Abstract
Vibrio cholerae O139 Bengal initially appeared in the southern coastal region of Bangladesh and spread northward, causing explosive epidemics during 1992 and 1993. The resurgence of V. cholerae O139 during 1995 after its transient displacement by a new clone of El Tor vibrios demonstrated rapid changes in the epidemiology of cholera in Bangladesh. A recent outbreak of cholera in two north-central districts of Bangladesh caused by V. cholerae O139 led us to analyze strains collected from the outbreak and compare them with V. cholerae O139 strains isolated from other regions of Bangladesh and neighboring India to investigate their origins. Analysis of restriction fragment length polymorphisms in genes for conserved rRNA (ribotype) revealed that the recently isolated V. cholerae O139 strains belonged to a new ribotype which was distinct from previously described ribotypes of toxigenic V. cholerae O139. All strains carried the genes for toxin-coregulated pili (tcpA and tcpI) and accessory colonization factor (acfB), the regulatory gene toxR, and multiple copies of the lysogenic phage genome encoding cholera toxin (CTXΦ) and belonged to a previously described ctxA genotype. Comparative analysis of the rfb gene cluster by PCR revealed the absence of a large region of the O1-specific rfb operon downstream of the rfaD gene and the presence of an O139-specific genomic region in all O139 strains. Southern hybridization analysis of the O139-specific genomic region also produced identical restriction patterns in strains belonging to the new ribotype and those of previously described ribotypes. These results suggested that the new ribotype of Bengal vibrios possibly originated from an existing strain of V. cholerae O139 by genetic changes in the rRNA operons. In contrast to previously isolated O139 strains which mostly had resistance to trimethoprim, sulfamethoxazole, and streptomycin encoded by a transposon (SXT element), 68.6% of the toxigenic strains analyzed in the present study, including all strains belonging to the new ribotype, were susceptible to these antibiotics. Molecular analysis of the SXT element revealed possible deletion of a 3.6-kb region of the SXT element in strains which were susceptible to the antibiotics. Thus, V. cholerae O139 strains in Bangladesh are also undergoing considerable reassortments in genetic elements encoding antimicrobial resistance.
Vibrio cholerae O139 Bengal emerged as a second etiologic agent of cholera in 1992 and caused explosive epidemics throughout Bangladesh, India, and neighboring countries (4, 25, 26). In Bangladesh, this new serogroup of epidemic V. cholerae was first detected in the southern coastal districts and offshore islands (29). The spread of the epidemic initially remained largely confined to the coastal districts, where the aquatic environment is saline and typical of the Bay of Bengal. Subsequently, V. cholerae O139 spread to the northeastern and north-central regions of the country and caused outbreaks of cholera (29). However, during 1994 and until the middle of 1995, in most northern and central areas of Bangladesh, including the capital city Dhaka, the O139 vibrios were replaced by a new clone of V. cholerae O1 of the El Tor biotype, whereas in the southern coastal regions V. cholerae O139 continued to exist (7, 9, 29). During the second half of 1995 and in 1996, nearly 4 years after the initial detection of O139 vibrios, cases due to both V. cholerae O1 and V. cholerae O139 were detected in various regions of Bangladesh. Analysis of these isolates revealed the emergence of a new clone of V. cholerae O139, and epidemiological assessment suggested that the new clone probably originated in the northern region of the country and spread toward the south (9). Recent surveillance by the Epidemic Control Preparedness Program of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B), during November 1997 revealed an outbreak of cholera due to V. cholerae O139 in Mymensingh and Kishoreganj, two rural districts of Bangladesh situated north of Dhaka. A preliminary estimate revealed that over 50,000 cases of cholera and at least 34 deaths occurred during this outbreak (28a). Details of the surveillance will be published elsewhere. In the present study, we used molecular techniques to characterize V. cholerae O139 strains isolated from the recent outbreak and compared these with V. cholerae O139 strains isolated from other parts of Bangladesh and neighboring countries between 1995 and 1998 to study clonal relationships and understand the origins of these recently emerged epidemic strains.
MATERIALS AND METHODS
V. cholerae strains.
A total of 68 clinical isolates of V. cholerae O139 were included in this study. Strains isolated in Bangladesh consisted of 19 isolates from the cholera outbreak in 1997 in two north-central districts of Bangladesh and 39 strains isolated in other regions of Bangladesh between 1995 and 1998. Other strains consisted of six strains isolated in India in 1997 (courtesy of G. B. Nair, National Institute of Cholera and Enteric Diseases, Calcutta), three strains isolated in Thailand (Armed Forces Research Institute of Medical Sciences, Bangkok), and a single nontoxigenic strain isolated in Argentina in 1993. Strains were stored either in a lyophilized form or in sealed deep nutrient agar at room temperature at the culture collection of the ICDDR,B. Before use, the identities of the cultures were confirmed by biochemical reaction and serology (36). Details of the strains are presented in Table 1.
