacteria. B. infantis and C. difficile were co-cultured in GAM-wos supplemented with human fecal suspension and Gal-β1,4-Rha, and C. difficile toxin (CD toxin) was quantified (Figure 4). Paired one-way ANOVA with Bonferroni correction was performed on the results obtained using fecal suspensions from five donors. For all fecal suspensions, no CD toxin was detected without inoculation of C. difficile in the culture (Data not shown). When C. difficile was inoculated into the fecal cultures, CD toxin was detected in all cultures (black bars in Supplemental Figure S5A). When B. infantis was added to the fecal cultures inoculated by C. difficile, the amount of CD toxin did not decrease significantly (gray bar in Figure 4). When Gal-β1,4-Rha was added to the fecal cultures inoculated by C. difficile and B. infantis, the amount of CD toxin decreased significantly (checkered bar in Figure 4). The CD toxin was drastically reduced to less than one-third in fecal cultures using feces A, C, and D (checkered bars in Supplemental Figure S5A). CD toxin levels in fecal cultures using feces B and E, which had lower amounts of the toxin when inoculated with C. difficile (black bars in Supplemental Figure S5), hardly decreased (checkered bars in Supplemental Figure S5A). Taken together, despite the presence of interindividual differences, Gal-β1,4-Rha significantly suppressed the production of CD toxin in the presence of B. infantis. The amount of CD toxin did not decrease significantly when B. infantis was not inoculated (Diagonal bar in Figure 4). These results suggested that the suppression of the production of CD toxin occurred most strongly when Gal-β1,4-Rha and B. infantis were present simultaneously. However, there was no difference between the bacterial counts of C. difficile in the fecal cultures (Supplemental Figure S5B).
Figure 4.
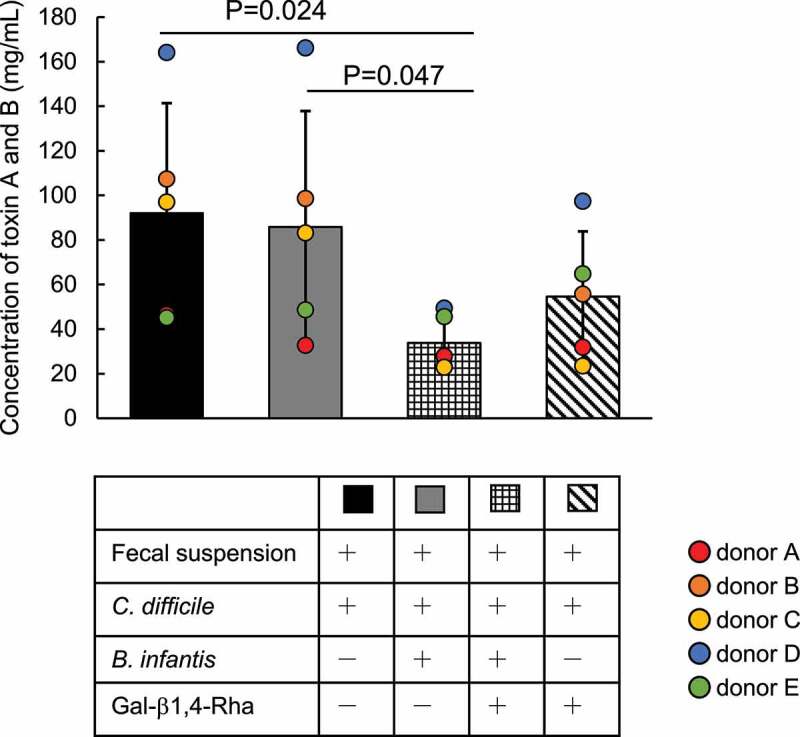
Inhibition of the production of CD toxins A and B by the combination of Gal-β1,4-Rha and B. infantis in fecal culture
C. difficile, B. infantis, and/or human fecal suspension from 5 donors were cultured in GAM-wos supplemented with or without 0.5% Gal-β1,4-Rha for 48 h, and CD toxins A and B in the fecal cultures were quantified by ELISA. Black bar, fecal suspension, and C. difficile were cultured in GAM-wos; gray bar, C. difficile, B. infantis, and fecal suspension were co-cultured in GAM-wos; checkered bar, C. difficile B. infantis, and fecal suspension were co-cultured in GAM-wos supplemented with 0.5% Gal-β1,4-Rha; diagonal bar, C. difficile and fecal suspension were cultured in GAM-wos supplemented with Gal-β1,4-Rha. These culture conditions are summarized in the table below the Figure (+, when the components were added to the medium; -, when not added). For all fecal suspensions, CD toxin was not detected when C. difficile was not inoculated into the culture and is not shown in the figure. Data represent means ± standard deviation. Paired one-way ANOVA followed by multiple comparison with Bonferroni correction was performed. Individual data points are represented by circles.
