Abstract
Understanding the evolutionary history of crops, including identifying wild relatives, helps to provide insight for conservation and crop breeding efforts. Cultivated Brassica oleracea has intrigued researchers for centuries due to its wide diversity in forms, which include cabbage, broccoli, cauliflower, kale, kohlrabi, and Brussels sprouts. Yet, the evolutionary history of this species remains understudied. With such different vegetables produced from a single species, B. oleracea is a model organism for understanding the power of artificial selection. Persistent challenges in the study of B. oleracea include conflicting hypotheses regarding domestication and the identity of the closest living wild relative. Using newly generated RNA-seq data for a diversity panel of 224 accessions, which represents 14 different B. oleracea crop types and nine potential wild progenitor species, we integrate phylogenetic and population genetic techniques with ecological niche modeling, archaeological, and literary evidence to examine relationships among cultivars and wild relatives to clarify the origin of this horticulturally important species. Our analyses point to the Aegean endemic B. cretica as the closest living relative of cultivated B. oleracea, supporting an origin of cultivation in the Eastern Mediterranean region. Additionally, we identify several feral lineages, suggesting that cultivated plants of this species can revert to a wild-like state with relative ease. By expanding our understanding of the evolutionary history in B. oleracea, these results contribute to a growing body of knowledge on crop domestication that will facilitate continued breeding efforts including adaptation to changing environmental conditions.
Keywords: cabbage, domestication, crop wild relatives, Mediterranean, origin, ecological niche
Introduction
“Greek legend has it that the cabbage sprung from where Zeus’ sweat hit the ground.”
–N.D. Mitchell (1976)
A key tenet of evolutionary and plant biology is understanding how plants respond and adapt to changes in environmental conditions, which can be better understood by leveraging genotypic diversity and investigating the connections between genotype and phenotype. Crop wild relatives (CWRs) provide pools of allelic diversity that at one time were shared through a common ancestor with cultivated relatives. Although Vavilov recognized the potential of CWRs in the early 1900s (Vavilov 1926), advances in genomics and genome editing techniques have enabled scientists to better realize the potential of CWRs as a source of diversity and novel traits for the improvement of cultivated populations (Prohens et al. 2017; Li et al. 2018; Fernie and Yan 2019; Khoury et al. 2020; Turner-Hissong et al. 2020). Yet these scientific advancements are hindered in that we still have not identified the CWRs of many important crop species. Although cabbage may not have exactly formed from Zeus’ sweat, its evolutionary history, including identifying the closest living wild relative and origin of domestication, is still left unclear due to taxonomic confusion and the lack of genetic and archaeological evidence.
The horticultural crop Brassica oleracea L. has played an important role in global food systems for centuries, providing a source of leaf and root vegetables, fodder, and forage (Shyam et al. 2012). When first introduced to the species, Darwin drew many parallels between his theory of natural selection and the cultivation practices that led to the varied forms of this plant (Darwin 1868). Although many people may recognize that various dog breeds are all part of the same species, they are often surprised to learn that the domesticated forms of B. oleracea, broccoli (var. italica), Brussels sprouts (var. gemmifera), cabbage (var. capitata), cauliflower (var. botrytis), kale (var. acephala), and kohlrabi (var. gongylodes) are all one species as well. The global market for B. oleracea crops was around 70.1 million metric tons, in terms of production for 2019 (The Food and Agriculture Organization; www.fao.org). Although just six major crop types comprise the majority of the U.S. market (Agricultural Marketing Service, Market News Reports; www.ams.usda.gov), outside of these six major cultivars there exists at least 12 additional cultivated crop types (supplementary table S1, Supplementary Material online). These include lesser known varieties such as Chinese white kale or Cantonese gai-lan (Mandarin Jiè lán ; var. alboglabra), a leafy vegetable with florets, romanesco (var. botrytis) with unique fractal patterned curds, and walking stick kale (var. longata), which grows 6–12 feet (1.8–3.7 m) in height.
; var. alboglabra), a leafy vegetable with florets, romanesco (var. botrytis) with unique fractal patterned curds, and walking stick kale (var. longata), which grows 6–12 feet (1.8–3.7 m) in height.
Compared with other crops, surprisingly little is known about the progenitor species and origin of domesticated B. oleracea. Primary challenges in identifying the progenitor species include the number of wild species that share a single cytodeme and are interfertile with B. oleracea (2n=18 chromosomes; similar genomic organization; referred to as the “C genome”), the corresponding confusion surrounding taxonomic relationships, and conflicting evidence regarding the center of origin. Wild relatives that share the C genome with domesticated B. oleracea include Brassica bourgeaui, Brassica cretica, Brassica hilarionis, Brassica incana, Brassica insularis, Brassica macrocarpa, Brassica montana, Brassica rupestris, and Brassica villosa. Throughout the literature, many of these species have been referred to by alternative names or have multiple subspecies. For example, B. cretica is described as having either three subspecies (subsp. aegea, cretica, and laconica; Snogerup et al. 1990) or only two (subsp. cretica and nivea; Gustafsson et al. 1976). The taxonomic confusion is perhaps best highlighted by Bailey (1930), who stated that “Some of these plants appear to be more confused in literature than in nature.” The progenitor species of B. oleracea is further obscured by the presence of weedy, cabbage-like plants along the coastline of western Europe (England, France, and Spain), which have also been referred to as B. sylvestris (Mitchell 1976) or B. oleracea var. sylvestris (Gladis and Hammer 2001). The role of these weedy populations in the domestication of B. oleracea is unclear, with some studies suggesting these coastal wild populations represent the progenitor species (Snogerup et al. 1990; Song et al. 1990), and others identifying these wild forms as plants that escaped cultivation (Mitchell 1976; Mitchell and Richards 1979).
Given the uncertainty surrounding wild relatives and weedy populations, researchers have proposed numerous hypotheses for the progenitor species of B. oleracea (table 1). Hypotheses range from a single domestication with a single progenitor species (Song et al. 1990; Allender et al. 2007) to multiple domestications arising from multiple progenitor species (de Candolle 1855; Neutrofal 1927; Lizgunova 1959; Helm 1963; Snogerup 1980; Heaney et al. 1987; Song et al. 1988; Swarup and Brahmi 2005). Findings that point to a single origin of domestication have proposed different wild species as the progenitor (Snogerup et al. 1990; Song et al. 1990; Hodgkin 1995; Maggioni et al. 2018). For instance, Neutrofal (1927) suggested that B. montana was the progenitor of cabbages and that B. rupestris was the progenitor of kohlrabi, whereas Schulz (1936) identified B. cretica as the progenitor of only cauliflower and broccoli. Helm (1963) proposed a triple origin in which a single progenitor species gave rise to cauliflower, broccoli, and sprouting broccoli, whereas kale and Brussels sprouts were derived from another unknown wild species, and that all other crop forms were derived from a third unknown wild species. Snogerup (1980) proposed that cabbages were derived from wild B. oleracea, kales were derived from both B. rupestris and B. incana, and that Chinese white kale was derived specifically from B. cretica subsp. nivea.
Table 1.
Wild Species Which Have Been Proposed as Progenitor Species for Brassica oleracea Crop Types.
| Cultivar | Wild Relative | Author |
|---|---|---|
| Broccoli | B. oleracea | Linnaeus |
| B. oleracea | Hedrick (1919) a | |
| B. oleracea | Giles (1941) b | |
| B. montana | Hegi (1919) | |
| B. oleracea (from Italy) | Giles (1941) | |
| B. cretica | Gates (1953) | |
| B. oleracea and B. alboglabra | Song et al. (1990) | |
| Brussels sprouts | B. oleracea | Linnaeus |
| B. oleracea (western Europe) | Gates (1953) | |
| B. oleracea (western Europe) | Snogerup (1980) | |
| B. oleracea and B. alboglabra | Song et al. (1990) | |
| Cabbage | B. oleracea | Linnaeus |
| B. oleracea | de Candolle (1824) | |
| B. oleracea | Hedrick (1919) a | |
| B. oleracea | Bailey (1930) | |
| B. montana | Hegi (1919) | |
| B. oleracea (western Europe) | Gates (1953) | |
| B. oleracea (western Europe) | Snogerup (1980) | |
| B. oleracea and B. alboglabra | Song et al. (1990) | |
| Cauliflower | B. oleracea | Linnaeus |
| B. oleracea | de Candolle (1824) | |
| B. oleracea | Bailey (1930) | |
| B. montana | Hegi (1919) | |
| B. cretica | Schulz (1936) | |
| B. oleracea (from Cypus) | Giles (1941) | |
| B. cretica | Gates (1953) | |
| B. oleracea and B. alboglabra | Song et al. (1990) | |
| B. cretica | Tutin et al. (1964) | |
| Kale | B. oleracea | Linnaeus |
| B. oleracea | Hedrick (1919) a | |
| B. oleracea | Bailey (1930) | |
| B. montana | Hegi (1919) | |
| B. montana | Netroufal (1927) | |
| B. oleracea (western Europe) | Gates (1953) | |
| B. cretica, B. incana, B. rupestris | Snogerup (1980) | |
| B. incana and B. insularis | Hosaka et al. (1990) | |
| B. oleracea and B. alboglabra | Song et al. (1990) | |
| Kohlrabi | B. oleracea | Linnaeus |
| B. rupestris | Netroufal (1927) | |
| Unknown Mediterranean species | Gates (1953) | |
| B. oleracea and B. alboglabra | Song et al. (1990) |
Note.—Specific location is included in parentheses if indicated by the author. Brassica oleracea sometimes referred to as B. oleracea var. sylvestris.
Edited observations by Sturtevant in the late 19th century.
Referring to Prof Buckman’s experiment.
Due to the lack of consensus on the progenitor species, the center of origin for B. oleracea has also remained obscure. One hypothesis is that domesticated B. oleracea originated in England from weedy B. oleracea populations, with early cultivated forms brought to the Mediterranean, where selection for many of the early crop types occurred (Hodgkin 1995). Other studies point specifically to Sicily, which boasts a large diversity of wild relatives, as the center of domestication (Schiemann 1932; Lizgunova 1959). This conforms with the observations of Vavilov (1951) that plants tended to be domesticated in a finite number of global centers of diversity, which includes the Mediterranean. Most recently, linguistic and literary evidence provided support for domestication in the Eastern Mediterranean, where there is a rich history of expressions related to the usage and cultivation of B. oleracea crop types in early Greek and Latin literature (Maggioni et al. 2010, 2018).
Using newly generated RNA-seq data for a diversity panel of 224 accessions that includes 14 cultivar types and nine wild relatives, representing the largest and most diverse collection of this species and its wild relatives to date, we integrate phylogenomics, population genomics, ecological niche modeling, archaeological, and literary evidence to clarify the taxonomy, identify the closest living wild relative, and provide insight on the origin of domestication for B. oleracea.