TABLE 1.
Analysis of genes for rRNA, CT (ctxA), and the SXT element in clinical strains of toxigenic V. cholerae O139 isolated between 1995 and 1998a
| Yr of isolation | Place of isolation | No. of isolates | rRNA gene restriction pattern | ctxA genotype | SXT genotype | Antimicrobial resistance patternb |
|---|---|---|---|---|---|---|
| 1995 | Matlab, Bangladesh | 5 | I | A | 1 | Sr SXTr |
| 1995 | Matlab, Bangladesh | 6 | II | A | 1 | Sr SXTr |
| 1997 | Matlab, Bangladesh | 5 | II | B | 2 | Susceptible |
| 1996 | Jhalokati, Bangladesh | 2 | II | B | 2 | Susceptible |
| 1995 | Dhaka, Bangladesh | 5 | II | A | 1 | Sr SXTr |
| 1996 | Dhaka, Bangladesh | 7 | II | B | 2 | Susceptible |
| 1996 | Sunamganj, Bangladesh | 2 | II | B | 1 | Sr SXTr |
| 1997 | Bakerganj, Bangladesh | 1 | II | B | 2 | Susceptible |
| 1998 | Bakerganj, Bangladesh | 6 | II | B | 2 | Susceptible |
| 1997 | Mymensingh, Bangladesh | 8 | III | B | 2 | Susceptible |
| 1997 | Kishoreganj, Bangladesh | 11 | III | B | 2 | Susceptible |
| 1997 | Bangkok, Thailand | 3 | II | B | 1 | Sr SXTr |
| 1997 | Calcutta, India | 6 | II | A | 2 | Susceptible |
All strains were negative in PCR assays for chromosomal regions representing rfbDEG, rfbNO, ompX, orf2, and orf3 of the rfb gene cluster of V. cholerae O1. Ribotypes, ctxA genotypes, and SXT genotypes are based on BglI restriction patterns of the respective genes and their flanking chromosomal sequences.
Strains designated “Susceptible” were susceptible to tetracycline, ampicillin, chloramphenicol, gentamicin, ciprofloxacin, norfloxacin, nalidixic acid, streptomycin, and SXT. Sr, streptomycin resistant; SXTr, SXT resistant.
Probes and hybridization.
Colony blots were prepared with nylon filters (Hybond; Amersham International PLC, Ayelesbury, United Kingdom) and processed by a standard method (16). Briefly, colonies were lysed with denaturing solution (0.5 M NaOH, 1.5 M NaCl) and neutralized in neutralizing solution (0.5 M Tris-HCl [pH 8.0], 1.5 M NaCl), and the liberated DNA was fixed to the nylon membrane by exposure to UV light for 3 min in accordance with the supplier’s instructions. For preparation of DNA blots, total cellular DNA was isolated from overnight cultures as described previously (32). Five-microgram aliquots of DNA were digested with the appropriate restriction enzymes (Bethesda Research Laboratories [BRL], Gaithersburg, Md.), electrophoresed in 0.8% agarose gels, and blotted onto nylon membranes (Hybond; Amersham) by Southern blotting (30).
The gene probe used in this study to detect the CTX genetic element was a 0.5-kb EcoRI fragment of pCVD27 (12) containing part of the ctxA gene. The toxR gene probe was a 2.4-kb BamHI fragment of pVM7 (18). The rRNA gene probe consisted of a 7.5-kb BamHI fragment of the Escherichia coli rRNA clone pKK3535 (3, 32). The O139-specific DNA probe was a 1.3-kb EcoRI fragment of pCRII-A3 (21), and the SXT probe was a NotI fragment of pSXT1 (35). The probes were labeled by random priming (11) with a random primer DNA labeling kit (BRL) and [α-32P]dCTP (3,000 Ci/mmol) (Amersham). Southern blots and colony blots were hybridized with the labeled probes, and autoradiographs were developed as described by us previously (7–9).
PCR assays.
