Abstract
Horizontal gene transfer (HGT) is a major driving force for bacterial evolution. To avoid the deleterious effects due to the unregulated expression of newly acquired foreign genes, bacteria have evolved specific proteins named xenogeneic silencers to recognize foreign DNA sequences and suppress their transcription. As there is considerable diversity in genomic base compositions among bacteria, how xenogeneic silencers distinguish self- from nonself DNA in different bacteria remains poorly understood. This review summarizes the progress in studying the DNA binding preferences and the underlying molecular mechanisms of known xenogeneic silencer families, represented by H-NS of Escherichia coli, Lsr2 of Mycobacterium, MvaT of Pseudomonas, and Rok of Bacillus. Comparative analyses of the published data indicate that the differences in DNA recognition mechanisms enable these xenogeneic silencers to have clear characteristics in DNA sequence preferences, which are further correlated with different host genomic features. These correlations provide insights into the mechanisms of how these xenogeneic silencers selectively target foreign DNA in different genomic backgrounds. Furthermore, it is revealed that the genomic AT contents of bacterial species with the same xenogeneic silencer family proteins are distributed in a limited range and are generally lower than those species without any known xenogeneic silencers in the same phylum/class/genus, indicating that xenogeneic silencers have multifaceted roles on bacterial genome evolution. In addition to regulating horizontal gene transfer, xenogeneic silencers also act as a selective force against the GC to AT mutational bias found in bacterial genomes and help the host genomic AT contents maintained at relatively low levels.
Keywords: horizontal gene transfer, xenogeneic silencer, DNA-binding mechanism, H-NS/MvaT/Lsr2/Rok, bacterial genomic AT content, bacterial genome evolution
Introduction
Horizontal gene transfer (HGT), also known as lateral gene transfer, refers to the exchange of genetic material between distantly related strains or species, in contrast to the vertical transmission from parent to offspring (Brown 2003). In bacteria that reproduce by asexual reproduction, HGT is an important driving force for bacterial evolution and speciation, and large numbers of horizontally acquired genes have been found in bacterial genomes (Ochman et al. 2000). More importantly, HGT promotes the rapid spread of the genetic materials responsible for virulence and antibiotic resistance among bacteria, contributing to the pathogenesis of many infectious diseases and making the existing drugs less effective in treatment (Freeman 1951; Akiba et al. 1960; Groisman and Ochman 1996).
Although horizontally acquired foreign genes enhance the competitiveness of bacteria under certain conditions, newly acquired genes are likely to function inappropriately when incorporated into a new genetic environment. In such situations, they are more likely to decrease rather than increase the fitness of the recipient bacteria (Baltrus 2013). For example, unregulated expression of the horizontally transferred genes may disrupt the existing regulatory networks or waste metabolic resources (Cohen et al. 2011), even though these genes may provide new beneficial traits. Therefore, the success of any given HGT event will be dictated by a balance between the fitness costs associated with a new gene against its potential benefits (Park and Zhang 2012). In order to mitigate these possible deleterious effects, it is suggested that many of the horizontally transferred genes are initially expressed at low levels (Gogarten and Townsend 2005; Park and Zhang 2012), and these sequences may confer a selective advantage for the recipient bacteria when expressed in an appropriate manner.
In the past decades, it has been shown that many bacteria genera have evolved specific proteins, known as “xenogeneic silencers,” that selectively repress the expression of foreign sequences (Navarre et al. 2006; Castang et al. 2008; Gordon et al. 2010; Smits and Grossman 2010; Pfeifer et al. 2016) that display distinct base compositions from the host genome, often with higher adenine and thymine (AT) contents (Lawrence and Ochman 1997; Karlin 2001; Rocha and Danchin 2002). Xenogeneic silencing enhances the ability of bacteria to tolerate the acquisition of foreign sequences and improves the probability that these sequences will be retained in the genome. Silencing thus facilitates HGT by protecting the functional and regulatory integrity of the host bacteria while still allowing the cell to acquire new DNA (Navarre et al. 2007; Ali et al. 2012; Will et al. 2015; Singh et al. 2016). All known xenogeneic silencers prefer to bind to AT-rich sequences, which may explain why horizontally acquired DNA exhibits elevated AT content, as AT-rich foreign sequences are better tolerated and more likely to be retained by recipient cells (Will et al. 2015; Navarre 2016). As a result of their functions, xenogeneic silencers are key regulators of horizontally acquired foreign sequences, including those that are involved in virulence and antibiotic resistance in a large number of bacterial pathogens, such as Enteropathogenic Escherichia coli (EPEC) and Enterohemorrhagic Escherichia coli (EHEC) (Mellies et al. 2007), Salmonella typhimurium (Navarre et al. 2006), Vibrio cholera (Ayala et al. 2017), Shigella spp. (Picker and Wing 2016), Mycobacterium tuberculosis (Liu and Gordon 2012), and Pseudomonas aeruginosa (Castang et al. 2008). Many other AT-rich genes, not of foreign origin, can also be recognized and regulated by xenogeneic silencers (Navarre et al. 2006; Castang et al. 2008; Gordon et al. 2010; Seid et al. 2017). In addition, all known xenogeneic silencers also belong to the nucleoid-associated proteins (NAPs), which play important roles in chromosome organization and transcription regulation (for recent reviews, see Dame et al. 2020; Hołówka and Zakrzewska-Czerwińska 2020).
Known xenogeneic silencers were previously classified into four families based on the sequence similarities and target specificities of their DNA binding domains. These families are represented by the H-NS protein of E. coli, Lsr2 of M. tuberculosis, MvaT of P. aeruginosa, and Rok of Bacillus subtilis (Perez-Rueda and Ibarra 2015). All four families of xenogeneic silencers share a common domain organization pattern, with an N-terminal oligomerization domain and a C-terminal DNA binding domain (Qin et al. 2019). Both domains are essential for their gene silencing function. This structural organization provides the mechanistic basis for gene silencing (Tendeng et al. 2003; Gordon et al. 2008). It was revealed that in a large stretch of DNA, the high-affinity binding sites of H-NS function as the nucleation sites, resulting in the cooperative binding of more proteins to the adjacent low-affinity sites (Rimsky et al. 2001; Bouffartigues et al. 2007). Later studies found that MvaT (Winardhi et al. 2012) and Lsr2 (Qu et al. 2013) also bind to DNA through a highly cooperative process. Although the binding affinity of a single DNA binding domain of these xenogeneic silencers to most sites along the DNA is relatively weak, with an equilibrium dissociation constant (Kd) of about 10−5–10−6 M, the oligomerized silencer protein complex can bind to long stretches of DNA with much higher affinity through its multivalent binding (Bouffartigues et al. 2007; Ruthenburg et al. 2007; Gordon et al. 2011; Qu et al. 2013; Ding et al. 2015). This cooperative binding promotes the formation of nucleoprotein filaments, which results in bridging (Dame et al. 2000; Dame et al. 2005; Chen et al. 2008) or stiffening (Amit et al. 2003; Lim, Lee et al. 2012; Lim, Whang et al. 2012; Winardhi et al. 2012; Qu et al. 2013; Winardhi et al. 2014; Winardhi et al. 2015) of DNA segments, and play important roles in bacterial chromosome compaction and gene silencing (Fang and Rimsky 2008; Ali et al. 2012; Lim, Lee et al. 2012; Kotlajich et al. 2015; Grainger 2016). Besides the core genome, homologs of these proteins are also widely found in plasmids, phages, and genomic islands (Pina-Iturbe et al. 2020). The architects of these xenogeneic silencers, and their functions in response to environmental changes or in phage–host interactions have been reviewed recently (Pfeifer et al. 2019; Qin et al. 2019).
Bacterial genomes display a wide range of base compositions, with AT contents ranging from ∼25% to 80%, depending on the species. Escherichia coli, for example, has a genome AT content of ∼48%, while Pseudomonas sp. are more GC-rich, with genomic AT contents of ∼34%. Although the xenogeneic silencers currently known all prefer to bind AT-rich foreign DNA sequences, it is clear that a segment of DNA that is “AT-rich” in Pseudomonas, for example with an AT content of 43%, could be viewed as “GC-rich” in the context of the E. coli genome. It is likely, then, that xenogeneic silencers selectively recognize and silence DNA sequences that appear foreign only with respect to their particular genomic background. It also suggests that these silencers might, in turn, drive the evolution of the genome through the gradual elimination of high-affinity silencing binding sites within core genes. This would be analogous to how specific restriction enzyme target sequences are under-represented in the genomes of bacteria that encode those enzymes (Aras et al. 2002).
In this review, we compare the DNA binding preferences of the known xenogeneic silencer family proteins, and the molecular mechanisms determining their specificities in DNA recognition, as it relates to the genomic backgrounds of their species. Not surprisingly, we observe that the DNA binding preferences and the distributions of different families of xenogeneic silencers are highly correlated with the bacterial genomic AT contents, which implies that xenogeneic silencers may play important roles in the evolution of bacterial genomes, not merely through affecting horizontal gene transfer per se but also on the base compositions of the core genomes.
