Abstract
UV irradiation induces the formation of cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts in DNA. These two types of lesions can be directly photorepaired by CPD photolyases and 6-4 photolyases, respectively. Recently, a new class of 6-4 photolyases named iron–sulfur bacterial cryptochromes and photolyases (FeS-BCPs) were found, which were considered as the ancestors of all photolyases and their homologs—cryptochromes. However, a controversy exists regarding 6-4 photoproducts only constituting ∼10–30% of the total UV-induced lesions that primordial organisms would hardly survive without a CPD repair enzyme. By extensive phylogenetic analyses, we identified a novel class of proteins, all from eubacteria. They have relatively high similarity to class I/III CPD photolyases, especially in the putative substrate-binding and FAD-binding regions. However, these proteins are shorter, and they lack the “N-terminal α/β domain” of normal photolyases. Therefore, we named them short photolyase-like. Nevertheless, similar to FeS-BCPs, some of short photolyase-likes also contain four conserved cysteines, which may also coordinate an iron–sulfur cluster as FeS-BCPs. A member from Rhodococcus fascians was cloned and expressed. It was demonstrated that the protein contains a FAD cofactor and an iron–sulfur cluster, and has CPD repair activity. It was speculated that this novel class of photolyases may be the real ancestors of the cryptochrome/photolyase family.
Keywords: photolyase, cryptochrome, cyclobutane pyrimidine dimer, iron–sulfur cluster, phylogenetic analysis
Introduction
The early Earth was anaerobic and iron and sulfur was abundant (Imlay 2006). Based on the coacervate theory of Oparin (Oparin 1938), researchers further proposed that the iron–sulfur complexes played an important role in the emergence of life (Huber and Wachtershauser 1997; Russell and Hall 1997; Koonin and Martin 2005). Many ancient proteins might possess iron–sulfur clusters as cofactors to facilitate their structure folding and to perform electron transferring or other catalytic functions (Imlay 2006). Meanwhile, due to the lack of oxygen and thus ozone in atmosphere, the surface of the early Earth was exposed to high doses of UV irradiation (Cockell 1998). To survive on the early Earth, the primordial organisms must have the efficient ability to repair UV-induced lesions, such as cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts in DNA (Beukers et al. 2008). Photorepair was a good choice for the primordial organisms, which could cure the UV-induced lesions directly by photolyases using external light as the energy source (Sancar 2003; Vechtomova et al. 2020). Indeed, photolyases are widely distributed in all kingdoms of living organisms, indicating that they have been evolved very early (Lucas-Lledo and Lynch 2009; Vechtomova et al. 2020). About 2.5 billion years ago, the oxygen concentration in the atmosphere began to rise by the production of photosynthesis (Lyons et al. 2014). This change caused the oxidation of environmental iron and the limitation of its bioavailability. Furthermore, the stability of the iron–sulfur clusters in proteins were threatened by oxygen (Imlay 2006). Therefore, many organisms abandoned the use of the iron–sulfur clusters in most of their proteins. Only a small number of crucial iron–sulfur cluster containing proteins were retained and protected by various antioxidant strategies (Imlay 2006). Meanwhile, a good result of oxygen accumulation was the formation of the ozone layer, which greatly attenuated the UV irradiation reached the surface of the Earth (Cockell 1998). Thereafter, the importance of photolyases might be lowered, allowing some of them to evolve to their functionally diverse homologs, called cryptochromes (Chaves et al. 2011; Lopez et al. 2021).