Results
Sequencing Depth and SNP Identification
RNA sequencing of 224 samples resulted in an average of 88,598,754 reads per sample, with a range of 59,543,560–151,814,032 reads. The minimum per-sample sequencing depth recovered was 9X, with a maximum depth of 12X. After mapping reads to the B. oleracea TO1000 genome (Parkin et al. 2014), SNPs were filtered to exclude those with a Fisher strand (FS) value greater than 30 and quality depth (QD) less than 2.0. This recovered 942,357 variants in total, with 879,865 variants on chromosomes 1–9 and 62,492 variants on remaining scaffolds. Chromosomal SNPs were then filtered to exclude sites with greater than 60% missing data, sites with mean per-sample depth values less than 5, and indels, resulting in a total of 103,525 SNPs. After a final filtering step for linkage disequilibrium (LD), a conservative final data set of 36,750 SNPs was generated. For all samples, no mapping bias was detected when comparing the percentage of uniquely mapped reads across cultivar groups, species, and sequencing lane (supplementary fig. S1, Supplementary Material online).
Phylogeny and Population Clustering Distinguish Wild and Feral Populations
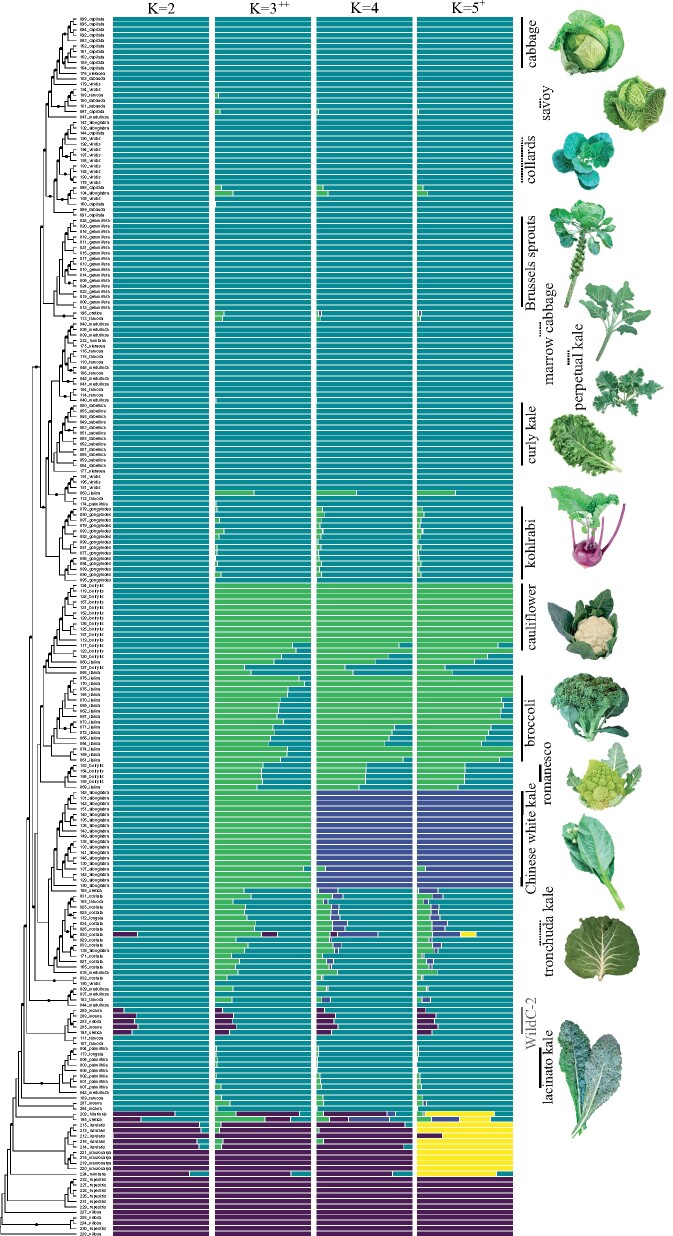
Sampling of B. oleracea cultivars included eight types of kales, five types of cabbages, Brussels sprouts, broccoli, cauliflower, Romanesco (var. botrytis), and kohlrabi (supplementary table S2, Supplementary Material online). Together, these cultivated types accounted for 188 of the 224 total samples. The remaining 36 samples included previously identified wild relatives: putatively wild B. oleracea, B. cretica, B. incana, B. montana, B. hilarionis, B. insularis, B. macrocarpa, B. rupestris, and B. villosa. The phylogenetic reconstruction of all 224 samples using SNPhylo (Lee et al. 2014) recovered several well-supported clades with greater than 70% bootstrap support, although overall support was generally poor (less than 70% bootstrap support), especially along the backbone. Chinese white kale, broccoli, cauliflower, romanesco, kohlrabi, curly kale, Brussels sprouts, B. rupestris, B. macrocarpa, and B. insularis were all recovered as monophyletic. Aside from red cabbages, cabbages were also monophyletic, but with only 55% bootstrap support. Seven cultivars (collards, tronchuda kale, savoy cabbage, perpetual kale, red cabbage, and marrow cabbage) were found throughout the tree as polyphyletic assemblages. Several wild samples were recovered within the cultivar clade, including two samples of B. cretica (196, 199), one sample of B. montana (222), and all samples of putatively wild B. oleracea (175, 176, 177; sample names in bold text; fig. 1). We also recovered a group in the cultivar clade consisting of five samples of three wild species, B. incana (205, 208, 209), B. villosa (233), and B. cretica (195), labeled “WildC-2” (for wild samples with the C genome). Many of these “wild” samples also share most or all of their ancestry with cultivars. At K = 2, in our fastSTRUCTURE analyses (Raj et al. 2014), samples clustered as either cultivars or wild (fig. 1). We find that two samples of B. incana (204, 207; likely both from Crimea), which are sister to all cultivated samples, share 100% of their ancestry with cultivated types, as do two samples of B. cretica (196, 199), one sample of B. montana (222), and all three samples of putatively wild B. oleracea (175, 176, 177). Together with the placement in the phylogeny, these analyses indicate that these are not truly wild samples, but represent feral types, defining feral here as either exoferal (a domesticated population derived from admixture with either a divergent population, a wild conspecific, another domesticated species, or another wild species) or endoferal (a population of domesticated plants that has escaped from cultivation without the aid of introgression/hybridization with wild conspecifics; Gering et al. 2019). Our newly identified WildC-2 shows mixed wild and cultivar ancestry, which was also observed for one sample of tronchuda kale (30). The marginal likelihood was maximized at K = 3, in which a cluster comprised of broccoli, cauliflower, and Chinese white kale separated from other cultivated types. At K = 4, Chinese white kale was distinct from broccoli and cauliflower. The structure in the data was best explained by K = 5, in which the clade comprised of B. insularis and B. macrocarpa was separated and had shared ancestry with Brassica cretica (198), B. hilarionis, B. montana (224), and one sample of tronchuda kale (30). Additional K values showed similar patterns (supplementary fig. S2, Supplementary Material online).
Fig. 1.
Demographics and population structure for 224 samples of cultivated Brassica oleracea (n = 188) and wild C genome species (n = 36). (Left) Individual sample phylogeny with putatively wild samples labeled in bold and black dots indicating bootstrap values less than 70%. (Middle) Ancestry proportions for K = 2 to K = 5 as inferred from fastSTRUCTURE; K = 3 maximizes marginal likelihood (++) and K = 5 best explains structure in the data (+). (Right) Monophyletic clades indicated by a solid line, largest cluster of paraphyletic groups indicated by dashed lined. Illustrations of corresponding crop types by Andi Kur.
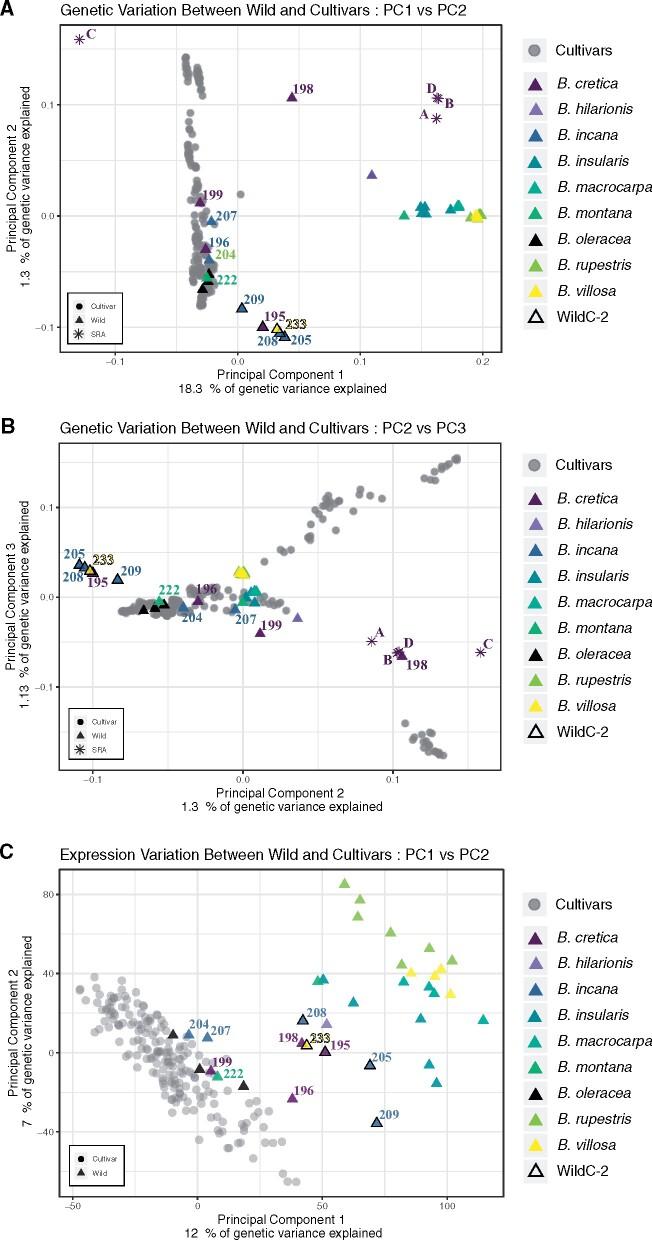
Principal component analysis (PCA) also separated cultivars from most wild samples (supplementary fig. S3D–F, Supplementary Material online). The PC1 axis distinguishes wild species from cultivars and the PC2 axis separates WildC-2 from all other wild species (triangles with black outlines). Although one sample of B. cretica (198) clusters closest to cultivated types, samples of B. incana, which were not in WildC-2, along with one sample of B. montana (222), two samples of B. cretica (196, 199), and all three samples of B. oleracea (175, 176, 177) cluster with the cultivars, corroborating the phylogenetic analyses. To further investigate the clustering patterns of B. cretica to cultivars, we included four additional wild-collected samples of two B. cretica subspecies (A and B=subsp. nivea, C and D=subsp. cretica; fig. 2A and B; supplementary fig. S3A, Supplementary Material online; labeled SRA in figure legend; Kioukis et al. 2020). Adding these samples supports the results of other studies that B. cretica, as a species, is very diverse. Although sample C does not group with other B. cretica samples using the PC1 axis, the PC2, PC3, and PC4 axes show much tighter clustering among the four wild-collected samples and one of our samples of B. cretica (198), indicating that our B. cretica (198) sample is an informative representative of wild-collected B. cretica (fig. 2A and B; supplementary fig. S3A, Supplementary Material online).