Presence of the toxin-coregulated pilus (TCP) pathogenicity island was determined by PCR assays specific for the tcpA, tcpI, and acfB genes. All oligonucleotides used either as probes or PCR primers were synthesized commercially by Oswel DNA Service (University of Edinburgh, Edinburgh, United Kingdom), and PCR reagents and kits were purchased from Perkin-Elmer Corporation (Norwalk, Conn.). Presence of tcpA genes specific for the classical and El Tor biotypes was determined by a multiplex PCR assay (14). The tcpI gene was detected by a specific PCR assay described by us previously (7). The acfB gene was detected by a PCR assay based on the published sequence of acfB (6) with two primers having the following sequences: 5′GGACCAAGCATTATTATCTCT and 5′AATGATAAACTTACTGATTAA. The PCR assay amplified a 1.9-kb region of the acfB gene. Amplification was performed for 25 cycles consisting of denaturation at 94°C for 2 min, annealing of primers at 50°C for 2 min, and primer extension at 72°C for 3 min.
PCR assays for defined regions of the rfb gene cluster and adjoining sequences were performed with six different sets of primers derived from the sequence of the V. cholerae O1 rfb gene cluster, as described by us previously (9). The expected sizes of the amplicons were ascertained by electrophoresis in agarose gels, and the identity of each PCR product was also verified by Southern blot hybridization. PCR-negative strains were further confirmed for the absence of the relevant genes by colony blot hybridization with the corresponding PCR-generated amplicons from a positive control strain, 569B, as specific probes.
Antimicrobial resistance.
All V. cholerae isolates were tested for antimicrobial resistance by the method of Bauer et al. (2) with standard antibiotic disks (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) at the following antibiotic concentrations (in micrograms per disk): ampicillin, 10; chloramphenicol, 30; streptomycin, 10; tetracycline, 30; trimethoprim-sulfamethoxazole (SXT), 1.25 and 23.75, respectively; kanamycin, 30; gentamicin, 10; ciprofloxacin, 5; norfloxacin, 10; nalidixic acid, 30.
RESULTS AND DISCUSSION
The epidemiology of cholera in the flood plains of the northern region of Bangladesh had previously been shown to differ considerably from that in the southern coastal region (29), probably due to differences in the water ecologies between the two regions. Soon after the emergence of V. cholerae O139, the existing strains of V. cholerae O1 were almost completely displaced and the O139 vibrios continued to dominate until the emergence of a new clone of El Tor vibrios (7). The new clone of El Tor vibrios displaced the O139 vibrios during 1994 and 1995 in the central and northern areas of Bangladesh, but the O139 vibrios continued to exist and became endemic in the southern coastal region, where this serogroup was initially detected in 1993 (4, 29). During 1995 and 1996, after the reemergence of V. cholerae O139, molecular epidemiological studies suggested that strains isolated from the southern regions were remnants of the initial clones whereas those isolated in the northern districts belonged to a new clone (9). The present study was designed to ascertain whether strains isolated from the recent outbreak in the two north-central districts of Bangladesh also represented a new clone of V. cholerae O139.
Ribotype analysis.
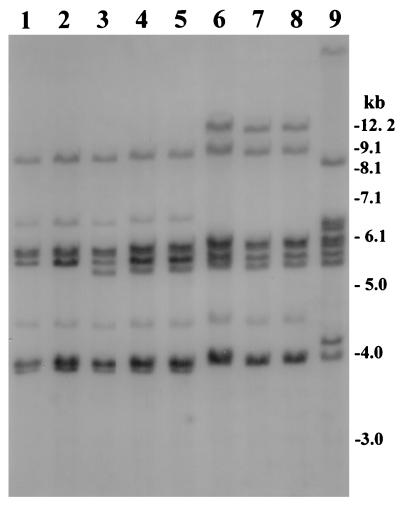
We have previously examined restriction patterns of conserved rRNA genes (ribotypes) and cholera toxin genes or chromosomal DNA flanking these genes to differentiate among clones of toxigenic V. cholerae which were otherwise phenotypically identical (7–9). These studies have also shown that BglI was the most discriminatory restriction enzyme used for ribotyping. In the present study, analysis of rRNA genes with BglI revealed three different restriction patterns (patterns I through III) among the toxigenic strains (Fig. 1). The nontoxigenic O139 strain from Argentina produced a restriction pattern which was widely different from those of the toxigenic strains described by us previously (9). All of the 19 strains isolated from the recent outbreak produced restriction pattern III (Fig. 1), which was different from the previously reported ribotype patterns of V. cholerae isolates belonging to the O139 or O1 serogroup (7–9, 23, 24). Strains isolated in India and Thailand analyzed in the present study produced either restriction pattern I or II (Table 1). Thus, V. cholerae O139 strains isolated from the recent epidemic in the north-central region of Bangladesh belonged to a new ribotype. The rRNA gene restriction patterns were reproducible in repeated assays and consisted of seven to eight major bands of between 12.5 and 3.9 kb in size. The restriction