Photolyases are generally composed of two structural domains, an N-terminal α/β domain and a C-terminal α-helical domain (Sancar 2003; Muller and Carell 2009; Vechtomova et al. 2020). A reduced FAD (FADH−) cofactor is required for the activity of all photolyases, which is bound in the C-terminal α-helical domain. Most photolyases also have an antenna cofactor, such as 5,10-methenyltetrahydrofolate (MTHF) or 8-hydroxy-5-deazaflavin (8-HDF), located in a cleft between the two domains, which can increase the catalytic efficiency under light-limiting conditions (Sancar 2003; Muller and Carell 2009; Kritsky et al. 2010, 2012). Photolyases can be divided into two types, CPD photolyases and 6-4 photolyases, which repair CPDs and 6-4 photoproducts in DNA, respectively (Sancar 2003; Yamamoto et al. 2017). Recently, a new class of 6-4 photolyases named iron–sulfur bacterial cryptochromes and photolyases (FeS-BCPs) were found in some prokaryotes. These proteins generally contain a unique 6,7-dimethyl 8-ribityl-lumazine (DMRL) as an antenna cofactor, and an iron–sulfur cluster [4Fe–4S] bound to the C-terminal domain (Oberpichler et al. 2011; Geisselbrecht et al. 2012; Zhang et al. 2013; Dikbas et al. 2019). Based on phylogenetic analyses, it was proposed that a 6-4 photolyase with an iron–sulfur cluster was the first common ancestor of the cryptochrome/photolyase family (CPF) (Zhang et al. 2013). This hypothesis was reasonable because FeS-BCPs were the only known members of CPF that contain an iron–sulfur cluster, which is an ancient feature. However, 6-4 photoproducts constitute only a small part (∼10–30%) of the total UV-induced DNA lesions, whereas CPDs constitute the large part (Sancar 2003; Weber 2005; Vechtomova et al. 2020). One cannot imagine that the primordial organisms would survive under high-UV conditions without a CPD repair enzyme. This discrepancy needs to be reconciled by new findings.
In this work, we performed extensive phylogenetic analyses of CPF proteins. From 574 CPF sequences of 271 organisms, a novel class of CPF was identified. The similarity of these proteins to class I/III CPD photolyases is relatively high, especially in the putative substrate-binding and FAD-binding regions. However, they are ∼250–400 residues in length, which are shorter than most CPF proteins. Therefore, we named this class of proteins short photolyase-like (SPL). To obtain more information about this novel class, a larger number of SPL sequences (191 in total) were retrieved. All the protein sequences in this class were found in eubacterial genomes. These sequences were divided into five subgroups by phylogenetic analysis. It is remarkable that all these proteins lack the “N-terminal α/β domain” of normal photolyases, implying that they do not have an antenna cofactor. Nevertheless, about one-third of SPLs contain four conserved cysteines, the arrangements of which resemble those of FeS-BCPs, indicating these SPLs may also contain an iron–sulfur cluster as FeS-BCPs. A representative SPL from Rhodococcus fascians (RfSPL) was cloned and expressed. It was demonstrated that RfSPL is a CPD photolyase with a FAD cofactor and an iron–sulfur cluster. We speculate that this novel class of photolyases may be the real ancestors of all CPF proteins.
Results
Identification of a Novel Class of the Cryptochrome/Photolyase Family
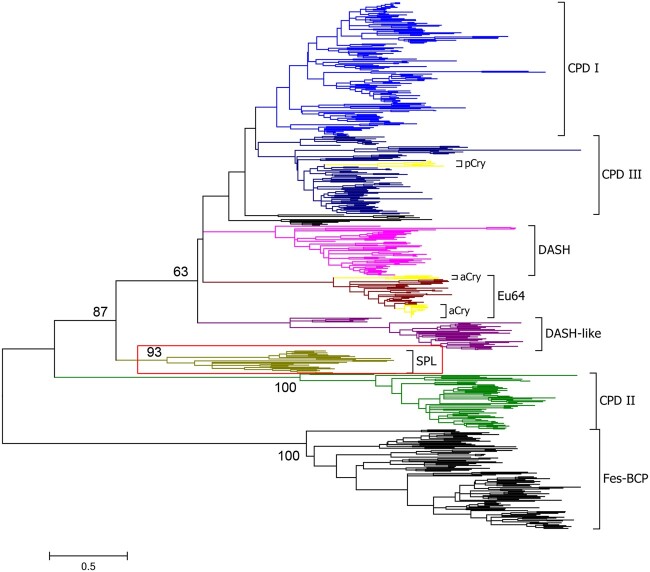
Photolyases are important DNA repair enzymes that repair UV-induced lesions such as CPDs and 6-4 products in DNA (Sancar 2003; Vechtomova et al. 2020). Cryptochromes are homologs of photolyases that play key roles in regulating circadian clock, development, and growth of organisms (Chaves et al. 2011; Lopez et al. 2021). These proteins form a large protein family called cryptochrome/photolyase family (CPF). To get a deeper insight into the category and evolutionary history of CPF, a total of 574 CPF sequences of 271 organisms from all three kingdoms were retrieved, and extensive phylogenetic analyses were performed.