DNA Binding Preferences of Xenogeneic Silencers
The AT-rich DNA binding preference is one of the characteristics that were noted when Lsr2 (Gordon et al. 2010), H-NS (Grainger et al. 2006; Navarre et al. 2006; Oshima et al. 2006; Kahramanoglou et al. 2011), MvaT (Castang et al. 2008), and Rok (Seid et al. 2017) were initially identified as xenogeneic silencers. The properties of the DNA sequences bound by Lsr2 of M. tuberculosis, H-NS of Salmonella enterica, MvaT of P. aeruginosa, and Rok of B. subtilis were revealed from protein binding microarray (PBM) experiments (Gordon et al. 2011; Ding et al. 2015; Duan et al. 2018). These PBM experiments assessed the relative binding preferences of each protein towards all possible 8-mer DNA sequences, assigning each sequence an enrichment score (E-score), ranging from 0.5 (most favored) to −0.5 (most disfavored) (Berger et al. 2008).
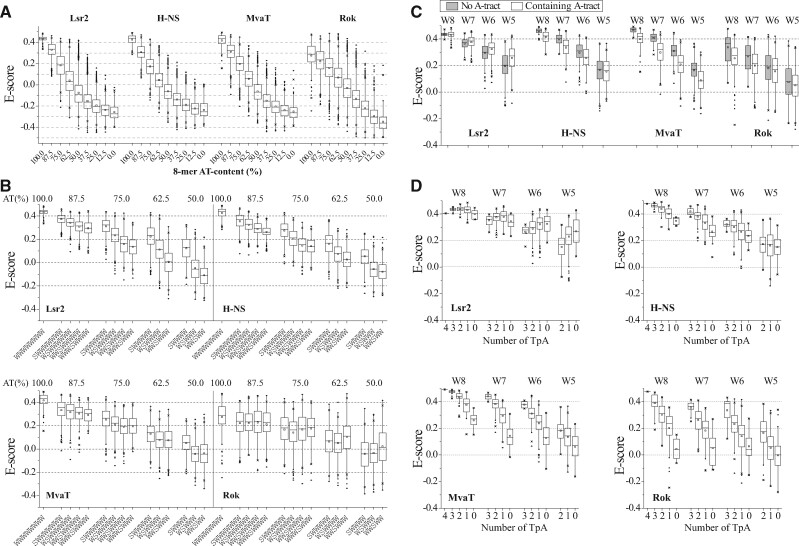
In general, the DNA binding preferences of Lsr2, H-NS, MvaT, and Rok are all positively correlated with 8-mer sequences that are rich in A or T bases (fig. 1A) (Gordon et al. 2010; Gordon et al. 2011; Ding et al. 2015; Duan et al. 2018). However, a more granular analysis of the PBM data indicated that AT content is not the only determinant for their binding affinities. We observed that the E-scores of 8-mers with the same AT content could differ significantly and were often influenced by other factors such as the presence of A-tracts, TpA steps, interruption by GC base-pairs, and even specific sequence motifs. Notably, binding by each of the four families of xenogeneic silencers was affected differently by various structural features within the target DNA sequence.
Fig. 1.
Influence of AT contents, GC insertions, A-tracts, and TpA steps on the binding of Lsr2, H-NS, MvaT, and Rok. (A) Box plot of the E-scores of 8-mers with different AT-content. Bands at the bottom, top, and inside of the box represent the first quartile, the third quartile, and the median, respectively. The small box represents the mean value. The whiskers indicate the highest and lowest points within the 1.5 interquartile range. The black dots represent the outliers. The crosses represent 99% and 1%. (B) Influence of GC insertions on the binding of Lsr2, H-NS, MvaT, and Rok to AT-rich sequences. E-scores of 8-mers that contain different AT-rich motifs are compared. W: A/T; S: G/C. The other positions of these 8-mers which are not shown are GC base pairs. (e.g., SWWWWWW includes SSWWWWW and SWWWWWS.) (C) Distribution of the E-scores of 8-mers with 5–8 contiguous AT base pairs containing A-tracts or not. Wj (j = 5/6/7/8) represents 8-mers that contain j contiguous A/T and 8-j G/C. (D) Distribution of the E-scores of 8-mers with 5–8 contiguous AT base pairs containing different numbers of TpA steps.
In figure 1B, the positional effects of G base-pairs within an AT-rich DNA sequence are shown with respect to binding by the different types of silencers. As expected, for all four xenogeneic silencers, the median E-scores of 8-mer sequences with 5–8 contiguous AT sequences (W: A/T; S: G/C; SiWjSk, i ≥ 0, j ≥ 5, k ≥ 0, i + j + k = 8) are lower when a terminal AT base pair is substituted with a GC base pair (e.g., WWWWWWWW > SWWWWWWW). When the position of the GC base pair shifts closer to the center of the binding site, the median E-scores for Lsr2 and H-NS are further reduced (e.g., SWWWWWWW > WSWWWWWW > WWSWWWWW > WWWSWWWW) (fig. 1B), indicating that both Lsr2 and H-NS prefer contiguous AT sequences without interruption by a G or C (Gordon et al. 2011). However, the presence of a centrally located G/C base within the target sequence only has a minor negative effect on MvaT binding, indicating that MvaT has a higher tolerance for G or C insertion (Ding et al. 2015). Contrary to Lsr2 and H-NS, Rok is insensitive to the position of G/C substitution for 7 or 8 contiguous AT base pairs, and even displays a slight preference for a G or C located in the middle of 5 or 6 contiguous AT base pairs (Duan et al. 2018) (fig. 1B).
Even with a stretch of contiguous AT DNA sequences, the binding preferences of xenogeneic silencers are affected differently by the arrangement of base steps, which is known to affect the structure and flexibility of DNA. A TpA step, due to disruptions in base stacking, causes DNA to be locally flexible and forms a wider minor groove compared with ApA and ApT steps (Travers 2004). In contrast, A-tracts (AnTm, n ≥ 0, m ≥ 0, n + m ≥ 4) are more rigid and have the most narrow minor grooves of any DNA sequence (Haran and Mohanty 2009). For Lsr2, the presence of A-tracts in 8-mers generally has a positive effect on its binding (fig. 1C), and TpA steps are not preferred (fig. 1D), especially for 8-mers containing shorter (5 or 6) contiguous AT base pairs flanked by GC. However, the length of A-tracts may also affect Lsr2 binding, as the presence of 7 or 8-mer A-tracts (e.g., W8 or W7 sequences with no TpA steps) have a negative effect (fig. 1D). Conversely, H-NS, MvaT, and Rok all prefer TpA steps to A-tracts, and DNA sequences with more TpA steps tend to have higher median E-scores (Ding et al. 2015; Duan et al. 2018) (fig. 1C and D).
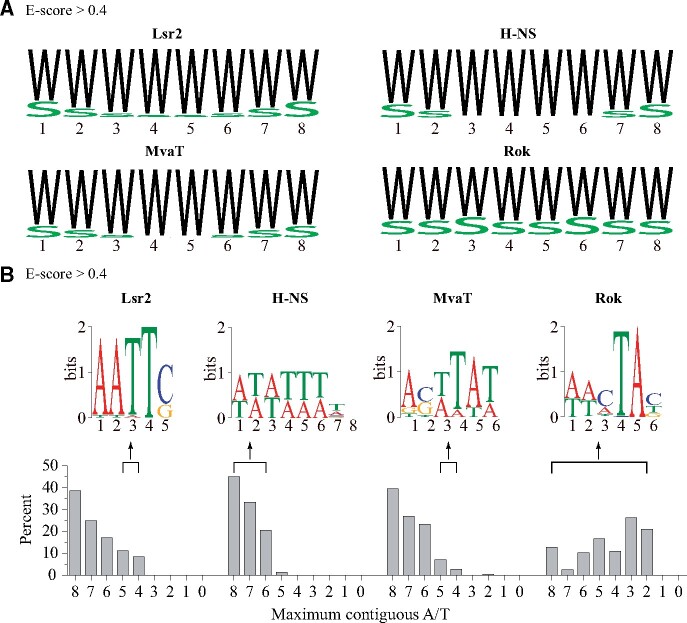
Although Rok generally prefers to bind AT-rich sequences, there are many GC-containing sequences among its high-affinity 8-mers (E-scores > 0.40), as evident from the WebLogo (Crooks et al. 2004) analysis (fig. 2A). Many of these high-affinity 8-mers only have 2 or 3 contiguous AT sequences (fig. 2B). A 6-mer MEME (Bailey et al. 2009) motif was found in 139 of the 157 8-mers with E-scores > 0.40, which contains a TpA step with a preceding C or A (fig. 2B). The sixth position of the motif is less conserved, and the most preferred 5-mers by Rok are “AACTA,” “TACTA,” and “ATATA” (Duan et al. 2018). This sequence specificity is critical to enable Rok to recognize foreign genes, which will be discussed later.
Fig. 2.
Base compositions of high-affinity 8-mers with E-scores > 0.4 for Lsr2, H-NS, MvaT, and Rok. (A) Frequency logos for 8-mers with E-scores > 0.40 generated with the WebLogo program. W: A/T; S: G/C. (B) 8-mers are classified into different groups according to their maximum length of contiguous A/T. The percentage of each group among high-affinity 8-mers with E-scores > 0.40 was calculated and compared. The features of certain groups of 8-mers are analyzed using MEME.
