Abstract
The immune system protects us from enormously diverse microbial pathogens but needs to be tightly regulated to avoid deleterious immune-mediated inflammation and tissue damage. A wide range of molecular determinants and cellular components work in concert to control the magnitude and duration of a given immune response. In the past decade, microRNAs (miRNAs), a major class of small non-coding RNA species, have been extensively studied as key molecular players in immune regulation. In this chapter, we will discuss how miRNAs function as negative regulators to restrict innate and adaptive immune responses. Moreover, we will review the current reports regarding miRNAs in human immunological diseases. Finally, we will also address the emerging roles of other non-coding RNAs, long non-coding RNAs (lncRNAs) in particular, in the regulation of the immune system.
1. Introduction
Since the first microRNA (miRNA), lin-4, was discovered in Caenorhabditis elegans by the laboratories of Victor Ambros and Gary Ruvkun in 1993 (Lee et al., 1993, Wightman et al., 1993), these small non-coding RNA species have been extensively studied for their roles in post-transcriptional regulation of gene expression. Like other protein encoding genes, primary miRNA transcripts are first transcribed in the nucleus but are sequentially processed by a microprocessor complex, which contains the RNase III Drosha and the double-stranded RNA binding protein DGCR8 in mammals and Pasha in other species, into a characteristic ~60 nucleotide stem-loop structure, named precursor-miRNA (pre-miRNA) (Lee et al., 2003, Han et al., 2006, Denli et al., 2004, Gregory et al., 2004). The pre-miRNAs are then exported via exportin 5 (Lund et al., 2004, Yi et al., 2003) into the cytoplasm, where the hairpin of pre-miRNA is further processed by the evolutionarily conserved RNase III enzyme Dicer to generate mature miRNAs (Bernstein et al., 2001, Hutvagner et al., 2001). These mature miRNAs are capable of being incorporated into the RNA-induced silencing complex (RISC) where they interact with the core component Argonaute protein (Hutvagner and Zamore, 2002, Lingel et al., 2003, Liu et al., 2004). Once assembling with mature miRNA and engaging the targets, active RISCs recognize complementary messenger RNA (mRNA) transcripts for degradation or translational silencing (Jing et al., 2005, Song et al., 2004, Liu et al., 2004). By modulating the expression level of their target genes, miRNAs have been found to regulate almost all biological processes including cell growth, differentiation, and apoptosis in both plants and animals (Ameres and Zamore, 2013). As is the case for non-immune cells, the role of miRNA-mediated gene regulation in the immune system has in recent years become a focus of intense investigation.
2. miRNAs in the immune system
Even though our understanding of miRNA biogenesis and its overall cellular function was initially established based on discoveries in nematodes and plants (Lee et al., 1993, Baulcombe, 2004), accumulating evidence has demonstrated that miRNAs also have a crucial role in controlling all aspects of immune responses (Mehta and Baltimore, 2016). The importance of miRNA in immune cells was first demonstrated in studies where key components in miRNA biogenesis were disrupted. To this end, conditional deletion of Dicer during early B cell development leads to a dramatic block at the pro- to pre-B transition (Koralov et al., 2008). Subsequent studies using mice with Dicer ablated specifically in peripheral activated B cells further demonstrated that miRNAs are also required for germinal center B-cell formation and the generation of the antibody diversity (Xu et al., 2012). Interestingly, unlike what has been reported in B cell differentiation, Dicer ablation in early T cell progenitors did not exhibit any substantial alterations in thymic T cell development with the exception of reduced thymic cellularity (Cobb et al., 2005). Nevertheless, the specificity of the role of miRNA in peripheral T cells is much more apparent, as T cells lacking Dicer failed to differentiate into multiple helper T cell lineages and exhibited aberrant effector T cell function (Muljo et al., 2005). Moreover, when Dicer or Drosha ablation was restricted to regulatory T (Treg) cell lineage, mice developed highly aggressive autoimmunity comparable to those devoid of a functional Foxp3 gene (Zhou et al., 2008, Liston et al., 2008, Chong et al., 2008), suggesting an indispensable role of miRNA in controlling Treg cell biology.
To date, more than hundreds of miRNAs have been reported to be differentially expressed in immune cells. Distinct miRNA signatures not only were found in individual immune cell lineages, but could also be detected in the same cell subsets that are in different developmental stages. For example, whereas miR-139 is highly expressed in Pro- and Pre-B cells, elevated levels of miR-28, miR-320 and miR-148a are detected in germinal center (GC) B cells and plasma cells, respectively (Kuchen et al., 2010). Moreover, expression of miRNAs in immune cells can also be dynamically regulated in response to a variety of stimuli, such as antigens recognized by T or B cell receptors, proinflammatory cytokines, and microbial components that trigger Toll-like receptors (O’Connell et al., 2007, Taganov et al., 2006, Cobb et al., 2006). To this end, a recent study reported that T cell activation induces proteasome-mediated degradation of Argonaute, and subsequently causes a global down-regulation of mature miRNAs (Bronevetsky et al., 2013). It was suggested that activation-induced miRNA down-regulation confers effector functions to helper T cells via relaxing the repression of genes that direct T cell differentiation. Finally, hierarchical clustering analysis of miRNA profiling clearly separated cells of the immune system from other tissues. Taken together, these results implied certain miRNAs might play a specific role in controlling development and effector functions of the immune system (Kuchen et al., 2010), and that miRNAs need to be tightly regulated as aberrant expression of miRNAs often leads to dysregulated innate and adaptive immunity.
3. miRNAs as negative regulators of immune responses
3.1. miRNA in adaptive immunity
While studies of mice with B or T cell-specific deletion of the entire miRNA pathway seemed to suggest that miRNAs generally play a positive role in promoting adaptive immunity as discussed in the previous section, many miRNAs have been identified as important negative regulators in restricting B and T cell responses (Figure 1). For example, miR-150, a miRNA that is predominantly expressed in mature B cells was shown to control multiple B cell populations through regulating the expression level of transcription factor c-Myb (Xiao et al., 2007). Genetic ablation of miR-150 resulted in the expansion of B1 cells, one of the subsets of mature B cells, in spleen and peritoneal cavity. While the numbers of follicular B cells were not significantly altered, upon immunization elevated antibody responses could be easily detected (Xiao et al., 2007). Compared to miR-150, miR-210, a miRNA that is highly induced upon B cell activation, appears to play an even larger role in functioning as a negative feedback regulator to restrain B cell responses; deletion of miR-210 leads to the development of age-associated autoantibodies (Mok et al., 2013). In contrast, while miR-142 was shown to target B-cell-activating factor receptor (BAFF-R), a molecule that is critical for B cell proliferation and survival, hypogammaglobulinemia phenotype was detected in mice devoid of miR-142 despite having increased follicular B cell numbers (Kramer et al., 2015). Similarly, despite the fact that activation-induced cytidine deaminase (AID), a potent enzyme critical for somatic hypermutation and class-switch recombination (CSR), has been shown to be a bona fide miR-155 target where lack of AID regulation by miR-155 led to defective affinity maturation (Teng et al., 2008, Dorsett et al., 2008), miR-155-deficient mice actually exhibited reduced germinal center function and failed to generate high-affinity IgG1 antibodies (Rodriguez et al., 2007). Together, these results suggested that other miR-142 or miR-155 targets are likely responsible for their respective effects on humoral immunity and further demonstrate the complex nature of miRNA-mediated immune regulation.
Figure 1. MiRNAs negatively regulate adaptive immune cells.