The body weight loss was suppressed by a combination of B. infantis and Gal-β1,4-Rha in a mouse model of C. difficile colitis
To investigate the growth inhibitory effect on C. difficile in vivo, C. difficile, B. infantis, and Gal-β1,4-Rha were administered in various combinations to antibiotic-treated mice for 5 days (Figure 5a), and change in body weight (Figure 5b and 5c) along with bacterial count of B. infantis (Figure 5d) and C. difficile (Figure 5e) in feces were measured. After infection of C. difficile, body weight began to decrease significantly in all groups. However, in mice administered with a combination of Gal-β1,4-Rha, C. difficile, and wild-type B. infantis, the weight significantly decreased from day 1 to 5, but recovered to the level of day 0 between days 5 and 7 (Group D in Figure 5b). Group D showed the smallest reduction in weight on day 7 compared to that on the first day, and its recovery from day 0 was significantly greater than that in group A, to which neither Gal-β1,4-Rha nor B. infantis was administered (Figure 5c). Group D, to which both Gal-β1,4-Rha and B. infantis was administered, had significantly higher weight recovery than group B, to which only Gal-β1,4-Rha was administered, and group C, to which only B. infantis was administered. Administration of Gal-β1,4-Rha alone did not cause weight loss (Supplemental Figure S6). In contrast, in mice administered with a combination of Gal-β1,4-Rha, C. difficile, and the B. infantis BLIJ_2090 mutant, the weight kept decreasing (Group E in Figure 5b). This suggests that B. infantis imports Gal-β1,4-Rha via the transporter identified in this study, and alleviates weight loss owing to C. difficile infection in vivo. In addition, the bacterial count of B. infantis in the feces of the mice administered with Gal-β1,4-Rha and wild-type B. infantis was significantly higher than that with the administration of wild-type B. infantis alone (Figure 5d). These suggest that Gal-β1,4-Rha may promote the growth of B. infantis in the intestinal lumen of mice. However, there was no difference between bacterial counts of C. difficile in mice feces, and the inhibitory effect on the growth of C. difficile was not observed in vivo (Figure 5e).
Figure 5.
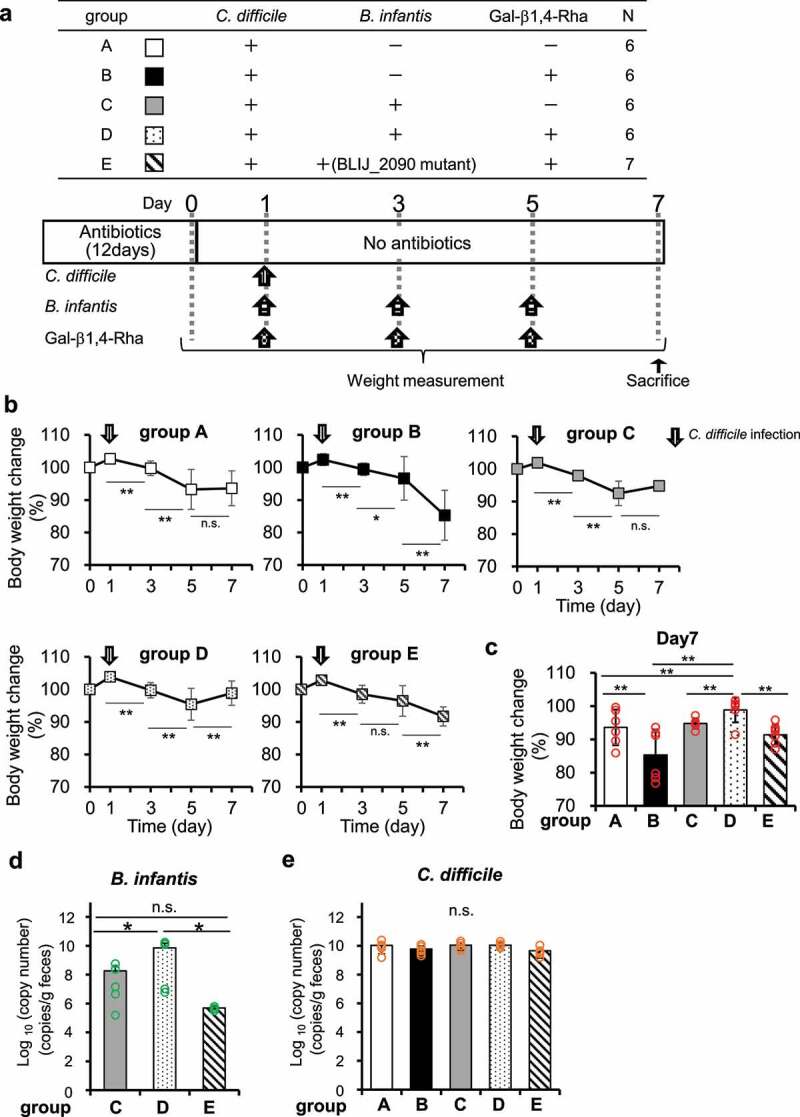
Effects of Gal-β1,4-Rha and B. infantis on weight loss in mice owing to C. difficile infection
(a) Experimental outline. The experimental groups are shown in the table (+, administered; −, not administered). The experimental schedule is shown below the table. The arrows indicate the timing of Gal-β1,4-Rha or bacterial administration. (b-c) Changes in body weights. b: Asterisks indicate statistical significance versus the same group on the day of the previous measurement after C. difficile infection. c: Asterisks indicate statistical significance versus groups A and D on day 7. (*p <.05 and **p <.01, two-way ANOVA with repeated measurement followed by Tukey multiple comparison test) Data represent means ± standard deviation. Individual data points are represented by circles. (d-e) Copy number of beta-galactosidase 42B gene of B. infantis (d) and 16S rRNA gene of C. difficile (e) in feces on day 7 quantified by qPCR. B. infantis has 1 copy of beta-galactosidase 42B gene per cell, and C. difficile has 12 copies of the 16S rRNA gene per cell. Asterisks indicate statistical significance (*p <.05, one-way ANOVA with Tukey-Kramer multiple comparison test). Data represent means ± standard deviation. Individual data points are represented by circles.