For comparison, we also analyzed the high-affinity 8-mers (E-scores > 0.40) for Lsr2, H-NS, and MvaT from the PBM data. 98.7% of the high-affinity 8-mers (E-scores > 0.40) of H-NS have at least 6 contiguous AT base pairs, much higher than that of Lsr2 (74.4%), MvaT (71.8%), or Rok (25.0%) (fig. 2B). WebLogo analysis showed that among the high-affinity 8-mers, there is no GC base pair at the central four positions for H-NS and the central two positions for MvaT, while GC base pairs may appear at all positions for Lsr2. These findings indicate that Lsr2 may also have a certain degree of tolerance for the presence of a GC base pair, which might be even higher than that of MvaT.
The MEME program did not produce any specific feature other than AT-richness using these high-affinity sequences for H-NS, MvaT, or Lsr2 with ≥ 6 contiguous AT base pairs (e.g., the MEME motif shown for H-NS) (fig. 2A). However, MEME analysis of the sequences with 4 or 5 contiguous AT base pairs of Lsr2 revealed a conserved “AATTG/C” sequence, indicating that Lsr2 tolerates GC base pair when there is an adjacent “AATT” motif (fig. 2B). This motif can also be found in 41.3% of the sequences with 6–8 contiguous AT base pairs. MEME analysis of the highly preferred sequences with 4 or 5 contiguous AT base pairs of MvaT did not reveal any specific pattern, except a 6-bp AT-rich motif with a GC base pair at the second position (fig. 2B), consistent with the higher tolerance of MvaT towards GC insertion.
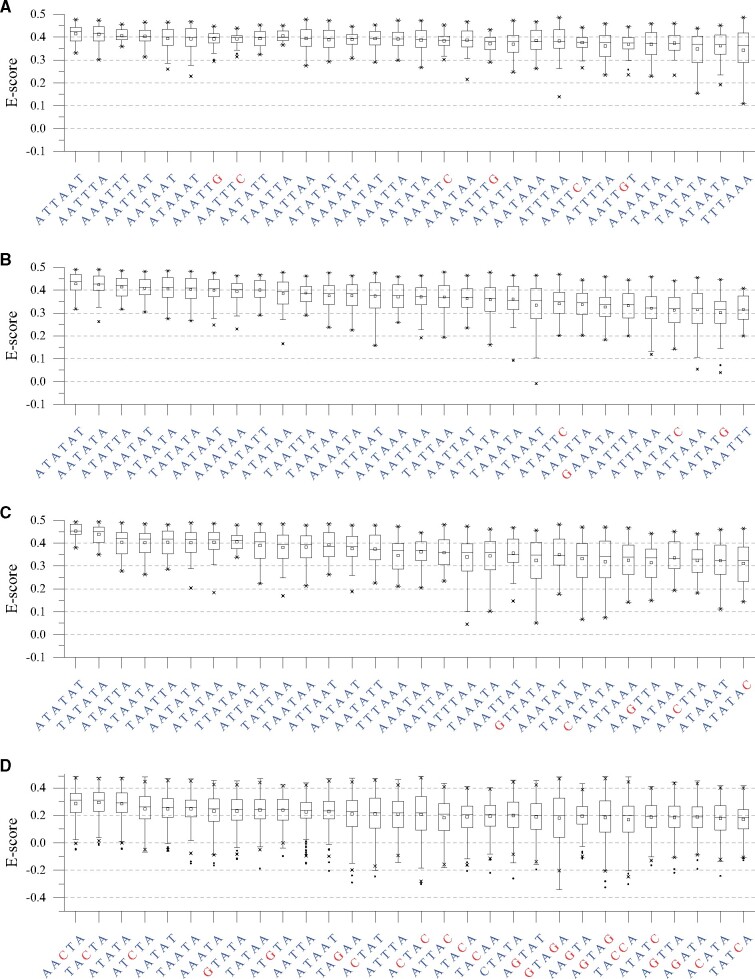
MEME-based determination of preferred binding sites, however, averages several sequences together without retaining information as to how the presence of specific bases at specific positions correlates to specific bases at other positions. A more granular analysis of DNA binding sites is presented to compare the preferences of xenogeneic silencers towards each 6/5-bp sequence, according to the E-score distribution of 8-mers containing it. Ranked by the median E-scores, the top 30 6-bp sequences preferred by Lsr2, H-NS, and MvaT, as well as the top 30 5-bp sequences for Rok, are shown in fig. 3, which reflect the DNA preferences discussed above more directly. Among the top sequences, AATT appears more frequently for Lsr2 (fig. 3A), and TpA steps are favored by H-NS/MvaT/Rok (fig. 3B–D), while the top two sequences for Rok are T/AACTA (fig. 3D). Lsr2 shows the highest compatibility towards different AT-rich 6-bp sequences (slow declining rate of median E-score) and displays a higher tolerance towards GC base pairs when AATT is present (fig. 3A), while MvaT shows a tolerance for G/C interruptions in the middle (fig. 3B and C). These sequences may provide a guidance for finding potential binding sites of xenogeneic silencers in bacterial genomes.
Fig. 3.
Top 30 6-bp sequences preferred by Lsr2 (A), H-NS (B), and MvaT (C), as well as top 30 5-bp sequences preferred by Rok (D) ranked by the median E-score of 8-mers. Preferences towards all 6- or 5-bp sequences could be found in supplementary data set S1, Supplementary Material online.
DNA Recognition Mechanisms of Xenogeneic Silencers
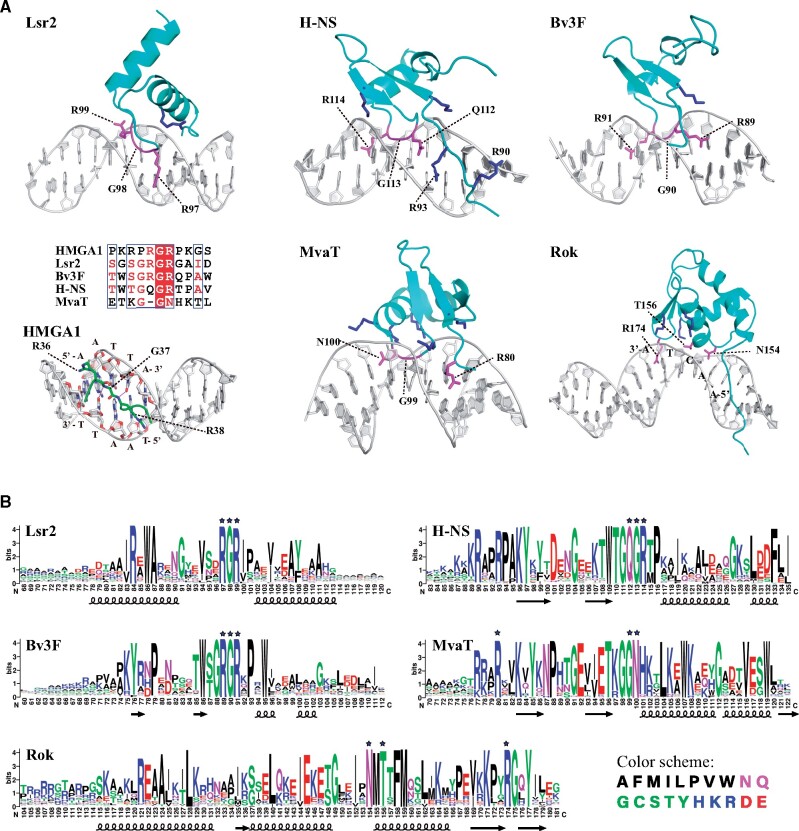
The reason why different AT-rich sequences are preferred by each of the C-terminal DNA binding domains of Lsr2, Bv3F, H-NS, MvaT, and Rok, can be explained by an understanding of their structures and how they bind their DNA targets. All four silencers bind AT-rich DNA at the minor groove and are outcompeted by the minor groove binding drug netropsin (Gordon et al. 2011; Ding et al. 2015; Duan et al. 2018). However, subtle but important differences in how these proteins bind the minor groove underlie their unique specificities.
Lsr2 is the first xenogeneic silencer for which the DNA recognition mechanism was elucidated. The C-terminal domain of Lsr2 (Lsr2CTD) mainly consists of two α-helices linked by a loop (fig. 4A and B). Molecular docking based on solution NMR titration data revealed that a trio of conserved residues (RGR) within the loop insert into the DNA minor groove, with the sidechains of the two arginine residues residing along the floor of the groove and pointing away from each other (Gordon et al. 2010) (fig. 4A). The substitution of these two arginine residues with alanine almost completely abolishes the DNA binding ability of Lsr2CTD (Gordon et al. 2011). Yet another conserved arginine residue (R84) stabilizes the binding by interacting with the DNA phosphate groups. This structural model explains why Lsr2 binding is so sensitive to disruption by the presence of GC base pairs. Namely, that the 2-NH2 group of G that protrudes into the minor groove can sterically prevent the insertion of the Lsr2 “RGR” motif. Interestingly, the conformation of DNA binding loop of Lsr2 highly resembles the DNA-bound “AT-hook” structure of HMGA1, which also has the “RGR” signature motif (Huth et al. 1997; Fonfria-Subiros et al. 2012), and both Lsr2 and HMGA1 share a common DNA binding preference towards AT-rich DNA containing “AATT” (Bustin and Reeves 1996; Cui and Leng 2007). Work on the structures of HMGA1/DNA complexes can help us better understand the DNA recognition mechanism of Lsr2.