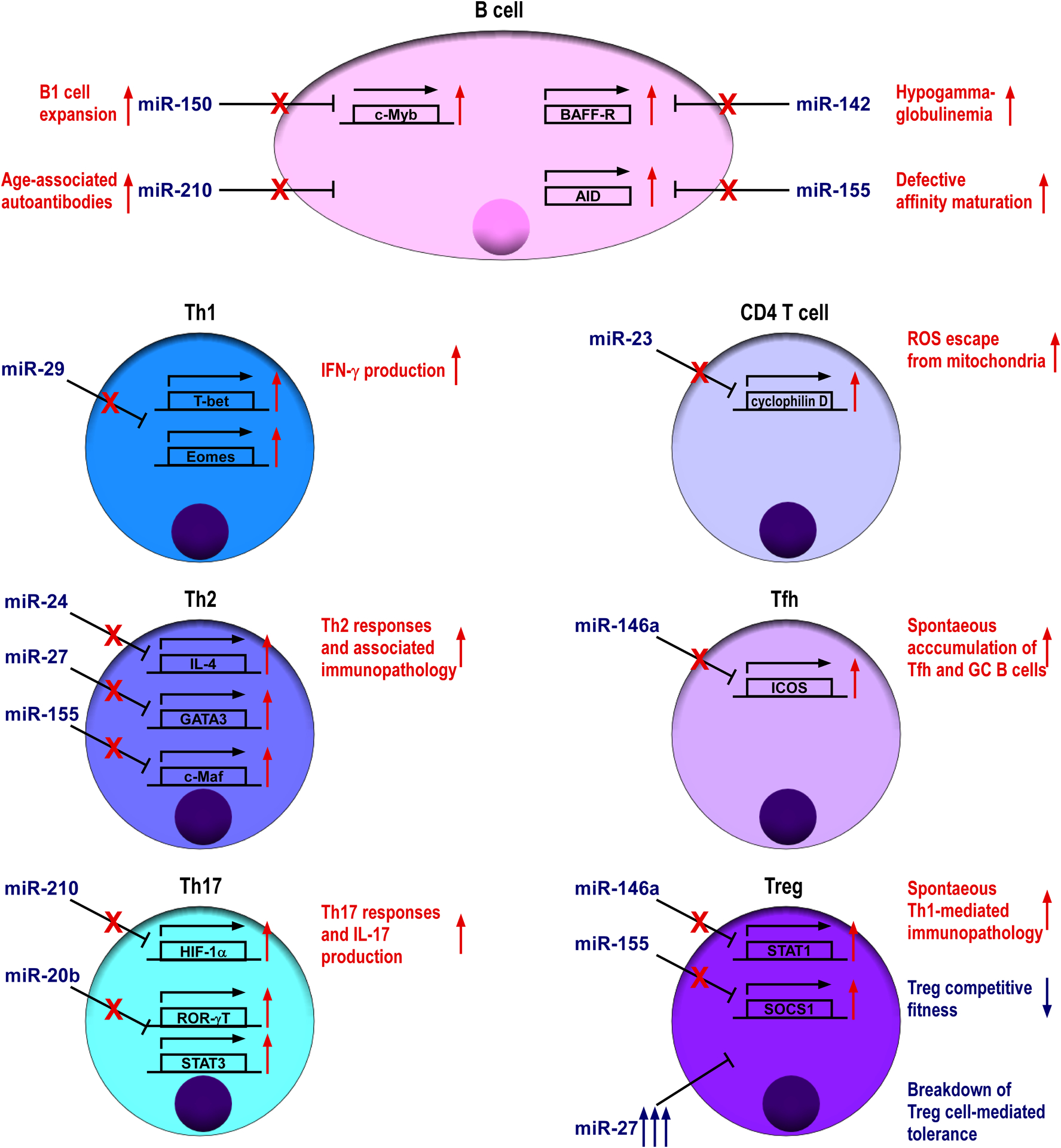
Specific miRNAs expressed by B or T cells repress key target genes that are involved in adaptive immune responses. Tfh cell, T follicular helper cell; Th cell, T helper cell; Treg cell, regulatory T cell.
Like B cell, activated T cells also express increased level of miR-155; mice devoid of miR-155 displayed increased lung airway remodeling, and miR-155-deficient CD4+ T cell cells are intrinsically biased toward Th2 differentiation in vitro. Mechanistically, it was shown that miR-155 can inhibit Th2 responses through modulating the level of c-Maf, a transcription factor known to promote Th2 immunity (Rodriguez et al., 2007). In addition to miR-155, we and others have recently demonstrated that miR-24 and miR-27, two members of the miR23 cluster family, collaboratively limit Th2 responses and associated immune pathology through targeting IL-4, GATA3 as well as other Th2-related genes in both direct and indirect manners (Cho et al., 2016, Pua et al., 2016). While miR-23 does not seem to play any role in Th2 regulation, it is indispensable for restraining activation-induced necrosis of CD4+ T cells by enforcing intracellular reactive oxygen species (ROS) equilibrium through targeting cyclophilin D, a regulator of ROS escape from mitochondria (Zhang et al., 2016). In addition to Th2 regulation, miRNAs have also been implicated in regulating other Th lineages. To this end, miR-29 was shown to control Th1 responses by repressing multiple genes associated to Th1 differentiation and function including both T-bet and Eomes, two transcription factors known to induce IFNγ production and IFNγ itself (Ma et al., 2011, Steiner et al., 2011). On the other hand, hypoxia-induced miR-210 was reported to negatively regulate Th17 responses through restricting the expression of HIF-1α, a key transcription factor that promotes Th17 polarization under limited oxygen (Wang et al., 2014). Similarly, Th17 differentiation and the resultant pathogenesis of experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis, could be suppressed by miR-20b via targeting RORγt and STAT3, two key Th17 transcription factors (Zhu et al., 2014). Finally, miR-146a is highly induced in T follicular helper (Tfh) cells, a specialized Th cell subset required for humoral immunity, and can act as a post-transcriptional brake to control Tfh cell and corresponding GC B cell responses by regulating ICOS-ICOSL axis (Pratama et al., 2015).
In addition to its role in Tfh cells, miR-146a has also been shown to function as a key molecular regulator to confer suppressor function to Treg cells. In the absence of miR-146a-mediated regulation of STAT1, mice succumbed to spontaneous IFN-γ-dependent Th1-mediated immunopathology (Lu et al., 2010). Moreover, our recent work also demonstrated that miR-27 controls multiple aspects of Treg cell biology and suggests that excessive expression of miR-27 in Treg cells resulted in a breakdown of Treg cell-mediated immunological tolerance (Cruz et al., 2017). Together with the aforementioned studies in which the entire miRNA pathway was ablated in Treg cells, these results suggested that miRNAs are able to mediate their regulatory effects on the immune system indirectly through maintaining optimal Treg cell function and homeostasis (Zhou et al., 2008, Liston et al., 2008, Chong et al., 2008).
3.2. miRNA in innate immunity
Similar to what was described in the adaptive immune system, significant progress has been made over the past decade in characterizing individual miRNAs that control the function of innate immune cells (Figure 2). Among them, miRNA-mediated regulation of myeloid cell function is best characterized. Both miR-146 and miR-155 were identified in macrophages in response to LPS activation (Taganov et al., 2006, O’Connell et al., 2007). Between these two miRNA, miR-146a serves as a negative regulator to limit inflammatory responses by targeting TRAF6 and IRAK1 (Taganov et al., 2006, Boldin et al., 2011). During virus infection, miR-146a expression is also upregulated in macrophages in RIG-I-dependent manner, and its function negatively regulates type I interferon (IFN) production through repressing IRAK2 (Hou et al., 2009). While miR-155 is generally considered as a positive player in mediating inflammatory responses as it can directly repress SOCS1 and SHIP1 (Androulidaki et al., 2009, O’Connell et al., 2009), two known inhibitory molecules affecting multiple signaling pathways, miR-155 can also down-modulate TLR/IL-1 inflammatory pathway via targeting TAB2 (Ceppi et al., 2009). Moreover, protein kinase Akt1, which is activated by LPS in macrophages, positively regulates let-7e, a miRNA that can inhibit TLR4 expression to control endotoxin sensitivity and tolerance (Androulidaki et al., 2009). In addition to direct TLR4 targeting, miR-21, another miRNA induced by LPS can also act as a negative feedback regulator of TLR4 signaling by targeting PDCD4, a proinflammatory protein required for LPS-induced death (Sheedy et al., 2010). Finally, miRNAs can also restrict DC function by directly suppressing the production of proinflammatory cytokines. To this end, NOD2-induced miR-29 expression in human dendritic cells (DCs) was shown to inhibit IL-23 expression by repressing IL-12p40 directly and IL-23p19 indirectly via targeting ATF2 (Brain et al., 2013).
