Abstract
A 298-bp region of the Cryptosporidium parvum 18S rRNA gene and a 390-bp region of the acetyl coenzyme A synthetase gene were sequenced for a range of Cryptosporidium isolates from wild house mice (Mus domesticus), a bat (Myotus adversus), and cattle from different geographical areas. Previous research has identified a distinct genotype, referred to as the “mouse”-derived Cryptosporidium genotype, common to isolates from Australian mice. Comparison of a wider range of Australian mouse isolates with United Kingdom and Spanish isolates from mice and cattle and also an Australian bat-derived Cryptosporidium isolate revealed that the “mouse” genotype is conserved across geographic areas. Mice are also susceptible to infection with the “cattle” Cryptosporidium genotype, which has important implications for their role as reservoirs of infection for humans and domestic animals.
Cryptosporidium parvum is now recognized as an important cause of diarrheal infections in animals and humans (4). Wild rodents are thought to provide an important reservoir of infection of C. parvum for farm animals because the oocysts are environmentally resistant and C. parvum has been detected in wild brown rats (Rattus norvegicus), wild house mice (Mus domesticus), wild wood mice (Apodemus sylvaticus), and wild bank voles (Clethrionomys glareolus) (2, 20). Recent research that has genetically characterized isolates of C. parvum from wild Australian mice (Mus domesticus) has revealed that mice carry a distinct genotype (10, 11). This “mouse” genotype has smaller oocysts than C. parvum (4.5 by 4.0 μm) and is genetically different from genotypes carried by cattle and humans (10, 11). Until recently, it was assumed that C. parvum was a uniform species, but there is now strong evidence that C. parvum is composed of numerous distinct genotypes: a “human” genotype found only in humans, a “cattle” genotype found in many domestic animals and also humans, and a number of other genotypes, some of which appear to be host specific (1, 7–17, 19). The aim of this study was to genetically characterize Cryptosporidium isolates from mice from diverse locations in order to determine if the “mouse” genotype is conserved in mouse-derived Cryptosporidium isolates from different geographical areas.
MATERIALS AND METHODS
Sources of parasite isolates, DNA purification, and PCR.
The sources of the parasite isolates are listed in Table 1, and DNA was purified as described previously (8). Primers and PCR conditions were as described previously (8).
TABLE 1.
Isolates of Cryptosporidium used in this study
| Code | Host | Geographical origin | Sourcea |
|---|---|---|---|
| H1 | Human | Perth, Western Australia | PMH |
| WB1 | Wildebeest (C. taurinus taurinus) | Barcelona, Spain | LPUB |
| C1 | Calf | Switzerland | CVL |
| UKC1 | Calf | United Kingdom | SNES |
| UKC2 | Calf | United Kingdom | SNES |
| UKC3 | Calf | United Kingdom | SNES |
| UKC4 | Calf | United Kingdom | SNES |
| UKC5 | Calf | United Kingdom | SNES |
| UKM7 | Mouse (M. musculus) | United Kingdom | SNES |
| UKM8 | Mouse (M. musculus) | United Kingdom | SNES |
| UKM9 | Mouse (M. musculus) | United Kingdom | SNES |
| UKM10 | Mouse (M. musculus) | United Kingdom | SNES |
| UKM11 | Mouse (M. musculus) | United Kingdom | SNES |
| UKM12 | Mouse (M. musculus) | United Kingdom | SNES |
| M7 | Mouse (M. musculus) | Victoria | CSIRO |
| M11 | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| M24 | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| M26 | Mouse (M. musculus) | Victoria | CSIRO |
| M27 | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| M4c | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| M6c | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| M8c | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| M23c | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| M27c | Mouse (M. musculus) | Walpeup, Victoria | CSIRO |
| SM1 | Mouse (M. musculus) | Barcelona, Spain | LPUB |
| SM2 | Mouse (M. musculus) | Barcelona, Spain | LPUB |
| SM4 | Mouse (M. musculus) | Barcelona, Spain | LPUB |
| Bat 1 | Bat (M. adversus) | New South Wales, Australia | TPZ |
PMH, Princess Margaret Hospital, Perth, Western Australia, Australia; CSIRO, Commonwealth Scientific and Industrial Research Organisation, Victoria, Australia; CVL, Central Veterinary Laboratories, Adelaide, South Australia, Australia; SNES, School of Natural and Environmental Sciences, Coventry University, Coventry, United Kingdom; LPUB, Laboratori de Parasitologica, University of Barcelona, Barcelona, Spain; TPZ, Taronga Park Zoo.