Fig. 4.
DNA binding mechanisms of Lsr2, HMGA1, H-NS, Bv3F, MvaT, and Rok. (A) Structures/Models showing the DNA binding mechanisms of the C-terminal domains of Lsr2, H-NS, Bv3F, MvaT (PDB code: 2MXF), Rok (PDB code: 5ZUX), and the AT-hook motif of HMGA1 (PDB code: 3UXW). Residues responsible for base recognition and conserved arginine/lysine residues interacting with DNA phosphate groups are colored magenta and blue, respectively. (B) Sequence logos of the C-terminal domains of H-NS, Lsr2, MvaT, Bv3F, and Rok family xenogeneic silencers. The homologs of xenogeneic silencers were obtained by searching the non-redundant database of NCBI using the blast program. For each family of xenogeneic silencers, several different proteins from distantly related species were used as seeds in order to get more comprehensive information. The sequences (supplementary data set S2, Supplementary Material online) were then aligned using Clustal Omega (Madeira et al. 2019) and analyzed using the WebLogo (Crooks et al. 2004) program. Residues involved in DNA recognition are marked with stars.
HMGA1 is one of the high mobility group A family proteins (HMGA, formerly named HMG-I/Y), which are nonhistone chromosomal proteins found in eukaryotes and act as architectural transcription factors (Reeves 2001). HMGA1 is composed of three DNA binding domains (DBD1, DBD2, and DBD3), and each contains an AT-hook motif with the consensus sequence “RGRP.” Solution structures of HMGA1 DBD2/3 in complex with a double-stranded DNA (GGGAAATTCCTC) were determined in 1997 (Huth et al. 1997). In 2012, the crystal structure of HMGA1 DBD3 in complex with a DNA molecule d (CGAATTAATTCG)2 was reported (Fonfria-Subiros et al. 2012). In each of these structures, the AT-hook motif binds the minor groove of the AATT sequence. This sequence specificity is partially due to the hydrogen bonds formed between the NH groups of the glycine and the following arginine residues in the “RGRP” motif with the O2 atoms of two thymine bases on opposite strands (Fonfria-Subiros et al. 2012). It was also suggested that the floor of the minor groove of the AATT binding site may provide optimal hydrophobic interactions with the side chains of two arginine residues, and the two guanine groups may also form hydrogen bonds with DNA residues next to “AATT” (fig. 4A) (Huth et al. 1997). Analogous to HMGA1, the “RGR” signature of the AT-hook-like motif in Lsr2 also makes contact with 6 base pairs of DNA, while the central “AATT” is expected to be important for high-affinity binding.
H-NS is the most extensively studied xenogeneic silencer. In the early 1990s, Shindo et al. (1995, 1999) determined the solution structure of the C-terminal DNA binding domain of E. coli H-NS and studied its interaction with DNA using NMR spectroscopy. But the structure is of low quality and how H-NS bind to DNA had remained unclear until our study (Gordon et al. 2011). The DNA binding mechanism of the Lsr2CTD inspired a further analysis of the structural parameters that dictate binding by H-NS. It was found that the primary sequence “SGRGR” of the Lsr2 AT-hook-like motif is structurally similar and functionally analogous to the conserved “TGQGR” sequence motif in the DNA binding domain of H-NS from E. coli or Salmonella. In the H-NS example, the “RGR” signature sequence has been replaced by “QGR.” However, in several other H-NS homologous proteins, including Bv3F from Burkholderia vietnamiensis, the sequence is “RGR” (fig. 4A and B). Solution structures of the C-terminal domains of Salmonella H-NS (H-NSCTD) and Bv3F (Bv3FCTD) revealed that the conserved sequences also adopt an AT-hook like conformation resembling that of the Lsr2CTD, even though the overall structures of the H-NS and Lsr2 DNA binding domains are quite different (fig. 4A) (Gordon et al. 2011). Structural models generated with HADDOCK based on the NMR DNA titration data indicate that H-NSCTD and Bv3FCTD recognize AT-rich DNA by inserting the “QGR” or “RGR” residues of the AT-hook-like motif into the DNA minor groove (fig. 4A). Mutations that change either the “QGR” or “RGR” motifs into “AGA” almost completely abolished the DNA binding ability of these proteins (Gordon et al. 2011). This binding model was further supported by a later molecular dynamics simulation study that also found the “QGR” residues can fully insert into the minor groove and form hydrogen bonds to the bases with both backbone and sidechain groups (Riccardi et al. 2019).
Why do some H-NS-like proteins have the QGR sequences instead of an RGR sequence in their critical binding motif? It is likely that this difference contributes to important but subtle differences in the DNA binding preferences between H-NS, which has the QGR motif, and Lsr2 and Bv3F, which have the RGR motif. H-NS prefers to bind sequences with multiple TpA steps instead of the “AATT” sequence. It is possible that the DNA binding preference of Bv3F, which is more structurally similar to H-NS but has the Lsr2-like “RGR” signature within its binding motif, will display binding preferences closer to that of Lsr2. Protein sequence alignments also show that H-NS and Bv3F have differences in other conserved residues outside of the QGR/RGR motif (fig. 4B). Hence, we suggest that H-NS and Bv3F like proteins should be classified as two distinct subgroups of silencers. In the following sections, we will show that the distributions of H-NS and Bv3F family proteins, as well as the AT contents of their resident genomes, also exhibit different features, which may reflect important functional differences between the silencers that contain a “QGR” and “RGR” motif.
In addition to the C-terminal domain, a less-ordered region (residues 87–94) between the N-terminal and C-terminal domains of H-NS from Salmonella or E. coli also plays a direct role in DNA binding (Fernandez-de-Alba et al. 2013; Gao et al. 2017). Two arginine residues (R90 and R93) within this region are highly conserved among H-NS homologs with the “QGR” signature, but not among Bv3F and other H-NS homologs (fig. 4B). Mutation of R90 or R93 can reduce the DNA binding affinity of H-NS (Gao et al. 2017), and the recent MD simulation study also suggested that residue R93 aids DNA binding by interacting with the DNA phosphate backbone (Riccardi et al. 2019). The detailed molecular mechanisms whereby H-NS-like and Bv3F-like proteins recognize DNA remain to be explored by structural studies.
MvaT was characterized as an H-NS functional analog in Pseudomonas (Tendeng et al. 2003). In 2015, the solution structure of MvaTCTD and DNA complex was determined and this is the first experimental structure of a xenogeneic silencer in complex with DNA (Ding et al. 2015). Although MvaTCTD and H-NSCTD have similar secondary and tertiary structures, there is no “QGR” or “RGR” signature in the loop of MvaTCTD corresponding to the AT-hook-like motif of H-NSCTD. Instead, the structural comparison revealed that the loop in MvaTCTD has a sequence of “KG-GN,” in which the arginine residue of “TGQGR” in the AT-hook-like motif of H-NS is substituted by an asparagine, and the glutamine residue between two glycine residues is missing (fig. 4A). In the complex structure of MvaTCTD/DNA, residues G99 and N100 of the “KG-GN” loop, together with the sidechain of an upstream R80 from the N-terminal region of MvaTCTD, are inserted into the DNA minor groove, an arrangement we term an “AT-pincer” motif (fig. 4A). The backbone amides of G99 and N100, as well as the sidechains of R80 and N100, form hydrogen bonds with DNA bases in the minor groove, covering the “ATATAT” base pairs (Ding et al. 2015). The cavity between R80 and G99/N100 in the DNA binding interface of the AT-pincer motif likely explains the higher tolerance of MvaT for G/C interruptions observed in the PBM data (fig. 1B). In addition, several lysine residues form a network that interacts with DNA phosphate groups on both strands. Mutating residues R80, N100, or these lysine residues individually into alanine all significantly reduced the DNA binding affinity of MvaT and impaired its functions in vivo, indicating that both the AT-pincer and the lysine network are critical for DNA binding (Ding et al. 2015). Correspondingly, these residues are also highly conserved among MvaT homologs (fig. 4B). The binding of MvaTCTD distorts the DNA molecule, both expanding the minor groove and rearranging the local base-stacking geometry. This requirement for local DNA flexibility is reminiscent of the TATA-binding protein (Juo et al. 1996) and may account for MvaT’s preference towards TpA steps over A-tracts. The structural rigidity and narrow minor groove of A-tracts would make these sequences far less likely to achieve the distorted conformation most conducive for MvaT binding (Ding et al. 2015).