Discussion
This study showed that some existing prebiotics promote the growth of opportunistic bacteria as well as beneficial bacteria in vitro, suggesting that some specific prebiotics promote the growth of opportunistic bacteria of unknown function in the human intestinal lumen. These opportunistic bacteria, which are predominant in the human intestinal lumen,3 previously were not considered harmful to human health. However, it has been suggested recently that some opportunistic bacteria may adversely affect health.7,8,26 In the present study, we screened various β-galactosides to develop a next-generation prebiotic that specifically supports the growth of bifidobacteria and lactic acid bacteria without boosting the growth of opportunistic or pathogenic bacteria present in the intestinal lumen. Gal-β1,4-Rha was successfully identified using our developed high-throughput culturing system.15 Reportedly, the abundance of Bacteroides ovatus was successfully controlled by calibrating the amount of administered porphyran, a non-digestible polysaccharide, using gnotobiotic mice.27 Therefore, by tailoring the dosage, Gal-β1,4-Rha could potentially revolutionize prebiotic therapy by the desired increase in numbers of the target beneficial bacteria.
We successfully screened Gal-β1,4-Rha as a next-generation prebiotic candidate. However, the guidelines for the use of health claims vary from country to country, and further tests are required for Gal-β1,4-Rha to be recognized as a prebiotic. Therefore, for next-generation prebiotics to be marketed, human trials must be conducted to demonstrate that they increase the abundance of bifidobacteria in the human gut.
In addition, we directly demonstrated using a gene-disrupted strain of B. longum 105-A that utilization of Gal-β1,4-Rha depends on BL105A_0502, a solute-binding protein of ABC transporter. Since homologs of BL105A_0502 are found specifically in Bifidobacterium, BL105A_0502 could be the key factor for the mechanism of Gal-β1,4-Rha-mediated, selective promotion of growth in bifidobacteria. BL105A_0502 has been reported as a solute-binding protein for prebiotic lactulose.13 In this study, we showed that Gal-1,4-Rha is another substrate of proteins encoded by BL105A_0502 and BLIJ_2090 by using gene-disruption strains. Therefore, BL105A_0502 has multiple substrates, suggesting that it is an important protein for bifidobacteria to utilize a variety of sugars, i.e., lactulose and Gal-1,4-Rha.
It has been reported that Gal-β1,4-Rha (Kd was not available28) and lactulose (Kd: (4.9 ± 0.3) × 10−4 M 28) binds to the solute-binding protein BALAC_0483 in B. animalis subsp. lactis Bl-04.28 However, the amino acid sequence identity of BALAC_0483 to BL105A_0502 and BLIJ_2090 is 51.2% and 48.1%, respectively, and therefore, BALAC_0483 is hardly considered a BL105A_0502 homolog.
Gal-β1,4-Rha is found in nature as part of the pectin rhamnogalacturonan I structure,29 which is found in tansy, Tanacetum vulgare L.30 Several studies have shown that bifidobacteria can utilize plant-derived oligosaccharides31,32 as nutrient sources. Therefore, it is considered that plant-derived dietary fibers reach the large intestine of the animal host in an undigested state. Bifidobacteria may occupy a niche by employing an ABC transporter like a BL105A_0502 homolog as a solute-binding protein, conferring the ability to use an oligosaccharide containing the Gal-β1,4-Rha structure as a carbon source. Notably, Gal-β1,4-Rha structure is reportedly included in the extracellular polysaccharide produced by Bifidobacterium longum JBL05.33 The intestinal opportunistic strain Bacteroides thetaiotaomicron degrades and utilizes glycans from yeast cell walls,34 suggesting that intestinal bacteria may utilize polysaccharides from other organisms in the gut. Assuming that bifidobacteria share the adjacent niche in the large intestinal lumen, although the amount of Gal-β1,4-Rha there is unknown, it is conceivable that bifidobacteria may utilize Gal-β1,4-Rha derived from extracellular polysaccharides as a nutrient source produced by the same genus.
Bifidobacteria are expected to degrade Gal-1,4-Rha by some enzymes after uptake via the ABC transporter. B. infantis possesses three types of GH42 β-galactosidases (Bga42A, Bga42B, and Bga42C), and it has been reported that these β-galactosidases are capable of hydrolyzing Gal-β1,3-Gal and Gal-β1,3-Glc, which are human milk oligosaccharides.35,36 In the genomic DNA of B. infantis, BLIJ_2092, a gene encoding GH42 β-galactosidase, is located in the vicinity of the BLIJ_2090 gene. In addition, there are similarities in the steric structures of Gal-β1,4-Rha, Gal-β1,3-Gal, and Gal-β1,3-Glc. Therefore, we considered GH42 β-galactosidase as a candidate for the Gal-β1,4-Rha-degrading enzyme but further elucidation of this assumption requires additional studies in the future.
