Abstract
We studied the SmaI and SstII macrorestriction patterns of 54 Staphylococcus epidermidis strains isolated from 14 patients infected following the implantation of joint prostheses. Multiple strains from pus and infected tissue specimens of each patient were selected on the basis of different colony morphologies and drug resistance patterns. The same criteria were used to select 23 S. epidermidis strains from hand swabs of eight healthy individuals. For 10 of the 14 patients, all the intrapatient strains appeared to be closely or possibly related, whereas related strains were detected in the skin flora of only one of the eight healthy individuals. This observation suggests that, in most cases, the patients were infected by a single S. epidermidis clone which subsequently underwent rearrangements that yielded derivatives with divergent phenotypes and, occasionally, divergent macrorestriction patterns. The four patients whose specimens contained unrelated S. epidermidis strains were probably infected with several polyclonal strains.
Infection is one of the most devastating complications of prosthetic joint surgery (3, 12, 14, 15, 17, 27, 33). Staphylococcus epidermidis, which can adhere to implants, has increasingly been identified as a cause of such infections (6, 16, 21, 23, 24, 26, 35). S. epidermidis is a normal commensal organism of the skin which may contaminate specimens during collection, and it is therefore difficult to assess the clinical significance of its detection, particularly when the bacterial colonies obtained have diverse aspects and drug resistance phenotypes. Detection of gram-positive cocci by direct inspection and/or repeated isolation of the same mixtures of strains from a series of specimens from the same patient provides good evidence of infection causality (17, 19, 36). The presence in pus and tissue specimens of multiple S. epidermidis strains, frequent in infections following implantation of joint prostheses (17), may be due either to the genomic instability of a single infectious clone or to an infection caused by a polyclonal mixture of strains such as the mixture of strains found in the skin flora. To examine these two possibilities, we compared the genomes of the various phenotypically distinct S. epidermidis strains detected in pus and tissue specimens from 14 patients with chronic prosthetic joint infections. Diverse S. epidermidis colonies isolated from the skin flora of eight healthy individuals were used for comparative analysis of the intraindividual colonies that were selected on the basis of their different colony morphologies and drug resistance phenotypes.
MATERIALS AND METHODS
Patients and healthy individuals.
The 14 patients included in this study were infected following the implantation of joint prostheses (see Table 1). Joint replacements were carried out in diverse hospitals, and there was no epidemiological link between the patients. The files of all patients except patients B and N were consulted. The 12 patients whose files were available had pain without fever. The delay between surgery and pain was not regularly registered in the files; therefore, we report (see Table 1) the delay between the implantation of the prosthesis and its replacement (3 to 84 months). The replacement was not necessarily done immediately after the first signs of infection. Five of the 12 patients (patients A, E, I, L, and P) had abscesses, which had fistulized in patients A, L, and P, and 6 patients (patients E, F, I, L, O, and P) presented with edema. Bacteriological samples were obtained at the time of intraoperative assessment and surgery for excisional arthroplasty. No antibiotic was administered to any patient for at least 1 week before joint surgery. Patients whose infected specimens yielded only S. epidermidis were included in this study.
TABLE 1.
Relevant characteristics of the S. epidermidis strains isolated from infected patients
| Patient (sex, age [yr]) | Pathology specimena | Date of sampling (day/mo/yr or mo/yr) | Strain designa-tion | Common and additional drug resistance phenotype markerb | Genotype based on restriction patterns obtained with the followingc:
|
|
|---|---|---|---|---|---|---|
| SmaI | SstII | |||||
| A (male, 70) | Infected total knee prosthesis (36) | Pc Su Gm Tm Km Rf Pf Fa | ||||
| Intraoperative joint samples | 8/2/1996 | 96182 | Nm Sm MLSc As | A1 | 1a | |
| 8/2/1996 | 96183 | Nm Sm MLSc SgA Cm Fm As | A2 | 1a | ||
| 8/2/1996 | 96184 | MLSc As | A3 | 1a | ||
| 19/2/1996 | 96187 | Sm Sp MLSc SgA Cm As | A3 | 1a | ||
| 19/2/1996 | 96188d | Ox Nm Sp MLSc Cm As | A3 | 1a | ||
| Intraoperative tissue samples from the tibia | 20/2/1996 | 96189 | Ox Nm Sm LSgA Cm Cd As Eb | A2 | 1b | |
