Abstract
To investigate novel patterns and processes of protein evolution, we have focused in the metallothioneins (MTs), a singular group of metal-binding, cysteine-rich proteins that, due to their high degree of sequence diversity, still represents a “black hole” in Evolutionary Biology. We have identified and analyzed more than 160 new MTs in nonvertebrate chordates (especially in 37 species of ascidians, 4 thaliaceans, and 3 appendicularians) showing that prototypic tunicate MTs are mono-modular proteins with a pervasive preference for cadmium ions, whereas vertebrate and cephalochordate MTs are bimodular proteins with diverse metal preferences. These structural and functional differences imply a complex evolutionary history of chordate MTs—including de novo emergence of genes and domains, processes of convergent evolution, events of gene gains and losses, and recurrent amplifications of functional domains—that would stand for an unprecedented case in the field of protein evolution.
Keywords: metallothionein domains, modular proteins, Chordata, Tunicata, ascidians/thaliaceans/appendicularians, metallothionein evolution
Introduction
Heavy metals such as zinc (Zn) and copper (Cu) are integral components of numerous proteins involved in multiple cellular functions (reviewed in Vallee and Falchuk [1993]; Maret [2013]; Vest et al. [2013]), whereas other metals such as cadmium (Cd) have no known biological functions and are highly toxic for living beings. Organisms have developed different molecular mechanisms that regulate the homeostasis of the physiological metals and counteract the harmful effects of the toxic ones. One of such mechanisms is based on an extremely diverse group of metal-binding proteins known as metallothioneins (MTs), which are engaged in the physiological control of metals operating as ion reservoirs, metal transporters and/or metal deliverers to target metalloproteins, but also in radical scavenging, oxidative stress protection, and antiapoptotic defense (reviewed in Capdevila et al. [2012]; Ziller and Fraissinet-Tachet [2018]).
Most MTs are low molecular weight (<100 amino acids) and cysteine-rich (≈15–30%) proteins. Their cysteine (Cys, C) residues are arranged in CxC, CC, and CCC motifs, whose number and distribution define functional domains. Many MTs have a bimodular organization made of two functional domains, in which each domain enables the coordination of a number of metal ions through metal–thiolate bonds (reviewed in Blindauer [2014]). MTs have been classified in a stepwise gradation, from extreme Zn/Cd-thioneins to extreme Cu-thioneins depending on their metal-binding preference (Bofill et al. 2009; Palacios, Atrian, et al. 2011). There have been attempts to predict the metal preference of a given MT from the analysis of their non-Cys amino acids, and in the case of gastropod MTs, for instance, it has been proposed that the lysine-asparagine (K/N) ratio could be important for metal preference, so that a preponderance of K over N residues would be a feature of Cd selectivity, whereas a predominance of N over K would be typical of Cu-MTs (Perez-Rafael et al. 2014; Pedrini-Martha et al. 2020). Due to the high sequence and structural diversity of MTs, however, it is unclear whether the rule of K/N ratio applies to nongastropod MTs, and thereby, metal-binding assays are needed to experimentally determine the metal preference of newly discovered MTs.
Comparative analyses of the MTs across diverse species have been useful to better understand the function and evolution of these proteins, and to illustrate unusual modes of molecular evolution of these genes and proteins. MT research has exposed, for instance, pervasive events of internal sequence duplications (Tanguy and Moraga 2001; Palacios, Espart, et al. 2014; Iturbe-Espinoza et al. 2016; Jenny et al. 2016; Calatayud et al. 2018), de novo emergence of domains (Calatayud, Garcia-Risco, Pedrini-Martha, et al. 2021), and a high bias in the amino acid composition probably as a result of the 3D structural requirements for metal binding. MT research has also provided useful information for many other research fields. In the field of Evolutionary Ecology and Environmental Sciences, for instance, MTs are considered a good model to investigate the mechanisms by which different organisms—from protists and fungi to insects, mollusks, and mammals—have adapted to diverse metal bioavailability and other environmental stress conditions. These mechanisms include duplications of MT genes (Maroni et al. 1987; Adamo et al. 2012; Luo et al. 2020), increases in the levels of MT expression (Timmermans et al. 2005; Janssens et al. 2008, 2009; Costa et al. 2012; Catalan et al. 2016; de Francisco et al. 2018), and changes in metal specificity (Tio et al. 2004; Palacios, Pagani, et al. 2011; de Francisco et al. 2017; Dallinger et al. 2020; García-Risco et al. 2020). In the fields of Bioinorganic Chemistry and Metallomics, MTs have been used to analyze metal–protein interactions, analyzing the qualitative and quantitative characteristics of the metal-binding capacities of these proteins (Blindauer and Leszczyszyn 2010; Palacios, Pagani, et al. 2011; Beil et al. 2019) and by extension, of a broad range of metalloproteins (Waldron et al. 2009).
In this work, we aim to explore the MTs found in a large number of species of the three tunicate classes—Ascidiacea, Thaliacea, and Appendicularia—in order to investigate the intriguing origin and evolution of MTs in chordates. Tunicates (Tunicata subphylum, a.k.a. Urochordata) are the sister group of vertebrates, and together with basally branching cephalochordates form the phylum Chordata. Tunicates have undergone a rapid and “liberal” pattern of evolution in comparison with the “conservative” pattern of vertebrates and cephalochordates (Somorjai et al. 2018; Ferrández-Roldán et al. 2019), yielding a group of ecologically diverse filter-feeding marine animals—including planktonic and benthic specimens as well as solitary and colonial forms–, which have adapted to different conditions of metal bioavailability. In addition, the filtering lifestyle makes tunicates prone to accumulate metals from the seawater (Papadopoulou and Kanias 1977; Tzafriri-Milo et al. 2019; and references therein), and thereby, molecular mechanisms such as the MTs that regulate metal homeostasis and detoxification are physiologically relevant to them.
Our understanding of the MTs in tunicates is, however, scarce, limited to two MT sequences from the ascidian Ciona robusta (formerly C. intestinalis) and Herdmania curvata (Franchi et al. 2011), and two sequences from the appendicularian Oikopleura dioica recently described by our group (Calatayud et al. 2018). To overcome this situation, we have conducted an exhaustive survey of the MTs in tunicate databases, identifying 168 MTs in 37 species of ascidians, 4 of thaliaceans, and 3 of appendicularians, representing planktonic and benthic species, as well as solitary and colonial forms. We have also functionally characterized the metal-binding properties of eight tunicate MTs, which were selected on the basis of their taxon representativeness, sequence composition, degree of multiplicity, or modular organization. Comparative analyses of tunicate MTs with those of cephalochordates and vertebrates have exposed a high degree of structural diversity between lineages, which would not have been accompanied, however, by differences in their metal preferences. This diversity seems the result of numerous and intricate evolutionary events, and therefore, chordate MTs would represent one of the most complex and enigmatic examples of protein evolution.
Results
The biological interest for tunicates in the last decades has given rise to many genomic and transcriptomic resources, facilitating the in silico identification of virtually any gene in a wide set of species. This advantageous situation allowed us to perform an extensive survey of the MT system in tunicate databases.
MTs in Ascidians
Ascidians, commonly known as sea squirts, are the most studied group of tunicates. Adults are sessile, either solitary or colonial, usually found in shallow water worldwide. We surveyed ascidian databases and identified 145 MT sequences from 37 species distributed among Aplousobranchia (three species), Phlebobranchia (eight species), and Stolidobranchia (3 molgulidae + 5 pyuridae + 18 styelidae species) orders (fig. 1; see supplementary table S1, Supplementary Material online, for the species list). Sequence comparison showed that most ascidian MTs are 38–45 amino acids long with 12 cysteines (31–27%) organized in a single functional domain. In this domain, the C-motifs were distributed as [CxC]x5[CxC]x2Cx2Cx2[CxC]x4[CxC]x3-6CC, which would represent the prototypical organization of the mono-modular ascidian MTs (supplementary fig. S1, Supplementary Material online, and table 1).
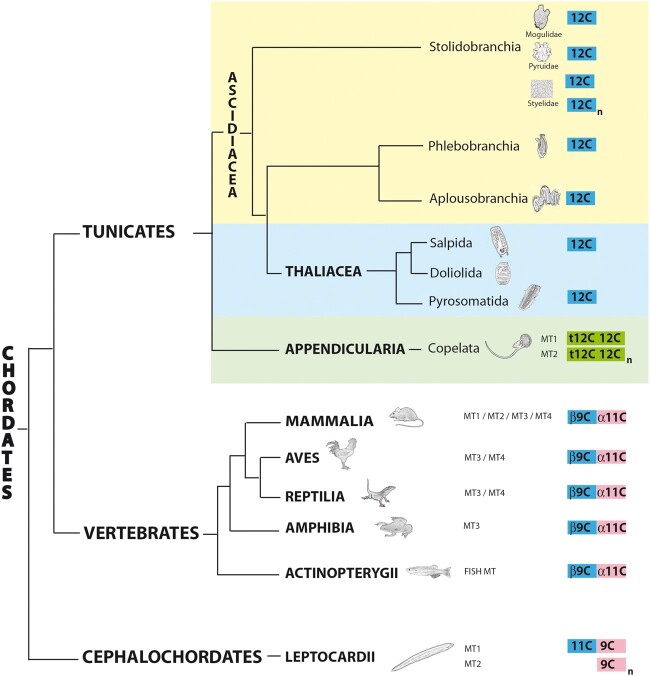
Fig. 1.