Figure 2. MiRNAs negatively regulate innate immune cells.
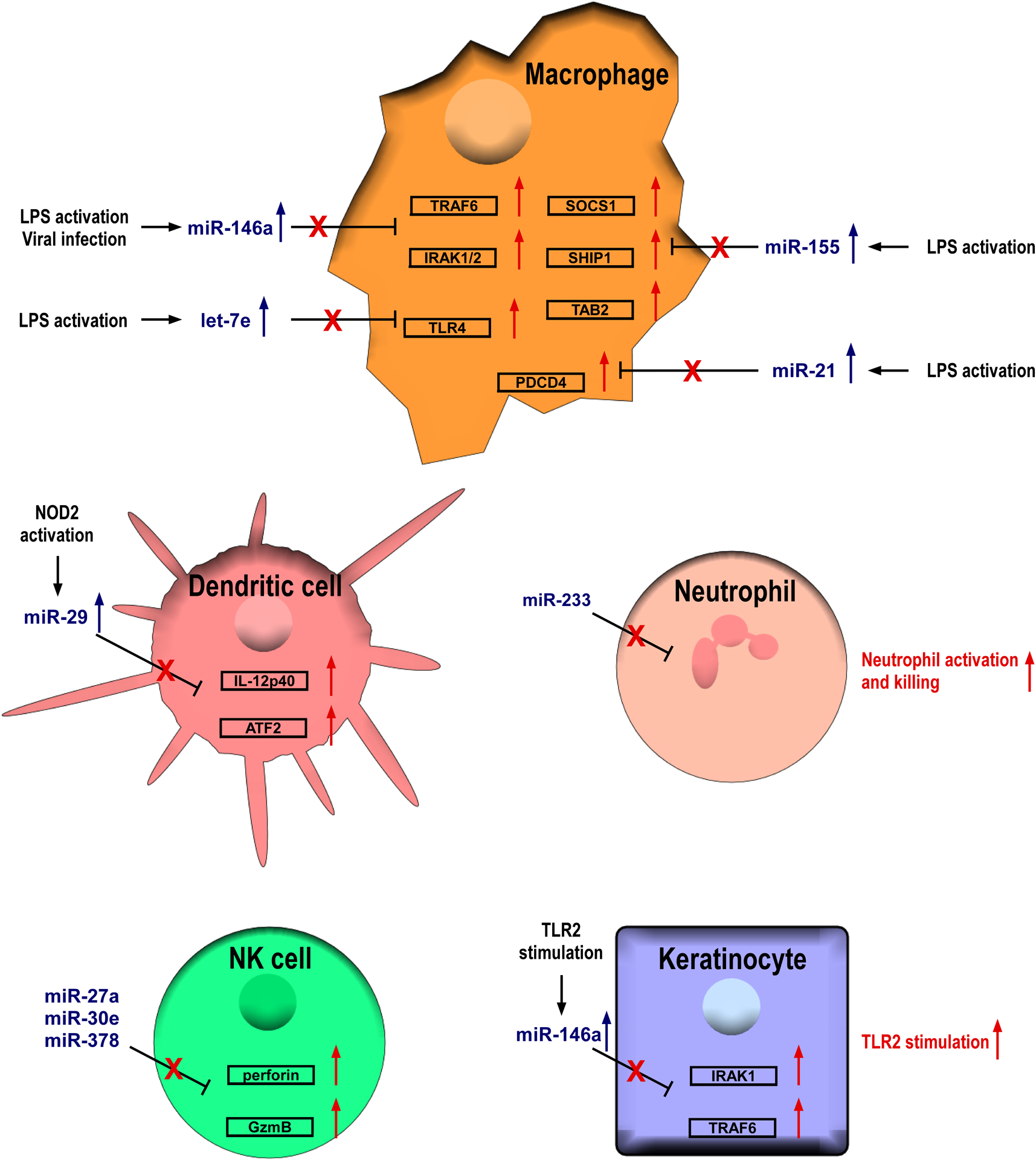
For miRNAs expressed in different innate immune cell populations, their key target genes are shown. NK, natural killer.
Beside myeloid cell subsets, much experimental evidence has also pointed to miRNAs as important negative regulators for other innate immune cells. For example, miR-223 has been shown to function as a cell intrinsic negative modulator of neutrophil activation and killing. Mice lack of miR-223 developed spontaneous inflammatory lung pathology and exhibited exaggerated tissue damage upon LPS treatment (Johnnidis et al., 2008). Moreover, miR-27a*, miR-378, and miR-30e have all been shown to act as negative regulators of NK-cell cytotoxicity by silencing perforin and GzmB expression (Kim et al., 2011, Wang et al., 2012). Finally, miRNAs can also regulate immune responses by targeting in non-innate immune cells. To this end, miR-146a expression was shown to be induced by TLR2 stimulation in human keratinocytes. MiR-146a can then serve as a potent negative feedback regulator to prevent further TLR2-induced inflammatory responses (Meisgen et al., 2014). Collectively, the aforementioned studies not only provide important insights into miRNA-mediated immune regulation but also offer the molecular basis to understand the precise role of miRNAs in the pathogenesis of human immunological diseases.
4. miRNAs in human immunological diseases
As summarized in the previous sections, studies employing genetically manipulated mice with in vitro experimental approaches and in vivo disease models have helped us to gain valuable knowledge of miRNA function in regulating immune responses. However, it is also important to study miRNA in the context of human immunological diseases. Indeed, numerous studies have shown the correlation between the expression of several miRNAs and immune related disorders, such as autoimmunity, hypersensitive diseases and hematopoietic malignancies.
4.1. miRNA in autoimmunity and hypersensitivity diseases
Abnormal expression of miRNAs has been associated with many autoimmune diseases (Table 1). One of the best examples is systemic lupus erythematosus (SLE), a multifaceted autoimmune disease with a strong genetic predisposition, characterized by enhanced type I interferon signaling. To this end, it has been reported that peripheral blood mononuclear cells (PBMCs) isolated from SLE patients express reduced miR-146a. The amount of miR-146a was shown to negatively correlate with the clinical disease activity and type I interferon levels in patients. It was suggested that lack of miR-146a-mediated regulation of STAT1 and IRF5 led to the excessive production of type I interferon (Tang et al., 2009). Moreover, sequencing analysis of single-nucleotide polymorphisms (SNPs) in SLE patients identified a genetic variant in the miR-146a promoter region that is functionally significant in downregulating the expression of miR-146a by altering its binding affinity for Ets-1 (Luo et al., 2011). In addition to miR-146a, diminished expressions of both miR-125a and miR-155 were also reported in patients with SLE (Zhao et al., 2010, Lashine et al., 2015). While elevated levels of RANTES in the absence of optimal miR-125a-mediated regulation was considered to promote the disease, increased expression of protein phosphatase 2A (PP2A) in juvenile SLE patients with reduced miR-155 was thought to be responsible for enhanced pathogenesis. On the other hand, elevated levels of miR-21 and miR-148a were detected in circulating CD4+ T cells from SLE patients. Both of these two miRNAs are able to upregulate autoimmune-associated methylation-sensitive genes such as CD70 and LFA-1 through promoting DNA hypomethylation and by repressing the expression of RASGRP1 and DNA methyltransferase 1 (DNMT1), respectively (Pan et al., 2010).