Fig. 2.
Principal component analysis (PCA) of SNPs and expression profiles. (A) Genetic variation PCA of PC1 versus PC2, (B) Genetic variation PC2 versus PC3, and (C) Expression profile PCA for PC1 versus PC2 of wild and cultivar samples. Triangles, wild samples; circles, cultivars; triangles with black outlines, WildC-2 samples with species identification indicated by color. Wild-collected B. cretica samples from Kioukis et al. (2020) indicated by asterisks, labeled as SRA.
For crop samples, estimates of inbreeding coefficients from PCAngsd (Meisner and Albrechtsen 2018) roughly matched expectations for the frequency of heterozygotes under Hardy–Weinberg equilibrium, whereas inbreeding coefficients for wild species suggest excess homozygosity (supplementary fig. S4, Supplementary Material online), possibly reflecting cultivation practices for germplasm management and the relative isolation of wild populations (i.e., small effective population size), respectively. Feral samples, those which were identified as wild taxa, but were found more closely related to cultivars than to wild taxa in our phylogeny and clustered with cultivated samples in our PCA (B. cretica—196, 199; B. incana—204, 207; B. montana—222; and wild B. oleracea—175, 176, 177), show patterns of heterozygosity that are similar to crop samples, as do the four samples of B. cretica from Kioukis et al. (2020). Our WildC-2 exhibited patterns of excess homozygosity more similar to other wild taxa.
Domestication Is Also Reflected in the Transcriptome
Using expression profiles (transcript abundances) of 51,438 genes for our original 224 samples, we tested if cultivars and wild samples would still cluster separately based on the transcriptome. Overall, results and clustering patterns were similar to analyses using SNPs, with the axes of PC1 and PC2 separating most wild species from cultivars (fig. 2C; supplementary fig. S3B and C, Supplementary Material online). We again found the same samples of B. incana (204, 207), B. cretica (196, 199), B. montana (222), and B. oleracea (175, 176, 177) clustering with the cultivars, but in expression analyses WildC-2 clustered with the other wild samples, rather than separately as in our SNP based PCA. Hierarchical clustering of the expression profiles recovered similar patterns with two major groups: wild and cultivated, again with WildC-2 clustering with the other wild samples (supplementary fig. S5, Supplementary Material online). Although most cultivar groups were not recovered as unique clusters, there were a few exceptions. Brussels sprouts, Chinese white kale, and curly kale all formed distinct clades, which corresponds to what we know about their growth habit. Since RNA was collected at the 7th leaf-stage, before substantial morphological differentiation occurs between cultivars, it is not too surprising that they do not cluster distinctively by cultivar. Curly kale is almost immediately visually distinguishable from other cultivars in that the first true leaves have margins which are already undulate and/or frilled, in contrast to the more lanceolate (i.e., long, widest in the middle, with tapered tips) leaves observed in most cultivars. Brussels sprouts are also easily identifiable at this early growing stage as they have short, oblong to nearly circular leaves. Although Chinese white kale leaves look more similar to the lanceolate shape of other cultivars, they grow more rapidly and plants in this group are annual instead of biennial, which may explain why these accessions cluster separately from other cultivars.
To identify modules of genes that might be driving the observed clustering patterns, we used weighted correlation network analysis (WGCNA; Langfelder and Horvath 2008). We found that 48 modules, ranging in size from 34 to 35,981 genes, provided the best fit for the data (supplementary table S3, Supplementary Material online). To assess what types of biological processes were overrepresented in these modules, we used syntenic Arabidopsis thaliana genes and performed a GO analysis through PANTHER v. 16.0 (Mi et al. 2021). Overlap of B. oleracea with A. thaliana genes ranged from 17% to 98.3%, perhaps indicating that some modules are more conserved, whereas others are unique to B. oleracea. Modules which were more conserved between the two species included genes related to herbivory defense compound production (secondary metabolite biosynthetic process, phenylpropanoid biosynthetic and metabolic processes), wound formation (suberin biosynthetic processes), and wax formation (wax biosynthetic and metabolic processes), the latter of which may be correlated to the characteristic glaucous leaves of cultivated B. oleracea (supplementary table S4, Supplementary Material online). Within the top five conserved modules, the transcript abundance (TPM) was significantly different among the different groups (P value≤2e-16 for modules 7, 13, 31, & 34; P value=2.19e-11 for module 30). Post hoc comparisons using Tukey’s honestly significant difference (HSD) revealed that transcript abundance in cultivars was significantly different compared with that of wild relatives across conserved modules, except for B. hilarionis, which was not recovered as significantly different from cultivars for any module. WildC-2 along with other identified feral samples had significantly different transcript abundance compared with cultivars for modules 7, 13, 31, and 34, but not for module 30. Significant differences were also found between WildC-2 and feral samples compared with wild relatives for several modules, with no obvious patterns across modules (supplementary fig. S6 and table S5, Supplementary Material online).
Species Tree and Admixture Inference Indicate Brassica cretica Is the Closest Living Wild Relative
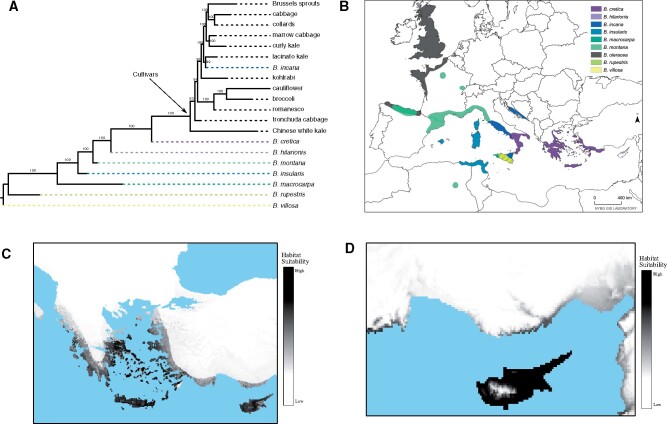
Given the results of population clustering using both SNPs and expression profiles, we further interrogated the species level relationships between wild relatives and cultivar groups by resolving the backbone of the phylogeny. Using the PoMo model (Schrempf et al. 2016) as implemented in IQ-Tree (Nguyen et al. 2015) and only including samples representing monophyletic groups as determined in the sample-level phylogeny, we found strong support for B. cretica as the closest living wild relative to cultivated B. oleracea (fig. 3A). Notably, for our species tree analyses, we included only one sample of B. cretica (198). This sample was used for species reconstruction due to its placement near other wild taxa in the sample level phylogeny and its clustering with wild-collected B. cretica from Kioukis et al. (2020) in the PCA. The current distribution of B. cretica occurs throughout the Eastern Mediterranean, primarily in Greece, highlighting a potential origin of domestication (fig. 3B). Another suggested wild relative, B. incana, is strongly supported as belonging to the cultivar clade, sister to lacinato kale. Although our sampling is limited in regard to the distribution of B. incana as a whole, this result supports our other findings that B. incana is not a completely wild assemblage, but that at least some populations are feral. Within cultivars, several expected relationships were recovered: collards and cabbage as sister lineages (Song et al. 1988; Farnham 1996), with Brussels sprouts sister to both; cauliflower and broccoli as sister clades (Song et al. 1988; Stansell et al. 2018), with romanesco sister to both; and Chinese white kale as sister to all other cultivars, agreeing with recent literature (Cheng et al. 2016; Stansell et al. 2018).
Fig. 3.
Species tree with current distribution and historical environmental niche modeling. (A) Species tree of wild and cultivar samples. Bootstrap support indicated above branches. (B) Current species distribution of wild relatives. (C) Suitable habitat for B. cretica and (D) B. hilarionis during the late-Holocene. Map of current distrubution provided by Elizabeth Gjieli, the Geographical Information Manager at the New York Botanical Garden GIS Laboratory.
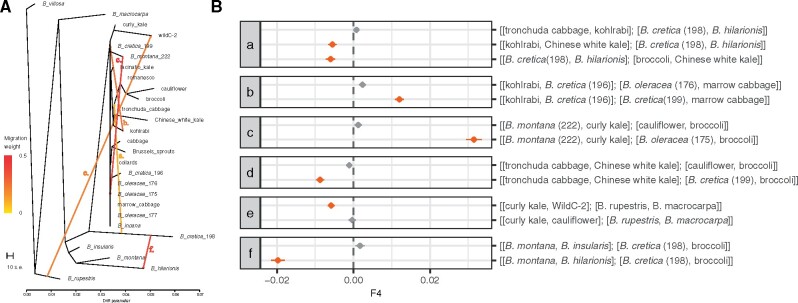
With the overall species relationships resolved, we aimed to tease apart the evolutionary history of the wild samples that clustered within the cultivar clade. Specifically, we asked if any of the identified feral samples were the products of admixture using TreeMix (Pickrell and Pritchard 2012). Although the tree model without any migration edges explained 87.3% of the variance in the data set, sequentially adding migration events to the tree resulted in five migrations events explaining 92% of the variation (fig. 4A and supplementary fig. S7, Supplementary Material online). Adding a single migration edge resulted in an admixture event from B. cretica (198) to a clade of [Chinese white kale+tronchuda cabbage]. To further test this event, we used four-population (f4) tests for treeness as implemented in TreeMix, where a significant nonzero value indicates the presence of gene flow (Reich et al. 2009; Pickrell and Pritchard 2012; fig. 4B). Although the tree [[tronchuda cabbage, kohlrabi],[B. cretica (198), B. hilarionis]] showed no significant evidence of gene flow (f4 = 0.0008, Z = 1.094), replacing tronchuda cabbage with Chinese white kale indicated significant gene flow from B. cretica (198) to Chinese white kale (f4=−0.0055, Z =−5.113). This result was further verified when adding a second migration edge, as the migration edge only included Chinese white kale, but the direction was reversed (from Chinese white kale to B. cretica [198]). The second event, from kohlrabi to a presumably feral sample of B. cretica (199), was supported by f4 tests, with the tree [[kohlrabi, B. cretica (196)],[B. cretica (199), marrow cabbage]] indicating significant evidence of gene flow from kohlrabi to B. cretica (199) (f4 = 0.012, Z = 10.5). This migration event is also seen phenotypically, as B. cretica (199) has a swollen stem when grown to maturity. No significant evidence of gene flow was found when substituting B. cretica (199) with B. oleracea (175), which is not expected to be involved in the admixture event (f4 = 0.00023, Z = 2.68). Two admixture events provide evidence of potential exoferal origins for at least two samples, B. oleracea (175) and B. cretica (199). The four-population tree of [[B. montana (222), curly kale],[B. oleracea (175), broccoli]] suggests significant gene flow from B. montana (222) to B. oleracea (175) (f4 = 0.315, Z = 15.77), as does the tree of [[tronchuda cabbage, Chinese white kale],[B. cretica (199), broccoli] for gene flow from Chinese white kale to B. cretica (199) (f4 =−0.009, Z =−7.98). The fifth added migration edge from B. rupestris to WildC-2 explains the shared ancestry recovered in the fastSTUCTURE results. The test for treeness with [[curly kale, WildC-2],[B. rupestris, B. macrocarpa]] indicated significant admixture from B. rupestris to WildC-2 (f4 =−0.006, Z=−6.50), but was nonsignificant when substituting WildC-2 with cauliflower (f4 =−0.0003, Z =−0.338). In general, these analyses highlight that the evolutionary history of B. oleracea is characterized by many admixture events and lineages of exoferal origins.