| 20/2/1996 | 96190 | Nm Sm Sp MLSc Cm Cd Tp Tc | A3 | 1a | ||
| 20/2/1996 | 96191 | Ox Nm Sp MLSc SgA Cm Cd As | A4 | 1c | ||
| 23/2/1996 | 96192 | Nm Sp MLSc Cm As | A2 | 1a | ||
| 23/2/1996 | 96193 | Nm Sp MLSc SgA Cm Fm As | A3 | 1b | ||
| 23/2/1996 | 96194 | Nm Sp MLSc SgA Cm As | A3 | 1b | ||
| B (male, 63) | Septic nonunion of the tibia with a centro-medullary nail | Pc Su As MLSi | ||||
| Intraoperative tissue samples | 5/1995 | 95160d | Ox | B | 2 | |
| 5/1995 | 95161d | Tc Fa | C | 3 | ||
| C (male, 67) | Infected total hip prosthesis inserted with gentamicin-loaded cement (84) | As | ||||
| Intraoperative joint samples | 3/1996 | 96229d | No additional marker | D | 4 | |
| Recess of the acetabulum | 3/1996 | 96230 | Su | D | 4 | |
| D (male, 70) | Infected total hip prosthesis inserted with gentamicin-loaded cement (80) | Pc Ox (mecA+) As | ||||
| Intraoperative tissue samples | 6/1994 | 94308d | Su | E | 5 | |
| 6/1994 | 94314 | No additional marker | E | 5 | ||
| 6/1994 | 94315 | Su Tp | E | 5 | ||
| E (male, 61) | Infected total shoulder prosthesis (19) | Pc Ox Pf Rf Su Tp As Hg Sm Pf | ||||
| Intraoperative tissue samples | 7/1994 | 94304 | Gm Tm Km Nm MLSc Cd Fa | F1 | 6 | |
| 7/1994 | 94305d | Cd | F2 | 7 | ||
| 7/1994 | 94306 | Gm Tm Km Nm | F3 | 7 | ||
| F (female, 73) | Infected total knee prosthesis (8) | Pc Pf Su Tp Fa | ||||
| Intraoperative tissue samples | 7/1994 | 94348d | MLSc Tc As | G | 8 | |
| 8/1994 | 94349 | MLSc | G | 8 | ||
| 8/1994 | 94350 | MLSc Tc | G | 8 | ||
| 9/1994 | 94351d | Ox Gm Tm Km MLSc Tc Cd As Hg Eb | H1 | 9a | ||
| 9/1994 | 94352 | Ox Gm Tm Km MLSc Tc Cd As Hg Ba | H2 | 9b | ||
| 9/1994 | 94353 | Ox Gm Tm Km Cd As Hg | H2 | 9b | ||
| Variants of strain 94351 obtained by subculture | Pc Pf Su Tp Fa Gm Tm Km Tc Cd As Hg Eb | |||||
| Rep1 | No additional marker | V | NDe | |||
| Rep2 | MLSc | H1 | ND | |||
| H (female, 49) | Infected total hip prosthesis (16) | Pc Su As Hg | ||||
| Intraoperative pus and tissue samples | 8/1994 | 94328d | Tmf MLSc Fa Tc Eb Af Pi | I1 | 10a | |
| 8/1994 | 94329 | Tmf Fa Tc Eb Af Pi | I1 | 10a | ||
| 8/1994 | 94330d | MLSc Tc Eb | I2 | 10b | ||
| 8/1994 | 94331 | Tc | I2 | 10b | ||
| 8/1994 | 94332 | MLSc Tc | I1 | 10c | ||
| 8/1994 | 94333 | Tmf MLSc Fa Eb Af Pi | I1 | 10a | ||
| I (male, 58) | Infected total knee prosthesis inserted with gentamicin-loaded cement (16) | As | ||||
| Intraoperative pus and tissue samples | 9/1996 | 96386d | Pc Ox Gm Tm Km Nm Sp MLSc SgA Cm Rf Fa Pf | J | 11 | |
| 9/1996 | 96388d | Pc Ox Pf Su Eb | K | 12 | ||
| 9/1996 | 96389d | Km Nm L SgA Su Hg | L | 13 | ||
| 9/1996 | 96390d | Pc Ox Tm Km Nm Fa Af Pi | M | 14 | ||
| 9/1996 | 96391 | No additional marker | N | 15 | ||
| K (female, 72) | Infected total hip prosthesis inserted with gentamicin-loaded cement (30) | Su As | ||||
| Intraoperative bone samples | 9/1996 | 96394d | Gm Tm Km Tp Hg | O | 16 | |
| 9/1996 | 96395d | Ox Tc Eb | P | 17 | ||
| L (female, 73) | Infected total shoulder prosthesis (3) | Pc Ox Su Eb As Tm Km Nm Sp MLSc Cd Rf Pf Tp Hg Pi | ||||
| Intraoperative bone samples from the upper extremity of the humerus | 7/1996 | 96396 | Gm | F4 | 18 | |
| 7/1996 | 96397d | No additional marker | F4 | 18 | ||
| 7/1996 | 96398 | Gm Fa | F4 | 18 | ||
| M (male, 72) | Infected total hip prosthesis (10) | Pc Ox Sm Sp Gm Tm Km MLSc Su Tp Pf | ||||
| Intraoperative tissue samples from the acetabulum | 7/1996 | 96400d | Fm | Q | 19 | |
| 7/1996 | 96401 | No additional marker | Q | 19 | ||
| N (female, 63) | Infected total hip prosthesis | Fa As | ||||
| Intraoperative joint capsule samples | 7/1996 | 96408d | Pc | R | 20 | |
| 7/1996 | 96409 | No additional marker | R | 20 | ||
| 7/1996 | 96412d | Pc Ox Tm Km Nm MLSi Cd Su Eb Ba | S | 21 | ||
| O (female, 77) | Infected total hip prosthesis (18) | Fa As | ||||
| Intraoperative tissue samples | 9/1996 | 96402 | Pc Ox Gm Tm Km Nm Cd Hg | T1 | 22a | |
| 9/1996 | 96403d | No additional marker | T2 | 22b | ||
| 9/1996 | 96406 | Pc Ox Gm Tm Km Nm MLSc Cd Hg | T1 | 22a | ||
| P (female, 78) | Infected total hip prosthesis (79) | Pc | ||||
| Intraoperative tissue samples | 3/1997 | 97055 | Ox Gm Tm Km Sm MLSc Cm Su Tc | U | 23 | |
| 3/1997 | 97058d | Ox Gm Tm Km Sm MLSc Cm | U | 23 | ||
| 3/1997 | 97060 | Ox Gm Tm Km Sm MLSc Cm Su Tp Tc | U | 23 | ||
| Cutaneomucous samples | ||||||
| Nasal mucous samples | 3/1997 | narL1 | Tc Mn Fa | W | ND | |
| 3/1997 | narL2 | Tc Fa | X | ND | ||
| 3/1997 | narL3 | OxTm Km Gm Fa Fm As Hg | Y | ND | ||
| 3/1997 | narR | Mn | Z | ND | ||
| Sample from contralateral groin | 3/1997 | groin L | Ox Tc Pf Rf Fa Fm As Cd Hg Ba | AA | ND | |
The time (in months) between the implantation of the prosthesis and its removal because of infection is indicated in parentheses.