During retrieving the homologous sequences of CPF from available genome data, we noticed that there are some short sequences with ∼250–400 residues that have relatively high similarity to the C-terminal α-helical domain of class I/III CPD photolyases. Furthermore, in the phylogenetic analysis, they constituted one monophyletic group containing 24 members with a high bootstrap value (93%) supporting, together with seven known groups formed by other CPF sequences: class I CPD photolyases, class III CPD photolyases (including plant cryptochromes), DASH proteins, DASH-like proteins (or named the prokaryote-specific group); eukaryotic 6-4 photolyases (including animal cryptochromes), class II CPD photolyases, and FeS-BCPs (Lucas-Lledo and Lynch 2009; Scheerer et al. 2015; Xu et al. 2017) (fig. 1 and supplementary fig. S1 and table S1, Supplementary Material online). We named this new class of proteins SPL due to the short nature of the sequences in the class. The main topological structures of the phylogenetic trees were not altered when the sequences of the large subunits of the archaeal/eukaryotic primases (PriLs) that have structural similarity with photolyases (Sauguet et al. 2010; Zhang et al. 2013) were included in the analysis (supplementary fig. S2, Supplementary Material online).
Fig. 1.
Phylogenetic tree of the cryptochrome/photolyase family (CPF). The evolutionary history was inferred by using the maximum likelihood method based on the Le_Gascuel_2008 with Freq. (+F) model (Le and Gascuel 2008) and 500 bootstrap iterations. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter=1.3083]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 2.0619% sites). Numbers at nodes represent the bootstrap support percentage values. The CPF proteins were divided into eight main groups: class I CPD photolyases (CPD I), class III CPD photolyases (CPD III), DASH proteins (DASH), eukaryotic 6-4 photolyases (Eu64), DASH-like proteins (DASH-like), short photolyase-likes (SPL), class II CPD photolyases (CPD II), and FeS-BCP proteins (FeS-BCP). Plant cryptochromes (pCry) and animal cryptochromes (aCry) were clustered as the subgroups of CPD III and Eu6-4, respectively. The SPL group was highlighted with a red box, which was identified in this report.
To get further information of SPLs, we retrieved more SPL sequences (191 in total) and analyzed them in detail. These sequences were found in 187 eubacterial genomes, including those of the classes Alphaproteobacteria (71/187), Betaproteobacteria (19/187), Gammaproteobacteria (15/187) of the phylum Proteobacteria, and the phyla Acidobacteria (1/187), Actinobacteria (12/187), Bacteroidetes (20/187), Balneolaeota (1/187), Chloroflexi (4/187), Cyanobacteria (41/187), Deinococcus–Thermus (4/187), Planctomycetes (1/187), and Spirochaetes (1/187) (supplementary table S2, Supplementary Material online). Four organisms (Thalassobacter stenotrophicus, Sulfitobacter guttiformis, Methylobacterium sp. 77, and Hassallia byssoidea) contain two copies of SPL sequences, which may be due to gene duplication events. It was observed that the organisms harboring a SPL sequence often contain a class I/III CPD photolyase sequence (94%, (94 + 82)/187), and/or a FeS-BCP sequence (85%, 159/187). Less than half of the SPL containing organisms contain a DASH-like sequence (36%, 67/187). Only a few of them contain a DASH sequence (4%, 8/187) or a class II CPD photolyase sequence (5%, 10/187). None of them have a eukaryotic 6-4 photolyase sequence, a plant, or an animal cryptochrome sequence (supplementary table S3, Supplementary Material online). It was Interesting that in a number of organisms, such as Cyclobacterium marinum DSM 745, a SPL coding sequence and a FeS-BCP coding sequence were predicted to be in one operon (Taboada et al. 2012), implying that their functions may be closely related.