Fig. 4.
Inferred admixture events. (A) Phylogeny five migrations labeled a–f. (B) Corresponding four-population tests for treeness.
Archaeological and Literary Evidence Point to a Late-Holocene Domestication
To further investigate the origins of domesticated B. oleracea, we surveyed archaeological, literary, and artistic evidence (supplementary tables S6 and S7, Supplementary Material online). The earliest reported claim of B. oleracea comes from an archaeological collection from the Austrian Alps. This collection comprises three seeds dated to the Middle Bronze Age (ca. 3550–3350 years before present or BP; Schmidl and Oeggl 2005). However, the lack of illustrations and discussion of separation criteria from other Brassica species makes us question the reliability of this species-level identification, as seeds of Brassica species are difficult to tell apart. The only other find of similar antiquity is B. oleracea seeds from the Late Bronze Age/Early Iron Age, identified by scanning electron microscopy and radiocarbon dated directly between ca. 3250–2970 BP (Kaniewski et al. 2011). These finds are associated with destruction levels at Gibala, Tell Tweini in western Syria on the Mediterranean coast. Although most of the archaeological finds are of seeds (supplementary table S6, Supplementary Material online), there is at least one documentation of pottery residues where lipids of Brassica leaf waxes were identified and dated to 850–750 BP (Evershed et al. 1992, 1994). The authors attribute this to the boiling of leaves of B. oleracea, and given the lack of evidence for other commonly eaten Brassica leaves in England at this time, this would appear a likely identification.
The earliest literary references to B. oleracea date to Greek scholars 2500–2000 BP (supplementary table S7, Supplementary Material online). Hipponax’s writing refers to a seven-leaf cabbage in an iambic verse (West 2011), whereas Hippocrates On the Nature of Women, written around 2410–2320 BP, refers to the use of cabbage, or krambe, in a few recipes (Totelin 2009). As early as 2320 BP, there is evidence for cultivar diversity. Theophrastus refers to three varieties: a curly-leaved type, a smooth-leaved type, and a wild type with a bitter taste, many branches, and many small round leaves (Yonge 1854). Pliny in his Natural History writing some 200 years later describes at least ten varieties in addition to those seen in the previous classical works (The Elder and Rackham 1950). However, whereas most scholars accept that the Greek or Latin translations of “cabbage” refer to B. oleracea, it is important to note that “cabbage” is not a Greek word and that the word “raphanos” is translated as both cabbage and B. cretica in the Greek-English Lexicon (Liddell and Scott 1940) and in Hort’s (1916) translation of Theophrastus’ Historia Plantarum. Certainly, there are differences between the subspecies of B. cretica that might be reflective of the varieties described by Theophrastus and Pliny, and which may explain the diversity we observed among B. cretica samples in our PCA results. Further, the description by Nicander (quoted by Athenaeus; Yonge 1854; p. 582) indicates that wild or perhaps feral forms of B. cretica were known in Ionia, the western coast of present-day Turkey, ca. 2150–2050 BP.
Late-Holocene Environmental Niche Modeling Highlights Wild Relatives’ Ranges
Based on archaeological information, the oldest relatively reliable occurrence for B. oleracea cultivation is dated 3250–2970 BP in Gibala NW Syria (Kaniewski et al. 2011). To predict what would be a suitable habitat for the wild relatives during the late-Holocene, we compiled occurrence records from GBIF (www.gbif.org) and (Snogerup et al. 1990), along with environmental data, to perform environmental niche modeling using MaxEnt 3.4.1 (Phillips et al. 2017). Notably, we find that B. cretica has an expanded Eastern Mediterranean habitat suitability (fig. 3C) that includes Cyprus. Presently, only B. hilarionis is known to occur in Cyprus (fig. 3B), however modeling predicts that in the late-Holocene it would also have had an expanded habitat suitability in the surrounding mainland coastal regions (fig. 3D). Since most of these wild species are narrow island endemics (Snogerup et al. 1990), species are generally estimated to have little change from current day distributions (supplementary fig. S8 and table S8, Supplementary Material online).
Discussion
Multiple Lines of Evidence Support a Single Eastern Mediterranean Origin
Our evidence from genome-scale, multilocus data along with archeology, literature, and environmental niche modeling best support a single Eastern Mediterranean domestication origin for B. oleracea, corroborating the conclusions of Maggioni et al. (2018) based on literary sources and (Maggioni et al. 2010) using linguistics. When modeling phylogeny and population structure, two Eastern Mediterranean species, B. cretica and B. hilarionis, are found as sister species to cultivars and are assigned ancestry from all cultivar populations for values of K from 2 to 5 (fig. 1), consistent with these species being likely progenitor species of B. oleracea cultivars. In our species tree reconstructions, we find just B. cretica as sister to all cultivars, specifically sample 198, which clusters with wild-collected B. cretica samples from Kioukis et al. (2020) in our PCA (fig. 2A and B), lending further support for B. cretica as the progenitor species. This same sample of B. cretica (198) as well as our sample of B. hilarionis are recovered as fairly homozygous, therefore they would likely be good starting material for future research related to de novo domestication via selective breeding or gene editing.
Although we do recover evidence of admixture between B. cretica (198) and both wild and cultivated taxa, the placement of B. cretica (198) as the closest living wild relative does not change. However, an inferred admixture event from B. cretica (198) to B. hilarionis does result in a topological change in the placement of B. hilarionis as sister to B. montana (224; originally collected in Spain) (supplementary fig. S7, Supplementary Material online). This novel relationship has not been identified before and warrants additional study with greater taxon sampling. The second migration event involving B. cretica (198) is from Chinese white kale. This event lends further evidence of admixture with wild germplasm during the domestication process, consistent with other examples demonstrating that domestication is not a single event, but a series of events characterized by continuous gene flow between wild and cultivated populations (Beebe et al. 1997; Wang et al. 2017). Together with the phylogeographic discontinuity of wild B. oleracea samples and their Eastern Mediterranean progenitors (fig. 3B), the more distant phylogenetic placement of B. insularis, B. macrocarpa, and B. villosa (fig. 3A), and strong patterns of shared ancestry between B. incana and cultivars (fig. 1), these results lead us to support the hypothesis of domestication in the Eastern Mediterranean with B. cretica as the closest living wild relative.
The Role of Ferality in the Domestication of Brassica oleracea
Multiple lines of evidence highlight the role of wild and feral populations as pools of diversity that contributed to crop diversification during domestication (Beebe et al. 1997; Allaby 2010; Fuller et al. 2014; Wang et al. 2017). Our data support a similar phenomenon in the domestication of B. oleracea: it appears that introgression from wild or feral populations contributed to the genetic composition of particular crops, and vice versa, which is revealed by in-depth analyses of admixture using population structure and tree-based methods (figs. 1 and 4; supplementary fig. S2 and S7, Supplementary Material online). Several samples of wild relatives, including B. cretica, as well as wild B. oleracea, B. incana, B. montana, and B. villosa, are recovered as feral in all analyses.
Although we find one sample of B. cretica (198) as the closest living wild relative, we also identify two samples of B. cretica (196 and 199) as likely feral and fall within the cultivar clade (fig. 1; see supplementary fig. S9, Supplementary Material online, for photos). Interestingly, Song et al. (1988) also recovered a polyphyletic B. cretica using RFLPs. Results presented here support previous findings that B. cretica was at one point at least partially domesticated. Snogerup et al. (1990) state that wild B. cretica was consumed as late as 1962 and, as noted in our literary results, some early references to B. oleracea in the literature could be translated as B. cretica, meaning the vast amount of described morphology in these works, which may be the result of cultivation, could now be reflected in the multiple named subspecies and described genetic diversity of modern B. cretica (Snogerup et al. 1990; Widén et al. 2002; Allender et al. 2007; Edh et al. 2007). Further, B. cretica was known to occur in Ionia (western coast of present day Turkey) ca. 2150–2050 BP and the evidence of B. cretica populations today in Lebanon, which are morphologically similar to B. cretica subsp. nivea, suggests widespread trade of these species by the earliest Mediterranean civilizations (Dixon 2006). However, these plants may have been introduced into these localities without cultivation as was proposed by Snogerup et al. (1990). Previous researchers have noted that B. cretica populations are typically found in coastal locations associated with ancient seaports, occupying their preferred ecological niche on chalk cliffs undisturbed by grazing (Mitchell 1976; Snogerup et al. 1990). We believe that these early forms of B. cretica may have played underappreciated roles in the domestication of B. oleracea crops and to fully understand the evolutionary history of B. oleracea, the demographic history and domestication story of B. cretica must be resolved.