We hypothesized that the mechanism of the growth inhibition of C. difficile was related to the decrease in pH during the growth of B. infantis, nutrient deprivation by B. infantis, and the release of growth inhibitors into the environment by B. infantis against C. difficile. Cell-free supernatants with a low pH inhibited the growth of C. difficile (Supplemental Figure S4G) but cell-free supernatants with a pH of 7.1 did not (Supplemental Figure S4H). Therefore, we inferred that a decrease in pH is a part of the mechanism of C. difficile growth inhibition that is independent of the ability of B. infantis to utilize Gal-β1,4-Rha. In the co-culture study, regardless of the presence or absence of sugars available for B. infantis or the level of C. difficile suppression, the cell density of C. difficile reached a maximum of 108 colony-forming units (CFU)/mL after 9 h under all culture conditions (Figure 3a, 3c, 3e and 3g), suggesting that nutrient deprivation by B. infantis was not related to the suppression of C. difficile. Because cell-free supernatants with a pH of 7.1 were ineffective in inhibiting the growth of C. difficile (Supplemental Figure S4H), it is unlikely that there are substances in the culture supernatant that inhibit the growth of C. difficile. In the co-culture study, the growth of C. difficile was considerably inhibited when co-cultured with wild-type B. infantis in the presence of Gal-1,4-Rha (Figure 3a), and the growth of C. difficile was slightly inhibited when co-cultured with the BLIJ_2090 mutant strain of B. infantis in the presence of Gal-β1,4-Rha (Figure 3g). Apart from the difference in the degree of inhibition, the pH after incubation in the co-culture was low under both the conditions (Supplemental Figure S4E and S4F). Therefore, we believed that factors other than pH are involved in the inhibition of C. difficile by B. infantis that took up Gal-β1,4-Rha through BLIJ_2090.
In co-culture with fecal suspensions, Gal-β1,4-Rha suppressed the production of CD toxin in some samples when B. infantis was not inoculated (Diagonal bar in Supplemental Figure S5A). It has been reported that bifidobacteria are the dominant species in the Japanese gut microbiome.37 Because all fecal donors in this study were Japanese, endogenous bifidobacteria were likely present in the feces. Gal-β1,4-Rha may have promoted the growth of endogenous bifidobacteria and reduced the amount of CD toxin.
In fecal culture, CD toxin levels decreased (Figure 4) but the cell density of C. difficile quantified by qPCR did not decrease (Supplemental Figure S5B). In the mice experiments, weight loss by infection with C. difficile was also alleviated (Figure 5b) but the cell density of C. difficile (Figure 5e) quantified by qPCR did not decrease. On the other hand, in vitro co-culture analysis of the number of viable cells showed that C. difficile cells, once increased to 108 CFU/mL, decreased to 104 CFU/mL at 48 h in a medium supplemented with Gal-β1,4-Rha. However, the bacterial cell density of C. difficile sampled in the same culture quantified by qPCR did not decrease at 48 h (9.2 × 109 ± 6.0 × 109 copies/mL). This is probably because qPCR may also count the number of dead cells, it is possible that the number of viable C. difficile cells was actually reduced in fecal culture and mice experiments.
In the mice model in this study, group D administered with both B. infantis and Gal-β1,4-Rha showed the strongest alleviation of weight loss due to C. difficile, indicating promising results of this combination. On the other hand, the maximum weight loss was observed in group B, which was administered with Gal-β1,4-Rha in addition to C. difficile (black symbols in Figure 5b). Administration of Gal-1,4-Rha alone to mice did not cause diarrhea (data not shown) and weight loss (Supplemental Figure S6), although the influx of Gal-β1,4-Rha into the intestinal wall damaged by C. difficile infection may have caused greater weight loss in group B owing to the changes in osmotic pressure and other factors. On the other hand, there was a significant difference in the weight loss recovery of groups B and D (Figure 5c), and this difference was attributed to the effect of bifidobacteria. Group D administered with wild-type B. infantis also showed significantly better recovery compared to group E administered with the BLIJ_2090 mutant (Figure 5c), suggesting that Gal-β1,4-Rha utilization by bifidobacteria may be responsible for the suppression of weight loss in mice.
Our study represents the first step in the research for the therapeutic application of Gal-β1,4-Rha. We believe that by adjusting the dosage and method of administration, it may become a therapeutic option in the future. Human clinical trials are also required to demonstrate the efficacy of our method. In addition, there is no known enzyme capable of specifically extracting Gal-β1,4-Rha from natural materials at present. Therefore, we produced this carbohydrate by enzymatic synthesis. If a method to produce large amounts of Gal-β1,4-Rha from natural products can be developed, we will be closer to conducting human clinical trials.
Until now, no probiotics or synbiotics have been clinically used for the treatment of C. difficile infection. In this study, we showed that administration of a combination of bifidobacteria and Gal-β1,4-Rha was able to suppress the growth of C. difficile in vitro, and that the body weight loss was suppressed by the administration with a combination of bifidobacteria and Gal-β1,4-Rha in a mouse model of C. difficile colitis. Therefore, our results may pave the way toward the establishment of a new treatment method for C. difficile colitis based on a combination of next-generation prebiotics and probiotics.