Sequencing.
PCR products were sequenced with an ABI Prism Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) according to the manufacturer’s instructions, except that the annealing temperature was raised to 60°C. Sequences were analyzed with SeqEd, version 1.0.3. (Applied Biosystems), and were aligned with the Clustal V (6) sequence alignment program.
Phylogenetic analysis of sequence information.
A phylogenetic analysis based on the nucleotide sequences of 18S rRNA gene (rDNA) and acetyl coenzyme A (acetyl-CoA) synthetase gene regions from different isolates was conducted by using PHYLIP 3.5p (5). A similarity index among Cryptosporidium isolates was created by using the formula for Kimura’s distance. Phylograms were constructed from genetic distance matrices by the unweighted pair group method of analysis and with the DRAWGRAM programs available in PHYLIP 3.5p (5).
RESULTS
Sequence analysis of 18S rDNA.
Sequence analysis of the 298-bp 18S rDNA product (data not shown) revealed the “mouse” genotype (10, 11) to be highly conserved between three mouse-derived Cryptosporidium isolates from Australia sequenced previously (10) (GenBank accession no. AF099667), six mouse-derived Cryptosporidium isolates from the United Kingdom, two mouse-derived Cryptosporidium isolates from Spain, and a bat isolate from New South Wales in Australia. Five mouse isolates from Australia (isolates M4c, M6c, M8c, M23c, and M27c) (GenBank accession no. AF099668) exhibited the “cattle” genotype and one mouse isolate from Spain (isolate SM4) was identical to Cryptosporidium muris (GenBank accession no. L19069). Cattle isolates from the United Kingdom and Australia were all identical and displayed the “cattle” genotype. Sequence analysis of a wildebeest-derived Cryptosporidium isolate from Spain (isolate WB1) revealed that it was of the “cattle” genotype.
Sequence analysis of the acetyl-CoA synthethase gene.
As with the rDNA sequencing results, sequence analysis of the acetyl-CoA synthetase gene revealed distinct differences between cattle and mouse isolates. All cattle isolates analyzed exhibited a common genotype. Mouse isolates from the United Kingdom were identical to five mouse isolates from Australia sequenced previously (10) and to two mouse isolates from Spain (isolates SM1 and SM2) (GenBank accession no. AF102768). Isolate SM4 was not amplified with the acetyl-CoA primers. Australian mouse isolates M4c, M6c, M8c, M23c, and M27c exhibited the “cattle” genotype, as did the wildebeest isolate (isolate WB1) from Spain (GenBank accession no. AF102767). It was not possible to amplify the bat isolate with the acetyl-CoA synthetase gene primers due to the low amount of DNA present.
Phylogenetic analysis of rDNA sequencing results.
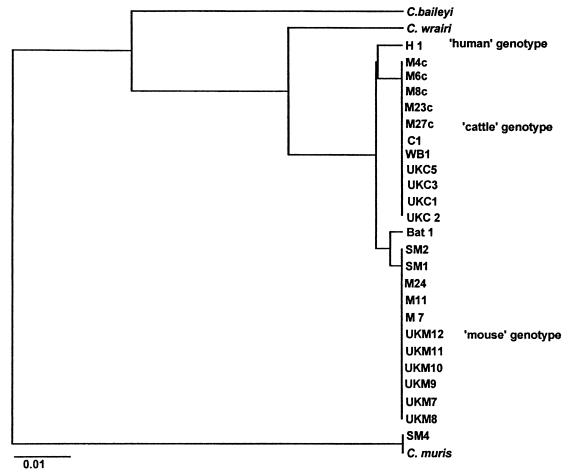
Additional isolates previously sequenced were also analyzed and included a human isolate (H1) and a cattle isolate (C1) (8), and additional Cryptosporidium isolates retrieved from the rRNA WWW Server on the World Wide Web (18). This extended phylogenetic analysis resulted in three distinct groups within C. parvum (Fig. 1): “human” group; a “cattle” group, which contained the cattle isolates, five Australian mouse isolates, and a wildebeest isolate (isolate WB1); and a “mouse” group, which contained all the Australian mouse isolates, all the mouse isolates from the United Kingdom, two Spanish mouse isolates, and the bat isolate from New South Wales. The remaining Spanish mouse isolate (isolate SM4) grouped with C. muris.
FIG. 1.