pattern representing the new ribotype contained a unique band of 12.5 kb which was not present in any of the other restriction patterns (Fig. 1). Hence, the O139 strains isolated from the recent epidemic were distinguishable from previously described O139 vibrios based on the ribotype pattern. The other two patterns detected among toxigenic O139 strains in the present study (patterns I and II) have been described previously (9), and these strains are likely to be remnants of the original clones of V. cholerae O139 which emerged during 1992 and 1993. However, another clone of toxigenic V. cholerae (9) which appeared transiently in 1996 was not found among the isolates analyzed in the present study. The detection of strains belonging to a new ribotype in the present study suggested that V. cholerae O139 strains are undergoing genetic changes leading to increased ribotype diversity. At present there is no standardized systematic nomenclature for designating ribotypes of V. cholerae O139 Bengal; hence, strains belonging to similar ribotypes have been designated differently in previous reports (8, 9, 24). With the prospect of emerging ribotype diversity among V. cholerae O139 isolates, it is important to establish a systematic nomenclature for identifying and monitoring the emergence and spread of epidemic O139 strains. We propose that the V. cholerae O139 Bengal strains producing the three different ribotype patterns, I through III (Fig. 1), be designated ribotypes B-I, B-II, and B-III (Table 1). Since previous studies have suggested that the nontoxigenic non-Bengal O139 strain (9) isolated in Argentina has an origin widely different from the epidemic Bengal strains, we propose the ribotype designation NB-1 for this strain. Analysis of a large number of V. cholerae O139 strains isolated in different countries is being carried out in our laboratory to further develop a standardized system for ribotype designations of toxigenic V. cholerae.
FIG. 1.
Southern hybridization analysis of rRNA genes in V. cholerae O139 strains isolated between 1995 and 1998 in Bangladesh, India, and Thailand. Genomic DNA was digested with BglI and probed with a 7.5-kb BamHI fragment of the E. coli rRNA clone pKK3535. Restriction pattern I, produced by two strains belonging to designated ribotype B-I isolated from Bangladesh and Thailand, respectively, are shown in lanes 1 and 2; restriction pattern II, produced by three strains belonging to ribotype B-II isolated in Bangladesh, India, and Thailand, respectively, are shown in lanes 3 through 5; and lanes 6 through 8 show restriction pattern III, produced by the new ribotype of V. cholerae O139 designated B-III, isolated from a recent outbreak in two north-central districts of Bangladesh. The pattern produced by the nontoxigenic O139 strain isolated in Argentina is shown in lane 9. Numbers indicating molecular sizes of bands correspond to a 1-kb DNA ladder (BRL) used as a molecular size marker.
Analysis of virulence genes.
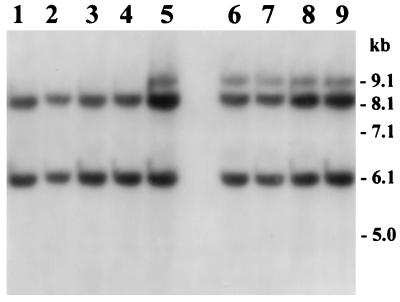
Previous studies have demonstrated that the rRNA gene restriction patterns (ribotypes) could be considered fairly stable markers for identifying different clones, and generally different ribotypes have also shown differences in the restriction patterns of several other genes studied (7, 9). We subjected the strains belonging to the new ribotype to further analysis to determine whether major pathogenic genes carried by strains of the new ribotype showed genetic dissimilarities with strains of the other ribotypes studied. The major colonization factor of toxigenic V. cholerae is TCP, the expression of which is under genetic control of the transcriptional activator ToxR and is coregulated with the expression of cholera toxin (CT) (18, 33). Although the major subunit of TCP is encoded by the tcpA gene, the formation and function of the pilus assembly requires the products of a number of other genes located on a large DNA region referred to as the TCP pathogenicity island, which includes the tcp and acf gene clusters (13, 15). Analysis of genes for the TCP pathogenicity island with PCR assays for the tcpA, tcpI, and acfB genes showed that all strains carried the TCP pathogenicity island. The PCR assay for tcpA amplified a 0.47-kb portion of the tcpA gene in all strains. The PCR assays for the tcpI and acfB genes produced amplicons of 2.1 and 1.9 kb, respectively, from all strains tested. Colony blot hybridization revealed that all strains in the present study also carried the toxR gene. Like previously described O139 Bengal vibrios, the tcpA amplicon derived from strains belonging to the new ribotype of V. cholerae O139 was identical to that produced by El Tor strains of V. cholerae (9). The ctxAB operon, which encodes the A and B subunits of