Materials and methods
Mice
C57BL/6 JJcl mice were obtained from CLEA Japan Inc. The animals were kept under conditions of 50% humidity and a 12:12 h light:dark cycle in specific pathogen-free conditions. They were fed a standard pellet diet (CE-2, CLEA Japan Inc.) and tap water ad libitum. Animal care and experiments conformed to the Guidelines for Animal Experiments of the University of Tokyo and were approved by the Animal Research Committee of the Institute of Medical Science at the University of Tokyo.
Human fecal samples
The experiment was approved by the Human Rights and Ethics Committee of Ishikawa Prefectural University. Five adults participated in the study (3 males, 2 females; age: 25.6 ± 8.14, range 21–40 years). Fecal samples were collected using a stool collection tube and placed in a closed container together with an Aneropack Kenki (Mitsubishi Gas Chemical Company, Inc.) to keep the samples anaerobic before the analysis.
Microbe strains
Bacteria were obtained from American Type Culture Collection (ATCC), German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), and Japan Collection of Microorganisms (JCM). Bacteria were cultured at 37°C in an anaerobic chamber (10% CO2, 10% H2, and 80% N2; InvivO2 400, Ruskinn Technology).
Mouse colonization
Male mice (n = 31) aged 8 weeks were given vancomycin (0.5 g/L) and doripenem (0.25 g/L) in their drinking water for 12 days. The mice were divided into 5 groups, and oral administration of spores of C. difficile (1.0 × 105 CFU/day), B infantis (1.0 × 106 CFU/day), and/or Gal-β1,4-Rha (2 g/kg bw/day) was performed one day after the end of antibiotic administration. Administration schedule was shown in Figure 5. The body weight of the mice was measured, and fecal samples were collected every two days for a week. Then the mice were sacrificed by cervical dislocation under anesthesia with isoflurane.
Preparation of oligosaccharides
LNB,18 Gal-β1,3-Glc,16 Gal-β1,3-Gal,16 GNB19 and Gal-β1,4-Rha16 were synthesized as previously described. Lactose was purchased from Nacalai Tesque. Lactulose, raffinose, GOS, and FOS were purchased from Wako Pure Chemicals. 1- Kestose was purchased from Tokyo Chemical Industry.
Preparation of sugar solution
MilliQ water was autoclaved, placed in a closed container together with the Aneropack Kenki, and let stand overnight to remove oxygen. Sugars were dissolved in the anaerobically stored water, and the sugar solution was filter-sterilized using Millex (Merck KGaA) in the anaerobic chamber.
Preparation of GAM without sugar medium (GAM-wos)
A GAM semisolid without dextrose broth (Nissui Pharmaceutical) was dissolved following the manufacturer’s instructions and filtered to remove agar. After autoclave sterilization, the medium was immediately placed in a closed container together with the Aneropack Kenki and let stand overnight to remove oxygen.
Preparation of modified MRS without sugar medium (mMRS-wos)
mMRS-wos was prepared as follows: a medium where glucose was removed from the composition of the de Man, Rogosa, and Sharpe medium38 was autoclaved and supplemented with 0.34% (w/v) sodium ascorbate and 0.02% (w/v) cysteine-HCl. Then the medium was placed in a closed container together with the Aneropack Kenki and let stand overnight to remove oxygen.
Screening of next-generation prebiotics
The bacterial culture for the screening of the next-generation prebiotic was conducted using previously described procedures.15 Briefly, intestinal bacterial strains were inoculated from frozen glycerol stock in (GAM; Nissui Pharmaceutical) in 96-deep well plates and incubated at 37°C for 1–2 days and used as pre-cultures. The pre-cultures were inoculated in 500 μL of GAM-wos supplemented with 0.5% (w/v) of each sugar in 96-deep well plates using a copy plate stand (Tokken). After 24 h of anaerobic incubation, growth was measured as optical density at 600 nm (OD600). To compare the ability of bacterial species to utilize sugars, the ratio of OD600 was obtained by dividing the bacterial growth (OD600) in the medium supplemented with sugar by that without sugar.
Generation of insertion mutants of bifidobacteria
In addition to the previously constructed gene disruptants of B. longum 105-A,13,14 Bifidobacterium mutants were further generated by inserting suicide plasmids into the genes encoding a putative solute-binding protein (Table 1). Insertional mutation in B. longum 105-A and B. infantis JCM 1222 was conducted by single-crossover recombination using previously described procedures.39 Briefly, plasmids for the introduction of mutations were constructed by cloning internal fragments of target genes encoding putative solute-binding proteins with 2.0-kb BamHI–digested fragment of pBS423.21 The plasmid sequence was integrated into the chromosome of bifidobacteria by homologous recombination. The genome sequences of B. longum 105-A (GenBank accession no. AP014658.1)40 and B. infantis JCM 1222T (accession no. AP010889.1)41 were used as a reference. The primers used are listed in Table S2.