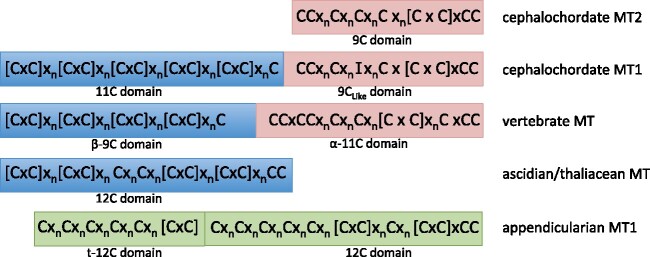
Phylogenetic relationships in the Chordata phylum. Regarding tunicates, traditional classifications divided this subphylum in three classes, Appendicularia (green background), Ascidiacea (yellow background), and Thaliacea (blue background), but recent analyses propose Thaliacea species (salps, doliolids, and pyrosomes) are nested within the Ascidiacea class, closer to the Aplousobranchia and Phlebobranchia orders than to the Stolidobranchia order (Delsuc et al. 2018; Kocot et al. 2018). A schematic representation of the MTs of each taxonomic group, with a mono-, bi-, or multimodular organization with distinct domains (color coded) containing seven, nine, 11, or 12 cysteines, is showed at the right (see fig. 7 and text for additional details).
Table 1.
Cysteine Motifs in 11/12-Cys Domains from Different Animal Taxa.
| Taxon | 11/12-Cys Domains | Cysteine Motifs | Metal Ions |
|---|---|---|---|
| Ascidians/thaliaceans | Full-length MT | [CxC]x5[CxC]x2Cx2Cx2[CxC]x4[CxC]x3–6CC | 4 (this work) |
| Appendicularians | 12C domain | Cx3Cx2Cx2Cx3Cx2[CxC]x2Cx4[CxC]xCC | 4 (Calatayud, Garcia-Risco, Capdevila, et al. 2021) |
| Vertebrates | α-domaina | CCxCCx3Cx2Cx3[CxC]x4–12Cx1–3CC | 4 (Stillman et al. 1987) |
| Cephalochordates | 11C domain | [CxC]x5[CxC]x3–4[CxC]x3[CxC]x3[CxC]x2C | nd |
| Mollusks | α-domain | [CxC]x4–6[CxC]x3Cx4–5[CxC]x3[CxC]x3[CxC]x2C | 4 (Digilio et al. 2009) |
| Echinoderms | α-domain | CxCCx5[CxC]x4CCx4CCx4CC | 4 (Tomas et al. 2013) |
| Insects | MTnB/C/D | Cx2Cx3[CxC]x4Cx3[CxC]x3[CxC]xC]x7CC | 4 (Egli et al. 2006) |
Note.—nd, not determined.
Notice that although vertebrate, echinoderm, and mollusk MTs have α-domains, the origin and evolutionary relationship of these α-domains remains uncertain.
Twenty-seven out of the 37 analyzed species showed multiple MT genes: seven species with two sequences, six with three, two with four, three with six, two with seven, four with eight, one with nine, and two species with up to ten sequences. Despite in some cases, due to the nature of the data source—mainly sequence read archive (SRA) projects–, allelic variants of a particular gene could not be discriminated from gene duplicates, overestimating thereby the degree of MT multiplicity in our catalog, our results suggested that, overall, the MT multiplicity in ascidians is pervasive, although a definitive evaluation of the degree of multiplicity must await more complete and deeper genomic databases.
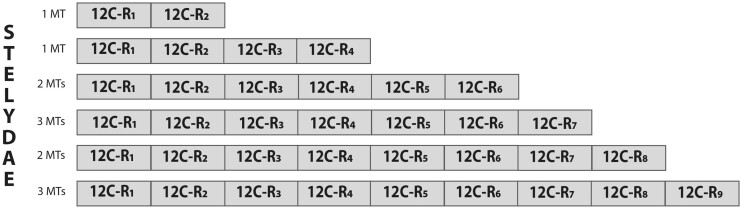
Interestingly, there were some exceptions to the prototypical organization, ranging from MTs that have lost or gained some cysteines or small protein fragments (supplementary fig. S1, Supplementary Material online) to multimodular forms containing tandem repeats of the prototypical domain (supplementary figs. S2 and S3, Supplementary Material online). We were able to reconstruct 12 multimodular MTs (five partial and seven full-length sequences), whose number of repeats ranged from two to nine (fig. 2). Noteworthy, all the species with multimodular MTs belonged to the styelidae family in the stolidobranchia order (fig. 1), suggesting that multimodular MTs arose by a lineage-specific evolutionary event.
Fig. 2.
Schematic representation of multimodular Stelydae MTs. Internal duplications of a 12-Cys domain (grey box) generated multimodular MTs with variable number of repeats (R), from 2 to 9. The number of MTs with different number of repeats (R1–R9) is indicated (DgrMT7 = 2 repeats; EtiMT8part ≥4 repeats; ScaMT6part and SsoMT8part ≥6 repeats; PmiMT10part, BleMT1, and BleMT2 ≥7 repeats; PauMT10part and PspMT7 ≥8 repeats; BscMT1, PmamiMT3, and PpoMT7 = 9 repeats) (see supplementary fig. S2, Supplementary Material online, for further details).
Metal-Binding Features of Mono-Modular Ascidian MTs
In order to analyze the metal-binding capacity and preference of the ascidian MTs, we selected three MTs of two species belonging to distinct orders within the Ascidiacea class: CroMT1 from C. robusta (phlebobranchia order), and HroMT1 and HroMT2 from Halocynthia roretzi (stolidobranchia order). These MTs were chosen because CroMT1 was the only MT in C. robusta, whereas HroMT1 and HroMT2 represented duplicate forms in H. roretzi. These MTs were also selected because of their K/N ratios, which in the CroMT1 was 3:5, opposite of that in HroMT1 and HroMT2 sequences, 6:3 and 8:2, respectively. HroMT2 showed indeed one of the highest K/N ratios among tunicate MTs (supplementary table S1, Supplementary Material online). The K/N ratio seems important for metal preference of gastropod MTs (Perez-Rafael et al. 2014; Pedrini-Martha et al. 2020), and the study of the metal selectivity of the CroMT1, HroMT1, and HroMT2 would allow us to experimentally evaluate the K/N rule in the ascidian system.
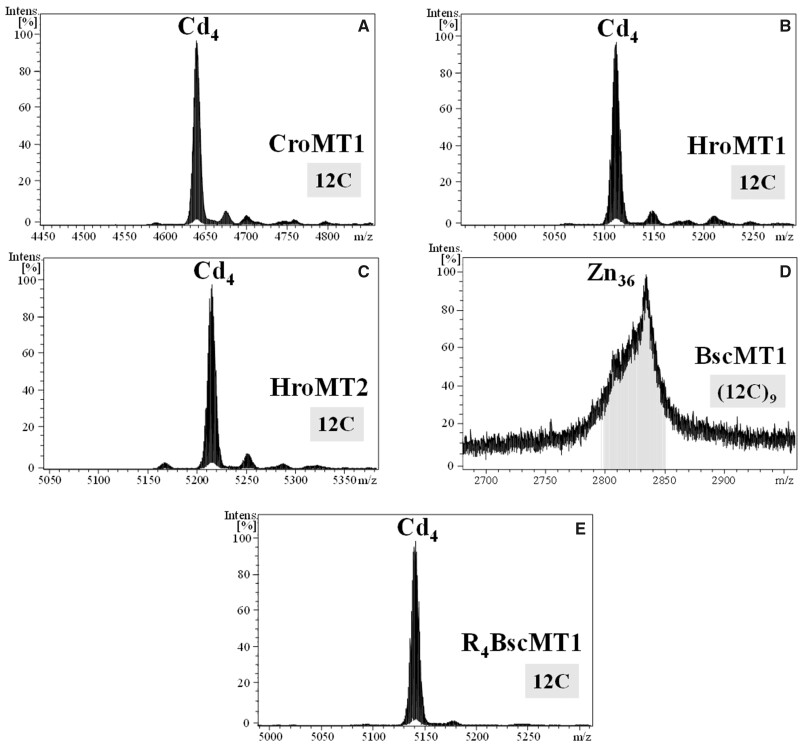
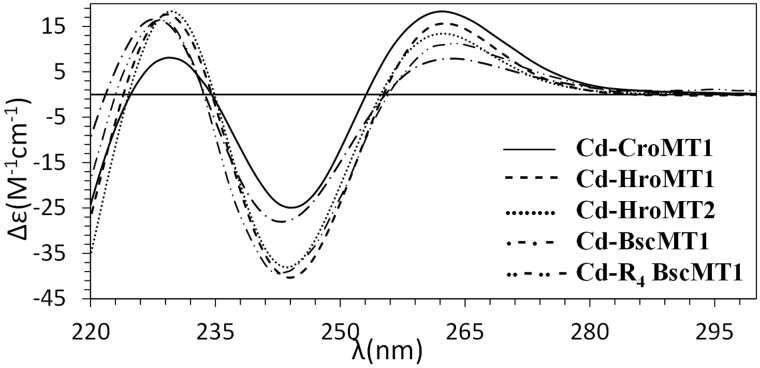
We characterized the formation of metal–MT complexes of CroMT1, HroMT1, and HroMT2 proteins heterologously expressed in Escherichia coli cultures supplemented with Cu, Cd, or Zn salts by inductively coupled plasma atomic emission spectroscopy (ICP-AES) and electrospray ionization mass spectrometry (ESI-MS). ICP-AES was used for protein quantification and metal-to-protein stoichiometry determination through the measurement of element composition of the samples (S, Zn, Cd, and Cu) (Bongers et al. 1988), and ESI-MS allowed us to determine the molecular mass of the species formed, that is, the speciation of the samples (Capdevila et al. 2012). Single well-folded Cd4-MT species were recovered from CroMT1, HroMT1, and HroMT2 Cd-productions (fig. 3A–C), and a major Zn4-MT species were found in their recombinant Zn productions (supplementary fig. S4A, C, and E, Supplementary Material online), which contrasted with the mixtures of homometallic Cu-MT species purified from Cu-supplemented E. coli cultures (supplementary fig. S4B, D, and F, Supplementary Material online). It is also noteworthy the resemblance of the CD spectra of the Cd–MT complexes of all ascidian MTs, denoting structurally equivalent metal clusters for these proteins (fig. 4). The exemplary exciton coupling centered at ∼250 nm exhibited in all the CD envelopes confirms high robustness and compactness of the Cd4S(Cys)12 clusters and, therefore, high Cd specificity. Altogether, these results showed that Zn4- or Cd4–MT complexes were energetically the most favorable forms of these proteins, suggesting that their function would be related to binding divalent metal ions, either as Zn- or Cd-thioneins [8]. Genuine Zn-thioneins, however, render heterometallic Cu, Zn–MT complexes when synthesized under Cu surplus, meaning that the ascidian MTs deviated toward a more specific Cd-thionein behavior (Palacios, Atrian, et al. 2011). Regarding the K/N rule used to predict the metal-binding preference in gastropod MTs, our results demonstrated that this rule did not apply to tunicate MTs, since the three of them displayed contrasted K/N ratios and their metal preferences were exactly the same.