CT, is part of a larger genetic element originally termed the CTX genetic element (22). Recent studies have shown that the CTX genetic element corresponds to the genome of CTXΦ, a lysogenic filamentous bacteriophage (34). In the present study, restriction fragment length polymorphism analysis of the ctxA gene and its flanking chromosomal sequence with the enzyme BglI revealed two different restriction patterns (A and B) for O139 strains. The ctxA restriction patterns consisted of either two or three bands between 9.1 and 6.1 kb (Fig. 2). Since the ctxA gene is known to have no internal BglI site (17), the number of bands represented the approximate number of copies of the integrated CTXΦ genome. Both of the ctxA restriction patterns produced by strains analyzed in the present study have been reported previously for V. cholerae O139 strains (9). All strains collected from the recent outbreak produced pattern B (Fig. 2) and carried three copies of the CTXΦ genome. We have previously proposed that origination of new toxigenic strains of V. cholerae is associated with the induction and propagation of CTXΦ (10). Diversification of the ctxA genotype among different strains may be a result of integration of CTXΦ into different regions of the host chromosome specified by the presence of a 17-bp attachment sequence referred to as attRS (34). Hence the new ribotype of V. cholerae O139 carries the attRS sequence at chromosomal sites similar to other previously described O139 vibrios. These results suggested that unlike the non-Bengal O139 strain from Argentina, which has been shown to have a non-O1 origin (9), the new ribotype of V. cholerae O139 is closely related other V. cholerae O139 Bengal vibrios.
FIG. 2.
Southern hybridization analysis of ctxA genes in V. cholerae O139 strains isolated from the epidemic in the north-central region of Bangladesh (lanes 6 through 9) in 1997 and from other districts between 1995 and 1998 (lanes 1 through 5). Genomic DNA was digested with BglI and probed with a 0.5-kb fragment of the ctxA gene. Restriction patterns corresponding to ctxA genotype A are shown in lanes 1 through 4, whereas those representing ctxA genotype B are shown in lanes 5 through 9. Numbers indicating molecular sizes of bands correspond to a 1-kb DNA ladder (BRL) used as a molecular size marker.
Antimicrobial resistance.
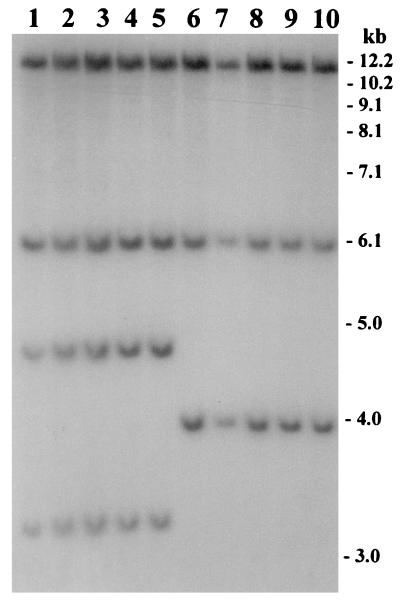
The O139 serogroup of V. cholerae which emerged during 1992 and 1993 was sensitive to tetracycline and showed a trend of increased resistance to SXT and streptomycin. Waldor and coworkers (35) reported the presence of a 62-kb self-transmissible transposon-like element (SXT element) encoding resistance to sulfamethoxazole, trimethoprim, and streptomycin in V. cholerae O139 strains isolated from this epidemic. The SXT element could be conjugally transferred from V. cholerae O139 to V. cholerae O1 and E. coli strains, where it integrated into recipient chromosomes in a site-specific recA-independent manner (35). In the present study, all strains of the new ribotype and 27 of 48 strains (56.25%) belonging to previously reported ribotypes were found to be sensitive to SXT and streptomycin (Table 1). All strains analyzed in the present study were also sensitive to tetracycline, ampicillin, chloramphenicol, gentamicin, ciprofloxacin, norfloxacin, and nalidixic acid. In keeping with the observations in Bangladesh, comparison of antibiotic resistance patterns between the O139 strains isolated during 1992 and 1993 and those isolated in 1996 and 1997 in India also showed that the latter strains were susceptible to SXT, unlike the O139 strains from 1992 and 1993 (19). To identify the genetic changes associated with the observed SXT sensitivity, we used a cloned SXT gene probe to study restriction fragment length polymorphisms in the SXT transposon. Two different BglI restriction patterns (patterns 1 and 2) of the SXT element were observed among the toxigenic O139 strains tested (Fig. 3). Strains producing pattern 2 were sensitive to SXT and streptomycin, whereas those producing pattern 1 were resistant to all three antibiotics. Further analysis of the restriction patterns suggested that the restriction site heterogeneity possibly occurred as a result of a deletion of an approximately 3.6-kb region of the SXT element in strains which were sensitive to SXT and streptomycin. Recent studies in India have also shown that O139 strains are becoming increasingly resistant to ampicillin and neomycin but increasingly susceptible to chloramphenicol and streptomycin (20). Considering the rapidly changing pattern of antibiotic resistance observed among V. cholerae O139, it appears that there is substantial mobility in other genetic elements encoding antimicrobial resistance in V. cholerae O139.