Culture conditions for bifidobacteria
Strains were inoculated on a GAM plate and cultured anaerobically at 37°C for a day to obtain the seed culture. Bacterial cells scraped from seed culture plates were inoculated into GAM broth and anaerobically incubated for 12 h at 37°C to obtain the pre-culture. The bacterial cells of pre-culture were washed twice with mMRS-wos. Washed cells were inoculated into mMRS-wos supplemented with or without 0.5% (w/v) sugars with initial OD600 of 0.02. Growth was tracked by measuring OD600.
Co-culture of B. infantis and C. difficile
C. difficile and B. infantis were streaked on a GAM plate, respectively, and anaerobically incubated at 37°C for 48 to 60 h. Single colonies were inoculated into GAM broth and incubated anaerobically for 24 to 26 h at 37°C to obtain pre-cultures. The pre-cultures were inoculated into 500 µL of GAM-wos supplemented with 0.5% (w/v) of Gal-β1,4-Rha or glucose with initial OD600 of 0.01 for C. difficile and B. infantis BLIJ_2090 mutant. For wild-type B. infantis, the initial OD600 was adjusted to 0.001. At 0, 9, 17, 24, and 48 h after inoculation, cultures were serially diluted from 10−1 to 10−6 in a PBS and spread on Clostridia Count Agar (Nissui) or GAM plate. Plates were incubated for 48 h at 37°C, and CFU were counted. On the Clostridia Count Agar, B. infantis and C. difficile formed white and black colonies, respectively. On the GAM plate, B. infantis and C. difficile formed white and cream-colored colonies, respectively. To confirm the estimation of bacterial species by colony morphology, four colonies with each characteristic were picked from these plates, then, 16S rDNA sequencing was performed using previously described procedures.15 The identity of bacterial species estimated by colony morphology was validated by 16S rDNA analysis. After 48 h of co-culture, the culture supernatant was collected and the pH was measured.
Culture using cell-free supernatants
B. infantis was streaked on a GAM plate and anaerobically incubated at 37°C for 48 h. Single colonies were inoculated into GAM broth and incubated anaerobically for 24 h at 37°C to obtain pre-cultures. The pre-cultures were inoculated into 5 mL of GAM-wos supplemented with or without 0.5% (w/v) of Gal-β1,4-Rha. For wild-type B. infantis, the initial OD600 was adjusted to 0.001. For the B. infantis BLIJ_2090 mutant, the initial OD600 was adjusted to 0.01. After 48 h of inoculation, the cultures were centrifuged (5,000 g for 10 min) and the supernatant was collected. A cell-free supernatant was obtained by passing through a 0.22 μm filter. A portion of the cell-free supernatant was adjusted to pH 7.1. C. difficile was inoculated into 500 µL of GAM-wos supplemented with 50% (v/v) cell-free supernatants with an initial OD600 of 0.01. Growth was determined by measuring OD600.
Fecal culture
C. difficile and B. infantis were respectively streaked on GAM plates and anaerobically incubated at 37°C for 48 h. Single colonies picked from plates were inoculated into tubes containing GAM broth and were anaerobically incubated for 24 h at 37°C to obtain pre-cultures. Feces were diluted 10-fold into PBS, autoclaved, and stored in the anaerobic chamber. The pre-cultures and fecal suspensions were inoculated in 500 µL of GAM-wos supplemented with or without 0.5% (w/v) Gal-β1,4-Rha in 96-deep well plates with initial OD600 of 0.01 for C. difficile and the fecal suspension and 0.001 for B. infantis. After 48 h of anaerobic incubation, the amount of CD toxin A and B in the supernatant was measured using an ELISA kit (TGC-E001-1; tgcBIOMICS) following the manufacturer’s instructions.
Clostridioides difficile spore formation
Spore formation of C. difficile was performed as previously described.42,43
Preparation of freeze-dried bacterial powder of B. infantis
B. infantis was streaked on a GAM plate and anaerobically incubated at 37°C for 2 days. Bacterial cells were scraped and inoculated into GAM broth and incubated anaerobically for 12 h at 37°C to obtain pre-cultures. 3.5 mL of pre-culture was inoculated in 200-mL GAM broth and incubated anaerobically for 12 h. The bacterial cell cultures were washed with PBS. The washed cells were re-suspended in 10 mL freeze-drying buffer containing 100 mM potassium phosphate buffer (pH 7.0), 3% (w/v) L-Glutamic Acid Monosodium Salt, 1.5% (w/v) ribitol, and 0.05% (w/v) cysteine-HCl and stored at −80°C before the freeze-drying process. After pre-freezing, freeze-drying was performed for 24 h with freeze dry system (FreeZone Plus 2.5 Liter Cascade Console Freeze Dry System; Labconco Corporation).
Quantification of bacterial genes in mice feces
Fecal samples were suspended in Tris-EDTA buffer, and lysozyme was added to a final concentration of 15 mg/mL. In addition, achromopeptidase was added to a final concentration of 2000 U/mL and incubated at 37°C for 15 min. SDS was added to a final concentration of 2% (w/v) and incubated at 60°C for 5 min. Fecal DNA was extracted using QIAamp Fast DNA Stool Mini Kit (QIAGEN). Then DNA was subjected to quantitative PCR. The copy numbers of bacterial genes were quantified using a SYBR Green system kit (TB Green Premix Ex Taq II; Takara Bio) and a thermal cycler for the real-time PCR (StepOneTM Real-Time PCR System; Applied Biosystems). The primers used are listed in table S2. The cycling conditions were as follows: 95°C for 1 min, followed by 40 cycles of 5 sec at 95°C and 30 sec at 60°C.