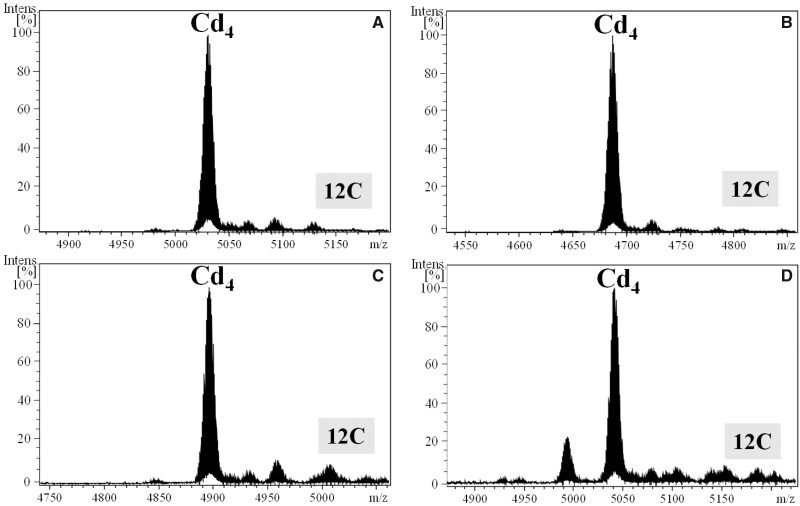
Fig. 3.
Deconvoluted ESI-MS spectra of the Cd(II)-productions of ascidian CroMT1 (A), HroMT1 (B), HroMT2 (C), and +14 charge state ESI-MS spectra of the Zn(II)-production of BscMT1 (D), as well as the deconvoluted ESI-MS spectra of the Cd(II)-production of its fragment R4 BscMT1 (E). Notice that BscMT1 is capable to bind up to 36 divalent ions, which nicely match with a multimodular organization made of nine repeated domains (12C)9, each one binding four divalent ions alike the mono-modular (12C) MTs (A–C) or the R4 (12C) domain (E).
Fig. 4.
Circular dichroism spectra of the Cd(II)-productions of CroMT1, HroMT1, HroMT2, BscMT1, and R4 BscMT1, all exhibiting very similar CD signals. The exciton coupling centered at ∼250 nm confirms high robustness and compactness of the clusters formed.
Metal-Binding Features of Multimodular Ascidian MTs
In order to determine the metal-binding features of multimodular MTs, we selected BscMT1 (ID: ATSW01006707.1; chromosome 6) of Botryllus schlosseri (stolidobranchia order) because this MT was not only identified in an accurately assembled genome (Voskoboynik et al. 2013), but also because it was supported by three EST sequences (supplementary table S1, Supplementary Material online). BscMT1 was a multimodular MT of 365 amino acids with 105 cysteines (29%) and a K/N ratio of 1:1 (31K vs. 31N) (supplementary fig. S2 and table S1, Supplementary Material online). BscMT1 was one of the longest MT described so far, and a detailed analysis of its sequence revealed that it was made of nine tandem repeats (R1–R9, the last one only partially complete) of a 12-Cys domain, each one resembling a prototypical ascidian form (supplementary fig. S3, Supplementary Material online).
The metal–BscMT1 complexes purified from the heterologous expression in E. coli cultures supplemented with Cu, Cd, or Zn salts were characterized by the IPC-AES and ESI-MS. The concentrations of metal–BscMT1 productions, despite the several attempts, were always low, probably due to the high molecular weight of BscMT1 and its structural complexity made of repeated domains. Only the samples recovered from the Zn(II)-enriched culture media rendered informative data through these techniques. The IPC-AES and ESI-MS analyses showed that this preparation contained Zn36–MT complexes (fig. 3D). The binding of 36 metal ions agreed with its multimodular organization made of nine repeated domains, each one similar to a prototypical M4-MT: 4 metal ions×9 domains = 36 metal ions. To further verify the functional correspondence between one of the 12-Cys tandem repeats of BscMT1 and the mono-modular ascidian MTs, we analyzed the ability of the R4 domain (R4-BscMT1) to form metal complexes when it was expressed alone. The election of the R4 domain was due to the fact that it was highly similar to the prototypical ascidian MTs, including some additional amino acids after the carboxyl-terminal CC doublet (supplementary fig. S3, Supplementary Material online). We heterologously expressed the R4 domain in E. coli grown in media supplemented with Cu, Cd, or Zn salts. The ICP-AES and ESI-MS analyses of the recovered samples confirmed the ability of this fragment to autonomously form metal complexes by itself, and revealed that the biochemical features of R4 domain were highly similar to the mono-modular ascidian forms (compare fig. 3A–C and E). Thus, this domain yielded major M4–R4 complexes with Zn(II) and Cd(II), being unique species only in the latter, whereas rendered a homometallic mixture of species in the presence of copper, as the ascidian MTs discussed above (supplementary fig. S4G and H, Supplementary Material online). Based on this R4 fragment as a representation of the whole protein, the multidomain BscMT1 would be a Cd-thionein capable to bind up to 36 divalent ions, and its stoichiometry—that is, M36-BscMT1—would nicely match with what would be expected from the repetition of nine domains, that is, M4-R1 to M4-R9. These results supported a direct relationship between the binding capacity and the number of the repeated domains in multimodular MTs.
MTs in Thaliaceans
We analyzed the thaliacean MTs by surveying the databases of the five available species. We identified 14 MTs from four thaliacean species, three from Salpida (Iasis cylindrica, Salpa fusiformis, and Salpa thompsoni), and one from Pyrosomata (Pyrosomella verticillata) orders. Sequence comparison showed that thaliacean MTs were 36–41 amino acids long, with 12 cysteines (33–29%) organized in a single functional domain with the C-motifs distributed as [CxC]x5[CxC]x2Cx2Cx2[CxC]x4[CxC]x3–4CC (noteworthy, we did not find multimodular MTs in thaliacean databases) (table 1). This motif arrangement was identical to that of the prototypical ascidian MTs, denoting a common origin of that thaliacean and ascidian MTs, and supporting the recently suggested phylogenetic relationship of these classes (Delsuc et al. 2018; Kocot et al. 2018) (fig. 1).
Importantly, MT multiplicity was found in the four thaliacean species although as in ascidians, it was challenging to determine whether the multiple sequences represented different genes or allelic variants. In S. thompsoni, for instance, we identified six MT sequences, five full-length (SthMT1, MT2, MT3, MT5, and MT6), and one partial (SthMT4), which coded for proteins 48% to 95% identical at amino acid level (supplementary table S2, Supplementary Material online). The highest identities were between SthMT1 and SthMT5 (88%) and SthMT2 and SthMT6 (95%) sequences, and therefore, these pairs were the most plausible cases of allelic variants. To investigate this possibility, we analyzed the conservation of 5′ and 3′ flanking regions, as well as of intron sequences in PCR-amplified genomic regions from two S. thompsoni specimens. Nucleotide identity (excluding gaps) in SthMT1–SthMT5 and SthMT2–SthMT6 alignments was 91–100%, 87–82% and 83–86% at 5′ regions, introns and 3′ regions, respectively (supplementary table S3, Supplementary Material online). These results would be compatible with that SthMT5 and SthMT6 were allelic variants of SthMT1 and SthMT2, respectively, although the possibility they may be recently duplicated genes cannot be fully discarded.
Metal-Binding Features of Thaliacean MTs
We analyzed the metal-binding capacity and metal preference of the thaliacean MTs, and compared them with those of ascidian MTs in order to investigate whether the differences in lifestyles—planktonic versus benthic—might have been key for MT evolution. We characterized the metal–MT complexes rendered for the four MTs from S. thompsoni, SthMT1 to SthMT4, heterologously expressed in Zn-, Cd-, or Cu-supplemented E. coli cultures. The clear Cd-thionein character of S. thompsoni MTs was reflected, once again, by the ICP-AES and ESI-MS analyses of the recovered samples. SthMTs yielded single Cd4-MT species under Cd(II) surplus (fig. 5), as well as major M4–MT complexes with Zn(II) (supplementary fig. S5A–D, Supplementary Material online). A mixture of species (for SthMT1 and SthMT3) or not detection of the metallated species (for SthMT2 and StMT4) were obtained in the presence of copper (supplementary fig. S5E and F, Supplementary Material online), evidencing that these proteins struggle to build metal clusters with Cu. Our results demonstrated that the four SthMTs were functionally very similar among them (compare panels A–D in fig. 5), as well as to the mono-modular ascidian MT (compare fig. 3 and fig. 5). The fact that thaliacean (SthMTs) and ascidian (CroMT1, HroMT1, HroMT2, and BscMT1) MTs exhibited a similar Cd-thionein character strongly supported that the prototypical ascidian/thaliacean MT (and most likely the ancestral tunicate MT, see below) was a mono-modular protein with a 12C domain capable to bind four metal ions with preference for Cd.
Fig. 5.
Deconvoluted ESI-MS of the Cd(II)-production of S. thompsoni SthMT1 (A), SthMT2 (B), SthMT3 (C), and to SthMT4 (D). Thaliacean MTs are mono-modular proteins made of a single 12C domain capable to bind four divalent ions, identically to ascidian MTs.