Table 1 |.
miRNAs involved in human immunological diseases
| Immune disease | miRNA | Target |
|---|---|---|
| Systemic lupus | miR-21 | RASGRP1 |
| erythematosus | miR-125a | KLF13 |
| miR-146a | IRF5, STAT1, Ets-1 | |
| miR-148a | DNMT1 | |
| MiR-155 | PP2A | |
| Multiple sclerosis | miR-27b | BMI1, TGFBR1, and SMAD4 |
| miR-326 | Ets-1 | |
| Type 1 diabetes | miR-326 | Ets-1, VDR |
| Rheumatoid arthritis | miR-146a | FAF1 |
| miR-155 | MMP-3 | |
| miR-223 | IGF-1 | |
| Psoriasis | MiR-203 | SOCS-3 |
| Asthma | miR-19a | PTEN, SOCS1, and NFAIP3 |
Like SLE, multiple sclerosis (MS) has also been linked to aberrant expression of many miRNAs. For example, the expression level of miR-326 has been shown to be highly correlated with disease severity in MS patients (Du et al., 2009). Mechanistically, miR-326 promotes Th17 cell induction through targeting Ets-1, a known negative regulator of Th17 differentiation. Moreover, increased miR-27, miR-128 as well as miR-340 were detected in CD4+ T cells of patients with MS. It was shown that through repressing BMI1, a molecule that stabilizes GATA3, miR-27 inhibits Th2 differentiation and promotes proinflammatory Th1 autoimmune responses (Guerau-de-Arellano et al., 2011). Following up this study, miR-27 was further shown to dampen TGFβ signaling, leading to impaired Treg cells and enhanced susceptibility to developing multiple sclerosis (Severin et al., 2016). Besides these two autoimmune diseases, abnormal miRNA expression has also been associated with other autoimmune inflammation including rheumatoid arthritis (RA), psoriasis and type I diabetes (Li et al., 2010, Stanczyk et al., 2008, Lu et al., 2014, Sonkoly et al., 2007, Sebastiani et al., 2011).
In addition to autoimmunity, dysregulated miRNAs also contribute to the development or pathogenesis of hypersensitivity diseases like asthma. MiRNA profiling analysis of human airway-infiltrating T cells revealed that miR-19a, a member of the miR-17~92 cluster, was greatly upregulated in CD4+ T cells isolated from asthmatic airways compared with cells from healthy subjects. Mechanistically, miR-19a was shown to repress multiple genes including PTEN, SOCS1 and TNFAIP3, leading to specific augmentation of Th2 responses and associated immune pathology (Simpson et al., 2014). Together, more and more studies have revealed a causative role of miRNA in the development of many human immunological diseases.
4.2. miRNA in blood cancer
As one of the major functions of miRNAs is to regulate cell differentiation and proliferation, it is not surprising that when dysregulated miRNAs can drive the development of malignancies of the immune system (Table 2). MiRNAs can act either as tumor suppressors or function as oncomirs to promote or prevent tumorigenesis. As tumor suppressors, miR-15a and miR-16 were shown to inhibit the development of B cell chronic lymphocytic leukemias (B-CLL) through targeting Bcl2, and that in more than 50% of B-CLL patients a region encoding miR-15a and miR-16 was found to be deleted (Cimmino et al., 2005, Calin et al., 2002). MiR-28 was also identified as a tumor suppressor and is significantly downregulated in Burkitt lymphoma. Oncogene Myc was shown to negatively regulate miR-28 expression leading to the uncontrolled proliferation of certain B cell subsets (Schneider et al., 2014). Another example is miR-29b whose expression is deregulated in primary acute myelogeneous leukemia (AML). Restoration of miR-29b in AML cell lines was able to induce apoptosis and dramatically reduce tumorigenicity, pointing to a clear tumor suppressor role (Garzon et al., 2009).
Table 2 |.
miRNAs involved in human blood cancer
| miRNA | Blood cancer | Target |
|---|---|---|
| Tumor suppressors | ||
| miR15a/miR16-1 | B cell chronic lymphocytic leukemia | Bcl-2 |
| miR-28 | Burkitt lymphoma | MAD2L1, BAG1 |
| miR-29b | Acute myelogeneous leukemia | MCL-1 |
| OncomiRs | ||
| miR-17-92 | B-cell lymphomas | c-myc |
| miR-125b | Myeloid and B-cell leukemia | IRF4 |
| miR-155 | Burkitt’s lymphoma, diffuse large B-cell lymphomas, Hodgkin’s lymphomas, and NK cell lymphoma | PTEN, PDCD4, and SHIP1 |
| miR-223 | T cell acute lymphoblastic leukemia | FBXW7 |
In contrast to the aforementioned roles of miRNA in preventing tumorigenesis, many miRNAs also exhibit oncogenic activity in hematologic malignancies. For example, the miR-17-92 cluster, which is located in a region of DNA that is frequently amplified in human B-cell lymphomas, has been shown to promote malignancy of immune cells (He et al., 2005, Tagawa and Seto, 2005, Inomata et al., 2009, Lu et al., 2010). In addition to miR-17-92 cluster, miR-155, another well-characterized oncomir, has also been shown to be expressed at higher levels in many different types of B cell lymphomas including Hodgkin’s lymphoma, DLBCL, Burkitt’s lymphoma as well as NK cell lymphoma (Eis et al., 2005, Kluiver et al., 2005, van den Berg et al., 2003, Metzler et al., 2004, Yamanaka et al., 2009). On the other hand, miR-223 was shown to be upregulated in human T cell acute lymphoblastic leukemia (T-ALL) in a TAL1-dependent manner (Mansour et al., 2013). It is thought that miR-223 promotes T-ALL through repressing a tumor suppressor, FBXW7. Finally, elevated levels of the oncomir, miR-125b, has been reported in a variety of human neoplastic blood disorders and could potentially induce myeloid- and B-cell leukemia by inhibiting IRF4 (So et al., 2014). The miRNA signatures identified from these clinical studies not only provide great value as prognostic parameters for cancer progression, but might also serve as potential novel therapeutic targets to treat human hematopoietic malignancies.
5. Other non-coding RNAs
Like miRNAs, many other non-coding RNA (ncRNA) species including long non-coding RNAs (lncRNAs) have also been identified as important gene regulators in the immune system (Figure 3). Since the first lncRNA H19 was reported in 1990 (Brannan et al., 1990), extensive investigation in lncRNA-mediated gene regulation has demonstrated that lncRNAs can regulate gene expression in various biological processes, including immune responses (Ponting et al., 2009). LncRNAs are transcribed by RNA polymerase II, 5’-capped, polyadenylated, and undergo splicing similar to that for mRNAs (Guttman et al., 2009). They can function both in cis to regulate the gene expression in close genomic proximity at the site of transcription, or in trans to target distant transcriptional activators or repressors (Ponting et al., 2009). Moreover, since lncRNAs usually contain multiple modular domains that can either interact with proteins or form complementary pairs with nucleotides, these molecules could connect DNA, RNA, and proteins and be involved in nearly all stages of gene regulation (Guttman and Rinn, 2012).
Figure 3. Non-coding RNAs in the immune system.