Abbreviations: Af, acriflavine; As, sodium arsenate; Ba, cetyltrimethylammonium bromide; Cd, cadmium acetate; Cm, chloramphenicol; Eb, ethidium bromide; Fa, fusidic acid; Fm, fosfomycin; Gm, gentamicin; Hg, mercuric nitrate; Km, kanamycin; L, lincosamide; MLSc, constitutive resistance to macrolides-lincosamides-streptogramin B; MLSi, inducible resistance to macrolides-lincosamides-streptogramin B; Mn, minocyclin; Nm, neomycin; Ox, oxacillin; Pc, β-lactam (penicillinase production); Pf, pefloxacin; Pi, propamidine isethionate; Rf, rifampin; SgA, streptogramin A; Sm, streptomycin; Sp, spectinomycin; Su, sulfonamide; Tc, tetracycline; Tm, tobramycin; Tp, trimethoprim. Markers common to all strains from a patient are in boldface type.
Major genotypes are designated by capital letters or arabic numerals according to the enzymes used, SmaI or SstII, respectively. The patterns of those clustered within the same major genotype differed by one to three restriction bands. When subtypes are detected in the same major genotype, they are designated by letters with number suffixes for SmaI subtypes and by numbers with letter suffixes for SstII subtypes. The strains clustered within the same subtype have the same macrorestriction pattern.
HindIII- and EcoRI-hybridization patterns with pBA2 (ribotypes) were determined. Analysis of these patterns enabled us to assign the strains to the species S. epidermidis.
ND, not determined.
The Tmr strains carried the gene aadD, but they were susceptible in vitro to kanamycin (MIC, <8 μg/ml) and to neomycin (MIC, <8 μg/ml).
Hand skin swabs were collected from healthy individuals including (i) six surgeons working in the same operating room in a Parisian hospital, but a hospital that was not one of those in which the patients of our study were hospitalized, and (ii) two individuals who did not work in a hospital (see Table 2).
TABLE 2.
Relevant characteristics of the 23 S. epidermidis strains isolated from the cutaneous flora of eight healthy individuals
| Healthy individual designation | Strain designation | Common and additional drug resistance phenotype markera | Genotype designation according to SmaI patternb |
|---|---|---|---|
| b | Pc As | ||
| b1 | Pf Hg | BB | |
| b2 | Km Nm Cd | CC | |
| b3 | No additional marker | DD | |
| c | Pc As Cd | ||
| c1 | Tc | EE | |
| c2 | Km Nm | FF | |
| c3 | km Nm Pf | GG | |
| f (surgeon) | Pc As Hg | ||
| f1 | Ox | HH1 | |
| f2 | Fa | II | |
| f7 | Ox Km Tm Gm Tc MLSc Tp Su Pf Fa Cd Ba | JJ | |
| i (surgeon) | Pc Ox As | ||
| j1 | Tc MLSc Pf Cd Hg | KK | |
| j2 | Km Nm Tm Gm MLSc Pf | HH2 | |
| j3 | Em Fa | LL | |
| l (surgeon) | Pc As | ||
| 13 | Ox | HH3 | |
| 14 | Ox Km Nm Tm Cl Em | MM | |
| 16 | Km Nm Tm Em | NN | |
| m (surgeon) | Pc As | ||
| m1 | Km Nm Tm Em | OO | |
| m2 | Ox Km Tm Gm MLSc SgA Su Pf Rf Fa Cd Hg | PP | |
| m3 | Ox Pf Eb Ba | QQ1 | |
| t (surgeon) | Pc Ox Km Tm Gm MLSc Hg Eb Af Ip | ||
| t4 | Cd Fm As Pf | RR1 | |
| t5 | As | SS | |
| t8 | Fm Cd Pf | RR2 | |
| w (surgeon) | Pc Ox As | ||
| w1 | Em | TT | |
| w3 | Pf Eb Af Ip | QQ2 |
Bacterial strains and plasmids.