Phylogram of Kimura’s distance generated from 18S rDNA sequence information among isolates of Cryptosporidium clustered by the unweighted pair group method of analysis.
Phylogenetic analysis of acetyl-CoA synthetase gene sequencing results.
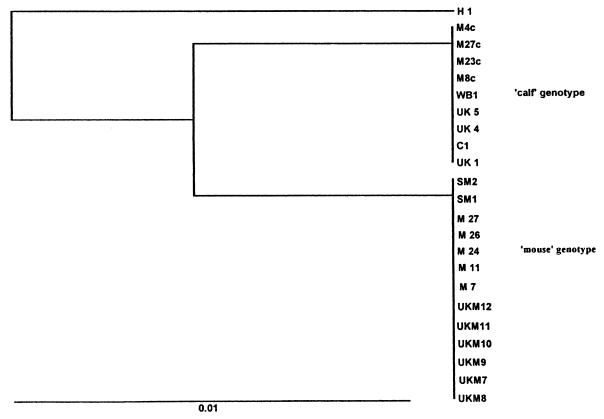
Phylogenetic analysis of the acetyl-CoA synthetase gene sequence information also produced three main groups (Fig. 2): A “human” group; a “cattle” group, which contained the cattle isolates, five Australian mouse isolates, and a wildebeest isolate (isolate WB1); and a “mouse” group, which contained the Australian mice isolates, all the mouse isolates from the United Kingdom, and two Spanish mouse isolates (isolates SM1 and SM2).
FIG. 2.
Phylogram of Kimura’s distance generated from acetyl-CoA synthethase gene sequence information among isolates of Cryptosporidium clustered by the unweighted pair group method of analysis.
DISCUSSION
Mice (M. musculus syn. domesticus) from different geographical areas were shown to carry a distinct genotype of C. parvum (referred to as the “mouse” genotype) by both rDNA and acetyl-CoA synthetase gene sequence analyses, indicating that this genotype is conserved across widely separated geographical areas.
The “mouse” genotype was also identified in a fecal sample from a large-footed mouse-eared bat (Myotus adversus), extending the host range of this genotype. Cryptosporidium has recently been reported in a big brown bat (Eptesicus fuscus) (3). In this report, cryptosporidial bodies (<5 μm) were found to be attached to the microvillar border of enterocytes of paraffin-embedded sections of the small intestine. This bat isolate was not genotyped, but the small sizes of the oocysts indicate that a C. parvum-like isolate was present. Future genotyping studies should examine bat isolates from different geographical locations in order to determine how common this genotype is among bats. The present report of Cryptosporidium in a large-footed bat is the only other report of a cryptosporidial infection in bats.
Cattle isolates from the United Kingdom all exhibited the “cattle” genotype by both rDNA and acetyl-CoA synthetase gene sequence analyses and were identical to an Australian cattle isolate (isolate C1), confirming the conserved and widespread nature of this genotype. The “cattle” genotype was also identified in an adult male wildebeest (Connochaetes taurinus taurinus). This is the first time that this genotype has been identified in this host.
Interestingly, five of the mouse isolates analyzed exhibited the “cattle” genotype, which is known to infect humans. These mice were trapped on farms in Victoria, Australia, where large numbers of sheep were grazing. Under the circumstances, sheep are the most likely source of infection for mice since sheep are known to carry isolates of the “cattle” genotype (10). A recent 2-year survey of wild mice and voles on a farm in Warwickshire, United Kingdom, reported prevalence rates of 22, 21, and 13% for C. parvum in M. musculus, A. sylvaticus, and C. glareolus, respectively (2). The apparent autumnal peak for C. parvum in all three rodent species coincided with the calving period at that farm, and it was concluded that “rodents may represent a significant reservoir of Cryptosporidium with a high potential for infection of man and livestock due to cohabitation” (2). The finding of the “cattle” genotype in Australian mice indicates that sheep and cattle may transmit the “cattle” genotype to mice, which may in turn transmit Cryptosporidium to other domestic animals and also to humans. However, the “mouse” genotype appears to be more common in mice, and as small rodent populations and Cryptosporidium prevalence are highest at the end of the summer, independent of the presence of cattle and sheep (2), it may be that mice are only occasionally infected with the “cattle” genotype during periods of heavy environmental contamination. Recent studies with pigs have shown that they are also capable of carrying two distinct genotypes: a “pig” genotype common to pigs from different geographical areas and the “cattle” genotype (10, 11, 13).