Sources have hypothesized that wild populations of B. oleracea in England are the progenitor(s) for modern cultivars (Snogerup et al. 1990; Song et al. 1990), whereas others have proposed that these are escaped cultivars (Mitchell 1976; Mitchell and Richards 1979). Consistent with these hypotheses, we find that the three wild B. oleracea samples in our study cluster with cultivars both phylogenetically and in PCA for both SNP data and expression profiles. Although these samples are from Canada (175), Denmark (176), and Germany (177), well outside the natural distribution range for B. oleracea—notably not from England, one of the hypothesized geographic origins—we suggest that an origin in England is unlikely given the archeological and literary data. Although the oldest archaeobotanical record for B. oleracea (Middle Bronze Age; ca. 3550–3350 BP) is from Austria, we regard this evidence with caution as wild populations of B. oleracea are not presently found in Austria and the major Brassica crops in this region include B. nigra (Tutin et al. 1964) or potentially cultivated turnip (B. rapa). Additionally, there is no compelling archaeological evidence to suggest the possible cultivation of cabbages in Europe prior to the Late Iron Age (2350–2050 BP) and Roman periods (1950–1650 BP), but there is evidence for knowledge of B. oleracea in Greece during this time (Maggioni et al. 2018; supplementary tables S6 and S7, Supplementary Material online). Overall, there are no records for B. oleracea from before this period within databases relating to the Eastern Mediterranean (Reihl 2014), Europe (Kroll 2001, 2005), Britain (Tomlinson and Hall 1996), the Czech Republic (Kreuz and Schäfer 2002), or within predynastic and Pharaonic Egypt (Murray 2000), despite having documentation for other Brassica species. Evidence for B. oleracea in Europe does not start appearing until ca. 1850 BP, when the appearance of seeds increased and can be attributed to the spread of crops both within and on the periphery of the Roman Empire (Van der Veen 2011). Additionally, several studies that sampled wild B. oleracea populations in the British Isles (Mitchell 1976; Mitchell and Richards 1979), South West England (Raybould et al. 1999), Atlantic coasts of western Europe (Mittell et al. 2020), and Atlantic coast of France (Maggioni et al. 2020) support that these wild B. oleracea populations are feral populations, typically with low levels of genetic diversity and some degree of isolation from other populations. Lanner-Herrera et al. (1996) sampled populations across Spain, France, and Great Britain, concluding that each population evolved independently, whereas more recently, Mittell et al. (2020) found that geographically close populations were more genetically different than distant populations. Our results provide additional evidence that feralization is commonplace for B. oleracea crops and that references to wild B. oleracea likely represent multiple, independent feralization events. Additional sampling of wild populations will enable opportunities to further investigate the relationships among these feral populations and cultivated crops.
Brassica incana, another suggested progenitor species (Snogerup 1980), is also supported as feral for the samples included in our analyses. Two of our five samples (204 and 207) are recovered as sister to all cultivars in our individual level phylogeny but are found to share 100% of their ancestry with cultivars rather than other wild taxa using fastSTRUCTURE when K = 2 (fig. 1). Further, these two samples were resolved as sister to lacinato kale in our species tree analysis, providing additional evidence that these samples represent a feral lineage, possibly of lacinato kale. This result may lend insight into why previous studies have found B. incana as sister to B. oleracea (Lázaro and Aguinagalde 1998; Mei et al. 2010; Arias and Pires 2012) and the observation by Snogerup et al. (1990) that samples of B. incana from the Crimea are more interfertile with cultivated B. oleracea than others. Although Snogerup et al. (1990) suggested that B. incana was more interfertile due to historical introgression, we do not find evidence of this for samples 204 and 207. However, the three other samples of B. incana (205, 208, 209), which belong to WildC-2, do show evidence of admixture with B. rupestris, likely explaining their clustering together both in the PCAs and phylogeny with B. cretica (195) and B. villosa (233) which also show admixture with B. rupestris (see supplementary, fig. S10, Supplementary Material online, for photos). All three B. incana were collected in Italy from two locations and therefore do not well represent the known B. incana range (fig. 3B), whereas the two other samples found in this clade, B. cretica (195) and B. villosa (233), were collected in Greece and Italy, respectively. Although all five WildC-2 samples share an introgression event from B. rupestris (figs. 1 and 4; supplementary fig. S7, Supplementary Material online), they are from different germplasm collections (IPK-gatersleben and USDA National Plant Germplasm System), ruling out the inferred migration being the result of current cultivation practices. It is possible that at least three of these samples (B. incana 205, 208, 209) are related to the wild kale of Crimea, which is posited as a B. rupestris–incana hybrid that was transferred to the Crimea via trade (Dixon 2006). This suggests that there was early widespread cultivation of these B. rupestris–incana types (Dixon 2006) and provides a plausible explanation for why B. incana and B. rupestris are closely related in previous studies (Lannér et al. 1997; Mei et al. 2010). The other two samples in WildC-2 (B. cretica 195 and B. villosa 233), possibly represent misidentifications, which is supported by their intermediate phenotypes (i.e., B. rupestris margins with varying amounts of trichomes; supplementary fig. S10, Supplementary Material online).
The last feral identification is that of B. montana, for which we find one sample as more closely related to wild taxa (224) and one more closely related to cultivars (222). The feral sample (222) is of unknown origin, but again the literature indicates that this may not be a surprising result. Many studies have previously indicated a close relationship between B. montana and B. oleracea. For example, Panda et al. (2003) concluded that B. montana may be a subspecies of B. oleracea, whereas Lannér et al. (1997) found that B. montana and B. oleracea clustered together using chloroplast data. Furthermore, several authors have suggested that some populations of B. montana were feral B. oleracea (Paolucci 1890; Onno 1933; Snogerup et al. 1990), which may be reflected in the overlapping ranges produced by our niche modeling of these two species (supplementary fig. S8, Supplementary Material online). Therefore, in combination with results from previous studies, our results support that at least some B. montana populations are of feral origin.
Taken together, it is clear that the current taxonomy of B. oleracea and its wild relatives is confounded by gene flow between wild and cultivated populations, resulting in confusion between wild and feral lineages and obscuring the true evolutionary history of this species. Additionally, although there is much interest in crop improvement using CWRs (Meyer et al. 2012; Khoury et al. 2020), feral lineages offer another, potentially more direct route to reintroducing genetic diversity into cultivated populations, as gene flow is less likely to be impeded by barriers such as reproductive isolation (Mabry et al. 2021). These feral populations may also provide additional avenues to explore the evolutionary capacity for range expansion and phenotypic plasticity.
Postdomestication Cultivar Relationships
Although our knowledge of the spread and diversification of B. oleracea crops after domestication is confounded by both the difficulties of identifying seeds of individual crop types and frequent introgression between crop types, we can infer some patterns using the species phylogeny. Like other studies (Cheng et al. 2016; Stansell et al. 2018), we find Chinese white kale sister to all other cultivars, representing the only Asian clade of crop types (fig. 3A). Although the spread of B. oleracea to eastern Asia is still undocumented archaeologically, recent pollen analysis has provided evidence for cultivation of other Brassica species, including B. rapa, in the Yangtze valley 3250–3350 BP, likely corresponding to movement across “Silk Road” trade routes (Zhang 2009). However, this only provides identification criteria, not archaeological evidence (Yang et al. 2018). A review of Chinese historical sources concluded that B. oleracea may have been introduced to China 1450–1350 BP and had evolved into Chinese white kale in Southern China by the period of the Tang Dynasty (1350–1250 BP; Zhang 2009). Due to its position as sister to all other cultivars and as the only Asian B. oleracea crop type, as well as its annual growth habit, this taxon warrants additional study to understand its own unique domestication story.
The dispersal of B. oleracea by human translocation westward, ultimately to the Atlantic coast of Europe, appears to have established both regional feral populations and the variety of modern crop types. Archaeological evidence suggests that this process may have begun with Late Bronze Age seafaring (3000–3300 years ago), when the whole Mediterranean became linked in trade perhaps for the first time (Broodbank 2015), and continued to provide a corridor for introgression and varietal diversification through the Iron Age (up to 2000 years ago). Trade links along the Atlantic seaboard from North Africa and Iberia through Britain and Ireland are clearly indicated in archaeology (Cunliffe 2004), and are associated with the first peopling of the Canary Islands from the north, where walking stick kale is endemic. Notably, many cultivars do not form monophyletic groups in our sample level phylogeny, likely indicative of admixture between crop types. This is supported by previous findings that broccoli is paraphyletic (Song et al. 1988; Stansell et al. 2018), as well as collards (Pelc et al. 2015), and by our findings that kale types such as tronchuda kale and perpetual kale are highly polyphyletic, suggesting that the kale morphotype has been selected for multiple times independently.
In conclusion, we confirm a single Eastern Mediterranean origin for B. oleracea and find B. cretica as the closest living wild relative. We highlight several feral samples that are not reflected by the current taxonomy but likely reflect important aspects of the domestication history for B. oleracea. Moving forward, it will be important to identify, collect, study, and preserve these feral samples as pools of allelic diversity, which may play an important role in future crop improvement, for example, as a source of potential pest and pathogen resistance (Mithen et al. 1987; Mithen and Magrath 1992; Mohammed et al. 2010). In clarifying the evolutionary history of B. oleracea and its wild relatives, we hope to enable this model system for additional studies on evolutionary phenomena such as parallel selection, polyploidy, and ferality. Additionally, since many of these wild species are very narrow endemics and are valuable for both crop improvement and for nature conservation, their identification and preservation are urgent. We hope this study can serve as a steppingstone, as the work before us has, for those who, like Darwin was, are intrigued by this group of plants and wish to further its study.
Materials and Methods
Taxon Sampling
Samples from cultivars accounted for 188 of the 224 total samples with the remaining 36 samples included being previously identified wild relatives (supplementary table S2, Supplementary Material online). These include accessions from the United States Department of Agriculture, Agriculture Research Service (USDA-ARS) Plant Genetic Resources Unit (PGRU; 114 accessions), The Leibniz Institute of Plant Genetics and Crop Plant Research (IPK; 71 accessions), Universidad Politécnica de Madrid (UPM; four accessions), The Nordic Genetic Resource Centre (NordGen; two accessions), Gomez Campo Collection (two accessions), John Innes Center (one accession), doubled haploid lines (17 samples, some accessions sampled twice), or from the Pires’ personal collection (13 accessions). Four replicates of each accession were grown from seed in a sterile growth chamber at the University of Missouri (MU; Columbia, MO) Bond Life Sciences Center in a randomized complete block design across two independent outgrowths. At the seventh leaf stage, leaf four was collected from each plant and immediately flash-frozen in liquid nitrogen for RNA extraction. Morphotype identity was validated in mature plants by growing all accessions twice over the span of 2 years (supplementary table S2, Supplementary Material online).
Whole-genome resequencing data for an additional four samples from Kioukis et al. (2020) of two varieties of B. cretica (var. cretica and var. nivea) was downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) to supplement our sampling of B. cretica. These samples are under the SRA accession as follows: A=SRR9331103, B=SRR9331104, C=SRR9331105, and D=SRR9331106. Samples of A and B are B. cretica var. nivea from mainland Greece and C and D are B. cretica var. cretica, one from the mainland (C) and one from the island of Crete (D).