A total of 82 S. epidermidis strains originating from 14 infected patients (54 strains from infected specimens and 5 strains from cutaneous samples of patient P) and from 8 healthy individuals (23 strains) were studied (see Tables 1 and 2, respectively). The infected specimens and the skin swabs were cultured on sheep blood agar. After at least 48 h of incubation at 37°C, the species of the colonies that were isolated from each sample and that had different morphologies (size, color which varied from white to grey, shape and outline, presence or absence of hemolytic activity, rough or smooth aspect) was determined as described previously (8), and the colonies were tested with the ID32 Staph system (BioMérieux, Marcy-l’Etoile, France). Patients whose infected specimens contained only S. epidermidis colonies were included in this study. The drug resistance pattern was determined for each different S. epidermidis colony isolated from the specimens of each patient. The strains that were distinguishable by at least one drug resistance marker were studied independently. Similarly, the S. epidermidis colonies isolated from skin flora were selected on the basis of their distinct morphologies and drug resistance patterns. Bacterial suspensions in brain heart infusion containing 30% glycerol were stored at 80°C before analysis.
Plasmid pBA2 (18) was used as a probe for ribotype determination. It consists of pBR322 carrying a 2.3-kb HindIII insert from Bacillus subtilis which encodes 16S rRNA.
Susceptibility to antimicrobial agents.
The pattern of resistance to antimicrobial agents was determined by the disk diffusion method (4). The markers tested were those which enabled us to detect the drug resistance phenotypes described to date among staphylococci. Commercially available disks loaded with the following antibiotics were used: penicillin G, oxacillin, spectinomycin, streptomycin, kanamycin, neomycin, gentamicin, tobramycin, chloramphenicol, erythromycin, lincomycin, trimethoprim, sulfonamide, tetracycline, minocycline, pefloxacin, rifampin, fusidic acid, fosfomycin, and vancomycin (Diagnostics Pasteur, Marne-la-Coquette, France) and mupirocin (Mast Diagnostics, Mast Group Ltd., Merseyside, United Kingdom). Additional disks prepared in our laboratory contained 20 μg of pristinamycin IIA, 40 μg of pristinamycin IB, 0.2 μmol of cadmium acetate, 0.2 μmol of sodium arsenate, 0.2 μmol of mercuric nitrate, 200 μg of ethidium bromide, 200 μg of acriflavine, 200 μg of propamidine isethionate, or 10 μg of cetyltrimethylammonium bromide.
Selective Mueller-Hinton agar media containing 4 μg of oxacillin per ml, 0.12 IU of penicillin per ml, 5 μg of erythromycin per ml, 3 μg of tetracycline per ml, or 20 μg of gentamicin per ml were used to screen for variants exhibiting distinct drug resistance patterns in the subcultures of strain 94351 (see Table 1).
Ribotype determination.
Cellular DNA was extracted, cleaved with HindIII and EcoRI (Pharmacia Biotech, Uppsala, Sweden) separately, electrophoresed, transferred onto Hybond N+ nylon membranes (Amersham International), and tested for hybridization under stringent conditions with pBA2 radiolabeled with [α-32P]dTCP (110 TBq/mmol) as described elsewhere (7, 8).
The sizes of the bands constituting the hybridization patterns (HPs) were introduced into our database (5, 7). Each of these HPs was compared with each of the EcoRI HPs and HindIII HPs previously obtained for validly classified staphylococci. Similarity was evaluated according to the Dice coefficient (11). The strains that had HPs that were indistinguishable from those detected previously could immediately be assigned to a species. For each new HP, as reported previously (5), the percent similarity with each EcoRI or HindIII HP in the database was calculated. An isolate exhibiting a new HP can be assigned to a known taxon if the highest percentages of similarity obtained are clustered within a single taxon.
Pulsed-field gel electrophoresis of macrorestriction fragments and comparative analysis of banding patterns.
The protocol used for the determination of SmaI or SstII restriction patterns was described previously (10). Concatameric bacteriophage lambda DNA molecules (48.5 kb; Bio-Rad) and the SmaI fragments of the cellular DNA from Staphylococcus aureus NCTC8325 were used as size standards. Macrorestriction fingerprints were compared visually and were scanned with GelCompar software (Applied Maths, Kortrijk, Belgium). A similarity matrix was created by using the band-based Dice similarity coefficient (11). The unweighted pair-group method with average linkages was used to cluster the strains on the basis of the patterns obtained with each of the two enzymes used.
The SmaI or SstII patterns of the isolates from the same patient or healthy individual were compared visually, in pairs, by using an enlarged photograph of the same gel. The strains were clustered according to the following criteria proposed by Tenover et al. (28): (i) Strains were grouped in the same major genotype if their patterns differed by no more than three bands (these strains were considered to be closely related and monoclonal); (ii) if the number of band differences between patterns was between four and six, the strains were scored as possibly related but were nevertheless classified into distinct genotypes to discriminate them clearly from the unambiguously closely related strains; and (iii) differences between patterns involving at least seven bands indicated different or unrelated strains. Major genotypes are designated by capital letters or arabic numerals according to the enzyme used, SmaI or SstII, respectively (see Table 1). The strains having indistinguishable patterns were classified within the same subtype. SmaI subtypes are designated by letters with number suffixes, and SstII subtypes are designated by numbers with letter suffixes. If the dendrograms revealed clusters that included strains from different patients with percentages of similarity of at least 80, the patterns of the strains grouped in the same cluster were compared visually on the same gel. Those strains whose patterns differed by no more than three macrorestriction fragments were assigned to the same genotype.