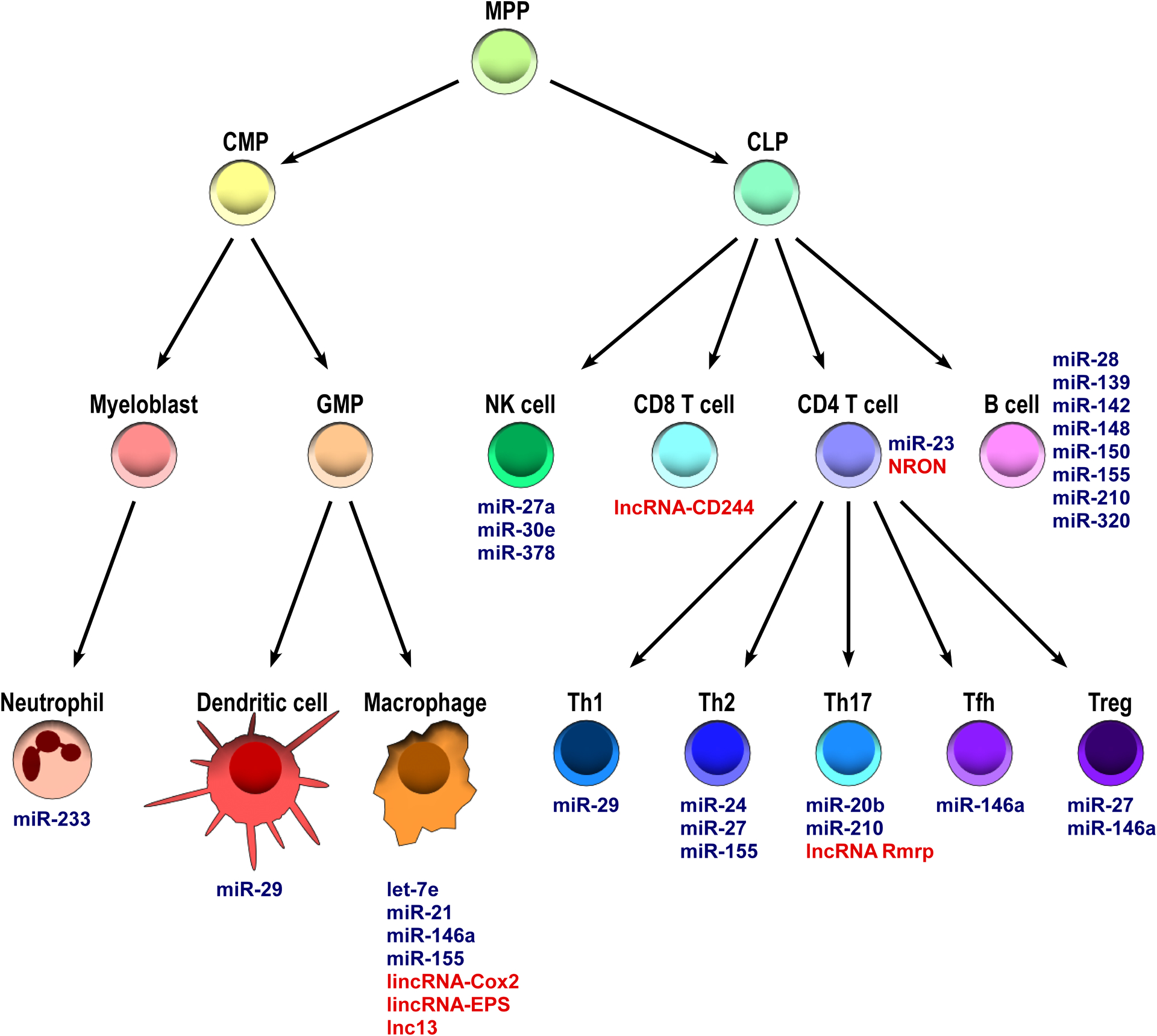
The schematic describes miRNAs (blue font) and lncRNAs (red font) that have key roles in controlling immune responses. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte–monocyte progenitor.
Recent report suggests that up to two third of transcribed genes across all cell types in humans are classified as lncRNAs (Iyer et al., 2015). In T cells, a lncRNA, NRON (noncoding RNA, repressor of NFAT) was shown to form a large cytoplasmic RNA-protein scaffold complex that can repress the transcriptional activation of NFAT-responsive genes via regulating NFAT nuclear trafficking (Willingham et al., 2005, Sharma et al., 2011). Moreover, another lncRNA, lncRNA-CD244, whose expression has been shown to be driven by CD244 signaling upon Tuberculosis (TB) infection, is able to inhibit cytokine production by CD8+ T cells by mediating histone H3K27 trimethylation at promoter regions of IFNγ and TNFα (Wang et al., 2015). On the other hand, in Th17 cells, rather than inhibiting their effector function, a lncRNA, lncRNA Rmrp was shown to interact with a complex of RORγt and a RNA helicase, DDX5, to activate RORγt-dependent Th17-relative gene transcription (Huang et al., 2015).
As for the role of lncRNAs in the innate immune cells, a TLR signaling induced intergenic lncRNA, lncRNA-Cox2, was reported to repress the expression of many critical inflammatory genes in macrophages through forming a complex with heterogeneous nuclear ribonucleoprotein (hnRNP) A/B and A2/B1 (Carpenter et al., 2013). In addition to lncRNA-Cox2, lncRNA-EPS was also recently identified as a key regulator in controlling macrophage inflammatory responses. Mice with lncRNA-EPS deficiency exhibited enhanced inflammation and lethality upon LPS challenges. Mechanistically, lncRNA-EPS was shown to limit the expression of immune response genes (IRGs) by controlling nucleosome positioning through interacting with hnRNP L (Atianand et al., 2016).
Finally, the link between lncRNA and human autoimmune diseases has also been demonstrated. Genome-wide association studies (GWAS) of celiac disease patients identified 5 SNPs in a region encoding a lncRNA, lnc13. Biopsies from celiac disease patients appeared to have substantially lower amounts of lnc13 compared with healthy donors. Further studies have shown that lnc13 is primarily expressed in the nucleus of human macrophages from the lamina propria. Like the aforementioned lncRNAs, lnc13 was also shown to repress many inflammatory genes through interaction with a hnRNP, hnRNP D in particular, as well as Hdac1 and chromatin. It was thus suggested that decreased levels of lnc13 in intestinal tissue from patients with celiac disease likely contributes to the observed inflammation in this autoimmune disorder (Castellanos-Rubio et al., 2016). Despite the great efforts made to study lncRNA biology, unlike miRNAs, it remains a major challenge to functionally evaluate a lncRNA as the sequence of the transcript lends no insight into how it may actually work within a given cell type. Nevertheless, it is evident that lncRNAs exhibit important regulatory functions in controlling immune responses as well as other biological processes.
6. Concluding remarks
Over a decade of intense scrutiny into the role of miRNAs in the immune system, there is little doubt that miRNAs function as crucial gene modulators that would impact almost all facets of immune responses in both physiological and pathological settings. While miRNAs do not completely turn off (or in some cases, turn on) the expression of their targets, they can act by repressing genes involved in positive-feedback regulatory circuits or by regulating a set of genes that are in a shared pathway or protein complex. As such, even relatively small changes in gene expression introduced by miRNAs could cause major biological consequences. From impairment of immune functions to the pathogenesis of a variety of immunological diseases, the fact that dysregulation of individual miRNAs in the immune system has repeatedly been demonstrated to have profound physiological effects further supports this notion. Moreover, beyond gaining further molecular insights into miRNA-mediated gene regulation in immunological research, recent advances in modulating miRNA function by miRNA mimics or antisense oligonucleotides have shown promise in miRNA-targeted therapeutics. With increasing knowledge of miRNA biology and the development of novel approaches for efficient delivery of miRNA modifying agents to specific immune cell subsets, we are confident that manipulating miRNA pathways will soon become a viable option to treat a wide array of human immunological diseases.