MTs in Appendicularians
We investigated the MTs in the Appendicularia (a.k.a. Larvacea) class. In a previous work, we had characterized the OdiMT1 and OdiMT2 genes (formerly OdMT1 and OdMT2) of O. dioica (Calatayud et al. 2018). We showed that both genes encoded MTs very different from all MTs described so far because their cysteines motifs were arranged in a novel pattern forming a “repeat unit” (RU) made of two C7 subunits (C7a and C7b) plus a carboxyl-terminal tail with five cysteines (C5) (Calatayud et al. 2018) (supplementary fig. S6A, Supplementary Material online). We concluded that OdiMT1 was made of a single RU, whereas OdiMT2 was a multimodular protein made of six direct tandem RU (Calatayud et al. 2018). We wondered whether these unconventional MTs were specific of O. dioica—gene and protein sequences are known to have considerably diverged during O. dioica evolution (reviewed in Ferrández-Roldán et al. [2019])—or they were widespread in the appendicularian lineage. We took advantage of recent sequence projects of several appendicularian species (Naville et al. 2019) and identified seven new MT sequences in two additional Oikopleura species: O. albicans (three sequences) and O. vanhoeffeni (four sequences) (supplementary table S1, Supplementary Material online). We did not identify any putative MT in databases of other oikopleurids such as O. longicauda, Bathochordaeus sp., and Mesochordaeus erythrocephalus, nor in the fritillarid species Fritillaria borealis, although any conclusion about the loss of the MTs in these lineages must await deeper sequence projects.
Regarding Oikopleura MTs, although most of the sequences were partial, they were clearly similar to O. dioica MTs. Sequence analyses showed that these new MTs were multimodular proteins made of repeats of a conserved 12-Cys domain with C-motifs distributed as CX3Cx2Cx2Cx3Cx2[CxC]x2Cx4[CxC]xCC (table 1 and supplementary fig. S6B, Supplementary Material online). This 12-Cys domain corresponded to the previously described C7b+C5 subunits (Calatayud et al. 2018). OdiMT1 consist, therefore, of two 12-Cys domains, one full-length (12C = C7b + C5), and one 12-Cys domain that has been trimmed (t-12C, formerly known as C7a) and lacks the five last cysteines (C5) (supplementary fig. S6A, Supplementary Material online and fig. 7). Interestingly, the trimmed domain when expressed alone did not efficiently bind divalent metals by itself, suggesting that it has lost its capacity to independently work (Calatayud, Garcia-Risco, Capdevila, et al. 2021). OdiMT2 would be a multimodular MT made of twelve 12-Cys domains, whose odd domains would have been also trimmed as t-12C. The identification of this conserved 12-Cys domain in all Oikopleura MTs suggested that this domain appeared in the ancestor of the Oikopleura genus, and that it was duplicated generating multimodular MTs independently in the diverse appendicularian species (supplementary fig. S6B, Supplementary Material online).
Fig. 7.
Comparison of the arrangement of Cys motifs in different MT domains. The arrangement of the Cys motifs in the 11-Cys amino-terminal domain of cephalochordate MT1s are somewhat similar to that in the vertebrate 9-Cys β domains and in the ascidian/thaliacean 12-Cys MTs (blue boxes), whereas the Cys motifs in the carboxyl-terminal domains of cephalochordate MT1s and MT2s (9C and 9CLike domains) are vaguely similar to those in vertebrate 11-Cys α domains (pink boxes). In contrast, the arrangement of the Cys motifs in the appendicularian MT1 seems totally different (green boxes).
MTs in Other Chordates
To provide a broad evolutionary perspective to our analyses we compared the tunicate MTs with those from the other chordate subphyla: cephalochordates and vertebrates.
In cephalochordates, MTs had been characterized in two amphioxus species, Branchiostoma floridae and Branchiostoma lanceolatum (Guirola et al. 2012). We extended the surveys of MTs to two additional amphioxus species, Branchiostoma belcheri (BbeMT1 and BbeMT2) and Branchiostoma japonicum (BjaMT1 and BjaMT2), and to Asymmetron lucayanum (AluMT1 and AluMT2), a cephalochordate distantly related to the other Branchiostoma species (Yue et al. 2014). We identified two MTs, MT1 and MT2, in all the cephalochordate species. B. lanceolatum MT2 showed two alternatively spliced forms, MT2-L (for the long form, with 67–70 amino acids) and MT2-S (for the short form, with 46–47 amino acids) (supplementary fig. S7, Supplementary Material online). Splice variants were also identified in B. floridae, B. japonicum, and A. lucayanum cDNA databases (i.e., BflMT2L and S, BjaMT2L and S, and AluMT2L and S), and alternative splicing was predicted from genomic sequence in B. belcheri species (BbeMT2L and S).
Sequence comparisons revealed that cephalochordate MT2s were multimodular proteins made of two (MT2S) or three (MT2L) 9-Cys domains (CCx2Cx2Cx3Cx[CxC]xCC) depending on an alternative splicing on exon 2 (supplementary figs. S7 and S8B, Supplementary Material online). In contrast, MT1s were bimodular proteins with two different domains: a 11-Cys, N-terminal domain with the C-motifs distributed as [CxC]x5[CxC]x3–4[CxC]x3[CxC]x3[CxC]x2C (table 1 and supplementary fig. S8A, Supplementary Material online) linked by 9–11 residues to a domain similar to the 9-Cys domain of MT2 (we named this domain 9-CysLike because an Ile substitutes the fourth Cys: CCx2Cx2Ix3Cx[CxC]xCC), followed by a carboxyl-terminal tail of 16–17 amino acids devoid of Cys residues. Interestingly, MT2 9-Cys and the MT1 9-CysLike domains showed a clear sequence similarity (supplementary fig. S8C, Supplementary Material online), which suggested that cephalochordate MT2s might have arisen from the duplication of an ancestral MT1 that lost the 11-Cys domain but amplified its 9-Cys domain.
In vertebrates, MTs had been classified in four different MT types: MT1, MT2, MT3, and MT4. Vertebrate MTs had been characterized in many species with significant differences of multiplicity in a species-dependent manner (Serén et al. 2014). Phylogenetic and synteny analyses had suggested the existence of mammalian MT1s and MT2s, amniote MT4s, tetrapod MT3s, and fish MTs (fig. 1) (Serén et al. 2014). The appearance of these four types would not be related with the two rounds of whole-genome duplication events that occurred during early vertebrate evolution since they are not localized in paralogous chromosomes but are located in tandem on a same chromosome (Serén et al. 2014). At protein level, all vertebrate MTs (even an MT we identified in the early divergent group of hagfish Eptatretus burgeri), showed a conserved bimodular organization made of a 9-Cys N-terminal domain (the so-called β-domain: [CxC]x5[CxC]x3[CxC]x2[CxC]x2Cx3) linked to a 11-Cys domain (the so-called α-domain: CCxCCx3Cx2Cx3[CxC]x6CxCC) at the carboxyl-terminal end (supplementary table S1 and fig. S9, Supplementary Material online). The arrangement of the C-motifs in the α and β domains did not show any obvious resemblance between them that might suggest of a common ancestry of these domains, though vertebrate β-domain appeared somewhat similar to cephalochordate and tunicate 11/12-Cys domains.
Discussion
Origin and Convergent Evolution of Tunicate MTs
Ascidian and thaliacean MTs share the same arrangement of their C-motifs (table 1) and are clearly similar in their amino acid sequence, also in the noncysteine residues: ascidian MTs are at least ≈40% identical in their noncysteine residues to any of the thaliacean MTs, and ≈60% identical when considering the full sequence. In contrast, their similarity with appendicularian MTs is negligible at the arrangement of the C-motifs (table 1) and in the composition of the noncysteine residues. For instance, OdiMT1 only is, at most, 13% identical to ascidian (e.g., CroMT1 and HroMT1) or thaliacean (e.g., SthMT1) MTs in noncysteine residues (notice that 25–30% sequence identity is generally taken as the minimal threshold for presumption of homology; Rost 1999). Therefore, although it cannot be ruled out that appendicularian MTs derive from ascidian and thaliacean MTs that have diverged too much for orthology to be recognized, their sequence and the C-motifs are so divergent that they seem to have had independent evolutionary origins. This possibility agrees with the idea that de novo emergence of MTs might be more habitual than previously thought because the only requirement for a peptide to function as a metal ion chelator would be a high content of coordinating residues (i.e., cysteines) and a relative small length that would favor its proper folding (Capdevila and Atrian 2011; Calatayud, Garcia-Risco, Pedrini-Martha, et al. 2021). The independent emergence of MTs in distant animal groups have been, indeed, implicitly suggested in diverse evolutionary studies (Capdevila and Atrian 2011; Blindauer 2014; Isani and Carpene 2014; Ziller and Fraissinet-Tachet 2018), and we have recently showed de novo evolution of MT domains within the Mollusca phylum (Calatayud, Garcia-Risco, Pedrini-Martha, et al. 2021).
Intriguingly, ascidian/thaliacean MTs and appendicularian MTs seem to have converged in the number of cysteines—12—of their basic functional organizations: the prototypical 12-Cys MTs of ascidians and thaliaceans, and the repeated 12-Cys domains of appendicularian MTs. Functional domains with 11/12 cysteines organized in different patterns of C-motifs but converging to similar 3D architectures (Beil et al. 2019) appear a recurrent evolutionary solution in MTs of phylogenetically distant animal clades, from vertebrates and tunicates to mollusks and insects (table 1). 3D structural constraints to efficiently coordinate four metal ions through the formation of metal–thiolate bonds might have led to the convergent evolution of 11/12-Cys domains in MTs of different animal phyla (Stillman et al. 1987; Egli et al. 2006; Digilio et al. 2009; Tomas et al. 2013; Calatayud, Garcia-Risco, Capdevila, et al. 2021; and this work).