Conservation analysis of BL105A_0502
Using the amino acid sequence of BL105A_0502 of B. longum 105-A as a query sequence for a BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi)25 search, the sequence conservation of BL105A_0502 was analyzed in the genomes of 12 human-associated bifidobacterial species23,24 and 56 prominent gut bacteria.3 Reference strains were analyzed for bifidobacterial species, and taxonomic units described in the report were analyzed for 56 prominent gut species. When the blast score was more than 500 bits, we determined that it was likely to be conserved.
Statistical analysis
Statistical analyses were performed using BellCurve for Excel (Social Survey Research Information Co., LTD.).
Supplementary Material
Funding Statement
This work was supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (25010A) and the Lotte Research Promotion Grant and by Grants-in-Aid from the Institute for Fermentation, Osaka (K-25-04, G2019-2-020);Grants-in-Aid from Institute for Fermentation, Osaka [K-25-04];Grants-in-Aid from Institute for Fermentation, Osaka [K-25-04];Grants-in-Aid from Institute for Fermentation, Osaka [G2019-2-020];
Data and materials availability
All data needed to evaluate the conclusions in this work are present in the paper and/or the Supplementary Materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Conceptualization, R.H., M.S., H.N., and S.K.; Methodology, R.H., M.S., S.M., S.F., A.Y., A.G., K.Y., T.Kato, H.O., and N.I.; Investigation, R.H., M.S., K.Y., Y.Y., Y.A., and S.E.; Resources, M.Nara, N.S., M.Nishimoto, M.K., and H.N.; Writing – Original Draft, R.H. and S.K.; Writing – Review & Editing, H.N., M.S., and K.Y; Visualization, R.H.; Funding Acquisition, H.N., M.K., M.S., T.Katayama, and S.K.; Project Administration, S.K.; Supervision, H.N., and S.K.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RDal. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–18. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R.. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164(3):337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14(8):491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 5.Mao B, Li D, Ai C, Zhao J, Zhang H, Chen W.. Lactulose differently modulates the composition of luminal and mucosal microbiota in C57BL/6J mice. J Agric Food Chem. 2016;64(31):6240–6247. doi: 10.1021/acs.jafc.6b02305. [DOI] [PubMed] [Google Scholar]
- 6.Mao B, Tang H, Gu J, Li D, Cui S, Zhao J,Hao Z, Wei C . In vitro fermentation of raffinose by the human gut bacteria. Food Funct. 2018;9(11):5824–5831. doi: 10.1039/C8FO01687A. [DOI] [PubMed] [Google Scholar]
- 7.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur T, Hämäläinen A-M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339–53 e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Ciaran Kelly, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66:e1–e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alang N, Kelly CR. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis. 2015;2(1):ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen Y-B, Hohmann EL, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med. 2019;381(21):2043–2050. doi: 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K, Hirano R, Sakai Y, Choi M, Sakanaka M, Kurihara S, Iino H, Xiao J-Z, Katayama T, Odamaki T, et al. Bifidobacterium response to lactulose ingestion in the gut relies on a solute-binding protein-dependent ABC transporter. Commun Biol. 2021;4(1):541. doi: 10.1038/s42003-021-02072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh T, Ojima MN, Sakanaka M, Ashida H, Gotoh A, Katayama T. Enzymatic adaptation of bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms. 2020;8(4):481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotoh A, Nara M, Sugiyama Y, Sakanaka M, Yachi H, Kitakata A, Nakagawa A, Minami H, Okuda S, Katoh T, et al. Use of Gifu Anaerobic Medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Biosci Biotechnol Biochem. 2017;81(10):1–9. doi: 10.1080/09168451.2017.1359486. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima M, Nishimoto M, Kitaoka M. Characterization of Three b-galactoside β-Galactoside Phosphorylases from Clostridium phytofermentans: discovery of d-galactosyl-beta1->4-l-rhamnose phosphorylase. J Biol Chem. 2009;284(29):19220–19227. doi: 10.1074/jbc.M109.007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima M, Nishimoto M, Kitaoka M. Practical preparation of D-galactosyl-beta1–>4-L-rhamnose D-Galactosyl-β1→4-L-rhamnose employing the combined action of phosphorylases. Biosci Biotechnol Biochem. 2010;74(8):1652–1655. doi: 10.1271/bbb.100263. [DOI] [PubMed] [Google Scholar]
- 18.Nishimoto M, Kitaoka M. Practical preparation of Lacto-N-biose Lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci Biotechnol Biochem. 2007;71(8):2101–2104. doi: 10.1271/bbb.70320. [DOI] [PubMed] [Google Scholar]