RESULTS
Identification to the species level.
Fifty-one of the 54 coagulase-negative staphylococci isolated from the infected specimens from patients were assigned to S. epidermidis by the ID32 Staph system. Analysis of the hybridization patterns with pBA2 enabled us to assign to S. epidermidis three strains, strains 95160, 96388, and 96390 (Table 1) not classified by the ID32 Staph system.
The ID32 Staph system was used to identify the S. epidermidis strains isolated from the flora of patient P (Table 1) and the healthy individuals (Table 2).
Drug resistance phenotype.
The intrapatient strains had in common 1 to 14 markers and were distinguishable by 1 to 10 additional markers (Table 1). Among the strains from healthy individuals, the intraindividual strains had in common one to three resistance markers and differed by 1 to 13 additional markers (Table 2).
Analysis of the macrorestriction patterns. (i) Comparison of the SmaI and SstII patterns of patient strains isolated from infected specimens.
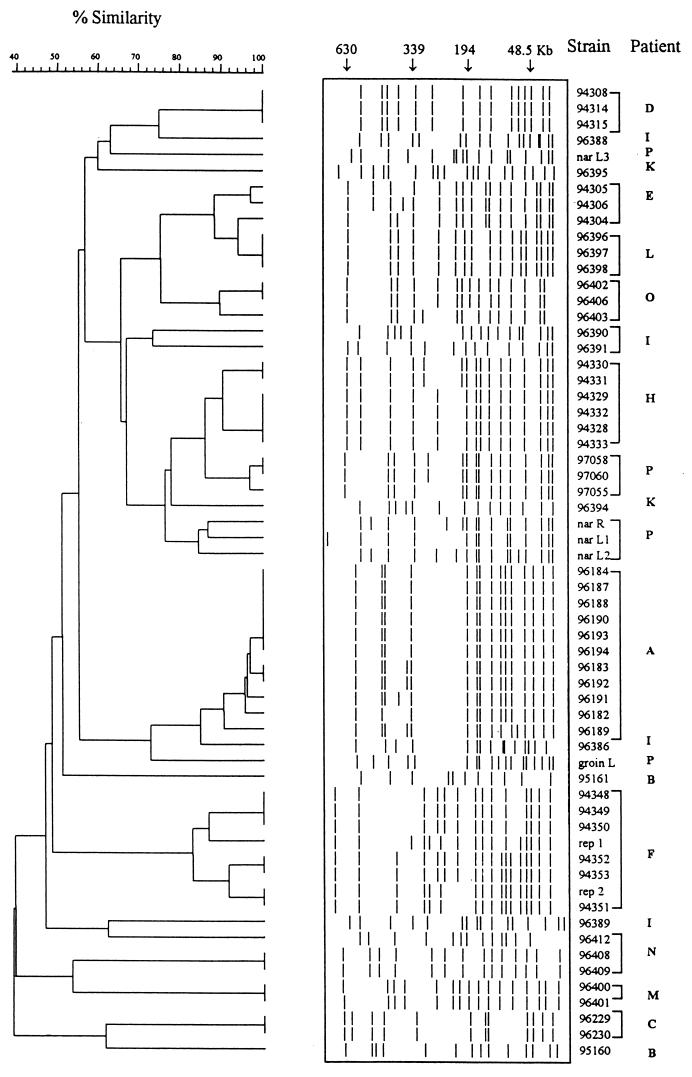
The 54 isolates of S. epidermidis tested (Table 1) gave a total of 30 different SmaI patterns and 29 different SstII patterns. Each SmaI profile included 12 to 17 fragments (Fig. 1), and each SstII profile included 12 to 21 fragments (data not shown).
FIG. 1.
Classification and schematic representation of the SmaI patterns of the S. epidermidis strains isolated from 14 patients (patients A to F, H, I, K and P) and of the two variants of 94351, Rep1 and Rep2, obtained by subculture.
The strains of each patient were clustered according to the similarities of the SmaI or SstII patterns. The clustering into genotypes of all intrapatient strains except strains 94304, 94305, and 94306, all of which were isolated from patient E, was the same for both enzymes (Table 1). On the basis of the SmaI patterns, these three strains were clustered into the same genotype. However, the SstII pattern of strain 94304 differed from those of strains 94305 and 94306 by five SstII fragments, suggesting that strain 94304 is in a separate genotype. Despite their distribution into two SstII genotypes, the three strains from patient E can be considered possibly related. S. epidermidis strains isolated from the infected samples from 8 of the 14 patients studied were monoclonal (Table 1; Fig. 1, patients A, C, D, H, L, M, O, and P), and their drug resistance patterns differed by one to nine markers. The six strains from patient F had five resistance markers in common but were distinguishable by one to eight additional markers and were possibly related since the differences between the SmaI or SstII patterns did not exceed six fragments. For patient N, strains 96408 and 96409, which differed from each other by only one drug resistance marker, had the same macrorestriction patterns with both enzymes; strain 96412 differed from strains 96408 and 96409 by seven and eight markers, respectively, and by more than seven SmaI and SstII bands. The different strains from each of the three patients B, I, and K appeared to be unrelated.
Surprisingly, the strains from patients E and L clustered in the same SmaI genotype, whereas the SstII patterns of the same strains differed by more than seven bands. Although patients E and L underwent prosthesis replacement in the same hospital, the interventions were 2 years apart and the first prostheses were implanted in different hospitals without any detectable link between the patients.