Phylogenetic analysis of these SPL sequences showed that they formed five subgroups (supplementary fig. S3 and table S2, Supplementary Material online). The largest subgroup was named “A,” because most members are from the class Alphaproteobacteria (68/83). The second large subgroup was named “C,” most members of which are from the phylum Cyanobacteria (39/62). The subgroups “Be” and “Ba” were mainly constituted with the members from the class Betaproteobacteria (13/15) and the phylum Bacteroidetes (20/24), respectively. The smallest subgroup was “Ac,” all members of which are from the phylum Actinobacteria (7/7).
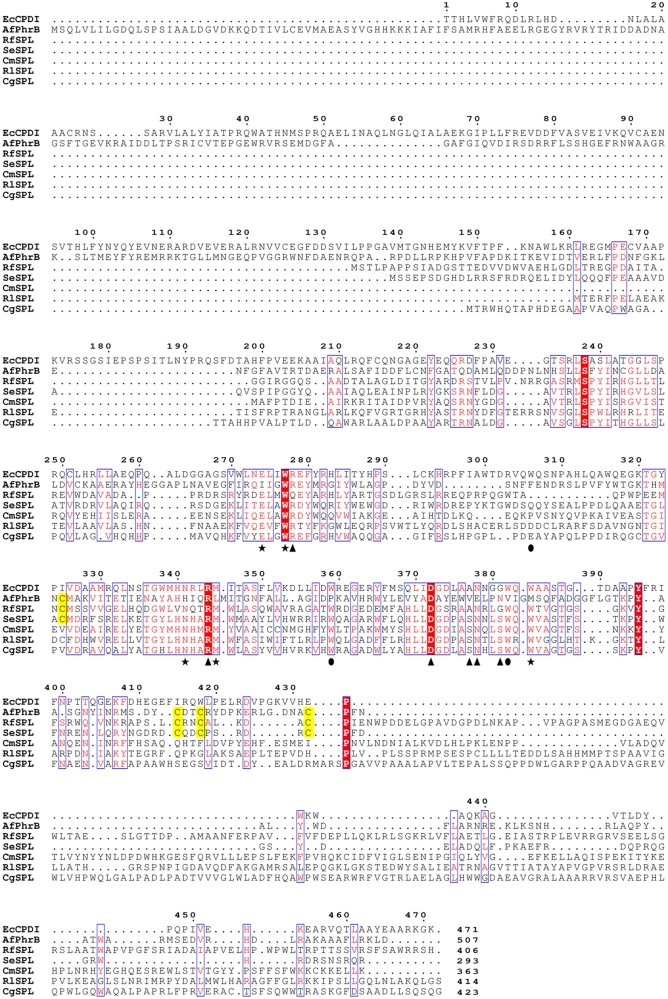
Structural Features of SPLs
The primary structures of five representative SPLs: R. fascians SPL (RfSPL, subgroup Ac), Synechococcus elongatus SPL (SeSPL, subgroup C), Cyclobacterium marinum SPL (CmSPL, subgroup Ba), Ruegeria lacuscaerulensis SPL (RlSPL, subgroup A), and Curvibacter gracilis SPL (CgSPL, subgroup Be) were aligned together with those of Escherichia coli class I CPD photolyase (EcCPDI, PDB ID: 1DNP) (Park et al. 1995) and Agrobacterium fabrum FeS-BCP (AfPhrB, PDB ID: 4DJA) (Zhang et al. 2013). The result was shown in figure 2. It was found that SPLs do not contain the homologous sequences of the N-terminal α/β domains of EcCPDI and AfPhrB, suggesting that SPLs may not have an antenna cofactor. The substrate-binding sites (Glu274, Trp277, Asn341, Met345, and Trp384) and FAD-binding sites (Arg344, Asp372, Ala377, Asn378, and Gly381) of EcCPDI are highly conserved in SPLs. Among the Trp triad (Trp306, Trp359, and Trp382) of EcCPDI, which act as an electron transfer chain during photoreduction of oxidized FAD (Aubert et al. 2000), only the latter two are conserved in SPLs. Most members of SPLs contain C-terminal extensions in various lengths. Intriguingly, about one-third (62 out of 191) of the retrieved SPL sequences, including RfSPL and SeSPL, have four conserved cysteines, the arrangements of which resemble those of AfPhrB (fig. 2 and supplementary table S4, Supplementary Material online). The SPLs that contain the four conserved cysteines are from subgroups C and Ac. The members of the subgroups A, Ba, and Be do not have the four conserved cysteines (supplementary table S4, Supplementary Material online).