References
- AMERES SL & ZAMORE PD 2013. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol, 14, 475–88. [DOI] [PubMed] [Google Scholar]
- ANDROULIDAKI A, ILIOPOULOS D, ARRANZ A, DOXAKI C, SCHWORER S, ZACHARIOUDAKI V, MARGIORIS AN, TSICHLIS PN & TSATSANIS C 2009. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity, 31, 220–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATIANAND MK, HU W, SATPATHY AT, SHEN Y, RICCI EP, ALVAREZ-DOMINGUEZ JR, BHATTA A, SCHATTGEN SA, MCGOWAN JD, BLIN J, BRAUN JE, GANDHI P, MOORE MJ, CHANG HY, LODISH HF, CAFFREY DR & FITZGERALD KA 2016. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell, 165, 1672–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAULCOMBE D 2004. RNA silencing in plants. Nature, 431, 356–63. [DOI] [PubMed] [Google Scholar]
- BERNSTEIN E, CAUDY AA, HAMMOND SM & HANNON GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–6. [DOI] [PubMed] [Google Scholar]
- BOLDIN MP, TAGANOV KD, RAO DS, YANG L, ZHAO JL, KALWANI M, GARCIA-FLORES Y, LUONG M, DEVREKANLI A, XU J, SUN G, TAY J, LINSLEY PS & BALTIMORE D 2011. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med, 208, 1189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAIN O, OWENS BM, PICHULIK T, ALLAN P, KHATAMZAS E, LESLIE A, STEEVELS T, SHARMA S, MAYER A, CATUNEANU AM, MORTON V, SUN MY, JEWELL D, COCCIA M, HARRISON O, MALOY K, SCHONEFELDT S, BORNSCHEIN S, LISTON A & SIMMONS A 2013. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity, 39, 521–36. [DOI] [PubMed] [Google Scholar]
- BRANNAN CI, DEES EC, INGRAM RS & TILGHMAN SM 1990. The product of the H19 gene may function as an RNA. Mol Cell Biol, 10, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRONEVETSKY Y, VILLARINO AV, EISLEY CJ, BARBEAU R, BARCZAK AJ, HEINZ GA, KREMMER E, HEISSMEYER V, MCMANUS MT, ERLE DJ, RAO A & ANSEL KM 2013. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med, 210, 417–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALIN GA, DUMITRU CD, SHIMIZU M, BICHI R, ZUPO S, NOCH E, ALDLER H, RATTAN S, KEATING M, RAI K, RASSENTI L, KIPPS T, NEGRINI M, BULLRICH F & CROCE CM 2002. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A, 99, 15524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARPENTER S, AIELLO D, ATIANAND MK, RICCI EP, GANDHI P, HALL LL, BYRON M, MONKS B, HENRY-BEZY M, LAWRENCE JB, O’NEILL LA, MOORE MJ, CAFFREY DR & FITZGERALD KA 2013. A long noncoding RNA mediates both activation and repression of immune response genes. Science, 341, 789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASTELLANOS-RUBIO A, FERNANDEZ-JIMENEZ N, KRATCHMAROV R, LUO X, BHAGAT G, GREEN PH, SCHNEIDER R, KILEDJIAN M, BILBAO JR & GHOSH S 2016. A long noncoding RNA associated with susceptibility to celiac disease. Science, 352, 91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CEPPI M, PEREIRA PM, DUNAND-SAUTHIER I, BARRAS E, REITH W, SANTOS MA & PIERRE P 2009. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A, 106, 2735–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHO S, WU CJ, YASUDA T, CRUZ LO, KHAN AA, LIN LL, NGUYEN DT, MILLER M, LEE HM, KUO ML, BROIDE DH, RAJEWSKY K, RUDENSKY AY & LU LF 2016. miR-23 approximately 27 approximately 24 clusters control effector T cell differentiation and function. J Exp Med, 213, 235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHONG MM, RASMUSSEN JP, RUDENSKY AY & LITTMAN DR 2008. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med, 205, 2005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIMMINO A, CALIN GA, FABBRI M, IORIO MV, FERRACIN M, SHIMIZU M, WOJCIK SE, AQEILAN RI, ZUPO S, DONO M, RASSENTI L, ALDER H, VOLINIA S, LIU CG, KIPPS TJ, NEGRINI M & CROCE CM 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A, 102, 13944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COBB BS, HERTWECK A, SMITH J, O’CONNOR E, GRAF D, COOK T, SMALE ST, SAKAGUCHI S, LIVESEY FJ, FISHER AG & MERKENSCHLAGER M 2006. A role for Dicer in immune regulation. J Exp Med, 203, 2519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COBB BS, NESTEROVA TB, THOMPSON E, HERTWECK A, O’CONNOR E, GODWIN J, WILSON CB, BROCKDORFF N, FISHER AG, SMALE ST & MERKENSCHLAGER M 2005. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med, 201, 1367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRUZ LO, HASHEMIFAR SS, WU CJ, CHO S, NGUYEN DT, LIN LL, KHAN AA & LU LF 2017. Excessive expression of miR-27 impairs Treg-mediated immunological tolerance. J Clin Invest, 127, 530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENLI AM, TOPS BB, PLASTERK RH, KETTING RF & HANNON GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature, 432, 231–5. [DOI] [PubMed] [Google Scholar]
- DORSETT Y, MCBRIDE KM, JANKOVIC M, GAZUMYAN A, THAI TH, ROBBIANI DF, DI VIRGILIO M, REINA SAN-MARTIN B, HEIDKAMP G, SCHWICKERT TA, EISENREICH T, RAJEWSKY K & NUSSENZWEIG MC 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity, 28, 630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DU C, LIU C, KANG J, ZHAO G, YE Z, HUANG S, LI Z, WU Z & PEI G 2009. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol, 10, 1252–9. [DOI] [PubMed] [Google Scholar]
- EIS PS, TAM W, SUN L, CHADBURN A, LI Z, GOMEZ MF, LUND E & DAHLBERG JE 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A, 102, 3627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARZON R, HEAPHY CE, HAVELANGE V, FABBRI M, VOLINIA S, TSAO T, ZANESI N, KORNBLAU SM, MARCUCCI G, CALIN GA, ANDREEFF M & CROCE CM 2009. MicroRNA 29b functions in acute myeloid leukemia. Blood, 114, 5331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREGORY RI, YAN KP, AMUTHAN G, CHENDRIMADA T, DORATOTAJ B, COOCH N & SHIEKHATTAR R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature, 432, 235–40. [DOI] [PubMed] [Google Scholar]
- GUERAU-DE-ARELLANO M, SMITH KM, GODLEWSKI J, LIU Y, WINGER R, LAWLER SE, WHITACRE CC, RACKE MK & LOVETT-RACKE AE 2011. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain, 134, 3578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUTTMAN M, AMIT I, GARBER M, FRENCH C, LIN MF, FELDSER D, HUARTE M, ZUK O, CAREY BW, CASSADY JP, CABILI MN, JAENISCH R, MIKKELSEN TS, JACKS T, HACOHEN N, BERNSTEIN BE, KELLIS M, REGEV A, RINN JL & LANDER ES 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature, 458, 223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUTTMAN M & RINN JL 2012. Modular regulatory principles of large non-coding RNAs. Nature, 482, 339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN J, LEE Y, YEOM KH, NAM JW, HEO I, RHEE JK, SOHN SY, CHO Y, ZHANG BT & KIM VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell, 125, 887–901. [DOI] [PubMed] [Google Scholar]
- HE L, THOMSON JM, HEMANN MT, HERNANDO-MONGE E, MU D, GOODSON S, POWERS S, CORDON-CARDO C, LOWE SW, HANNON GJ & HAMMOND SM 2005. A microRNA polycistron as a potential human oncogene. Nature, 435, 828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU J, WANG P, LIN L, LIU X, MA F, AN H, WANG Z & CAO X 2009. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol, 183, 2150–8. [DOI] [PubMed] [Google Scholar]