Dendrograms were constructed on the basis of the similarity of the SmaI or SstII patterns by the method of unweighted pair-group method with average linkages (see Fig. 1 for SmaI patterns). For the patterns with no more than three fragment differences, the percent similarities were 90 to 100.
(ii) SmaI profiles of the S. epidermidis strains isolated from the skin flora of patient P.
From patient P’s flora, five strains with distinct phenotypes were distributed into five different SmaI genotypes which did not include any of the three strains isolated from the infected specimens (Table 1; Fig. 1). The three strains isolated from both nares, narR, narL1, and narL2, were possibly related because their SmaI patterns differed by four or five fragments.
(iii) SmaI patterns of the hand flora strains isolated from healthy individuals.
Among the samples from the eight healthy individuals, only the hand swab of surgeon t gave monoclonal strains (Table 2). The SmaI patterns of strains t4 and t8 differed by only one SmaI fragment, whereas the pattern of the third strain (strain t5) from the same surgeon differed by at least eight fragments. For each of the seven other healthy individuals, the intraindividual colonies were considered unrelated, with at least seven SmaI band differences between the patterns (Table 2). Some of the S. epidermidis strains isolated from different surgeons working in the same operating room were clustered (Table 2), with percentages of similarity above 90 (data not shown). Indeed, the SmaI patterns of strains f1, l3, and j2 from surgeons f, l, and j, respectively, differed from each other by no more than two fragments, and those of strains w3 and m3 from surgeons w and m, respectively, differed from each other by three fragments. The related strains, which were resistant to oxacillin, were detected on the hands of several surgeons and were probably acquired in the hospital.
Analysis of HPs with pBA2 (ribotypes).
The HindIII and EcoRI HPs obtained with pBA2 were determined for the three coagulase-negative staphylococci which could not be classified by the ID32 Staph system (strains 95160, 96388, and 96390) and for a strain representative of each of the genotypes and subtypes of strains from patients (Table 1). For the latter strains, the assignment to S. epidermidis by the ID32 Staph System was confirmed by the comparative analysis of the ribotypes with those of the validly classified staphylococci already in our database.
Some strains considered to be different on the basis of their SmaI or SstII macrorestriction patterns had the same HPs for both enzymes, as follows: ribotype A for strains 96229, 96389, 96390, 96394, and 96397; ribotype B for strains 96160, 94305, and 96397; and ribotype C for strains 94348, 94351, 96386, and 96388. As reported previously (31), the discriminatory power of ribotyping is not satisfactory for the typing of S. epidermidis strains. Thus, analysis of ribotypes was used only to identify the strains to the species level.
Analysis of the stability of strain 94351 from patient F.
Although they were clustered in two genotypes, the six strains from patient F were scored as possibly related (fewer than six band differences). Strain 94351 had the largest number of drug resistance markers and was chosen for use in an evaluation of phenotypic and genomic stability after subculturing in brain heart infusion for 900 generations. Three hundred isolated colonies were studied, and two types of variants were detected: (i) those which had lost their oxacillin resistance, for example, strain Rep2, and (ii) those which had lost their resistance to oxacillin and to macrolides-lincosamides-streptogramin B, for example, Rep1 (Table 1). The loss of oxacillin resistance was associated with the loss of the mecA gene (data not shown). Neither of the two variants selected in vitro had a drug resistance pattern that was the same as those of any of the S. epidermidis strains isolated from the specimens from patient F. The SmaI patterns of strains Rep2 and 94351 were indistinguishable. The SmaI pattern of strain Rep1 differed from that of strain 94351 by five restriction fragments and from that of each of 94348, 94349, and 94350 by six fragments. The variant Rep2 strain was more closely related to the strains from patient F than to those from the 13 other patients (Fig. 1), suggesting that this variant was not a contaminant.
DISCUSSION
Variations in phenotypic characteristics, including virulence factors (2, 9, 32, 35) and drug resistance patterns (6, 13, 29), and in plasmid content (22) have often been reported for S. epidermidis strains. The detection of multiple S. epidermidis strains distinguishable by their drug resistance patterns in blood cultures, in pus, and in various other specimens from infected patients does not, however, result exclusively from the instability of phenotypic traits but results also from coinfection with unrelated S. epidermidis strains (1, 17, 20, 30, 34). Indeed, some cases of endocarditis following implantation of a prosthetic valve were recently shown to be attributable to polyclonal S. epidermidis populations (1, 30). Therefore, the detection in samples from the same patient of S. epidermidis strains with different antibiograms does not necessarily indicate contamination of the samples during collection. For the 14 patients in our study, contamination of the specimens by the polyclonal S. epidermidis strains of the skin flora is not likely, not only because most specimens were collected under the rigorously aseptic conditions required for surgery and in a sterile operating field but also because mixtures of phenotypically divergent colonies were detected in at least two specimens from the same patient. The proportion of each S. epidermidis strain in the mixtures could not be ascertained in our study because the drug resistance phenotype of every different strain was taken into consideration when the antibiotic therapy was chosen. This policy has probably contributed to the very high rate of successful eradication of infections following prosthesis replacements in the two hospitals that participated in the study.