Evolution of the Metal-Binding Preference
Ascidian/thaliacean MTs and appendicularian MTs also share the Cd-thionein character. The evolution of Cd-MTs seems, indeed, a common adaptive event in marine animals, including mollusk (Calatayud, Garcia-Risco, Pedrini-Martha, et al. 2021), crustacean (Narula et al. 1995; Valls et al. 2001; Munoz et al. 2002), echinoderm (Riek et al. 1999), and chordate species (Guirola et al. 2012; Calatayud et al. 2018). The preference of tunicate MTs for Cd further support the idea that ancestral MTs arose and evolved probably driven by the Cd(II) concentrations in the seawater because Cd is highly toxic by interfering in Zn-dependent cellular processes (Dallinger et al. 2020; Calatayud, Garcia-Risco, Pedrini-Martha, et al. 2021). In addition to sporadic episodes of increases in Cd emissions throughout geological eras (from Paleozoic to Mesozoic and Cenozoic; Dallinger et al. [2020] and references therein), Cd is frequently found with Zn in ore deposits of the earth crust. The natural Cd(II) concentrations are, therefore, higher where seawater comes into contact with the continental earth crust (≈0.8 nmol/kg) than in surface pelagic waters (≈0.002 nmol/kg) (Bruland 1980). Cd(II) concentrations are also higher in eutrophic coastal regions of the littoral and its close neritic zone (≈0.16 nmol/kg) (Bruland 1980), which are usual habitats for tunicates. It is noteworthy that all tunicate MTs analyzed in this work show a similar sequence and metal preference, either they are from benthic ascidians or from planktonic thaliaceans, or from solitary (C. intestinalis and H. roretzi) or colonial species (B. schlosseri). This similarity suggests that the different lifestyles of tunicates might have not been a significant factor in the evolution of metal-binding preference.
Evolution of MT Multiplicity and Multimodular MTs
Increasing the metal-binding capacity enhance the physiological capabilities of the organisms to adapt to diverse conditions of metal bioavailability and other environmental stress situations (Jenny et al. 2016; Baumann et al. 2017). In a simplified view, such increments might be attained through two strategies: increasing the number of MT genes (i.e., augmenting MT multiplicity) (Maroni et al. 1987; Adamo et al. 2012; Luo et al. 2020) or increasing the number of metal-binding domains in a given MT (i.e., generating multimodular MTs) (Jenny et al. 2016; Baumann et al. 2017). Tunicates have used both strategies.
Evolution of MT Multiplicity
Multiplicity in the MT system has been associated to neofunctionalization or subfunctionalization processes, in which different paralogs have distinct metal-binding preferences and/or expression patterns. For instance, in mollusks, several MTs can be found in a given species, each one with distinct metal selectivity—that is, Cd-MT, Cd/Cu-MT, and Cu-MT—and differential transcription patterns (Palacios, Perez-Rafael, et al. 2014; Dvorak et al. 2018; Dallinger et al. 2020). Similarly, in the mammalian system, ubiquitous metal-induced forms MT1 and MT2 have preference for Zn(II), whereas restrictedly expressed MT3 and MT4 have intermediate Zn/Cu- and Cu-thionein characters (Capdevila and Atrian 2011; Vasak and Meloni 2011; Artells et al. 2013). MT multiplicity has been observed in 36 tunicate species, patchily distributed among the three tunicate classes. However, in contrast with the observed in the MT system of other animal groups, the analyzed tunicate MT paralogs do not seem to have essentially diverged in their metal-binding selectivity. The two paralogs of the ascidian H. roretzi (HroMT1 and HroMT2), the four paralogs of the thaliacean S. thompsoni (SthMT1 to SthMT4), and the two paralogs of the appendicularian O. dioica (OdiMT1 and OdiMT2; Calatayud et al. 2018) have a conserved Cd selectivity. These results suggest that tunicate paralogs have undergone a subfunctionalization process partitioning the expression of the predecessor, rather than a neofunctionalization event acquiring new metal preferences. Future detailed spatiotemporal analyses of the transcription patterns of each paralog will provide a definitive answer to this question, and will allow us to understand the ecophysiological needs that led the evolution of multiple MTs in these species.
Evolution of Multimodular MTs
Evolution of multimodular MTs seems a recurrent solution for creating large MTs with high cysteine content and a high capacity of metal binding (Niederwanger et al. 2017; Palacios et al. 2017; Calatayud et al. 2018; and this work). Multimodular MTs have been found in very diverse organisms, from fungi (Palacios, Espart, et al. 2014; Iturbe-Espinoza et al. 2016) and mollusks (Calatayud, Garcia-Risco, Pedrini-Martha, et al. 2021; and references therein), to chordates (Guirola et al. 2012; Calatayud et al. 2018). Here, we have identified multimodular MTs in 20 species of ascidian and appendicularian classes (thaliaceans appear devoid of multimodular MTs). These multimodular MTs are made of tandem repeats of a prototypical organization within each tunicate class—[CxC]x5[CxC]x2Cx2Cx2[CxC]x4[CxC]x3–6CC for ascidian and CX3Cx2Cx2Cx3Cx2[CxC]x2Cx4[CxC]xCC for appendicularian MTs–, implying independent evolutionary origins for ascidian and appendicularian multimodular MTs. In ascidians, the multimodular MTs arose during the evolution of the Styelidae family in the Stolidobranchia order, after its split from the Pyuridae family, 277 Ma (Delsuc et al. 2018). In appendicularians, multimodular MTs arose after its divergence from the other tunicate classes, 447 Ma (Delsuc et al. 2018). The adaptive causes that favored the evolution of large MT in some benthic ascidian species but not in others, or in some planktonic tunicates such as appendicularians but not in thaliaceans, are still an unsolved question.
The Enigmatic Origin of the MTs in Chordates
The extensive catalog of tunicate MTs collected in this work together with the analysis of cephalochordate and vertebrate MTs has allowed us to investigate the origin and evolution of the MTs in the chordate phylum. This has been a particularly difficult challenge because when comparing distant taxonomic groups (e.g., at phylum, subphylum, or class level), one has to be cautious with the MT sequence alignments. The general cysteine richness of the MTs distorts the comparisons by forcing the alignment of Cys positions and inserting many gaps in the aligned sequences. This distortion results in a false impression of similarity between nonhomologous MT sequences, and if the alignments are used for phylogenetic reconstructions, they can lead to wrong evolutionary inferences. To circumvent this drawback, we focused our analyses on the patterns of the cysteine motifs (CxC, CC, and CCC) in the MT domains that, as it has been proved for other molecular markers (Rokas and Holland 2000), might have enormous potential as a tool for phylogenetic inferences.
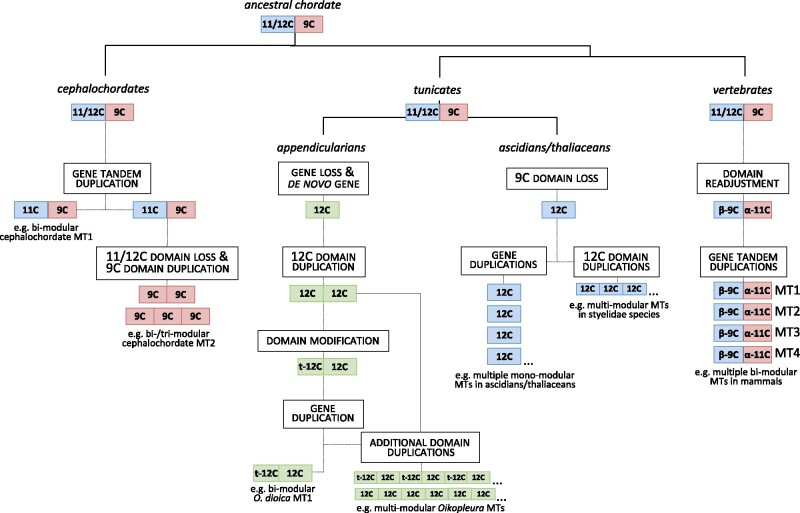
Regarding the ancestral chordate MT, it is generally accepted that due to their substantial genomic stasis (Cañestro and Albalat 2012; Somorjai et al. 2018; Coppola et al. 2019), cephalochordate genes and genomes are more similar to those of the chordate ancestor than those of tunicates and vertebrates (Louis et al. 2012). It can be argued, therefore, that the MT of the chordate ancestor was a bimodular protein with an amino-terminal 11/12-Cys domain and a carboxyl-terminal 9-Cys domain (fig. 6), resembling the organization of the current cephalochordate MT1s. The similarity of cephalochordate and vertebrate domains (fig. 7) supports the bimodular organization of the ancestral chordate MT. In addition, the fact that cephalochordate MT1s are also similar to the α and β1 domains of mollusks (Calatayud, Garcia-Risco, Pedrini-Martha, et al. 2021) would push back in evolutionary time this MT organization to the origins of the bilaterian animals.
Fig. 6.
Reconstruction of MT evolution in the Chordata phyla. The most parsimonious scenario would be that the ancestral chordate MT was a bimodular MT made of an N-terminal 11/12-Cys domain (in blue) and a C-terminal 9-Cys domain (in red), similar to the current cephalochordate MT1. In cephalochordates, this ancestral form was tandem duplicated. One copy lost the 11/12-Cys domain and expanded the 9-Cys domain, yielding the current bi/trimodular MT2 form. In vertebrates, the number of cysteines in each domain was readjusted to the current 9 Cys of the N-terminal β-domains, and 11 Cys of the C-terminal α domain. Successive lineage-specific tandem duplications of this primeval vertebrate MT led to different MT types (MT1, MT2, MT3, and MT4) with a variable degree of multiplicity in different species. In tunicates, the ancestral form lost the C-terminal 9-Cys domain in ascidian/thaliacean lineages, leading a mono-modular. This MT was frequently duplicated generating a high degree of MT multiplicity in many ascidian and thaliacean species. In contrast, internal duplications led to the emergence of multimodular MTs in the stelydae lineage. Finally, in the appendicularian clade, the ancestral MT appears to have been lost, and a new MT would have emerged de novo by the duplication of a 12-Cys domain (in green). During O. dioica evolution, the new bimodular MT lost five Cys in the N-terminal domain, yielding a 7-Cys/12-Cys arrangement (as it is found in current OdiMT1). Next, this 7-Cys/12-Cys MT was internal duplicated generating multimodular MTs (as it is OdiMT2).