FIG. 3.
Analysis of the SXT element in V. cholerae O139 strains isolated between 1995 and 1998 in Bangladesh. Genomic DNA was digested with BglI and probed with the SXT gene probe. Lanes 1 through 5 show restriction patterns corresponding to SXT genotype 1, whereas lanes 6 through 10 represent SXT genotype 2. Lane numbers, place, and year of isolation of strains are as follows: 1 and 2, Matlab, 1995; 3 and 4, Dhaka, 1995; 5, Sunamganj, 1996; 6 and 7, Mymensingh, 1997; 8 and 9, Kishoreganj, 1997; 10, Bakerganj, 1998. Numbers indicating molecular sizes of bands correspond to a 1-kb DNA ladder (BRL).
Origin of the new ribotype of V. cholerae O139.
Previous reports on the comparative analysis of V. cholerae O139 and V. cholerae O1 suggested that strains belonging to the O139 serogroup may have emerged from a genetic change in the serotype-specific genes of a toxigenic El Tor strain. These studies indicated that O139 strains resulted from the progenitor El Tor strain due to insertion of a large foreign genomic region encoding O139-specific genes and simultaneous deletion of most of the O1-specific rfb gene cluster (5, 31). The donor for the O139-specific DNA in this horizontal gene transfer event, however, has not been identified. In agreement with previous reports, a large region of chromosomal DNA representing the O1-specific rfb gene cluster, including regions representing rfbDEG, rfbNO, ompX, orf2, and orf3, was found to be absent in all O139 strains analyzed in the present study. PCR-based comparative analysis of the O139 vibrios with specific primers corresponding to six defined regions of the rfb gene cluster and flanking sequences of V. cholerae O1 showed that while all six regions could be amplified from control El Tor strains, the O139 strains showed positive amplification in two of the six PCR assays. These two assays amplified a 497-bp region of the rfaD locus and a 394-bp region corresponding to the rfbQRS locus in all of the toxigenic O139 vibrios. All O139 strains were negative in the PCR assays for chromosomal regions representing rfbDEG, rfbNO, ompX, orf2, and orf3 of the rfb gene cluster of V. cholerae O1. Southern hybridization analysis of genomic DNA with the O139-specific probe also produced identical restriction patterns for all O139 vibrios examined (data not shown). In addition, strains of the new ribotype belonged to a ctxA genotype which was identical to that of previously described O139 strains. Contrary to this, however, the recently reemerged O139 strains in India have been reported to show an altered organization of the CTX element (28).
The present study demonstrates the emergence of strains belonging to a new ribotype which are otherwise identical to previously described clones of O139 vibrios. Considering the ctxA genotypes, the organization of the rfb genes, and the presence of other virulence genes tested, it appears that the new ribotype probably did not emerge from a recent gene transfer event comprising horizontal transfer of O139-specific genes from an unidentified donor; rather, it seems more likely that the new ribotype originated from an existing O139 strain. This agrees with a previous study by Ruiting and Reeves (27) suggesting that recombination between rRNA operons can give rise to ribotype diversity.
Epidemiological significance of ribotype diversity.
Previously recorded events of appearance of new ribotypes of toxigenic V. cholerae in Bangladesh showed that the new ribotypes were also distinct in their ctxA genotypes, suggesting that the new ribotypes possibly emerged from nontoxigenic progenitors (7, 9). This would allow more phenotypic and genetic diversity among toxigenic V. cholerae strains. The present study, on the other hand, shows that strains belonging to the new ribotype are otherwise genetically similar to previously described O139 strains. Hence, strains belonging to the new ribotype have epidemic or pandemic potential similar to previously described O139 vibrios. However, the new ribotype pattern of the strains has epidemiological application as a marker in monitoring the spread of these pathogenic strains. In the present study, all 19 strains isolated from the recent cholera outbreak belonged to the new ribotype. This suggests that the recent outbreak in northern Bangladesh probably started from a point source and coincided with the origination of this new ribotype. Analysis of strains isolated from other areas of Bangladesh during the same period showed that these strains belong to previously described ribotypes, and hence the strains associated with the outbreak were largely confined to the two north-central districts of the country. During 1996, a new ribotype of V. cholerae O139 was detected in Bangladesh and in the same geographical region (9). However, in the present study we did not find the presence of any strain belonging to this ribotype. This suggests that in this region of Bangladesh the O139 vibrios are undergoing rapid genetic reassortment, resulting in transient appearance of different clones, whereas in the southern coastal region the original ribotypes still exist as endemic strains. The reasons for the rapid and transient emergence of different ribotypes of O139 vibrios in the northern region of Bangladesh are not clear.