The source of the delayed infections for the 14 patients in our study is not known. S. epidermidis of the skin flora or the environment may have been introduced into the operative wound. Alternatively, infection of the prosthesis may have been of blood origin and thus was not acquired during surgery. Collection of the S. epidermidis strains from the skin flora of the patient just before surgery, from the flora of the staff, and from the environment of the operating room would be required to trace the source of infection.
The comparative analysis of drug resistance patterns was insufficient to assess the degree of genomic relatedness of strains because some monoclonal strains in our study gave colonies that differed by up to nine drug resistance markers. Mapping of the genome regions carrying the drug resistance genes in the related strains from each patient would be required to elucidate the genetic changes responsible for the differences in drug resistance markers. When patients are infected with a single clone, the mutation, rearrangement, loss, or transposition of DNA may lead to the diversity of phenotypes (25). Genetic transfers, in particular, conjugation, may also occur when a polyclonal population is present at a single site. When phenotypic variations are not associated with detectable modifications of the macrorestriction profiles, it is likely that drug resistance phenotypic diversity results from divergence in plasmid content, but minor chromosomal modifications cannot be ruled out. This was probably the case for the Rep2 derivative whose SmaI pattern was indistinguishable from that of the parental strain, strain 94351, despite the loss of mecA, which is usually located in the chromosomes of staphylococci.
In our study, only 4 of the 14 patients appeared to be infected with a polyclonal population of strains. For the 10 other patients, all the intrapatient strains appeared to be closely or possibly related, despite the substantial diversity of the drug resistance patterns. Thus, for most patients, the multiple S. epidermidis strains found in the specimens were derivatives of a single clone. Such derivatives may have resulted from changes that occurred in situ during the infection process or may have preexisted in the intraoperative source of infection, which is often the skin flora. However, the latter possibility is not probable because closely related S. epidermidis colonies distinguishable by their morphologies and drug resistance patterns were detected in the skin flora of only one of the eight healthy individuals studied.
In conclusion, it is important to try to identify the largest number of colonies with distinct aspects in deep samples collected from patients whose prostheses are suspected of being infected so that antibiotic therapy can be directed at the most resistant isolates. Failure to eradicate these infections may, in some cases, be due to the failure to detect all the different S. epidermidis variants and clones present at the site of infection.
ACKNOWLEDGMENT
We thank C. Tran for secretarial assistance.
REFERENCES
- 1.Archer G L. Polyclonal Staphylococcus endocarditis: response. Clin Infect Dis. 1997;25:72–73. doi: 10.1086/514498. [DOI] [PubMed] [Google Scholar]
- 2.Baddour L M, Barker L P, Christensen G D, Parisi J T, Simpson W A. Phenotypic variation of Staphylococcus epidermidis in infection of transvenous endocardial pacemaker electrodes. J Clin Microbiol. 1990;28:676–679. doi: 10.1128/jcm.28.4.676-679.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brause B D. Infections associated with prosthetic joints. Clin Rheum Dis. 1986;12:523–536. [PubMed] [Google Scholar]
- 4.Chabbert Y A. Sensibilité bacterienne aux antibiotiques. In: Le Minor L, Véron M, editors. Bactériologie médicale. Médecine Science. Paris, France: Flammarion; 1982. pp. 204–212. [Google Scholar]
- 5.Chesneau O, Aubert S, Morvan A, Guesdon J L, El Solh N. Usefulness of the ID32 Staph system and a method based on rRNA gene restriction site polymorphism analysis for species and subspecies identification of staphylococcal clinical isolates. J Clin Microbiol. 1992;30:2346–2352. doi: 10.1128/jcm.30.9.2346-2352.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen G D, Baddour L M, Madison B M, Parisi J T, Abraham S N, Hasty D L, Lowrance J H, Josephs J A, Simpson W A. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to β-lactam antibiotics, and virulence. J Infect Dis. 1990;161:1153–1169. doi: 10.1093/infdis/161.6.1153. [DOI] [PubMed] [Google Scholar]
- 7.De Buyser M L, Morvan A, Aubert S, Dilasser F, El Solh N. Evaluation of a ribosomal RNA gene probe for the identification of species and subspecies within the genus Staphylococcus. J Gen Microbiol. 1992;138:889–899. doi: 10.1099/00221287-138-5-889. [DOI] [PubMed] [Google Scholar]
- 8.De Buyser M L, Morvan A, Grimont F, El Solh N. Characterization of Staphylococcus species by ribosomal RNA gene restriction patterns. J Gen Microbiol. 1989;135:989–999. doi: 10.1099/00221287-135-4-989. [DOI] [PubMed] [Google Scholar]