The evolutionary scenario of a chordate MT ancestor similar to the current cephalochordate MT1 implies a number of evolutionary events independently shaping the MTs in each chordate subphylum: (I) In cephalochordates, an ancient tandem duplication of the ancestral MT followed by the loss of the 11-Cys domain and the amplification of the 9-Cys domain in one of the copies would have led the MT2 forms of the subphylum (fig. 6); (II) In vertebrates, the ancestral MT would have readjusted its Cys motifs, leading to the current amino-terminal β- and the carboxyl-terminal α-domains (fig. 6). Successive lineage-specific tandem duplications would have led to a variable degree of the MT multiplicity in the different vertebrate species (Serén et al. 2014); (III) In tunicates, the ancestral MT would have lost the 9-Cys domain in the ascidian/thaliacean lineage. This mono-modular 12-Cys MT was patchily duplicated, giving rise to a variable number of MTs in different ascidian/thaliacean species, or internally repeated in the multimodular MTs of styelidae species (fig. 6); (IV) Finally, in the fast–evolving appendicularians, the ancestral chordate MT would have been lost, and a new MT with a different pattern of Cys motifs seems to have evolved de novo (fig. 6). The primeval appendicularian MT would have been a bimodular form made of two 12-Cys domains, apparently unrelated with the other 12-Cys domains of chordates. This ancestral appendicularian MT lost five Cys in the N-terminal domain in the O. dioica lineage (but remained intact in other Oikopleura lineages), yielding the t-12C + 12C organization of the current OdiMT1. This MT was later duplicated, and internal duplications in one of the copies of a repeat unit (RU) made of the t-12C + 12C domains gave rise to the large multimodular OdiMT2 (Calatayud et al. 2018).
In summary, our analyses have shown: 1) de novo emergence of MT genes and domains during chordate diversification; 2) convergent evolution to 12-Cys domains in chordate MTs, but also in other animals, probably favored by essential constraints of the metal coordination; 3) convergent evolution in the metal preference for Cd ions, probably triggered by an ancestral requirement for cadmium-selective forms in many marine animals; 4) lineage-specific events of gene duplications yielding significant levels of MT multiplicity; and 5) independent amplifications of domains that generated multimodular MTs with high metal-binding capacities. All these results have led us to reconstruct an intricate evolutionary history for the chordate MTs, from a bimodular, cadmium-selective form in the last common ancestor, to a high structural and functional MT diversity currently found in the diverse species of the phylum. To discover the adaptive causes that shaped this history and the molecular mechanisms that made it possible will certainly be a fascinating challenge.
Materials and Methods
Chordate MT Search in Databases
Tunicate and cephalochordate MT sequences were retrieved from public databases (NCBI database: https://www.ncbi.nlm.nih.gov/; and Octopus database: http://octopus.obs-vlfr.fr/public/botryllus/blastbotryllus.php) by BlastP and TBlastN searches using known chordate MTs as queries: tunicate CroMT1 (ACN32211), HcuMTa (AY314949), OdiMT1 (AYN64372), and OdiMT2 (AYN64376); cephalochordate BflMT1 (XP_035672826), BflMT2 (BW841405), BlaMT1 (BL11229), and BlaMT2 (JT872034); and vertebrate HsaMT1A (NP_005937), HsaMT2A (NP_005944), HsaMT3 (NP_005945), and HsaMT4 (NP_116324). In addition, RNA-SRA for each tunicate species deposited in NCBI were BLAST searched using as queries MT sequences from the nearest phylogenetic species as well as from different species covering the main tunicate clades. Raw sequence data were retrieved and assembled using SeqMan 8.0.2 (Pro Assembler) software from the DNASTAR Lasergene package, and manually inspected in order to reconstruct new MT sequences. The MT nature of each new identified sequence was evaluated by BlastX searches against metazoan NCBI nonredundant protein sequence database. The amino acid sequences and the accession numbers of the retrieved MTs are provided in supplementary table S1, Supplementary Material online. Sequence alignments were generated with Aliview program (Larsson 2014) and reviewed manually.
Characterization of S. thompsoni Genes
Salpa thompsoni specimens were kindly provided by Nora-Charlotte Pauli from the Bettina Meyer’s group at Alfred Wegener Institute for Polar and Marine Research (AWI). Genomic DNA was extracted using Quick-DNA/RNA Microprep Plus Kit Protocol (Zymo Research, CA) using 5 mg of tissue and following the manufacturer’s instructions. The DNA concentration, purity, and integrity were checked using Tecan Infinite M200 (Tecan Group Ltd, Switzerland) measuring the absorbance at 260 and 280 nm.
Based on S. thompsoni genome and transcriptome projects (PRJNA318929 and GFCC00000000.1, respectively) and the raw data from S. thompsoni SRA projects, several pairs of primers were designed in order to PCR amplify the 5′ and 3′ flanking regions, as well as of intron sequences of the different SthMTs (supplementary table S4, Supplementary Material online).
For SthMT1 we used the primer pairs FwMT1/MT5 with RvMT1/MT2; for SthMT5, FwMT1/MT5 with RvMT5, and finally, for SthMT6 we used FwMT2/MT6 with RvMT6.
For each PCR reaction, 1 ng of genomic DNA was amplified using selected pairs of primers and the Phusion High-Fidelity DNA Polymerase (Invitrogen, Thermo Fisher Scientific, Waltham, MA) in a final 25 μl reaction. PCR conditions were 98 °C 30 s (s); 35 cycles of 98 °C 10 s, 50–54 °C 30 s (supplementary table S4, Supplementary Material online) and 72 °C 3 min; and 72 °C 10 min. PCR products were visualized in 0.8% agarose gels, isolated with the GelElute Plasmid Miniprep Kit (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), and cloned with TOPO TA Cloning Kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA). Plasmid DNA was purified from bacteria using the GeneElute Plasmid Miniprep Kit (Sigma-Aldrich), screened for insert presence by digestion with Fast Digest EcoRI (Invitrogen, Thermo Fisher Scientific, Waltham, MA), and sequenced at the Scientific and Technological Centers of the University of Barcelona using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) in an automatic sequencer (ABIPRISM 310, Applied Biosystems). Sequences were analyzed with Unipro UGENE software (Okonechnikov et al. 2012).
Production and Purification of Recombinant Metal–MT Complexes
Synthetic cDNAs codifying the selected MTs (i.e., CroMT1, HroMT1, HroMT2, BscMT1, R4-BscMT1, SthMT1, SthMT2, SthMT3, and SthMT4) were provided by Synbio Technologies (Monmouth Junction, NJ), cloned in the pGEX-4T-1 expression vector (GE Healthcare, Chicago, IL), and transformed in protease-deficient E. coli BL21 strain for heterologous expression. For heterologous protein production, 500 ml of Luria–Bertani (LB) medium with 100 μg/ml ampicillin was inoculated with E. coli BL21 cells transformed with the corresponding recombinant plasmid. After overnight growth at 37 °C/250 rpm, the cultures were used to inoculate 5 l of fresh LB-100 μg/ml ampicillin medium. Gene expression was induced with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. After the first 30 min of induction, cultures were supplemented with ZnCl2 (300 μM), CdCl2 (300 μM), or CuSO4 (500 μM) in order to generate metal–MT complexes. Cells were harvested by centrifugation for 5 min at 9,100 × g (7,700 rpm), and bacterial pellets were suspended in 125 ml of ice-cold phosphate-buffered saline (PBS: 1.4 M NaCl, 27 mM KCl, 101 mM Na2HPO4, 18 mM KH2PO4, and 0.5% v/v β-mercaptoethanol). Resuspended cells were sonicated (Sonifier Ultrasonic Cell Disruptor) 8 min at voltage 6 with pulses of 0.6 s, and then centrifuged for 40 min at 17,200 × g (12,000 rpm) and 4 °C.
Soluble protein extracts containing GST-MT fusion proteins were incubated with glutathione sepharose beads (GE Healthcare) for 1 h at room temperature with gentle rotation. GST-MT fusion proteins bound to the sepharose beads were washed with 30 ml of cold 1xPBS bubbled with argon to prevent oxidation. After three washes, GST-MT fusion proteins were digested with thrombin (SERVA Electrophoresis GmbH Heidelberg, Germany, 25 U/l of culture) overnight at 17 °C, thus enabling separation of the metal–MT complexes from the GST that remained bound to the sepharose matrix. The eluted metal–MT complexes were concentrated with a 3 kDa Centripep Low Concentrator (Amicon, Merck-Millipore, Darmstadt, Germany), and fractionated on a Superdex-75 FPLC column (GE Healthcare) equilibrated with 20 mM Tris–HCl, pH 7.0. The protein-containing fractions, identified by their absorbance at 254 nm, were pooled and stored at −80 °C until use.
Analysis of Metal–MT Complexes
Protein quantification and element composition of all the samples were achieved by ICP-AES measurements performed in a Optima 4300DV apparatus (Perkin-Elmer, MA) (S, 182.040 nm; Zn, 213.856 nm; Cd, 228.802 nm; Cu, 324.803 nm) under conventional conditions following an already stablished method (Bongers et al. 1988). Molecular weights were determined by ESI-MS, in a MicroTof-Q instrument (Bruker Daltonics Gmbh, Bremen, Germany) connected to a Series 1100 HPLC pump (Agilent Technologies) controlled by the Compass Software. The instrument was calibrated with ESI-L Low Concentration Turning Mix (Agilent Technologies, Santa Clara, CA). Metallated forms were detected under native conditions: 20 µl of sample injected through a PEEK tube at 30–50 µl min−1 in a 3.5–5.0 kV capillary-counter voltage, at 90–110 °C of desolvation temperature, and with dry gas at 6 l min−1. The spectra were recorded between a m/z range from 800 to 3,000. The liquid carrier was a 90:10 mixture of 15 mM ammonium acetate and acetonitrile at pH 7.0. All molecular masses were calculated according to the bibliography (Fabris et al. 1996).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Nora-Charlotte Pauli and Bettina Meyer at Alfred Wegener Institute for Polar and Marine Research for providing Salpa thompsoni specimens, and all tunicate laboratories for sharing sequence results ahead of their publication. They thank Sebastian Artime for experimental support. They also thank the Centres Científics i Tecnològics (CCiT) de la Universitat de Barcelona (DNA sequencing) and the Servei d’Anàlisi Química (SAQ) de la Universitat Autònoma de Barcelona (ICP-AES, ESI-MS) for allocating instrument time. R.A. was supported by BIO2015-67358-C2-1-P, C.C. was supported by BFU2016-80601-P and PID2019-110562GB-I00, and M.C. and O.P. were supported by BIO2015-67358-C2-2-P, the Spanish Ministerio de Ciencia e Innovación. M.C. and O.P. are members of the “Grup de Recerca de la Generalitat de Catalunya,” ref. 2017SGR-864, and R.A. and CC. of ref. 2017SGR-1665. M.G.-R. acknowledges the UAB the PIF grant.