Rok from B. subtilis shows an entirely distinct DNA binding mechanism compared with Lsr2, H-NS, Bv3F, or MvaT. The C-terminal domain of Rok (RokCTD) adopts a typical winged helix fold with three α-helices and three anti-parallel β-strands (Duan et al. 2018). An NMR structure of RokCTD in complex with a DNA sequence of d (CTAATAACTAGTTATTAG)2 finds that the winged helix domain binds the DNA minor groove at the sequence “AACTA,” a sequence that was identified as a high-affinity 5-bp sequence in a PBM analysis (fig. 3D). Residues N154, T156 at the N-terminal of α3 helix and residue R174 of wing W1 composed of the loop between strands β2 and β3, are inserted in the minor groove of this region. These three residues are highly conserved among Rok homologs (fig. 4B). The NH2 group of N154 forms two hydrogen bonds with the O2 atoms of two consecutive thymine bases, and the OH group of T156 forms a hydrogen bond with the N2 atom of the guanine, while the guanidine group of R174 may form several hydrogen bonds with the TpA step. Like MvaT, the DNA binding of Rok is also assisted by four lysine residues that interact with DNA phosphate groups (fig. 4A). Similar to what was observed with MvaT, Rok binding also leads to large conformational changes of DNA, including minor groove widening and axis bending. Thus, Rok also prefers TpA steps, but dislikes A-tracts (fig. 1D). Consistent with these structural characteristics of Rok, the PBM median E-scores of 8-mers containing “AACTA” or “TACTA” are the highest (fig. 3D) (Duan et al. 2018).
Correlation between Xenogeneic Silencers and Bacterial Genomic AT Contents
It is apparent that xenogeneic silencers may serve as a “safety guard” of the host genome by silencing the expression of incoming foreign sequences as a default state. This reduces the fitness costs of such sequences and buys time for the host bacteria to rewire its genetic circuitry to possibly allow the safe integration of foreign genes into the genome. For this reason, it is essential for xenogeneic silencers to distinguish DNA that is self from nonself. Targeting foreign DNA based on the difference in base composition (AT content) rather than specific sequence features provides a solution to this problem, as it is widely found in bacteria that foreign genes tend to have higher AT contents than host genomes (Lawrence and Ochman 1997; Karlin 2001).
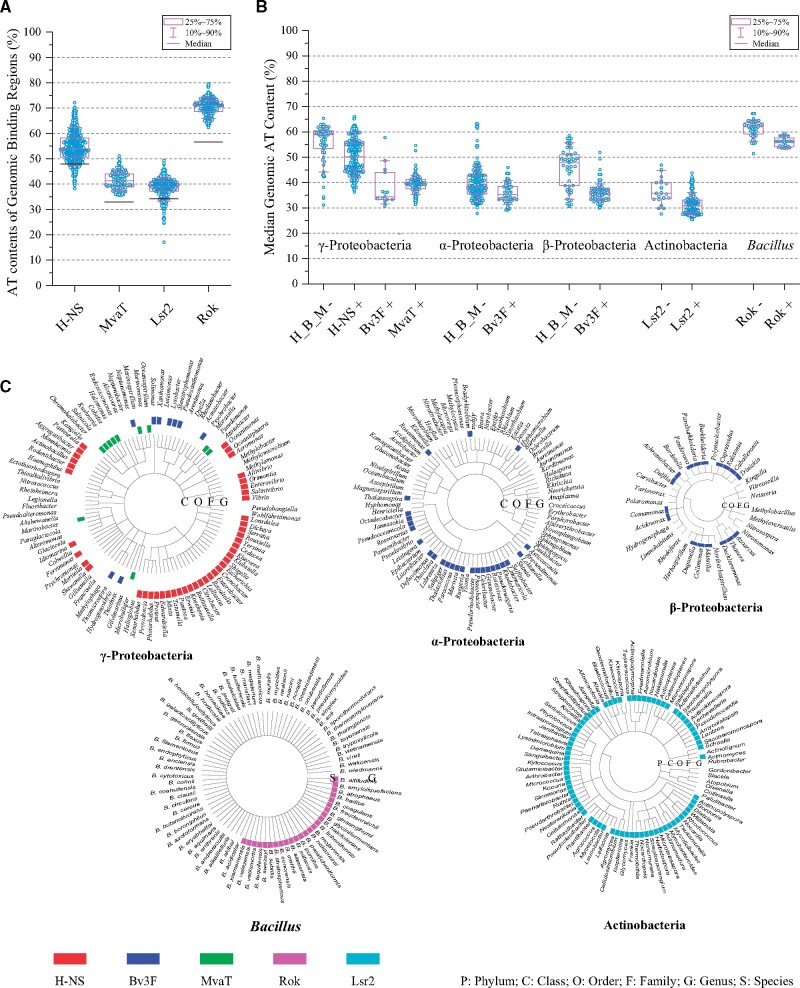
But it should also be noted that the genomic AT contents of various bacterial species differ from each other significantly, implying that the characteristics of a “xenogeneic gene” is not universal. Indeed, the reported genomic binding regions of H-NS (Navarre et al. 2006), MvaT (Castang et al. 2008), Lsr2 (Gordon et al. 2010), or Rok (Seid et al. 2017) identified from ChIP-chip or ChIP-seq experiments are not universally AT-rich (> 50%), but instead show higher AT contents relative to their respective genomic average (fig. 5A). For example, the genomic AT contents of P. aeruginosa (∼33%) and M. tuberculosis (∼34%) are much lower than that of S. typhimurium (∼48%), and accordingly, the AT contents of the MvaT and Lsr2 binding regions are significantly lower than that of H-NS, which is consistent with the PBM data showing that Lsr2 and MvaT have higher tolerances for the presence of GC base pairs than H-NS (fig. 2). A careful analysis of the ChIP-chip experiment data found that the fractions of the bound DNA sequences begin to increase quickly when the sequence AT contents reach ∼38% for Lsr2 and ∼50% for H-NS, which obviously correlate with the host genomic AT contents of M. tuberculosis (∼34%) and S. typhimurium (∼48%), respectively (Gordon et al. 2011) (see fig. S8 of the ref.). These findings suggest that the DNA binding preferences of a xenogeneic silencer evolve to avoid recognizing the core genome as foreign and are calibrated to function in the context of their own specific genetic milieu.
Fig. 5.
Relationship between xenogeneic silencers and bacterial genomic AT-contents. (A) Distributions of the AT contents of the genomic binding regions of H-NS (Navarre et al. 2006), MvaT (Castang et al. 2008), Lsr2 (Gordon et al. 2010), or Rok (Seid et al. 2017) revealed by ChIP-chip or ChIP-seq experiments. The black bar indicates the genomic AT contents of Salmonella typhimurium, Pseudomonas aeruginosa, Mycobacterium tuberculosis, and Bacillus subtilis. (B) Distributions of the genomic AT contents of species with or without known xenogeneic silencers. Species shown here have at least two sets of genomic sequencing data and median genomic sizes over 2 Mbp. “H_B_M –” represents species without any homologs of H-NS, Bv3F, or MvaT. (C) Taxonomy trees showing the distributions of known xenogeneic silencers. The trees were generated using the Common Tree tool of NCBI. Bacterial genera or species that contain one of the five kinds of xenogeneic silencers are marked with a color as indicated. Only those bacterial genera with three or more well-classified species are shown. The figures are produced with the TBtools program (Chen et al. 2020). Details about the distribution analysis are provided in the supplementary methods, Supplementary Material online.
Within each family of xenogeneic silencers, the key residues for DNA recognition are highly conserved (fig. 4B), indicating that they should have similar DNA binding preferences. Thus, it is expected that bacterial species with the same family of xenogeneic silencers may have similar genomic AT contents, and the distribution ranges of their genomic AT contents should be correlated with the DNA binding preferences of the xenogeneic silencers. To verify this, we conducted a more extensive analysis of the bacterial host distribution of different families of xenogeneic silencers. The results revealed that H-NS family proteins that contain the “QGR” DNA binding motif exist in bacterial species of the Gammaproteobacteria class with genomic AT contents mainly in the range of 42–59% (fig. 5B and C). Bv3F family proteins, which adopt the “RGR” type AT-hook-like DNA binding motif, can be found in species of Gamma-, Beta-, and Alphaproteobacteria classes (fig. 5C), with the genomic AT contents mainly ranging from ∼32% to 42% (fig. 5B). MvaT family proteins are also found in Gammaproteobacteria, but mainly in species with relatively lower genomic AT contents (∼35% to 44%). Based on their sequence similarities (fig. 4B), H-NS, Bv3F, and MvaT family proteins should have the same evolutionary origin. Considering that proteobacteria have evolved in the order of Alphaproteobacteria => Betaproteobacteria => Gammaproteobacteria (Gupta 2000), it is possible that Bv3F is ancestral to H-NS and MvaT, which may have emerged when Gammaproteobacteria diverged. Lsr2 family proteins are prevalent in the Actinobacteria phylum (fig. 5C), and the genomic AT contents of most species containing Lsr2 are between 27% and 35% (fig. 5B).
Therefore, the bacterial species containing Lsr2 have the lowest genomic AT content distribution range (median 29.5%), followed by those containing Bv3F (median 35.4%), MvaT (median 39.7%), and H-NS (median 50.2%). Correspondingly, the PBM and ChIP-chip/seq data revealed that Lsr2 shows the greatest ability to bind DNA sequences with relatively low AT contents, followed by MvaT, and then H-NS (figs. 2 and 5A).