RNA Isolation and Sequencing
RNA was isolated using the ThermoFisher Invitrogen PureLink RNA mini kit (Invitrogen, Carlsbad, CA) followed by TruSeq library preparation (Illumina, San Diego, CA) and sequencing on the NextSeq platform (Illumina, San Diego, CA) for 2×75 bp reads. Library preparation and sequencing were performed through the MU DNA Core Facility. For eight flow cells, 24 samples were multiplexed and sequenced in a single flow cell, followed by a ninth flow cell with 17 samples, and a tenth flow-cell with 16 samples.
Mapping and SNP Calling
Short reads were mapped to the B. oleracea TO1000 genome (Chinese white kale; Parkin et al. 2014; release-41) by first using the STAR v. 2.5.2 (Dobin et al. 2013) two-pass alignment to identify splice junctions, which were then used in the second pass to improve mapping (Engström et al. 2013). The TO1000 genome of Chinese white kale was chosen due to wild relatives having a more kale-like phenotype and its placement as sister to the other cultivars in recent studies (Cheng et al. 2016; Stansell et al. 2018). Mapped reads (BAM format) were then processed following the GATK v. 3.8 best practices for RNA-seq reads (McKenna et al. 2010; Van der Auwera et al. 2013; Poplin et al. 2017). To ensure that reads were mapping correctly, the GATK “Split’N’Trim” function was used to split reads into exon segments and trim any overhanging reads in intron segments. In total, 7,564,168 variants were called before any filtering was performed. The resulting variants were filtered to exclude those with a Fisher strand (FS) value greater than 30 and quality depth (QD) less than 2.0. The remaining 879,865 chromosomal variants were then filtered using vcftools v. 0.1.17 (Danecek et al. 2011) to exclude sites with greater than 60% missing data (–max-missing 0.4), sites with mean sample depth values less than 5 (–min-meanDP 5), and indels (–remove-indels;) resulting in a total of 103,525 SNPs. Finally, SNPs were filtered for linkage disequilibrium (LD) using using PLINK v. 1.90 with a window size of 80 kb, or about two times the estimated length for 80% LD decay (Cheng et al. 2016), a step size of 5 kb, and a variance inflation factor of 2 (–indep 80 kb 5 2; Purcell et al. 2007), for a final data set of 36,750 SNPs. The four B. cretica genome resequencing samples (Kioukis et al. 2020) were also mapped to the B. oleracea TO1000 genome (Chinese white kale; Parkin et al. 2014; release-41), using BWA (Li and Durbin 2009).
Phylogenetic and Introgression Inference
To test how the different populations are related to one another and which wild relative is most closely related to the cultivated types, we used three different phylogenetic programs; SNPhylo v. 20160204 (Lee et al. 2014) to assess individual sample relationships, IQ-Tree v. 1.6 (Nguyen et al. 2015) to test species level relationships, and TreeMix v. 1.13 (Pickrell and Pritchard 2012) to assess introgression. For SNPhylo (Lee et al. 2014), we ran analyses using an r2 cutoff of 0.1 for LD, minor allele frequency ≥0.01, proportion of missing sites ≤0.4, 1,000 bootstrap replicates, and rooted with sample 238 (B. villosa). For IQ-Tree, we used the Polymorphism-aware phylogenetic Models (PoMo) software (Schrempf et al. 2016; -m GTR+P) to perform phylogenetic comparisons using population genetic data, using 1,000 bootstrap replicates via the ultrafast bootstrap approximation method (Hoang et al. 2018) and B. villosa to root the tree. For our IQ-Tree analysis, we subsampled data to include only those samples which were recovered as monophyletic in our SNPhylo tree (supplementary table S2, Supplementary Material online; samples with asterisks). To test both the topology of relationships and for gene flow between populations, we used TreeMix with the following parameters: no sample size correction (-noss), rooted with B. villosa (-root villosa), bootstrapping over blocks of 500 SNPs (-bootstrap -k 500), and to incorporate between two and ten migration events (-m). TreeMix (Pickrell and Pritchard 2012) was run with samples of B. cretica, B. incana, B. montana, and B. oleracea as individuals, but used samples found in WildC-2, cultivars, and wild relatives as populations. Four-population (f4) tests for treeness (Reich et al. 2009; Pickrell and Pritchard 2012) were used to test the support of the inferred migration edges from Treemix (Pickrell and Pritchard 2012) via the fourpop method.
Population Structure and Variation
To test ancestry proportions and identify the likely genetic structure of described populations we used fastSTRUCTURE v. 1.0 (Raj et al. 2014). We tested K values from 2 to 8 using default convergence criteria and priors followed by the chooseK.py script to determine the appropriate number of model components that best explain structure in the data set.
ANGSD v. 0.925 (Korneliussen et al. 2014) was used to calculate genotype likelihoods for all samples, plus the four additional B. cretica samples from Kioukis et al. (2020), using the parameters -doGlf 2 -doMajorMinor 1 -doMaf 2 -minMapQ 30 -SNP_pval 1e-6, followed by analysis with PCAangsd v. 0.97 (Meisner and Albrechtsen 2018) to visualize population structure, estimate allele frequencies, and calculate individual inbreeding coefficients using the parameters -admix -selection 1 -inbreed 2.
Clustering Based on Expression Profiles
First, Salmon v. 1.2.1 (Patro et al. 2015) was used to acquire transcript abundances for each sample and the estimated number of reads originating from transcripts. The input for expression profile analysis was prepared using tximport (Soneson et al. 2015) with design=∼plantout+cultivar type. Correction for library size (estimateSizeFactors) and variance-stabilizing transformation (vst) was performed in DESeq2 v. 1.28.1 (Love et al. 2014). To test for clustering based on expression profiles, we ran a PCA on the normalized expression values and performed clustering based on Euclidean distance using the “prcomp” and “hclust” functions, respectively, in the “stats” v. 3.6.2 package for R v. 3.6.0 (R Core Team 2018). To assess networks of genes driving differences observed in the PCA, we used WGCNA v. 1.68 (Langfelder and Horvath 2008). Following (Zhang and Horvath 2005), we found that a soft-thresholding power of nine was best as it was the lowest power that satisfied the approximate scale-free topology criterion, resulting in 48 modules of genes.
To determine biological processes which were overrepresented in the resulting modules, A. thaliana orthologs of B. oleracea were determined using both synteny and BLAST. Synteny-based annotations were extracted from table S7 in Parkin et al. (2014), whereas the BLAST annotation was performed using blastn in BLAST v. 2.10.0+ (Camacho et al. 2009). The B. oleracea CDS database was downloaded from https://plants.ensembl.org/Brassica_oleracea/Info/Index (last accessed September 2020), and the A. thaliana CDS database from Araport11_genes.201606.cds.fasta from https://www.arabidopsis.org/ (last accessed Septemeber 2020). The blastn parameters were -evalue 1E-6 -max_target_seqs 1. Genes determined using synteny were then used to perform a GO analysis through PANTHER v. 16.0 (Mi et al. 2021). ANOVAs were used to test for differences in transcript abundance among cultivars, ferals (including WildC-2), and wild relatives by using the “aov” function in R v. 3.6.0 (R Core Team 2018) followed by multiple comparisons with Tukey’s HSD using the function “glht.”
Environmental Niche Modeling
We compiled occurrence records for wild relatives from the Global Biodiversity Information Facility (GBIF, www.gbif.org) data portal and data from Snogerup et al. (1990). From the GBIF data, we omitted records that were duplicated, lacked location data and/or vouchers, were collected from the grounds of botanical gardens, and that were clearly outside of the native range. From the Snogerup et al. (1990) data, we omitted records that could not be georeferenced to <5 km spatial uncertainty. Populations of B. cretica in Lebanon and Israel and of B. incana in Crimea are thought to be likely early human introductions (Snogerup et al. 1990) and records from these areas were omitted. Occurrences above 1200 m altitude were also omitted, as these species rarely occur above 1,000 m and observations above these altitudes may represent anthropogenic dispersals to disturbed areas or misidentifications. To minimize sampling bias due to clustered observations (Beck et al. 2014; Boria et al. 2014), we thinned the filtered occurrences to records greater than or equal to 10 km apart using the “spThin” package in R (Aiello-Lammens et al. 2015). After filtering and thinning, 172 records remained for B. cretica, 65 for B. incana, 57 for B. insularis, 101 B. montana, 15 for B. villosa, and seven and six for the narrow endemics B. macrocarpa and B. hilarionis respectively. Next, we obtained rasters for 19 bioclimatic variables at 2.5 minutes resolution based on contemporary climate data from WorldClim v. 2.0 (Fick and Hijmans 2017) and rasters for 19 bioclimatic variables at 2.5 minutes resolution based on late-Holocene climate projections using data derived from PaleoClim (Fordham et al. 2017; Brown et al. 2018). Rasters were clipped using QGIS v. 3.83 (Open Source Geospatial Foundation Project, QGIS Geographic Information System, http://qgis.org) to constrain the geographical background to windows slightly larger than the area circumscribed by contemporary observational data (Phillips et al. 2009; Acevedo et al. 2012). Although it is common practice to eliminate collinear environmental variables to avoid overfitting (Braunisch et al. 2013), recent simulations have shown that removing highly collinear variables has an insignificant impact on maximum entropy model performance (Feng et al. 2019) so all original variables were included. Projections for late-Holocene habitat suitability were generated using MaxEnt v. 3.4.1 (Phillips et al. 2017). Linear, quadratic, product, and hinge features and jackknife resampling were used to measure variable importance. Relative model performance was evaluated with the adjusted area under receiver operating characteristic (ROC) curve (AUC; DeLong et al. 1988). Although optimal performance cannot be determined with this approach using presence-only data, relative performance can still be assessed (Phillips et al. 2006).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Drs Bob Schnabel, Troy Rowan, Harly Durbin, and Paul Blischak for their assistance with computational analyses, the Mizzou DNA core, Nathan Bivens, Ming-Yi Zhou, and Karen Bromert, for their assistance in getting quality data for sequencing, and our computing resources, specifically the Research Computing Support Services (RCSS) and Informatics Research Core Facility (IRCF) at the University of Missouri. We are also grateful to Andi Kur for providing botanical illustrations of B. oleracea cultivar types and Elizabeth Gjieli, the Geographical Information Manager at the New York Botanical Garden GIS Laboratory, for providing the wild species range map. We thank Sarah Unruh for valuable feedback on early versions of this manuscript, Dr Jeff Ross-Ibarra for his help with interpreting admixture statistics, and two anonymous reviewers whose comments were extremely helpful in improving the manuscript. Funding for this project was provided by USDA-ARS Project No. 8060-21000-024-00D and the National Science Foundation Postdoctoral Fellowship in Biology (Award No. 1711347, S.T.-H.).