Data Availability
The data underlying this article are available in NCBI (https://www.ncbi.nlm.nih.gov/) and Octopus (http://octopus.obs-vlfr.fr/public/botryllus/blastbotryllus.php) databases. The amino acid sequences and the accession numbers of the MTs used in this work are available in supplementary table S1, Supplementary Material online.
References
- Adamo GM, Lotti M, Tamas MJ, Brocca S.. 2012. Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces cerevisiae. Microbiology 158(Pt 9):2325–2335. [DOI] [PubMed] [Google Scholar]
- Artells E, Palacios O, Capdevila M, Atrian S.. 2013. Mammalian MT1 and MT2 metallothioneins differ in their metal binding abilities. Metallomics 5(10):1397–1410. [DOI] [PubMed] [Google Scholar]
- Baumann C, Beil A, Jurt S, Niederwanger M, Palacios O, Capdevila M, Atrian S, Dallinger R, Zerbe O.. 2017. Structural adaptation of a protein to increased metal stress: NMR structure of a marine snail metallothionein with an additional domain. Angew Chem Int Ed Engl. 56(16):4617–4622. [DOI] [PubMed] [Google Scholar]
- Beil A, Jurt S, Walser R, Schonhut T, Guntert P, Palacios O, Atrian S, Capdevila M, Dallinger R, Zerbe O.. 2019. The solution structure and dynamics of Cd-metallothionein from helix pomatia reveal optimization for binding Cd over Zn. Biochemistry 58(45):4570–4581. [DOI] [PubMed] [Google Scholar]
- Blindauer CA.2014. Metallothioneins. In: Maret W, Wedd A, editors. Binding, transport and storage of metal ions in biological cells. Cambridge: The Royal Society of Chemistry. p. 594–653. [Google Scholar]
- Blindauer CA, Leszczyszyn OI.. 2010. Metallothioneins: unparalleled diversity in structures and functions for metal ion homeostasis and more. Nat Prod Rep. 27(5):720–741. [DOI] [PubMed] [Google Scholar]
- Bofill R, Capdevila M, Atrian S.. 2009. Independent metal-binding features of recombinant metallothioneins convergently draw a step gradation between Zn- and Cu-thioneins. Metallomics 1(3):229–234. [DOI] [PubMed] [Google Scholar]
- Bongers J, Walton CD, Richardson DE, Bell JU.. 1988. Micromolar protein concentrations and metalloprotein stoichiometries obtained by inductively coupled plasma atomic emission spectrometric determination of sulfur. Anal Chem. 60(24):2683–2686. [DOI] [PubMed] [Google Scholar]
- Bruland KW.1980. Oceanographic distributions of cadmium, zinc, nickel, and copper in the North Pacific. Earth Planetary Sci Lett. 47(2):176–198. [Google Scholar]
- Calatayud S, Garcia-Risco M, Capdevila M, Cañestro C, Palacios O, Albalat R. Forthcoming 2021. Modular evolution and population variability of Oikopleura dioica metallothioneins. Front Cell Dev Biol. doi: 10.3389/fcell.2021.702688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calatayud S, Garcia-Risco M, Pedrini-Martha V, Eernisse DJ, Dallinger R, Palacios Ò, Capdevila M, Albalat R.. 2021. Modularity in protein evolution: modular organization and de novo domain evolution in mollusk metallothioneins. Mol Biol Evol. 38(2):424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calatayud S, Garcia-Risco M, Rojas NS, Espinosa-Sanchez L, Artime S, Palacios O, Cañestro C, Albalat R.. 2018. Metallothioneins of the urochordate Oikopleura dioica have Cys-rich tandem repeats, large size and cadmium-binding preference. Metallomics 10(11):1585–1594. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Albalat R.. 2012. Transposon diversity is higher in amphioxus than in vertebrates: functional and evolutionary inferences. Brief Funct Genomics. 11(2):131–141. [DOI] [PubMed] [Google Scholar]
- Capdevila M, Atrian S.. 2011. Metallothionein protein evolution: a miniassay. J Biol Inorg Chem. 16(7):977–989. [DOI] [PubMed] [Google Scholar]
- Capdevila M, Bofill R, Palacios Ò, Atrian S.. 2012. State-of-the-art of metallothioneins at the beginning of the 21st century. Coordination Chem Rev. 256(1–2):46–62. [Google Scholar]
- Catalan A, Glaser-Schmitt A, Argyridou E, Duchen P, Parsch J.. 2016. An indel polymorphism in the MtnA 3′ untranslated region is associated with gene expression variation and local adaptation in Drosophila melanogaster. PLoS Genet. 12(4):e1005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola U, Ristoratore F, Albalat R, D’Aniello S.. 2019. The evolutionary landscape of the Rab family in chordates. Cell Mol Life Sci. 76(20):4117–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa D, Marien J, Janssens TK, van Gestel CA, Driessen G, Sousa JP, van Straalen NM, Roelofs D.. 2012. Influence of adaptive evolution of cadmium tolerance on neutral and functional genetic variation in Orchesella cincta. Ecotoxicology 21(7):2078–2087. [DOI] [PubMed] [Google Scholar]
- Dallinger R, Zerbe O, Baumann C, Egger B, Capdevila M, Palacios O, Albalat R, Calatayud S, Ladurner P, Schlick-Steiner B, et al. 2020. Metallomics reveal a persisting impact of cadmium on the evolution of metal-selective snail metallothioneins. Metallomics 12(5):702–720. [DOI] [PubMed] [Google Scholar]
- de Francisco P, Martin-Gonzalez A, Turkewitz AP, Gutierrez JC.. 2017. Extreme metal adapted, knockout and knockdown strains reveal a coordinated gene expression among different Tetrahymena thermophila metallothionein isoforms. PLoS One 12(12):e0189076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Francisco P, Martin-Gonzalez A, Turkewitz AP, Gutierrez JC.. 2018. Genome plasticity in response to stress in Tetrahymena thermophila: selective and reversible chromosome amplification and paralogous expansion of metallothionein genes. Environ Microbiol. 20(7):2410–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Philippe H, Tsagkogeorga G, Simion P, Tilak MK, Turon X, Lopez-Legentil S, Piette J, Lemaire P, Douzery EJP.. 2018. A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 16(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digilio G, Bracco C, Vergani L, Botta M, Osella D, Viarengo A.. 2009. The cadmium binding domains in the metallothionein isoform Cd7-MT10 from Mytilus galloprovincialis revealed by NMR spectroscopy. J Biol Inorg Chem. 14(2):167–178. [DOI] [PubMed] [Google Scholar]
- Dvorak M, Lackner R, Niederwanger M, Rotondo C, Schnegg R, Ladurner P, Pedrini-Martha V, Salvenmoser W, Kremser L, Lindner H, et al. 2018. Metal binding functions of metallothioneins in the slug Arion vulgaris differ from metal-specific isoforms of terrestrial snails. Metallomics 10(11):1638–1654. [DOI] [PubMed] [Google Scholar]
- Egli D, Domènech J, Selvaraj A, Balamurugan K, Hua H, Capdevila M, Georgiev O, Schaffner W, Atrian S.. 2006. The four members of the Drosophila metallothionein family exhibit distinct yet overlapping roles in heavy metal homeostasis and detoxification. Genes Cells 11(6):647–658. [DOI] [PubMed] [Google Scholar]
- Fabris D, Zaia J, Hathout Y, Fenselau C.. 1996. Retention of thiol protons in two classes of protein zinc ion coordination centers. J Am Chem Soc. 118(48):12242–12243. [Google Scholar]
- Ferrández-Roldán A, Martí-Solans J, Cañestro C, Albalat R.. 2019. Oikopleura dioica: an emergent chordate model to study the impact of gene loss on the evolution of the mechanisms of development. In: Tworzydlo W, Bilinski SM, editors. Evo-Devo: non-model species in cell and developmental biology. Cham: Springer International Publishing. p. 63–105. [DOI] [PubMed] [Google Scholar]
- Franchi N, Boldrin F, Ballarin L, Piccinni E.. 2011. CiMT-1, an unusual chordate metallothionein gene in Ciona intestinalis genome: structure and expression studies. J Exp Zool A Ecol Genet Physiol. 315A(2):90–100. [DOI] [PubMed] [Google Scholar]
- García-Risco M, Calatayud S, Niederwanger M, Albalat R, Palacios Ò, Capdevila M, Dallinger R.. 2020. Two unconventional metallothioneins in the apple snail Pomacea bridgesii have lost their metal specificity during adaptation to freshwater habitats. Int J Mol Sci. 22(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirola M, Perez-Rafael S, Capdevila M, Palacios O, Atrian S.. 2012. Metal dealing at the origin of the Chordata phylum: the metallothionein system and metal overload response in amphioxus. PLoS One 7(8):e43299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isani G, Carpene E.. 2014. Metallothioneins, unconventional proteins from unconventional animals: a long journey from nematodes to mammals. Biomolecules 4(2):435–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Espinoza P, Gil-Moreno S, Lin W, Calatayud S, Palacios O, Capdevila M, Atrian S.. 2016. The fungus Tremella mesenterica encodes the longest metallothionein currently known: gene, protein and metal binding characterization. PLoS One 11(2):e0148651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens TKS, Lopéz R. D R, Mariën J, Timmermans MJTN, Montagne-Wajer K, van Straalen NM, Roelofs D.. 2008. Comparative population analysis of metallothionein promoter alleles suggests stress-induced microevolution in the field. Environ Sci Technol. 42(10):3873–3878. [DOI] [PubMed] [Google Scholar]