Although Bv3F shows overall sequence homology to H-NS, bacteria with Bv3F family proteins have genomic AT contents closer to those with Lsr2, but not H-NS. This can be explained by the finding that both Bv3F and Lsr2 use the “RGR” AT-hook-like motif to recognize DNA, rather than the “QGR” of H-NS. Thus, it appears that bacterial species with lower genomic AT contents tend to have xenogeneic silencers with higher GC tolerance, as the AT contents of xenogeneic genes may also be lower.
Rok is different from H-NS, Bv3F, Lsr2, or MvaT in that it shows a stronger DNA sequence specificity. Bacillus subtilis has a high genomic AT content of about 56%, which means that a majority of its genomic regions are AT-rich. But the most preferred sequences of Rok, “AACTA” and “TACTA,” have dramatically low occurrence frequencies in the genome of B. subtilis, and the abundances of 5-bp sequences with 4 or 5 AT base pairs are negatively correlated with their PBM median E-scores (Duan et al. 2018). Again, this suggests that foreign genes may have base compositions different from the host genome, and those with a high abundance of Rok preferred sequences could be recognized and silenced. This correlation between the DNA binding preferences of Rok and the genomic characteristics of B. subtilis may well explain how Rok recognizes foreign DNA sequences selectively.
A distribution analysis of the Rok homologous proteins in the Bacillus genus revealed that Rok homologs are mostly found in those species with genomic AT contents between 53% and 59% (fig. 5B). In those Bacillus species with higher AT contents (59–68%), such as Bacillus anthracis, the “TACTA” and “AACTA” sequences are much more abundant. Therefore, if Rok were present, it would silence many “core” genes in the genome, very likely including essential genes (Duan et al. 2018). Like Rok, bacterial species containing Lsr2, H-NS, Bv3F, or MvaT also generally have relatively lower genomic AT contents, compared with those without these xenogeneic silencers in the same phylum or class, respectively (fig. 5B).
Within each class of bacteria, species that contain xenogeneic silencers appear to have lower genomic AT content than those that do not (fig. 5B). In addition, the copy number of xenogeneic silencer genes also appears to be correlated with the genomic AT content. For example, bacterial genomes containing two or more genes encoding H-NS family proteins generally have lower AT contents (<50%), while most species with genomic AT contents over 50% only have one H-NS gene (supplementary fig. S1, Supplementary Material online). The same is true for Lsr2. The correlations for Bv3F and MvaT are not significant, while only a few genomes have ≥ 2 rok genes (supplementary fig. S1, Supplementary Material online). Different xenogeneic genes in the same genome may have both overlapping and distinct functions, and may affect the expression level of each other. For example, H-NS and its paralog StpA in E. coli can repress the transcription of each other’s genes (Sonden and Uhlin 1996; Zhang et al. 1996; Qin et al. 2019). And similarly, deletion of mvaT resulted in increased expression of its paralog gene mvaU in P. aeruginosa, and vice versa (Vallet-Gely et al. 2005). It is possible that more copies of xenogeneic silencer genes do not always result in higher levels of xenogeneic silencer proteins in the cell. The overall effects of multiple xenogeneic silencer genes on bacteria remain to be understood.
Besides, for species containing the same kind of xenogeneic silencers, their genomic AT contents exhibit a weak negative correlation with their genomic sizes (supplementary fig. S2, Supplementary Material online), which is a general trend found in bacteria (Guo et al. 2009; Bohlin et al. 2014; Almpanis et al. 2018). Interestingly, most of the outlier species with significantly higher AT contents than the bulk in figure 5B have very small genomic sizes.
It is notable that AT-rich bacteria, including most Gram-positive Firmicutes, lack known xenogeneic silencers, suggesting that silencers would fail to distinguish self from non-self in the context of an AT-rich core genome. It is also worth mentioning that no xenogeneic silencer has yet been found for many bacteria with relatively GC-rich genomes, including most of the species in the Alphaproteobacterial class. It is still not clear whether these bacteria do not have a xenogeneic silencer, or their xenogeneic silencers are simply unrelated to currently known ones. Although some proteins have been proposed to share certain structural or functional similarities with H-NS, such as the Roc/MucR from Brucella abortus (Baglivo et al. 2018) and GapR from Caulobacter crescentus (Lourenço et al. 2020), but there is no clear evidence to show whether they indeed function as xenogeneic silencers.
Multifaced Roles of Xenogeneic Silencers on Bacterial Genome Evolution
The forces that shape the genome include not only horizontal gene transfer but also mutations and recombination (Koonin 2016). The evolutionary forces that have driven such a wide variation in bacterial genomic AT contents, however, have long been discussed and remain under debate (Muto and Osawa 1987; Rocha and Danchin 2002; Foerstner et al. 2005; Nishida 2013; Agashe and Shankar 2014; Reichenberger et al. 2015; Long et al. 2018). The effect of horizontal gene transfer may be underestimated as ancient horizontal acquired genes are likely to gradually acquire the molecular characteristics of the host genome over time due to gene amelioration (Marri and Golding 2008).
The existence of bacteria with highly GC-rich core genomes is, in fact, a bit curious. The DNA these bacteria accumulate via horizontal gene transfer tends to be more AT-rich than the core genome. Furthermore, with regard to point mutations, it has been found that in most bacteria, mutational biases push genomes toward increasing AT content (Rocha and Danchin 2002; Lind and Andersson 2008; Hershberg and Petrov 2010; Lee et al. 2012; Agashe and Shankar 2014; Bohlin et al. 2018). This drive toward AT-richness is particularly pronounced in endosymbionts that discard many of their DNA repair mechanisms (McCutcheon and Moran 2012). But as GC-rich genomes are widespread, and most species have not become overwhelmingly AT-rich during evolution, the AT bias must be opposed by selective forces favoring GC substitutions or purging AT mutations (Rocha and Feil 2010; Lassalle et al. 2015). However, the nature, strength, and source of the selective forces are still not well understood. Some studies found that E. coli or C. crescentus displayed a higher growth rate when GC-rich genes were expressed, compared with those strains harboring AT-rich gene variants. But the underlying mechanism is not clear, and no comparable results were observed in P. aeruginosa (Raghavan et al. 2012; Kelkar et al. 2015). In eukaryotes, GC alleles are more favored during homologous recombination, which was named GC-Biased Gene Conversion (gBGC). Lassalle et al. (2015) found that gBGC may also occur in bacteria and contribute to the evolution of AT contents of bacteria genomes. However, AT contents in bacteria evolve through a complex combination of factors, and recombination alone is unlikely to counteract the AT-enrichment of bacterial genomes (Bobay and Ochman 2017). More recently, Weissman et al. (2019) found that the non-homologous end-joining repair pathways of bacteria may elevate the genomic GC contents.
It was also hypothesized that the genomic AT contents of bacteria might be related to the variation in bacterial nucleoid-associated proteins (NAPs) (Nishida 2012a,b, 2013), which is supported by the evidence and arguments presented here. We show that bacterial species containing the same family of xenogeneic silencers have a limited distribution range of genomic AT contents, which is related to the DNA binding preferences of the xenogeneic silencers and the number of xenogeneic silencer genes per genome. Meanwhile, bacterial species generally tend to have lower genomic AT contents when xenogeneic silencers are present in their genomes, compared with those species without any known xenogeneic silencers from the same phylum/class/genus. These correlations highlight the role of xenogeneic silencers on bacterial genome evolution. On the one hand, by safely guiding the integration of foreign sequences into the host genome, xenogeneic silencers will promote the acquisition of foreign genes. On the other hand, xenogeneic silencers may also act as a selective force on the random mutation events against the GC to AT mutational bias, especially for core genomic genes.
It was found that the AT-to-GC substitution rate is higher than the GC-to-AT substitution rate for the core genes of Salmonella, while the two horizontally acquired islands SPI-1 and SPI-4 have a much higher GC-to-AT substitution rate than that of AT-to-GC substitution, consistent with the selective pressure to maintain their regulation by H-NS (Desai et al. 2013). In the presence of xenogeneic silencers, it is possible that some GC to AT mutations cannot be preserved during evolution, if they lead to the silencing of essential genes by xenogeneic silencers. Consistently, the core genes shared by Salmonella and E. coli strains have relatively lower AT content than the genomic averages. It was also found that noncoding regions of the bacterial genomes are more AT-rich than coding regions (Bohlin et al. 2008), and both the AT contents and its variations in coding regions are significantly lower in core genomes than accessory genomes (Bohlin et al. 2017).
Xenogeneic silencers belong to a larger group of nucleoid-associated proteins (NAPs), each of which may affect the rates of both mutation and recombination, two key processes for the evolution of bacterial genomes (Tavita et al. 2012; Warnecke et al. 2012; Kivisaar 2019). NAPs were found to have a significant growth phase-specific effect on mutation dynamics in E. coli, as they can not only shield DNA from certain mutagenesis processes during the stationary phase but also interfere with efficient DNA repair during the exponential phase (Warnecke et al. 2012). In addition, NAPs may also affect the frequency of homologous recombination, an essential process for horizontal gene transfer, as chromosomal DNA bound with NAPs might be less accessible to homologous recombinant processes (Tavita et al. 2012).