- 9.Deighton M, Pearson S, Capstick J, Spelman D, Borland R. Phenotypic variation of Staphylococcus epidermidis isolated from a patient with native valve endocarditis. J Clin Microbiol. 1992;30:2385–2390. doi: 10.1128/jcm.30.9.2385-2390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derbise A, Dyke K G H, El Solh N. Characterization of a Staphylococcus aureus transposon Tn5405, located within Tn5404 and carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid. 1996;35:174–188. doi: 10.1006/plas.1996.0020. [DOI] [PubMed] [Google Scholar]
- 11.Dice L R. Measures of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 12.Dougherty S H. Pathobiology of infection in prosthetic devices. Rev Infect Dis. 1988;10:1102–1117. doi: 10.1093/clinids/10.6.1102. [DOI] [PubMed] [Google Scholar]
- 13.Etienne J, Renaud F, Bes M, Brun Y, Greenland T B, Freney J, Fleurette J. Instability of characteristics amongst coagulase-negative staphylococci causing endocarditis. J Med Microbiol. 1990;32:115–122. doi: 10.1099/00222615-32-2-115. [DOI] [PubMed] [Google Scholar]
- 14.Garvin K L, Hanssen A D. Infection after total hip arthroplasty. Past, present, and future. J Bone Joint Surg. 1995;77:1576–1588. doi: 10.2106/00004623-199510000-00015. [DOI] [PubMed] [Google Scholar]
- 15.Gristina A G, Naylor P T, Myrvik Q N. Mechanisms of musculoskeletal sepsis. Orthop Clin N Am. 1991;22:363–371. [PubMed] [Google Scholar]
- 16.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 17.Hope P G, Kristinsson K G, Norman P, Elson R A. Deep infection of cemented total hip arthroplasties caused by coagulase-negative staphylococci. J Bone Joint Surg. 1989;71:851–855. doi: 10.1302/0301-620X.71B5.2584258. [DOI] [PubMed] [Google Scholar]
- 18.Iglesias A, Ceglowski P, Trautner T A. Plasmid transformation in Bacillus subtilis. Effects of the insertion of Bacillus subtilis rRNA genes into plasmids. Mol Gen Genet. 1983;192:149–155. doi: 10.1007/BF00327660. [DOI] [PubMed] [Google Scholar]
- 19.James P J, Butcher I A, Gardner E R, Hamblen D L. Methicillin-resistant Staphylococcus epidermidis in infection of hip arthroplasties. J Bone Joint Surg. 1994;76:725–727. [PubMed] [Google Scholar]
- 20.Khatib R, Riederer K M, Clark J A, Khatib S, Briski L E, Wilson F M. Coagulase-negative staphylococci in multiple blood cultures: strain relatedness and determinants of same-strain bacteremia. J Clin Microbiol. 1995;33:816–820. doi: 10.1128/jcm.33.4.816-820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludlam H A, Noble W C, Marples R R, Bayston R, Phillips I. The epidemiology of peritonitis caused by coagulase-negative staphylococci in continuous ambulatory peritoneal dialysis. J Med Microbiol. 1989;30:167–174. doi: 10.1099/00222615-30-3-167. [DOI] [PubMed] [Google Scholar]
- 23.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson M, Frykberg L, Flock J I, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect Immun. 1998;66:2666–2673. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen I T, Firth N, Skurray R A. Resistance to antimicrobial agents other than β-lactams. In: Crossley K B, Gordon G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 175–212. [Google Scholar]
- 26.Pfaller M, Herwaldt L. Laboratory, clinical and epidemiological aspect of coagulase-negative staphylococci. Clin Microbiol Rev. 1988;1:281–299. doi: 10.1128/cmr.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rupp M E, Archer G L. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994;19:231–243. doi: 10.1093/clinids/19.2.231. [DOI] [PubMed] [Google Scholar]
- 28.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toldos C M, Yagüe G, Ortiz G, Segovia M. Assessment of multiple coagulase-negative staphylococci isolated in blood cultures using pulsed-field gel electrophoresis. Eur J Clin Microbiol Infect Dis. 1997;16:581–586. doi: 10.1007/BF02447920. [DOI] [PubMed] [Google Scholar]
- 30.van Wijngaerden E, Peetermans W E, van Lierde S, van Eldere J. Polyclonal Staphylococcus endocarditis. Clin Infect Dis. 1997;25:69–71. doi: 10.1086/514499. [DOI] [PubMed] [Google Scholar]
- 31.Walcher-Salesse S, Monzon-Moreno C, Aubert S, El Solh N. An epidemiological assessment of coagulase-negative staphylococci from an intensive care unit. J Med Microbiol. 1992;36:321–331. doi: 10.1099/00222615-36-5-321. [DOI] [PubMed] [Google Scholar]
- 32.Williams P, Swift S, Modun B. Continuous ambulatory peritoneal dialysis-associated peritonitis as a model device-related infection: phenotypic adaptation, the staphylococcal cell envelope and infection. J Hosp Infect. 1995;30:35–43. doi: 10.1016/0195-6701(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi K, Adams R A, Morrey B F. Infection after total elbow arthroplasty. J Bone Joint Surg. 1998;80-A:481–491. doi: 10.2106/00004623-199804000-00004. [DOI] [PubMed] [Google Scholar]
- 34.Zaidi A K M, Harrell L J, Rost J R, Reller L B. Assessment of similarity among coagulase-negative staphylococci from sequential blood cultures of neonates and children by pulsed-field gel electrophoresis. J Infect Dis. 1996;174:1010–1014. doi: 10.1093/infdis/174.5.1010. [DOI] [PubMed] [Google Scholar]
- 35.Ziebuhr W, Heilmann C, Götz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziza J M, Desplaces N, Léonard P, Mamoudy P. Infections sur prothèses articulaires. Rev Méd Int. 1997;18:431s–434s. doi: 10.1016/s0248-8663(97)80150-6. [DOI] [PubMed] [Google Scholar]