- HUANG W, THOMAS B, FLYNN RA, GAVZY SJ, WU L, KIM SV, HALL JA, MIRALDI ER, NG CP, RIGO F, MEADOWS S, MONTOYA NR, HERRERA NG, DOMINGOS AI, RASTINEJAD F, MYERS RM, FULLER-PACE FV, BONNEAU R, CHANG HY, ACUTO O & LITTMAN DR 2015. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature, 528, 517–22. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- HUTVAGNER G, MCLACHLAN J, PASQUINELLI AE, BALINT E, TUSCHL T & ZAMORE PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–8. [DOI] [PubMed] [Google Scholar]
- HUTVAGNER G & ZAMORE PD 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–60. [DOI] [PubMed] [Google Scholar]
- INOMATA M, TAGAWA H, GUO YM, KAMEOKA Y, TAKAHASHI N & SAWADA K 2009. MicroRNA-17–92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood, 113, 396–402. [DOI] [PubMed] [Google Scholar]
- IYER MK, NIKNAFS YS, MALIK R, SINGHAL U, SAHU A, HOSONO Y, BARRETTE TR, PRENSNER JR, EVANS JR, ZHAO S, POLIAKOV A, CAO X, DHANASEKARAN SM, WU YM, ROBINSON DR, BEER DG, FENG FY, IYER HK & CHINNAIYAN AM 2015. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet, 47, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JING Q, HUANG S, GUTH S, ZARUBIN T, MOTOYAMA A, CHEN J, DI PADOVA F, LIN SC, GRAM H & HAN J 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell, 120, 623–34. [DOI] [PubMed] [Google Scholar]
- JOHNNIDIS JB, HARRIS MH, WHEELER RT, STEHLING-SUN S, LAM MH, KIRAK O, BRUMMELKAMP TR, FLEMING MD & CAMARGO FD 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature, 451, 1125–9. [DOI] [PubMed] [Google Scholar]
- KIM TD, LEE SU, YUN S, SUN HN, LEE SH, KIM JW, KIM HM, PARK SK, LEE CW, YOON SR, GREENBERG PD & CHOI I 2011. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood, 118, 5476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLUIVER J, POPPEMA S, DE JONG D, BLOKZIJL T, HARMS G, JACOBS S, KROESEN BJ & VAN DEN BERG A 2005. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol, 207, 243–9. [DOI] [PubMed] [Google Scholar]
- KORALOV SB, MULJO SA, GALLER GR, KREK A, CHAKRABORTY T, KANELLOPOULOU C, JENSEN K, COBB BS, MERKENSCHLAGER M, RAJEWSKY N & RAJEWSKY K 2008. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell, 132, 860–74. [DOI] [PubMed] [Google Scholar]
- KRAMER NJ, WANG WL, REYES EY, KUMAR B, CHEN CC, RAMAKRISHNA C, CANTIN EM, VONDERFECHT SL, TAGANOV KD, CHAU N & BOLDIN MP 2015. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood, 125, 3720–30. [DOI] [PubMed] [Google Scholar]
- KUCHEN S, RESCH W, YAMANE A, KUO N, LI Z, CHAKRABORTY T, WEI L, LAURENCE A, YASUDA T, PENG S, HU-LI J, LU K, DUBOIS W, KITAMURA Y, CHARLES N, SUN HW, MULJO S, SCHWARTZBERG PL, PAUL WE, O’SHEA J, RAJEWSKY K & CASELLAS R 2010. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity, 32, 828–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LASHINE YA, SALAH S, ABOELENEIN HR & ABDELAZIZ AI 2015. Correcting the expression of miRNA-155 represses PP2Ac and enhances the release of IL-2 in PBMCs of juvenile SLE patients. Lupus, 24, 240–7. [DOI] [PubMed] [Google Scholar]
- LEE RC, FEINBAUM RL & AMBROS V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75, 843–54. [DOI] [PubMed] [Google Scholar]
- LEE Y, AHN C, HAN J, CHOI H, KIM J, YIM J, LEE J, PROVOST P, RADMARK O, KIM S & KIM VN 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature, 425, 415–9. [DOI] [PubMed] [Google Scholar]
- LI J, WAN Y, GUO Q, ZOU L, ZHANG J, FANG Y, ZHANG J, ZHANG J, FU X, LIU H, LU L & WU Y 2010. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther, 12, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINGEL A, SIMON B, IZAURRALDE E & SATTLER M 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature, 426, 465–9. [DOI] [PubMed] [Google Scholar]
- LISTON A, LU LF, O’CARROLL D, TARAKHOVSKY A & RUDENSKY AY 2008. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med, 205, 1993–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU J, CARMELL MA, RIVAS FV, MARSDEN CG, THOMSON JM, SONG JJ, HAMMOND SM, JOSHUA-TOR L & HANNON GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science, 305, 1437–41. [DOI] [PubMed] [Google Scholar]
- LU LF, BOLDIN MP, CHAUDHRY A, LIN LL, TAGANOV KD, HANADA T, YOSHIMURA A, BALTIMORE D & RUDENSKY AY 2010. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell, 142, 914–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU MC, YU CL, CHEN HC, YU HC, HUANG HB & LAI NS 2014. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol, 177, 641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUND E, GUTTINGER S, CALADO A, DAHLBERG JE & KUTAY U 2004. Nuclear export of microRNA precursors. Science, 303, 95–8. [DOI] [PubMed] [Google Scholar]
- LUO X, YANG W, YE DQ, CUI H, ZHANG Y, HIRANKARN N, QIAN X, TANG Y, LAU YL, DE VRIES N, TAK PP, TSAO BP & SHEN N 2011. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet, 7, e1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA F, XU S, LIU X, ZHANG Q, XU X, LIU M, HUA M, LI N, YAO H & CAO X 2011. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol, 12, 861–9. [DOI] [PubMed] [Google Scholar]
- MANSOUR MR, SANDA T, LAWTON LN, LI X, KRESLAVSKY T, NOVINA CD, BRAND M, GUTIERREZ A, KELLIHER MA, JAMIESON CH, VON BOEHMER H, YOUNG RA & LOOK AT 2013. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med, 210, 1545–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEHTA A & BALTIMORE D 2016. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol, 16, 279–94. [DOI] [PubMed] [Google Scholar]
- MEISGEN F, XU LANDEN N, WANG A, RETHI B, BOUEZ C, ZUCCOLO M, GUENICHE A, STAHLE M, SONKOLY E, BRETON L & PIVARCSI A 2014. MiR-146a negatively regulates TLR2-induced inflammatory responses in keratinocytes. J Invest Dermatol, 134, 1931–40. [DOI] [PubMed] [Google Scholar]
- METZLER M, WILDA M, BUSCH K, VIEHMANN S & BORKHARDT A 2004. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer, 39, 167–9. [DOI] [PubMed] [Google Scholar]
- MOK Y, SCHWIERZECK V, THOMAS DC, VIGORITO E, RAYNER TF, JARVIS LB, PROSSER HM, BRADLEY A, WITHERS DR, MARTENSSON IL, CORCORAN LM, BLENKIRON C, MISKA EA, LYONS PA & SMITH KG 2013. MiR-210 is induced by Oct-2, regulates B cells, and inhibits autoantibody production. J Immunol, 191, 3037–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MULJO SA, ANSEL KM, KANELLOPOULOU C, LIVINGSTON DM, RAO A & RAJEWSKY K 2005. Aberrant T cell differentiation in the absence of Dicer. J Exp Med, 202, 261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’CONNELL RM, CHAUDHURI AA, RAO DS & BALTIMORE D 2009. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A, 106, 7113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’CONNELL RM, TAGANOV KD, BOLDIN MP, CHENG G & BALTIMORE D 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A, 104, 1604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN W, ZHU S, YUAN M, CUI H, WANG L, LUO X, LI J, ZHOU H, TANG Y & SHEN N 2010. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol, 184, 6773–81. [DOI] [PubMed] [Google Scholar]
- PONTING CP, OLIVER PL & REIK W 2009. Evolution and functions of long noncoding RNAs. Cell, 136, 629–41. [DOI] [PubMed] [Google Scholar]