Interplay of a variety of factors in the aquatic environment and genetic and phenotypic changes in V. cholerae, as well as the immune status of the human host, may contribute to the existence and dominance of different clones of toxigenic V. cholerae. We have previously reported different phenotypic and genetic changes in V. cholerae O139 in Bangladesh and the prevalence of different clones (1, 8). Although in the present study we did not detect any change in the major pathogenic genes carried by the strains belonging to the new ribotype, we did not rule out the possibility of simultaneous genetic changes occurring in the rRNA operons and other unidentified genes that might influence the prevalence of the O139 strains by interacting with environmental factors. In view of the fluctuation observed in the prevalence of V. cholerae O139 relative to that of V. cholerae O1 in human infection (7, 9) and rapid genetic and phenotypic changes, including changing patterns of antibiotic resistance, further studies are required to explain the appearance and disappearance of cholera epidemics and the mobility of genetic elements encoding virulence properties as well as antimicrobial resistance.
ACKNOWLEDGMENTS
This research was funded by the U.S. National Institutes of Health under grant RO1 AI39129-01A1 to the Department of International Health, Johns Hopkins University, and the ICDDR,B. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries. Current donors providing core support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, China, Japan, Saudi Arabia, Sri Lanka, Sweden, Switzerland, the United Kingdom, and the United States; international organizations including the Arab Gulf Fund, the European Union, the United Nations Children’s Fund, the United Nations Development Programme, and the World Health Organization; private foundations including the Aga Khan Foundation, the Child Health Foundation, the Ford Foundation, the Population Council, the Rockefeller Foundation, the Thrasher Research Foundation, and the George Mason Foundation; and private organizations including the East West Center, Helen Keller International, the International Atomic Energy Agency, the International Center for Research on Women, the International Development Research Center, the International Life Sciences Institute, Karolinska Institute, the London School of Hygiene and Tropical Medicine, Lederle Praxis, the National Institute of Health, New England Medical Center, Procter & Gamble, RAND Corporation, the Social Development Center of Philippines, the Swiss Red Cross, Johns Hopkins University, the University of Alabama at Birmingham, the University of Iowa, the University of Goteborg, UCB Osmotics Ltd., Wander AG, and others.
We thank John Mekalanos, Harvard Medical School, Boston, Mass., for the ToxR clone and the V. cholerae O139-specific probe and Matthew Waldor, New England Medical Center, Boston, Mass., for the SXT clone.
REFERENCES
- 1.Albert M J, Bhuiyan N A, Talukder K A, Faruque A S G, Nahar S, Faruque S M, Ansaruzzaman M, Rahman M. Phenotypic and genetic changes in Vibrio cholerae O139 Bengal. J Clin Microbiol. 1997;35:2588–2592. doi: 10.1128/jcm.35.10.2588-2592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer A W, Kirby W M M, Sherris J C, Turk M. Antibiotic susceptibility by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 3.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 4.Cholera Working Group, International Centre for Diarrhoeal Disease Research, Bangladesh. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet. 1993;342:387–390. [PubMed] [Google Scholar]
- 5.Comstock L E, Maneval D, Jr, Panigrahi P, Joseph A, Levine M M, Kaper J B, Morris J G, Jr, Johnson J A. The capsule and O-antigen in Vibrio cholerae O139 Bengal are associated with a genetic region not present in Vibrio cholerae O1. Infect Immun. 1995;63:317–323. doi: 10.1128/iai.63.1.317-323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everiss K D, Hughes K J, Kovach M E, Peterson K M. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect Immun. 1994;62:3289–3298. doi: 10.1128/iai.62.8.3289-3298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque S M, Ahmed K M, Alim A R M A, Qadri F, Siddique A K, Albert M J. Emergence of a new clone of toxigenic Vibrio cholerae O1 biotype El Tor displacing V. cholerae O139 Bengal in Bangladesh. J Clin Microbiol. 1997;35:624–630. doi: 10.1128/jcm.35.3.624-630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque S M, Roy S K, Alim A R M A, Siddique A K, Albert M J. Molecular epidemiology of toxigenic Vibrio cholerae in Bangladesh studied by numerical analysis of rRNA gene restriction patterns. J Clin Microbiol. 