- 19.Nishimoto M, Kitaoka M. One-pot enzymatic production of beta-D-galactopyranosyl-(1–>3)-2-acetamido-2-deoxy-D-galactose (galacto-N-biose) from sucrose and 2-acetamido-2-deoxy-D-galactose (N-acetylgalactosamine). Carbohydr Res. 2009;344(18):2573–2576. doi: 10.1016/j.carres.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura H, Takeuchi A, Kano Y. Construction of Escherichia coli-Bifidobacterium coli–Bifidobacterium longum Shuttle Vector Transforming B. longum 105-A and 108-A. Biosci Biotechnol Biochem. 1997;61(7):1211–1212. doi: 10.1271/bbb.61.1211. [DOI] [PubMed] [Google Scholar]
- 21.Hirayama Y, Sakanaka M, Fukuma H, Murayama H, Kano Y, Fukiya S, Yokota A. Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: functional analysis of the alpha-galactosidase gene for raffinose assimilation. Appl Environ Microbiol. 2012;78(14):4984–4994. doi: 10.1128/AEM.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parche S, Amon J, Jankovic I, Rezzonico E, Beleut M, Barutcu H, Schendel I, Eddy MP, Burkovski A, Arigoni F, et al. Sugar transport systems of Bifidobacterium longum NCC2705. J Mol Microbiol BiotechnolJournal of Molecular Microbiology and Biotechnology. 2007;12(1–2):9–19. doi: 10.1159/000096455. [DOI] [PubMed] [Google Scholar]
- 23.Bottacini F, Ventura M, van Sinderen D, O’Connell Motherway M. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact. 2014;13(Suppl 1):S4. doi: 10.1186/1475-2859-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi JH, Lee KM, Lee MK, Cha CJ, Kim GB. Bifidobacterium faecale sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2014;64(Pt_9):3134–3139. doi: 10.1099/ijs.0.063479-0. [DOI] [PubMed] [Google Scholar]
- 25.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinform. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575(7783):505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557(7705):434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theilmann MC, Fredslund F, Svensson B, Lo Leggio L, Abou Hachem M. Substrate preference of an ABC importer corresponds to selective growth on beta-(1,6)-galactosides in Bifidobacterium animalis subsp. lactis. J Biol Chem. 2019;294(31):11701–11711. doi: 10.1074/jbc.RA119.008843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schols HA, Visser R, Voragen A. Pectins and pectinases. 2009.
- 30.Polle AY, O RG, Shashkov AS, Ovodov YS. Some structural features of pectic polysaccharide from tansy, Tanacetum vulgare L. Carbohydr Polym. 2002;49(3):337–344. doi: 10.1016/S0144-8617(01)00346-0. [DOI] [PubMed] [Google Scholar]
- 31.Crociani F, Alessandrini A, Mucci MM, Biavati B. Degradation of complex carbohydrates by Bifidobacterium spp. Int J Food Microbiol. 1994;24(1–2):199–210. doi: 10.1016/0168-1605(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 32.Komeno M, Hayamizu H, Fujita K, Ashida H. Two Novel alpha-l-Arabinofuranosidases from Bifidobacterium longum subsp. longum Belonging to Glycoside Hydrolase Family 43 Cooperatively Degrade Arabinan. Appl Environ Microbiol. 2019;85(6):e02582-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohno M, Suzuki S, Kanaya T, Yoshino T, Matsuura Y, Asada M, Kitamura S. Structural characterization of the extracellular polysaccharide produced by Bifidobacterium longum JBL05. Carbohydr Polym. 2009;77(2):351–357. doi: 10.1016/j.carbpol.2009.01.013. [DOI] [Google Scholar]
- 34.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517(7533):165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H, et al. Bifidobacterium longum subsp. infantis uses two different b-galactosidases β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22(3):361–368. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 36.Viborg AH, Katayama T, Abou Hachem M, Andersen MC, Nishimoto M, Clausen MH, Urashima T, Svensson B, Kitaoka M. Distinct substrate specificities of three glycoside hydrolase family 42 b-galactosidases -galactosidases from Bifidobacterium longum subsp. infantis ATCC 15697. Glycobiology. 2014;24(2):208–216. doi: 10.1093/glycob/cwt104. [DOI] [PubMed] [Google Scholar]
- 37.Nishijima S, Suda W, Oshima K, Kim S-W, Hirose Y, Morita H, Hattori M. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23(2):125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De MAN JC, ROGOSA M, Sharpe ME. A medium for the cultivation of Lactobacilli. J Appl Bacteriol. 1960;23(1):130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 39.Sakanaka M, Hansen ME, Gotoh A, Katoh T, Yoshida K, Odamaki T, Yachi H, Sugiyama Y, Kurihara S, Hirose J, et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci Adv. 2019;5(8):eaaw7696. doi: 10.1126/sciadv.aaw7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanesaki Y, Masutani H, Sakanaka M, Shiwa Y, Fujisawa T, Nakamura Y, Yokota A, Fukiya S, Suzuki T, Yoshikawa H. Complete genome sequence of bifidobacterium longum 105-A, a strain with high transformation efficiency. Genome Announc. 2014;2(6):e01311-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469(7331):543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 42.Edwards AN, Suarez JM, McBride SM. Culturing and maintaining Clostridium difficile in an anaerobic environment. J Vis Exp. 2013;(79):e50787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinttila T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97(6):1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in this work are present in the paper and/or the Supplementary Materials.