Taken together, the roles of xenogeneic silencers on the evolution of bacterial genomes are multifaced, and of greater impact on both gene content and nucleotide compositions than previously expected. Bacteria appear to have evolved or acquired the right xenogeneic silencers according to their own core genomic sequence features during evolution, which in return may affect their acquisition of foreign genes, and act as a selective force on the random mutation events of genomes.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by grant 2016YFA0501203 from the Ministry of Science and Technology of China, and grant 31570734 from the National Natural Science Foundation of China to B.X., and grants from Canadian Institutes of Health Research (CIHR) PJT-156261 and PJT-173353 to J.L.
Data Availability
The PBM data for Lsr2, H-NS, MvaT, and Rok can be found in the supplementary data of our previously published works (Gordon et al. 2011; Ding et al. 2015; Duan et al. 2018). DNA binding preferences of Lsr2, H-NS, MvaT, and Rok towards 6- or 5-bp sequences can be found in supplementary data set S1, Supplementary Material online. C-terminal sequence alignments of different xenogeneic silencer family proteins are presented in supplementary data set S2, Supplementary Material online. Median genomic AT-contents and sizes of bacterial species with or without xenogeneic silencers are provided in supplementary data set S3, Supplementary Material online.
References
- Agashe D, Shankar N.. 2014. The evolution of bacterial DNA base composition. J Exp Zool B Mol Dev Evol. 322(7):517–528. [DOI] [PubMed] [Google Scholar]
- Akiba T, Koyama K, Ishiki Y, Kimura S, Fukushima T.. 1960. On the mechanism of the development of multiple-drug-resistant clones of Shigella. Jpn J Microbiol. 4(2):219–227. [DOI] [PubMed] [Google Scholar]
- Ali SS, Xia B, Liu J, Navarre WW.. 2012. Silencing of foreign DNA in bacteria. Curr Opin Microbiol. 15(2):175–181. [DOI] [PubMed] [Google Scholar]
- Almpanis A, Swain M, Gatherer D, McEwan N.. 2018. Correlation between bacterial G+C content, genome size and the G+C content of associated plasmids and bacteriophages. Microbial Genomics. 4(4):e000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit R, Oppenheim AB, Stavans J.. 2003. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 84(4):2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aras RA, Small AJ, Ando T, Blaser MJ.. 2002. Helicobacter pylori interstrain restriction-modification diversity prevents genome subversion by chromosomal DNA from competing strains. Nucleic Acids Res. 30(24):5391–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala JC, Silva AJ, Benitez JA.. 2017. H-NS: an overarching regulator of the Vibrio cholerae life cycle. Res Microbiol. 168(1):16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglivo I, Pirone L, Malgieri G, Fattorusso R, Roop Ii RM, Pedone EM, Pedone PV.. 2018. MucR binds multiple target sites in the promoter of its own gene and is a heat-stable protein: is MucR a H-NS-like protein? FEBS Open Bio. 8(4):711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS.. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue):W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA.2013. Exploring the costs of horizontal gene transfer. Trends Ecol Evol. 28(8):489–495. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133(7):1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay LM, Ochman H.. 2017. Impact of recombination on the base composition of bacteria and archaea. Mol Biol Evol. 34(10):2627–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlin J, Eldholm V, Brynildsrud O, Petterson JH, Alfsnes K.. 2018. Modeling of the GC content of the substituted bases in bacterial core genomes. BMC Genomics 19(1):589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlin J, Eldholm V, Pettersson JH, Brynildsrud O, Snipen L.. 2017. The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes. BMC Genomics 18(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlin J, Sekse C, Skjerve E, Brynildsrud O.. 2014. Positive correlations between genomic %AT and genome size within strains of bacterial species. Environ Microbiol Rep. 6(3):278–286. [DOI] [PubMed] [Google Scholar]
- Bohlin J, Skjerve E, Ussery DW.. 2008. Investigations of oligonucleotide usage variance within and between prokaryotes. PLoS Comput Biol. 4(4):e1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S.. 2007a. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol. 14(5):441–448. [DOI] [PubMed] [Google Scholar]
- Brown JR.2003. Ancient horizontal gene transfer. Nat Rev Genet. 4(2):121–132. [DOI] [PubMed] [Google Scholar]
- Bustin M, Reeves R.. 1996. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 54:35–100. [DOI] [PubMed] [Google Scholar]
- Castang S, McManus HR, Turner KH, Dove SL.. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A. 105(48):18947–18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R.. 2020. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 13(8):1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen JM, Ren H, Shaw JE, Wang YJ, Li M, Leung AS, Tran V, Berbenetz NM, Kocíncová D, Yip CM, et al. 2008. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 36(7):2123–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O, Gophna U, Pupko T.. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol. 28(4):1481–1489. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T, Leng F.. 2007. Specific recognition of AT-rich DNA sequences by the mammalian high mobility group protein AT-hook 2: a SELEX study. Biochemistry 46(45):13059–13066. [DOI] [PubMed] [Google Scholar]
- Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJL.. 2005. DNA bridging: a property shared among H-NS-like proteins. J Bacteriol. 187(5):1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Rashid F-ZM, Grainger DC.. 2020. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat Rev Genet. 21(4):227–242. [DOI] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Goosen N.. 2000. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 28(18):3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Bhonagiri-Palsikar V, Hallsworth-Pepin K, Clifton SW, Weinstock GM, McClelland M.. 2013. Evolutionary genomics of Salmonella enterica subspecies. MBio. 4(2):e00579–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P, McFarland KA, Jin S, Tong G, Duan B, Yang A, Hughes TR, Liu J, Dove SL, Navarre WW, et al. 2015. A novel AT-rich DNA recognition mechanism for bacterial xenogeneic silencer MvaT. PLoS Pathog. 11(6):e1004967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan B, Ding PF, Hughes TR, Navarre WW, Liu J, Xia B.. 2018. How bacterial xenogeneic silencer rok distinguishes foreign from self DNA in its resident genome. Nucleic Acids Res. 46(19):10514–10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC, Rimsky S.. 2008. New insights into transcriptional regulation by H-NS. Curr Opin Microbiol. 11(2):113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-de-Alba C, Berrow NS, Garcia-Castellanos R, Garcia J, Pons M.. 2013. On the origin of the selectivity of plasmidic H-NS towards horizontally acquired DNA: linking H-NS oligomerization and cooperative DNA binding. J Mol Biol. 425(13):2347–2358. [DOI] [PubMed] [Google Scholar]
- Foerstner KU, von Mering C, Hooper SD, Bork P.. 2005. Environments shape the nucleotide composition of genomes. EMBO Rep. 6(12):1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfria-Subiros E, Acosta-Reyes F, Saperas N, Pous J, Subirana JA, Campos JL.. 2012. Crystal structure of a complex of DNA with one AT-hook of HMGA1. PLoS One 7(5):e37120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman VJ.1951. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J Bacteriol. 61(6):675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Foo YH, Winardhi RS, Tang Q, Yan J, Kenney LJ.. 2017. Charged residues in the H-NS linker drive DNA binding and gene silencing in single cells. Proc Natl Acad Sci U S A. 114(47):12560–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP.. 2005. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 3(9):679–687. [DOI] [PubMed] [Google Scholar]
- Gordon BR, Imperial R, Wang L, Navarre WW, Liu J.. 2008. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J Bacteriol. 190(21):7052–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BR, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, Navarre WW, Xia B, Liu J.. 2011. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci U S A. 108(26):10690–10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BR, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J.. 2010. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 107(11):5154–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC.2016. Structure and function of bacterial H-NS protein. Biochem Soc Trans. 44(6):1561–1569. [DOI] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Goldberg MD, Busby SJ.. 2006. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34(16):4642–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Ochman H.. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 87(5):791–794. [DOI] [PubMed] [Google Scholar]
- Guo F-B, Lin H, Huang J.. 2009. A plot of G + C content against sequence length of 640 bacterial chromosomes shows the points are widely scattered in the upper triangular area. Chromosome Res. 17(3):359–364. [DOI] [PubMed] [Google Scholar]
- Gupta RS.2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev. 24(4):367–402. [DOI] [PubMed] [Google Scholar]
- Haran TE, Mohanty U.. 2009. The unique structure of A-tracts and intrinsic DNA bending. Q Rev Biophys. 42(1):41–81. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA.. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6(9):e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hołówka J, Zakrzewska-Czerwińska J.. 2020. Nucleoid associated proteins: the small organizers that help to cope with stress. Front Microbiol. 11: 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM.. 1997. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Mol Biol. 4(8):657–665. [DOI] [PubMed] [Google Scholar]
- Juo ZS, Chiu TK, Leiberman PM, Baikalov I, Berk AJ, Dickerson RE.. 1996. How proteins recognize the TATA box. J Mol Biol. 261(2):239–254. [DOI] [PubMed] [Google Scholar]
- Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM.. 2011. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39(6):2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S.2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9(7):335–343. [DOI] [PubMed] [Google Scholar]