Author Contributions
M.E.M., S.D.T.H., A.C.M., H.A., P.P.E., J.D.M., D.A.C.P., G.R.T., C.J.S., G.B., J.L., D.Q.F., T.B., R.G.A., J.E.D., M.A.G., and J.C.P designed the project. M.E.M., E.Y.G., H.A., and S.D.T.H. grew plants and collected tissue. M.E.M. and E.Y.G. extracted and isolated RNA. M.E.M. analyzed the genetic data. A.C.M. produced the species distribution models. C.J.S., D.Q.F., and R.G.A. researched archeology and written data. S.D.T.H., H.A., and J.E.D. assisted with processing and analyzing the data. M.E.M. wrote the original manuscript. S.D.T.H., A.C.M., H.A., P.P.E., J.D.M., D.A.C.P., G.R.T., C.J.S., G.B., J.L., D.Q.F., T.B., R.G.A., J.E.D., M.A.G., and J.C.P. provided critical feedback on manuscript drafts.
Data Availability
The sequences reported in this article have been deposited in the Sequence Read Archive database (accession number PRJNA544934; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA544934). The resulting transcript abundances and VCF file are deposited at DRYAD (https://doi.org/10.5061/dryad.5mkkwh763). Scripts used can be found on github (https://github.com/mmabry/Brassica-oleracea-Population-and-Phylogenetics).
References
- Acevedo P, Jiménez-Valverde A, Lobo JM, Real R.. 2012. Delimiting the geographical background in species distribution modelling. J Biogeogr. 39(8):1383–1390. [Google Scholar]
- Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, Anderson RP.. 2015. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38(5):541–545. [Google Scholar]
- Allaby R.2010. Integrating the processes in the evolutionary system of domestication. J Exp Bot. 61(4):935–944. [DOI] [PubMed] [Google Scholar]
- Allender CJ, Allainguillaume J, Lynn J, King GJ.. 2007. Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica oleracea L. and (n = 9) wild relatives. Theor Appl Genet. 114(4):609–618. [DOI] [PubMed] [Google Scholar]
- Arias T, Pires JC.. 2012. A fully resolved chloroplast phylogeny of the Brassica crops and wild relatives (Brassicaceae: Brassiceae): Novel clades and potential taxonomic implications. Taxon 61(5):980–988. [Google Scholar]
- Bailey LH.1930. The cultivated Brassicas second paper. Gentes Herbarum. v.2(5):209–267. [Google Scholar]
- Beck J, Böller M, Erhardt A, Schwanghart W.. 2014. Spatial bias in the GBIF database and its effect on modeling species’ geographic distributions. Ecol Inform. 19:10–15. [Google Scholar]
- Beebe S, Toro Ch O, González AV, Chacón MI, Debouck DG.. 1997. Wild-weed-crop complexes of common bean (Phaseolus vulgaris L., Fabaceae) in the Andes of Peru and Colombia, and their implications for conservation and breeding. Genet Resour Crop Evol. 44(1):73–91. [Google Scholar]
- Boria RA, Olson LE, Goodman SM, Anderson RP.. 2014. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol Modell. 275:73–77. [Google Scholar]
- Braunisch V, Coppes J, Arlettaz R, Suchant R, Schmid H, Bollmann K.. 2013. Selecting from correlated climate variables: a major source of uncertainty for predicting species distributions under climate change. Ecography 36(9):971–983. [Google Scholar]
- Broodbank C.2015. The making of the Middle Sea: a history of the mediterranean from the beginning to the emergence of the classical world. London: Thames & Hudson.
- Brown JL, Hill DJ, Dolan AM, Carnaval AC, Haywood AM.. 2018. PaleoClim, high spatial resolution paleoclimate surfaces for global land areas. Sci Data. 5:180254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Sun R, Hou X, Zheng H, Zhang F, Zhang Y, Liu B, Liang J, Zhuang M, Liu Y, et al. 2016. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet. 48(10):1218–1224. [DOI] [PubMed] [Google Scholar]
- Cunliffe B.2004. Facing the ocean: the Atlantic and its peoples, 8000 BC-AD 1500. Oxford, England: Oxford University Press. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C.1868. The variation of animals and plants under domestication. Vol. 2. Cambridge, United Kingdom. Cambridge University Press.
- de Candolle A.1855. Géographie botanique raisonnée ou exposition des faits principaux et des lois concernant la distribution géographique des plantes de l’époque actuelle. Paris, V. Masson
- DeLong ER, DeLong DM, Clarke-Pearson DL.. 1988. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3):837–845. [PubMed] [Google Scholar]
- Dixon GR.2006. Origins and diversity of Brassica and its relatives. Wallingford, UK: CABI. p. 1–33.
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edh K, Widén B, Ceplitis A.. 2007. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae). Mol Ecol. 16(23):4972–4983. [DOI] [PubMed] [Google Scholar]
- Engström PG, Steijger T, Sipos B, Grant GR, Kahles A, Rätsch G, Goldman N, Hubbard TJ, Harrow J, Guigó R, et al. 2013. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods. 10(12):1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evershed RP, Arnot KI, Collister J, Eglinton G, Charters S.. 1994. Application of isotope ratio monitoring gas chromatography–mass spectrometry to the analysis of organic residues of archaeological origin. Analyst 119(5):909–914. [Google Scholar]
- Evershed RP, Heron C, Charters S, Goad LJ.. 1992. The survival of food residues: new methods of analysis, interpretation and application. In: Proceedings of the British Academy. Vol. 77, No. 2. United Kingdom: Oxford University Press.
- Farnham MW.1996. Genetic variation among and within United States collard cultivars and landraces as determined by randomly amplified polymorphic DNA markers. Jashs 121(3):374–379. [Google Scholar]
- Feng X, Park DS, Liang Y, Pandey R, Papeş M.. 2019. Collinearity in ecological niche modeling: confusions and challenges. Ecol Evol. 9(18):10365–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Yan J.. 2019. De novo domestication: an alternative route toward new crops for the future. Mol Plant. 12(5):615–631. [DOI] [PubMed] [Google Scholar]
- Fick SE, Hijmans RJ.. 2017. WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. Int J Climatol. 37(12):4302–4315. [Google Scholar]
- Fordham DA, Saltré F, Haythorne S, Wigley TML, Otto-Bliesner BL, Chan KC, Brook BW.. 2017. PaleoView: a tool for generating continuous climate projections spanning the last 21 000 years at regional and global scales. Ecography 40(11):1348–1358. [Google Scholar]
- Fuller DQ, Denham T, Arroyo-Kalin M, Lucas L, Stevens CJ, Qin L, Allaby RG, Purugganan MD.. 2014. Convergent evolution and parallelism in plant domestication revealed by an expanding archaeological record. Proc Natl Acad Sci U S A. 111(17):6147–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering E, Incorvaia D, Henriksen R, Conner J, Getty T, Wright D.. 2019. Getting back to nature: feralization in animals and plants. Trends Ecol Evol. 34(12):1137–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladis T, Hammer K.. 2001. Nomenclatural notes on the Brassica oleracea-group. Genet Resour Crop Evol. 48(1):7–11. [Google Scholar]
- Gustafsson M, Bentzer B, Von Bothmer B., Snogerup. 1976. Meiosis in Greek Brassica of the oleracea group. Bot Not.129:73–84. [Google Scholar]
- Heaney RK, Fenwick RG, Mithen RF, Lewis BG.. 1987. Glucosinolates of wild and cultivated Brassica species. Phytochemistry 26(7):1969–1973. [Google Scholar]
- Helm J.1963. Morphologisch-taxonomische Gliederung der Kultursippen vonBrassica oleracea L. Kulturpflanze 11(1):92–210. [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin T.1995. Cabbages, kales, etc. Brassica oleracea (Cruciferae). In: Smartt J, Simmonds JW, editors. Evolution of crop plants. 2nd edn. Harlow: Longman Scientific & Technical. p. 76–82. [Google Scholar]
- Hort A.1916. Theophrastus: enquiry into plants. Cambridge (MA): Harvard University Press. [Google Scholar]
- HosakaK, , KianianSF, , McGrathJM, , Quiros CF.. 1990. Development and chromosomal localization of genome-specific DNA markers of Brassica and the evolution of amphidiploids and n = 9 diploid species. Genome 33(1):131–142. [Google Scholar]
- Kaniewski D, Van Campo E, Van Lerberghe K, Boiy T, Vansteenhuyse K, Jans G, Nys K, Weiss H, Morhange C, Otto T, et al. 2011. The Sea Peoples, from cuneiform tablets to carbon dating. PLoS One 6(6):e20232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury CK, Carver D, Greene SL, Williams KA, Achicanoy HA, Schori M, León B, Wiersema JH, Frances A.. 2020. Crop wild relatives of the United States require urgent conservation action. Proc Natl Acad Sci U S A. 117(52):33351–33357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioukis A, Michalopoulou VA, Briers L, Pirintsos S, Studholme DJ, Pavlidis P, Sarris PF.. 2020. Intraspecific diversification of the crop wild relative Brassica cretica Lam. using demographic model selection. BMC Genomics 21(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuz A, Schäfer E.. 2002. A new archaeobotanical database program. Veget Hist Archaeobot. 11(1–2):177–180. [Google Scholar]
- Kroll H.2001. Literature on archaeological remains of cultivated plants (1999/2000). Veget Hist Archaeobot. 10(1):33–60. [Google Scholar]
- Kroll H.2005. Literature on archaeological remains of cultivated plants 1981–2004. Available from: archaeobotany.de/database.html. Accessed January 2, 2016.