- Janssens TKS, Roelofs D, Van Straalen NM.. 2009. Molecular mechanisms of heavy metal tolerance and evolution in invertebrates. Insect Sci. 16(1):3–18. [Google Scholar]
- Jenny MJ, Payton SL, Baltzegar DA, Lozier JD.. 2016. Phylogenetic analysis of molluscan metallothioneins: evolutionary insight from Crassostrea virginica. J Mol Evol. 83(3–4):110–125. [DOI] [PubMed] [Google Scholar]
- Kocot KM, Tassia MG, Halanych KM, Swalla BJ.. 2018. Phylogenomics offers resolution of major tunicate relationships. Mol Phylogenet Evol. 121:166–173. [DOI] [PubMed] [Google Scholar]
- Larsson A.2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30(22):3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis A, Roest Crollius H, Robinson-Rechavi M.. 2012. How much does the amphioxus genome represent the ancestor of chordates? Brief Funct Genomics. 11(2):89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Finet C, Cong H, Wei HY, Chung H.. 2020. The evolution of insect metallothioneins. Proc Biol Sci. 287: 20202189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret W.2013. Zinc and the zinc proteome. In: Banci L, editor. Metallomics and the cell. Dordrecht: Springer Netherlands. p. 479–501. [Google Scholar]
- Maroni G, Wise J, Young JE, Otto E.. 1987. Metallothionein gene duplications and metal tolerance in natural populations of Drosophila melanogaster. Genetics 117(4):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz A, Forsterling FH, Shaw CF III, Petering DH.. 2002. Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms. J Biol Inorg Chem. 7(7–8):713–724. [DOI] [PubMed] [Google Scholar]
- Narula SS, Brouwer M, Hua Y, Armitage IM.. 1995. Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy. Biochemistry 34(2):620–631. [DOI] [PubMed] [Google Scholar]
- Naville M, Henriet S, Warren I, Sumic S, Reeve M, Volff JN, Chourrout D.. 2019. Massive changes of genome size driven by expansions of non-autonomous transposable elements. Curr Biol. 29(7):1161–1168.e1166. [DOI] [PubMed] [Google Scholar]
- Niederwanger M, Dvorak M, Schnegg R, Pedrini-Martha V, Bacher K, Bidoli M, Dallinger R.. 2017. Challenging the metallothionein (MT) gene of Biomphalaria glabrata: unexpected response patterns due to cadmium exposure and temperature stress. Int J Mol Sci. 18:1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M, UGENE Team. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167. [DOI] [PubMed] [Google Scholar]
- Palacios O, Atrian S, Capdevila M.. 2011. Zn- and Cu-thioneins: a functional classification for metallothioneins? J Biol Inorg Chem. 16(7):991–1009. [DOI] [PubMed] [Google Scholar]
- Palacios O, Espart A, Espin J, Ding C, Thiele DJ, Atrian S, Capdevila M.. 2014. Full characterization of the Cu-, Zn-, and Cd-binding properties of CnMT1 and CnMT2, two metallothioneins of the pathogenic fungus Cryptococcus neoformans acting as virulence factors. Metallomics 6(2):279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios O, Jimenez-Marti E, Niederwanger M, Gil-Moreno S, Zerbe O, Atrian S, Dallinger R, Capdevila M.. 2017. Analysis of metal-binding features of the wild type and two domain-truncated mutant variants of Littorina littorea metallothionein reveals its Cd-specific character. Int J Mol Sci. 18(7):1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios O, Pagani A, Pérez-Rafael S, Egg M, Höckner M, Brandstätter A, Capdevila M, Atrian S, Dallinger R.. 2011. Shaping mechanisms of metal specificity in a family of metazoan metallothioneins: evolutionary differentiation of mollusc metallothioneins. BMC Biol. 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios O, Perez-Rafael S, Pagani A, Dallinger R, Atrian S, Capdevila M.. 2014. Cognate and noncognate metal ion coordination in metal-specific metallothioneins: the Helix pomatia system as a model. J Biol Inorg Chem. 19(6):923–935. [DOI] [PubMed] [Google Scholar]
- Papadopoulou C, Kanias GD.. 1977. Tunicate species as marine pollution indicators. Mar Pollut Bull. 8(10):229–231. [Google Scholar]
- Pedrini-Martha V, Koll S, Dvorak M, Dallinger R.. 2020. Cadmium uptake, MT gene activation and structure of large-sized multi-domain metallothioneins in the terrestrial door snail Alinda biplicata (Gastropoda, Clausiliidae). Int J Mol Sci. 21(5):1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rafael S, Monteiro F, Dallinger R, Atrian S, Palacios O, Capdevila M.. 2014. Cantareus aspersus metallothionein metal binding abilities: the unspecific CaCd/CuMT isoform provides hints about the metal preference determinants in metallothioneins. Biochim Biophys Acta. 1844(9):1694–1707. [DOI] [PubMed] [Google Scholar]
- Riek R, Precheur B, Wang Y, Mackay EA, Wider G, Guntert P, Liu A, Kagi JH, Wuthrich K.. 1999. NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA. J Mol Biol. 291(2):417–428. [DOI] [PubMed] [Google Scholar]
- Rokas A, Holland PW.. 2000. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 15(11):454–459. [DOI] [PubMed] [Google Scholar]
- Rost B.1999. Twilight zone of protein sequence alignments. Protein Eng. 12(2):85–94. [DOI] [PubMed] [Google Scholar]
- Serén N, Glaberman S, Carretero MA, Chiari Y.. 2014. Molecular evolution and functional divergence of the metallothionein gene family in vertebrates. J Mol Evol. 78(3–4):217–233. [DOI] [PubMed] [Google Scholar]
- Somorjai IML, Martí-Solans J, Diaz-Gracia M, Nishida H, Imai KS, Escriva H, Cañestro C, Albalat R.. 2018. Wnt evolution and function shuffling in liberal and conservative chordate genomes. Genome Biol. 19(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman MJ, Cai W, Zelazowski AJ.. 1987. Cadmium binding to metallothioneins. Domain specificity in reactions of alpha and beta fragments, apometallothionein, and zinc metallothionein with Cd2. J Biol Chem. 262(10):4538–4548. [PubMed] [Google Scholar]
- Tanguy A, Moraga D.. 2001. Cloning and characterization of a gene coding for a novel metallothionein in the Pacific oyster Crassostrea gigas (CgMT2): a case of adaptive response to metal-induced stress? Gene 273(1):123–130. [DOI] [PubMed] [Google Scholar]
- Timmermans MJ, Ellers J, Roelofs D, van Straalen NM.. 2005. Metallothionein mRNA expression and cadmium tolerance in metal-stressed and reference populations of the springtail Orchesella cincta. Ecotoxicology 14(7):727–739. [DOI] [PubMed] [Google Scholar]
- Tio L, Villarreal L, Atrian S, Capdevila M.. 2004. Functional differentiation in the mammalian metallothionein gene family: metal binding features of mouse MT4 and comparison with its paralog MT1. J Biol Chem. 279(23):24403–24413. [DOI] [PubMed] [Google Scholar]
- Tomas M, Domènech J, Capdevila M, Bofill R, Atrian S.. 2013. The sea urchin metallothionein system: comparative evaluation of the SpMTA and SpMTB metal-binding preferences. FEBS Open Bio. 3:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafriri-Milo R, Benaltabet T, Torfstein A, Shenkar N.. 2019. The potential use of invasive Ascidians for biomonitoring heavy metal pollution. Front Mar Sci. 6:611. [Google Scholar]
- Vallee BL, Falchuk KH.. 1993. The biochemical basis of zinc physiology. Physiol Rev. 73(1):79–118. [DOI] [PubMed] [Google Scholar]
- Valls M, Bofill R, Gonzalez-Duarte R, Gonzalez-Duarte P, Capdevila M, Atrian S.. 2001. A new insight into metallothionein (MT) classification and evolution. The in vivo and in vitro metal binding features of Homarus americanus recombinant MT. J Biol Chem. 276(35):32835–32843. [DOI] [PubMed] [Google Scholar]
- Vasak M, Meloni G.. 2011. Chemistry and biology of mammalian metallothioneins. J Biol Inorg Chem. 16(7):1067–1078. [DOI] [PubMed] [Google Scholar]
- Vest KE, Hashemi HF, Cobine PA.. 2013. The copper metallome in eukaryotic cells. In: Banci L, editor. Metallomics and the cell. Dordrecht (The Netherlands: ): Springer Netherlands. p. 451–478. [DOI] [PubMed] [Google Scholar]
- Voskoboynik A, Neff NF, Sahoo D, Newman AM, Pushkarev D, Koh W, Passarelli B, Fan HC, Mantalas GL, Palmeri KJ, et al. 2013. The genome sequence of the colonial chordate, Botryllus schlosseri. Elife 2:e00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron KJ, Rutherford JC, Ford D, Robinson NJ.. 2009. Metalloproteins and metal sensing. Nature 460(7257):823–830. [DOI] [PubMed] [Google Scholar]
- Yue JX, Yu JK, Putnam NH, Holland LZ.. 2014. The transcriptome of an amphioxus, Asymmetron lucayanum, from the Bahamas: a window into chordate evolution. Genome Biol Evol. 6(10):2681–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller A, Fraissinet-Tachet L.. 2018. Metallothionein diversity and distribution in the tree of life: a multifunctional protein. Metallomics 10(11):1549–1559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in NCBI (https://www.ncbi.nlm.nih.gov/) and Octopus (http://octopus.obs-vlfr.fr/public/botryllus/blastbotryllus.php) databases. The amino acid sequences and the accession numbers of the MTs used in this work are available in supplementary table S1, Supplementary Material online.