1995;33:2833–2838. doi: 10.1128/jcm.33.11.2833-2838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque S M, Ahmed K M, Siddique A K, Zaman K, Alim A R M A, Albert M J. Molecular analysis of toxigenic Vibrio cholerae O139 Bengal strains isolated in Bangladesh between 1993 and 1996: evidence for the emergence of a new clone of the Bengal vibrios. J Clin Microbiol. 1997;35:2299–2306. doi: 10.1128/jcm.35.9.2299-2306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque S M, Asadulghani, Abdul Alim A R M, Albert M J, Islam K M N, Mekalanos J J. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg A, Volgelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 12.Kaper J B, Morris J G, Jr, Nishibuchi M. DNA probes for pathogenic Vibrio species. In: Tenover F C, editor. DNA probes for infectious disease. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 65–77. [Google Scholar]
- 13.Karaolis D K, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keasler S P, Hall R H. Detection and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 15.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Mekalanos J J, Swartz D J, Pearson G D N, Harford N, Groyne F, Wilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 18.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra R, Basu A, Dutta D, Nair G B, Takeda Y. Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet. 1996;348:1181. doi: 10.1016/s0140-6736(05)65326-3. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay A K, Basu A, Garg P, Bag P K, Ghosh A, Bhattacharya S K, Takeda Y, Nair G B. Molecular epidemiology of reemergent Vibrio cholerae O139 Bengal in India. J Clin Microbiol. 1998;36:2149–2152. doi: 10.1128/jcm.36.7.2149-2152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair G B, Bag P K, Shimada T, Ramamurthy T, Takeda T, Yamamoto S, Kurazono H, Takeda Y. Evaluation of DNA probes for specific detection of Vibrio cholerae O139 Bengal. J Clin Microbiol. 1995;33:2186–2187. doi: 10.1128/jcm.33.8.2186-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popovic T, Bopp C, Olsvic O, Wachsmuth K. Epidemiological application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popovic T, Fields P I, Olsvik O, Wells J G, Evins G M, Cameron D N, Farmer III J J, Bopp C A, Wachsmuth K, Sack R B, Albert M J, Nair G B, Shimada T, Feeley J C. Molecular subtyping of toxigenic Vibrio cholerae O139 causing epidemic cholera in India and Bangladesh, 1992–1993. J Infect Dis. 1995;171:122–127. doi: 10.1093/infdis/171.1.122. [DOI] [PubMed] [Google Scholar]
- 25.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. Emergence of a novel strain of Vibrio cholerae with epidemic potential in Southern and Eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 26.Rivas M, Toma C, Miliwebsky E, Caffer M I, Galas M, Varela P, Tous M, Bru A M, Binsztein N. Cholera isolates in relation to the “eighth pandemic.”. Lancet. 1993;342:926–927. [PubMed] [Google Scholar]
- 27.Ruiting L, Reeves P R. Recombination between rRNA operons created most of the ribotype variation observed in the seventh pandemic clone of Vibrio cholerae. Microbiology. 1998;144:1213–1221. doi: 10.1099/00221287-144-5-1213. [DOI] [PubMed] [Google Scholar]
- 28.Sharma C, Maiti S, Mukhopadhyay A K, Basu A, Basu I, Nair G B, Mukhopadhyaya R, Das B, Kar S, Ghosh R K, Ghosh A. Unique organization of the CTX genetic element in Vibrio cholerae O139 strains which reemerged in Calcutta, India, in September 1996. J Clin Microbiol. 1997;35:3348–3350. doi: 10.1128/jcm.35.12.3348-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Siddique, A. K. Unpublished data.
- 29.Siddique A K, Akram K, Zaman K, Mutsuddy P, Eusof A, Sack R B. Vibrio cholerae O139: how great is the threat of a pandemic? Trop Med Int Health. 1996;1:393–398. doi: 10.1046/j.1365-3156.1996.d01-54.x. [DOI] [PubMed] [Google Scholar]
- 30.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 31.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stull T L, LiPuma J J, Edlind T D. A broad spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988;157:280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- 33.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 35.Waldor M K, Tschape H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. World Health Organization guidelines for the laboratory diagnosis of cholera. Geneva, Switzerland: Bacterial Disease Unit, World Health Organization; 1974. [Google Scholar]