- Langfelder P, Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannér C, Bryngelsson T, Gustafsson M.. 1997. Relationships of wild Brassica species with chromosome number 2n= 18, based on RFLP studies. Genome 40(3):302–308. [DOI] [PubMed] [Google Scholar]
- Lanner-Herrera C, Gustafeson M, Filt AS, Bryngelsson T.. 1996. Diversity in natural populations of wild Brassica oleracea as estimated by isozyme and RAPD analysis. Genet Resour Crop Evol. 43(1):13–23. [Google Scholar]
- Lázaro A, Aguinagalde I.. 1998. Genetic diversity in Brassica oleracea L. (Cruciferae) and wild relatives (2n= 18) using isozymes. Ann Bot. 82(6):821–828. [Google Scholar]
- Lee T-H, Guo H, Wang X, Kim C, Paterson AH.. 2014. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C.. 2018. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol. 36(12):1160–1163. [DOI] [PubMed] [Google Scholar]
- Liddell HG, Scott R.. 1940. A Greek-English Lexicon Perseus. Oxford, United Kingdom: Oxford University Press. [Google Scholar]
- Lizgunova TV.1959. The history of botanical studies of the cabbage. Brassica oleracea L. Bull Appl Bot Genet Plant Breed. 32:37–70. [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabry ME, Rowan TN, Pires JC, Decker JE.. 2021. Feralization: confronting the complexity of domestication and evolution. Trends Genet. 37(4):302–305. [DOI] [PubMed] [Google Scholar]
- Maggioni L, von Bothmer R, Poulsen G, Aloisi KH.. 2020. Survey and genetic diversity of wild Brassica oleracea L. germplasm on the Atlantic coast of France. Genet Resour Crop Evol. 67(7):1853–1866. [Google Scholar]
- Maggioni L, von Bothmer R, Poulsen G, Branca F.. 2010. Origin and domestication of cole crops (Brassica oleracea L.): linguistic and literary considerations1. Econ Bot. 64(2):109–123. [Google Scholar]
- Maggioni L, von Bothmer R, Poulsen G, Lipman E.. 2018. Domestication, diversity and use of Brassica oleracea L., based on ancient Greek and Latin texts. Genet Resour Crop Evol. 65(1):137–159. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J, Li Q, Yang X, Qian L, Liu L, Yin J, Frauen M, Li J, Qian W.. 2010. Genomic relationships between wild and cultivated Brassica oleracea L. with emphasis on the origination of cultivated crops. Genet Resour Crop Evol. 57(5):687–692. [Google Scholar]
- Meisner J, Albrechtsen A.. 2018. Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210(2):719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, DuVal AE, Jensen HR.. 2012. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196(1):29–48. [DOI] [PubMed] [Google Scholar]
- Mi H, Ebert D, Muruganujan A, Mills C, Albou L-P, Mushayamaha T, Thomas PD.. 2021. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49(D1):D394–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell ND.1976. The status of Brassica oleracea L. subsp. oleracea (wild cabbage) in the British Isles. Watsonia 11:97–103. [Google Scholar]
- Mitchell ND, Richards AJ.. 1979. Brassica oleracea L. ssp. oleracea (B. sylvestris (L.) Miller). J Ecol. 67(3):1087–1096. [Google Scholar]
- Mithen RF, Lewis BG, Heaney RK, Fenwick GR.. 1987. Resistance of leaves of Brassica species to Leptosphaeria maculans. Trans Br Mycol Soc. 88(4):525–531. [Google Scholar]
- Mithen RF, Magrath R.. 1992. Glucosinolates and resistance to Leptosphaeria maculans in wild and cultivated Brassica species. Plant Breed. 108(1):60–68. [Google Scholar]
- Mittell EA, Cobbold CA, Ijaz UZ, Kilbride EA, Moore KA, Mable BK.. 2020. Feral populations of Brassica oleracea along Atlantic coasts in western Europe. Ecol Evol. 10(20):11810–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed A, Addo-Quaye A, Asare-bediako E.. 2010. Control of diamond back moth (Plutella xylostella) on cabbage (Brassica oleracea var capitata) using intercropping with non-host crops “E. Asare-Bediako,” AA Addo-Quaye and “A. Mohammed” Department of Crop Science, University of Cape Coast, Cape Coast, Ghana. Am J Food Technol. 5:269–274. [Google Scholar]
- Murray MA.2000. Fruits, vegetables, pulses and condiments. Cambridge, England: Cambridge University Press. [Google Scholar]
- Neutrofal F.1927. Zytologische Studien über die Kulturrassen von Brassica oleracea. Oesterr Bot Z. 76:105–115. [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onno M.1933. Die Wildformen aus dem Verwandtschaftskreis Brassica oleracea L. Sterr Bot Z. 82(4):309–334. [Google Scholar]
- Panda S, Martín J, Aguinagalde I.. 2003. Chloroplast and nuclear DNA studies in a few members of the Brassica oleracea L. group using PCR-RFLP and ISSR-PCR markers: a population genetic analysis. Theor Appl Genet. 106(6):1122–1128. [DOI] [PubMed] [Google Scholar]
- Paolucci L.1890. Flora marchigiana. Pesaro, Premiato Stab. Tipo-Lit. Federici.
- Parkin IAP, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE, Town CD, Nixon J, Krishnakumar V, Bidwell SL, et al. 2014. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 15(6):R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Kingsford C.. 2015. .Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14(4):417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelc SE, Couillard DM, Stansell ZJ, Farnham MW.. 2015. Genetic diversity and population structure of collard landraces and their relationship to other Brassica oleracea crops. Plant Genome 8(3):eplantgenome2015.04.0023. [DOI] [PubMed] [Google Scholar]
- Phillips SJ, Anderson RP, Dudík M, Schapire RE, Blair ME.. 2017. Opening the black box: an open-source release of Maxent. Ecography 40(7):887–893. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE.. 2006. Maximum entropy modeling of species geographic distributions. Ecol Modell. 190(3–4):231–259. [Google Scholar]
- Phillips SJ, Dudík M, Elith J, Graham CH, Lehmann A, Leathwick J, Ferrier S.. 2009. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol Appl. 19(1):181–197. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. 2017. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv, 201178. doi: 10.1101/201178. [Google Scholar]
- Prohens J, Gramazio P, Plazas M, Dempewolf H, Kilian B, Díez MJ, Fita A, Herraiz FJ, Rodríguez-Burruezo A, Soler S, et al. 2017. Introgressiomics: a new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 213(7):1–19. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
- Raj A, Stephens M, Pritchard JK.. 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197(2):573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould AF, Mogg RJ, Clarke RT, Gliddon CJ, Gray AJ.. 1999. Variation and population structure at microsatellite and isozyme loci in wild cabbage (Brassica oleracea L.) in Dorset (UK). Genet Resour Crop Evol. 46(4):351–360. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 461(7263):489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reihl S.2014. Archaeobotanical database of Eastern Mediterranean and Near Eastern sites. Available from: https://www.ademnes.de/.
- Schiemann E.1932. Entstehung der kulturpflanzen. Borntraeger. [Google Scholar]
- Schmidl A, Oeggl K.. 2005. Subsistence strategies of two Bronze Age hill-top settlements in the eastern Alps—Friaga/Bartholomäberg (Vorarlberg, Austria) and Ganglegg/Schluderns (South Tyrol, Italy). Veget Hist Archaeobot. 14(4):303–312. [Google Scholar]
- Schrempf D, Minh BQ, De Maio N, von Haeseler A, Kosiol C.. 2016. Reversible polymorphism-aware phylogenetic models and their application to tree inference. J Theor Biol. 407:362–370. [DOI] [PubMed] [Google Scholar]
- Schulz OE.1936. Die natuÈrlichen Panzenfamilien. In: Engler A, Harms H, editors. Cruciferae. 2nd ed. p. 176. [Google Scholar]
- Shyam P, Wu XM, Bhat SR.. 2012. History, evolution, and domestication of Brassica crops. Plant Breed Rev. 35:19–84. [Google Scholar]
- Snogerup S.1980. The wild forms of the Brassica oleracea group (2n= 18) and their possible relations to the cultivated ones. Brassica crops and wild allies.[I]. Tokyo, Japan: Japan Scientific Societies Press. p. 121–132.
- Snogerup S, Gustafsson M, Von Bothmer R.. 1990. Brassica sect. Brassica (Brassicaceae) I. Taxonomy and variation. Willdenowia 19:271–365. [Google Scholar]
- Soneson C, Love MI, Robinson MD.. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Osborn TC, Williams PH.. 1990. Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). Theor Appl Genet. 79(4):497–506. [DOI] [PubMed] [Google Scholar]
- Song KM, Osborn TC, Williams PH.. 1988. Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). Theor Appl Genet. 75(5):784–794. [DOI] [PubMed] [Google Scholar]
- Stansell Z, Hyma K, Fresnedo-Ramírez J, Sun Q, Mitchell S, Björkman T, Hua J.. 2018. Genotyping-by-sequencing of Brassica oleracea vegetables reveals unique phylogenetic patterns, population structure and domestication footprints. Hortic Res. 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup V, Brahmi P.. 2005. Cole crops. In: Dhillon BS, Tyagi RK, Saxena S, Randhawa GJ, editors. Plant Genetic Resources: Horticultural Crops. New Delhi: Narosa Publishing House Pvt. Ltd. p. 75–88.
- The Elder P, Rackham H.. 1950. Natural history with an English translation. Vol 5: libri XVII–XIX. Cambridge (MA): Harvard University Press. [Google Scholar]
- Tomlinson P, Hall AR.. 1996. A review of the archaeological evidence for food plants from the British Isles: an example of the use of the Archaeobotanical Computer Database (ABCD). Internet Archaeol. 1(1). doi: 10.11141/ia.1.5. [Google Scholar]
- Totelin LMV.2009. Hippocratic recipes: oral and written transmission of pharmacological knowledge in fifth- and fourth-century Greece. Leiden, Netherlands: BRILL.
- Turner-Hissong SD, Mabry ME, Beissinger TM, Ross-Ibarra J, Chris Pires J.. 2020. Evolutionary insights into plant breeding. Curr Opin Plant Biol. 54:93–100. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, Valentine DH, Walters SM, Webb DA.1964. Flora Europaea: lycopodiaceae to Platanaceae. London: Cambridge University Press. [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 43:11–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veen M.2011. Consumption, trade and innovation. Leiden, Netherlands: Brill publishers.
- Vavilov NI.1926. Studies on the origin of cultivated plants. Leningrad: Institut de Botanique Appliquee et d’Amelioration des Plantes. [Google Scholar]
- Vavilov N.1951. The origin, variation, immunity and breeding of cultivated plants. Soil Sci. 72(6):482. [Google Scholar]
- Wang H, Vieira FG, Crawford JE, Chu C, Nielsen R.. 2017. Asian wild rice is a hybrid swarm with extensive gene flow and feralization from domesticated rice. Genome Res. 27(6):1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML.2011. Studies in Greek Elegy and Iambus. Walter de Gruyter.
- Widén B, Andersson S, Rao G-Y, Widén M.. 2002. Population divergence of genetic (co)variance matrices in a subdivided plant species, Brassica cretica: g matrix variation in Brassica cretica. J Evol Biol. 15(6):961–970. [Google Scholar]
- Yang S, Zheng Z, Mao L, Li J, Chen B.. 2018. Pollen morphology of selected crop plants from southern China and testing pollen morphological data in an archaeobotanical study. Veget Hist Archaeobot. 27(6):781–799. [Google Scholar]
- Yonge CD.1854. The deipnosophists, or, Banquet of the learned of Athenæus. London: Henry G. Bohn. [Google Scholar]
- Zhang B, Horvath S.. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 4:Article 17. [DOI] [PubMed] [Google Scholar]
- Zhang P.2009. Studies on the origin of Brassica alboglabra Bailey. China Veg. (14):62–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences reported in this article have been deposited in the Sequence Read Archive database (accession number PRJNA544934; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA544934). The resulting transcript abundances and VCF file are deposited at DRYAD (https://doi.org/10.5061/dryad.5mkkwh763). Scripts used can be found on github (https://github.com/mmabry/Brassica-oleracea-Population-and-Phylogenetics).