- Kelkar YD, Phillips DS, Ochman H.. 2015. Effects of genic base composition on growth rate in G+C-rich genomes. G3 (Bethesda) 5:1247–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisaar M.2019. Mutation and recombination rates vary across bacterial chromosome. Microorganisms 8(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV.2016. Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Res. 5:1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlajich MV, Hron DR, Boudreau BA, Sun Z, Lyubchenko YL, Landick R.. 2015. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. Elife. 4:e04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, Perian S, Bataillon T, Nesme X, Duret L, Daubin V.. 2015. GC-Content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 11(2):e1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H.. 1997. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 44(4):383–397. [DOI] [PubMed] [Google Scholar]
- Lee H, Popodi E, Tang H, Foster PL.. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 109(41):E2774–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Lee SY, Kenney LJ, Yan J.. 2012. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci Rep. 2(1):509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Whang YR, Kenney LJ, Yan J.. 2012. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 40(8):3316–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Andersson DI.. 2008. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci U S A. 105(46):17878–17883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gordon BR.. 2012. Targeting the global regulator Lsr2 as a novel approach for anti-tuberculosis drug development. Expert Rev Anti Infect Ther. 10(9):1049–1053. [DOI] [PubMed] [Google Scholar]
- Long H, Sung W, Kucukyildirim S, Williams E, Miller SF, Guo W, Patterson C, Gregory C, Strauss C, Stone C, et al. 2018. Evolutionary determinants of genome-wide nucleotide composition. Nat Ecol Evol. 2(2):237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço RF, Saurabh S, Herrmann J, Wakatsuki S, Shapiro L.. 2020. The nucleoid-associated protein GapR uses conserved structural elements to oligomerize and bind DNA. MBio 11(3): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri PR, Golding GB.. 2008. Gene amelioration demonstrated: the journey of nascent genes in bacteria. Genome 51(2):164–168. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Mellies JL, Barron AM, Carmona AM.. 2007. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun. 75(9):4199–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Osawa S.. 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 84(1):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW.2016. The impact of gene silencing on horizontal gene transfer and bacterial evolution. Adv Microb Physiol. 69:157–186. [DOI] [PubMed] [Google Scholar]
- Navarre WW, McClelland M, Libby SJ, Fang FC.. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21(12):1456–1471. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC.. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313(5784):236–238. [DOI] [PubMed] [Google Scholar]
- Nishida H.2012a. Evolution of genome base composition and genome size in bacteria. Front Microbiol. 3:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H.2013. Genome DNA sequence variation, evolution, and function in bacteria and archaea. Curr Issues Mol Biol. 15:19–24. [PubMed] [Google Scholar]
- Nishida H.2012b. Nucleosome positioning. ISRN Mol Biol. 2012:245706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA.. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405(6784):299–304. [DOI] [PubMed] [Google Scholar]
- Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N.. 2006. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13(4):141–153. [DOI] [PubMed] [Google Scholar]
- Park C, Zhang J.. 2012. High expression hampers horizontal gene transfer. Genome Biol Evol. 4(4):523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rueda E, Ibarra JA.. 2015. Distribution of putative xenogeneic silencers in prokaryote genomes. Comput Biol Chem. 58:167–172. [DOI] [PubMed] [Google Scholar]
- Pfeifer E, Hünnefeld M, Popa O, Frunzke J.. 2019. Impact of xenogeneic silencing on phage-host interactions. J Mol Biol. 431(23):4670–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer E, Hunnefeld M, Popa O, Polen T, Kohlheyer D, Baumgart M, Frunzke J.. 2016. Silencing of cryptic prophages in Corynebacterium glutamicum. Nucleic Acids Res. 44(21):10117–10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picker MA, Wing HJ.. 2016. H-NS, its family members and their regulation of virulence genes in Shigella species. Genes (Basel) 7(12):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina-Iturbe A, Suazo ID, Hoppe-Elsholz G, Ulloa-Allendes D, Gonzalez PA, Kalergis AM, Bueno SM.. 2020. Horizontally acquired homologs of xenogeneic silencers: modulators of gene expression encoded by plasmids, phages and genomic islands. Genes (Basel) 11(2):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Erkelens AM, Ben Bdira F, Dame RT.. 2019. The architects of bacterial DNA bridges: a structurally and functionally conserved family of proteins. Open Biol. 9(12):190223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Lim CJ, Whang YR, Liu J, Yan J.. 2013. Mechanism of DNA organization by Mycobacterium tuberculosis protein Lsr2. Nucleic Acids Res. 41(10):5263–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan R, Kelkar YD, Ochman H.. 2012. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci U S A. 109(36):14504–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R.2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277(1-2):63–81. [DOI] [PubMed] [Google Scholar]
- Reichenberger ER, Rosen G, Hershberg U, Hershberg R.. 2015. Prokaryotic nucleotide composition is shaped by both phylogeny and the environment. Genome Biol Evol. 7(5):1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi E, van Mastbergen EC, Navarre WW, Vreede J.. 2019. Predicting the mechanism and rate of H-NS binding to AT-rich DNA. PLoS Comput Biol. 15(3):e1006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsky S, Zuber F, Buckle M, Buc H.. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol Microbiol. 42(5):1311–1323. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Feil EJ.. 2010. Mutational patterns cannot explain genome composition: are there any neutral sites in the genomes of bacteria? PLoS Genet. 6(9):e1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A.. 2002. Base composition bias might result from competition for metabolic resources. Trends Genet. 18(6):291–294. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD.. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 8(12):983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seid CA, Smith JL, Grossman AD.. 2017. Genetic and biochemical interactions between the bacterial replication initiator DNAA and the nucleoid-associated protein Rok in Bacillus subtilis. Mol Microbiol. 103(5):798–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo H, Iwaki T, Ieda R, Kurumizaka H, Ueguchi C, Mizuno T, Morikawa S, Nakamura H, Kuboniwa H.. 1995. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS Lett. 360(2):125–131. [DOI] [PubMed] [Google Scholar]
- Shindo H, Ohnuki A, Ginba H, Katoh E, Ueguchi C, Mizuno T, Yamazaki T.. 1999. Identification of the DNA binding surface of H-NS protein from Escherichia coli by heteronuclear NMR spectroscopy. FEBS Lett. 455(1-2):63–69. [DOI] [PubMed] [Google Scholar]
- Singh K, Milstein JN, Navarre WW.. 2016. Xenogeneic silencing and its impact on bacterial genomes. Annu Rev Microbiol. 70:199–213. [DOI] [PubMed] [Google Scholar]
- Smits WK, Grossman AD.. 2010. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 6(11):e1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonden B, Uhlin BE.. 1996. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 15(18):4970–4980. [PMC free article] [PubMed] [Google Scholar]
- Tavita K, Mikkel K, Tark-Dame M, Jerabek H, Teras R, Sidorenko J, Tegova R, Tover A, Dame RT, Kivisaar M.. 2012. Homologous recombination is facilitated in starving populations of Pseudomonas putida by phenol stress and affected by chromosomal location of the recombination target. Mutat Res. 737(1-2):12–24. [DOI] [PubMed] [Google Scholar]
- Tendeng C, Soutourina OA, Danchin A, Bertin PN.. 2003. MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiology (Reading). 149(11):3047–3050. [DOI] [PubMed] [Google Scholar]
- Travers AA.2004. The structural basis of DNA flexibility. Philos Trans A Math Phys Eng Sci. 362(1820):1423–1438. [DOI] [PubMed] [Google Scholar]
- Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL.. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 102(31):11082–11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Supek F, Lehner B.. 2012. Nucleoid-associated proteins affect mutation dynamics in E. coli in a growth phase-specific manner. PLoS Comput Biol. 8(12):e1002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman JL, Fagan WF, Johnson PLF.. 2019. Linking high GC content to the repair of double strand breaks in prokaryotic genomes. PLoS Genet. 15(11):e1008493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will WR, Navarre WW, Fang FC.. 2015. Integrated circuits: how transcriptional silencing and counter-silencing facilitate bacterial evolution. Curr Opin Microbiol. 23:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winardhi RS, Fu W, Castang S, Li Y, Dove SL, Yan J.. 2012. Higher order oligomerization is required for H-NS family member MvaT to form gene-silencing nucleoprotein filament. Nucleic Acids Res. 40(18):8942–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winardhi RS, Gulvady R, Mellies JL, Yan J.. 2014. Locus of enterocyte effacement-encoded regulator (Ler) of pathogenic Escherichia coli competes off histone-like nucleoid-structuring protein (H-NS) through noncooperative DNA binding. J Biol Chem. 289(20):13739–13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winardhi RS, Yan J, Kenney LJ.. 2015. H-NS regulates gene expression and compacts the nucleoid: insights from single-molecule experiments. Biophys J. 109(7):1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M.. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 15(6):1340–1349. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PBM data for Lsr2, H-NS, MvaT, and Rok can be found in the supplementary data of our previously published works (Gordon et al. 2011; Ding et al. 2015; Duan et al. 2018). DNA binding preferences of Lsr2, H-NS, MvaT, and Rok towards 6- or 5-bp sequences can be found in supplementary data set S1, Supplementary Material online. C-terminal sequence alignments of different xenogeneic silencer family proteins are presented in supplementary data set S2, Supplementary Material online. Median genomic AT-contents and sizes of bacterial species with or without xenogeneic silencers are provided in supplementary data set S3, Supplementary Material online.