- PRATAMA A, SRIVASTAVA M, WILLIAMS NJ, PAPA I, LEE SK, DINH XT, HUTLOFF A, JORDAN MA, ZHAO JL, CASELLAS R, ATHANASOPOULOS V & VINUESA CG 2015. MicroRNA-146a regulates ICOS-ICOSL signalling to limit accumulation of T follicular helper cells and germinal centres. Nat Commun, 6, 6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUA HH, STEINER DF, PATEL S, GONZALEZ JR, ORTIZ-CARPENA JF, KAGEYAMA R, CHIOU NT, GALLMAN A, DE KOUCHKOVSKY D, JEKER LT, MCMANUS MT, ERLE DJ & ANSEL KM 2016. MicroRNAs 24 and 27 Suppress Allergic Inflammation and Target a Network of Regulators of T Helper 2 Cell-Associated Cytokine Production. Immunity, 44, 821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RODRIGUEZ A, VIGORITO E, CLARE S, WARREN MV, COUTTET P, SOOND DR, VAN DONGEN S, GROCOCK RJ, DAS PP, MISKA EA, VETRIE D, OKKENHAUG K, ENRIGHT AJ, DOUGAN G, TURNER M & BRADLEY A 2007. Requirement of bic/microRNA-155 for normal immune function. Science, 316, 608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHNEIDER C, SETTY M, HOLMES AB, MAUTE RL, LESLIE CS, MUSSOLIN L, ROSOLEN A, DALLA-FAVERA R & BASSO K 2014. MicroRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc Natl Acad Sci U S A, 111, 8185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEBASTIANI G, GRIECO FA, SPAGNUOLO I, GALLERI L, CATALDO D & DOTTA F 2011. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev, 27, 862–6. [DOI] [PubMed] [Google Scholar]
- SEVERIN ME, LEE PW, LIU Y, SELHORST AJ, GORMLEY MG, PEI W, YANG Y, GUERAU-DE-ARELLANO M, RACKE MK & LOVETT-RACKE AE 2016. MicroRNAs targeting TGFbeta signalling underlie the regulatory T cell defect in multiple sclerosis. Brain, 139, 1747–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARMA S, FINDLAY GM, BANDUKWALA HS, OBERDOERFFER S, BAUST B, LI Z, SCHMIDT V, HOGAN PG, SACKS DB & RAO A 2011. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A, 108, 11381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEEDY FJ, PALSSON-MCDERMOTT E, HENNESSY EJ, MARTIN C, O’LEARY JJ, RUAN Q, JOHNSON DS, CHEN Y & O’NEILL LA 2010. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol, 11, 141–7. [DOI] [PubMed] [Google Scholar]
- SIMPSON LJ, PATEL S, BHAKTA NR, CHOY DF, BRIGHTBILL HD, REN X, WANG Y, PUA HH, BAUMJOHANN D, MONTOYA MM, PANDURO M, REMEDIOS KA, HUANG X, FAHY JV, ARRON JR, WOODRUFF PG & ANSEL KM 2014. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol, 15, 1162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SO AY, SOOKRAM R, CHAUDHURI AA, MINISANDRAM A, CHENG D, XIE C, LIM EL, FLORES YG, JIANG S, KIM JT, KEOWN C, RAMAKRISHNAN P & BALTIMORE D 2014. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood, 124, 1502–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONG JJ, SMITH SK, HANNON GJ & JOSHUA-TOR L 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science, 305, 1434–7. [DOI] [PubMed] [Google Scholar]
- SONKOLY E, WEI T, JANSON PC, SAAF A, LUNDEBERG L, TENGVALL-LINDER M, NORSTEDT G, ALENIUS H, HOMEY B, SCHEYNIUS A, STAHLE M & PIVARCSI A 2007. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One, 2, e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANCZYK J, PEDRIOLI DM, BRENTANO F, SANCHEZ-PERNAUTE O, KOLLING C, GAY RE, DETMAR M, GAY S & KYBURZ D 2008. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum, 58, 1001–9. [DOI] [PubMed] [Google Scholar]
- STEINER DF, THOMAS MF, HU JK, YANG Z, BABIARZ JE, ALLEN CD, MATLOUBIAN M, BLELLOCH R & ANSEL KM 2011. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity, 35, 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAGANOV KD, BOLDIN MP, CHANG KJ & BALTIMORE D 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A, 103, 12481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAGAWA H & SETO M 2005. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia, 19, 2013–6. [DOI] [PubMed] [Google Scholar]
- TANG Y, LUO X, CUI H, NI X, YUAN M, GUO Y, HUANG X, ZHOU H, DE VRIES N, TAK PP, CHEN S & SHEN N 2009. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum, 60, 1065–75. [DOI] [PubMed] [Google Scholar]
- TENG G, HAKIMPOUR P, LANDGRAF P, RICE A, TUSCHL T, CASELLAS R & PAPAVASILIOU FN 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity, 28, 621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DEN BERG A, KROESEN BJ, KOOISTRA K, DE JONG D, BRIGGS J, BLOKZIJL T, JACOBS S, KLUIVER J, DIEPSTRA A, MAGGIO E & POPPEMA S 2003. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer, 37, 20–8. [DOI] [PubMed] [Google Scholar]
- WANG H, FLACH H, ONIZAWA M, WEI L, MCMANUS MT & WEISS A 2014. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol, 15, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG P, GU Y, ZHANG Q, HAN Y, HOU J, LIN L, WU C, BAO Y, SU X, JIANG M, WANG Q, LI N & CAO X 2012. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J Immunol, 189, 211–21. [DOI] [PubMed] [Google Scholar]
- WANG Y, ZHONG H, XIE X, CHEN CY, HUANG D, SHEN L, ZHANG H, CHEN ZW & ZENG G 2015. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A, 112, E3883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIGHTMAN B, HA I & RUVKUN G 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell, 75, 855–62. [DOI] [PubMed] [Google Scholar]
- WILLINGHAM AT, ORTH AP, BATALOV S, PETERS EC, WEN BG, AZA-BLANC P, HOGENESCH JB & SCHULTZ PG 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science, 309, 1570–3. [DOI] [PubMed] [Google Scholar]
- XIAO C, CALADO DP, GALLER G, THAI TH, PATTERSON HC, WANG J, RAJEWSKY N, BENDER TP & RAJEWSKY K 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell, 131, 146–59. [DOI] [PubMed] [Google Scholar]
- XU S, GUO K, ZENG Q, HUO J & LAM KP 2012. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood, 119, 767–76. [DOI] [PubMed] [Google Scholar]
- YAMANAKA Y, TAGAWA H, TAKAHASHI N, WATANABE A, GUO YM, IWAMOTO K, YAMASHITA J, SAITOH H, KAMEOKA Y, SHIMIZU N, ICHINOHASAMA R & SAWADA K 2009. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood, 114, 3265–75. [DOI] [PubMed] [Google Scholar]
- YI R, QIN Y, MACARA IG & CULLEN BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev, 17, 3011–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG B, LIU SQ, LI C, LYKKEN E, JIANG S, WONG E, GONG Z, TAO Z, ZHU B, WAN Y & LI QJ 2016. MicroRNA-23a Curbs Necrosis during Early T Cell Activation by Enforcing Intracellular Reactive Oxygen Species Equilibrium. Immunity, 44, 568–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO X, TANG Y, QU B, CUI H, WANG S, WANG L, LUO X, HUANG X, LI J, CHEN S & SHEN N 2010. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum, 62, 3425–35. [DOI] [PubMed] [Google Scholar]
- ZHOU X, JEKER LT, FIFE BT, ZHU S, ANDERSON MS, MCMANUS MT & BLUESTONE JA 2008. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med, 205, 1983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU E, WANG X, ZHENG B, WANG Q, HAO J, CHEN S, ZHAO Q, ZHAO L, WU Z & YIN Z 2014. miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORgammat and STAT3. J Immunol, 192, 5599–609. [DOI] [PubMed] [Google Scholar]