More extensive characterization of rodent isolates of Cryptosporidium from a wider geographical distribution and from both urban and rural habitats is necessary before their role as reservoirs of infection in humans and domestic animals can be more fully determined.
ACKNOWLEDGMENTS
This study was supported by the Public Health Research and Development Committee (PHRDC) of the National Health and Medical Research Council of Australia and by the Vertebrate Biocontrol Centre, Australian National University, Canberra, Australia. U. M. Morgan is a PHRDC Research Fellow.
We thank A. Elliot for expert microscopy assistance and Micah Davies for assisting with the trapping in Victoria.
REFERENCES
- 1.Awad-El-Kariem F M, Robinson H A, Dyson D A, Evans D S, Wright S, Fox M T, McDonald V M. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology. 1995;110:129–132. doi: 10.1017/s0031182000063885. [DOI] [PubMed] [Google Scholar]
- 2.Chalmers R M, Sturdee A P, Bull S A, Miller A, Wright S E. The prevalence of Cryptosporidium parvum and C. muris in Mus domesticus, Apodemus sylvaticus and Clethrionomys glareolus in an agricultural system. Parasitol Res. 1997;83:478–482. doi: 10.1007/s004360050283. [DOI] [PubMed] [Google Scholar]
- 3.Dubey J P, Hamir A N, Sonn R J, Topper M J. Cryptosporidiosis in a bat (Eptesicus fuscus) J Parasitol. 1998;84:622–623. [PubMed] [Google Scholar]
- 4.Fayer R, Speer C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 1–42. [Google Scholar]
- 5.Felsenstein J. PHYLIP. Phylogeny interface package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 6.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1991;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 7.Morgan U M, Constantine C C, O’Donoghue P, Meloni B P, O’Brien P A, Thompson R C A. Molecular characterisation of Cryptosporidium isolates from humans and other animals using RAPD (random amplified polymorphic DNA) analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 8.Morgan U M, Constantine C C, Forbes D A, Thompson R C A. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 9.Morgan U M, Sargent K D, Elliot A, Thompson R C A. Cryptosporidium in cats—additional evidence for C. felis. Vet J. 1998;156:159–161. doi: 10.1016/s1090-0233(05)80047-4. [DOI] [PubMed] [Google Scholar]
- 10.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertzberg H, Elliot A, Thompson R C A. Molecular characterisation of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- 11.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertzberg H, Elliot A, Thompson R C A. Sequence and PCR-RFLP analysis of the internal transcribed spacers of the rDNA repeat unit in isolates of Cryptosporidium from different hosts. Parasitology. 1999;118:49–58. doi: 10.1017/s0031182098003412. [DOI] [PubMed] [Google Scholar]
- 12.Morgan U M, Pallant L, Dwyer B W, Forbes D A, Rich G, Thompson R C A. Comparison of PCR and microscopy for detection of Cryptosporidium in human fecal samples: clinical trial. J Clin Microbiol. 1998;36:995–998. doi: 10.1128/jcm.36.4.995-998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan U M, Buddle R, Armson A, Thompson R C A. Molecular and biological characterisation of Cryptosporidium in pigs. Aust Vet J. 1999;77:44–47. doi: 10.1111/j.1751-0813.1999.tb12428.x. [DOI] [PubMed] [Google Scholar]
- 14.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S L, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sargent K D, Morgan U M, Elliot A, Thompson R C A. Morphological and genetic characterisation of Cryptosporidium oocysts from domestic cats. Vet Parasitol. 1998;77:221–227. doi: 10.1016/s0304-4017(98)00122-8. [DOI] [PubMed] [Google Scholar]
- 16.Spano F, Putignani L, Mclauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 17.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blancq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van de Peer V, Van den Breock I, De Rijk P, De Wachter R. Database on the structure of small subunit RNA. Nucleic Acids Res. 1994;22:3488–3494. doi: 10.1093/nar/22.17.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasquez J R, Gooze L, Kim K, Gut J, Patersen C, Nelson R G. Potential antifolate resistance determinants and genotypic variation in the bifunctional reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol Biochem Parasitol. 1996;79:153–165. doi: 10.1016/0166-6851(96)02647-3. [DOI] [PubMed] [Google Scholar]
- 20.Webster J P, MacDonald D W. Parasites of wild brown rats (Rattus norvegicus) on UK farms. Parasitology. 1995;111:247–255. doi: 10.1017/s0031182000081804. [DOI] [PubMed] [Google Scholar]