Fig. 2.
Sequence alignment of the primary structures of five representative SPLs: Rhodococcus fascians SPL (RfSPL), Synechococcus elongatus SPL (SeSPL), Cyclobacterium marinum SPL (CmSPL), Ruegeria lacuscaerulensis SPL (RlSPL), and Curvibacter gracilis SPL (CgSPL), and those of Escherichia coli class I CPD photolyase (EcCPDI) and Agrobacterium fabrum FeS-BCP (AfPhrB). The putative the FAD-binding sites were marked with triangles. The putative substrate-binding sites were marked with stars. The sites of the putative electron transfer chain during photoreduction were marked with circles. The putative the iron–sulfur cluster coordination cysteines in SPLs and those in AfPhrB were highlighted by yellow blocks. The number at the end of each sequence represents the length of the protein.
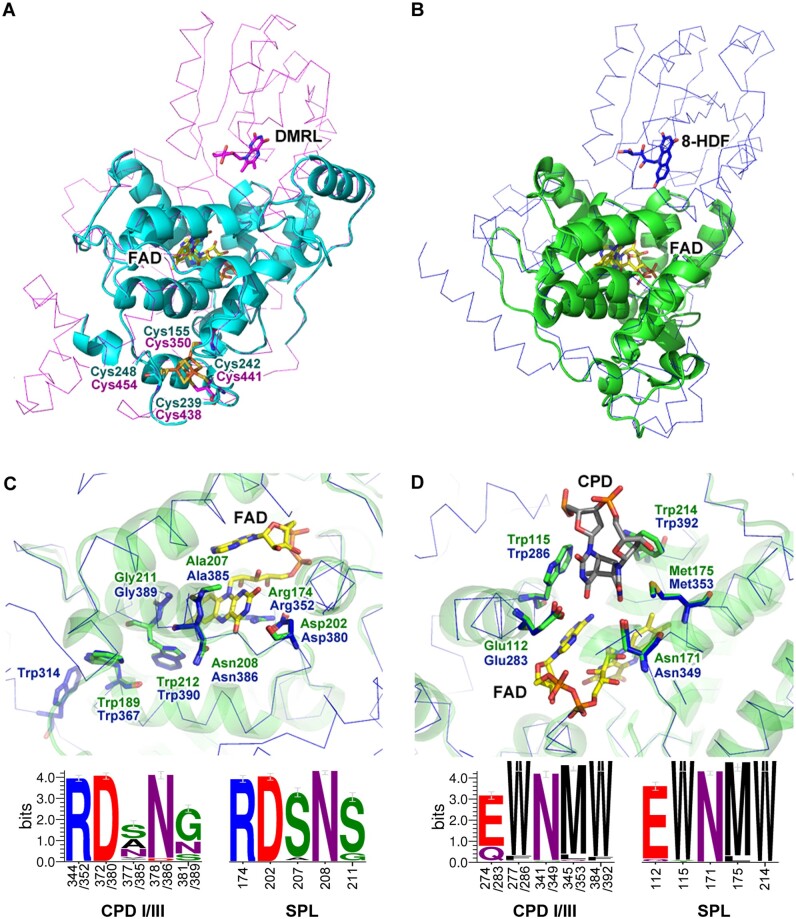
To get more structural information of SPLs, homology structural models of RfSPL were constructed using two of the top-ranked templates (AfPhrB, PDB ID: 4DJA and S. elongatus class I CPD photolyase, SeCPDI, PDB ID: 1TEZ). Among the total of 406 residues of RfSPL, fragments of 4-256 and 41-252 were modeled based on the structural templates of AfPhrB and SeCPDI, respectively (fig. 3A and B). It is clear that the modeled structures do not contain the homologous region of the N-terminal α/β domains of other CPF proteins. The C-terminal extension of RfSPL could not be modeled due to low similarities to the templates. In the first model, four cysteines of RfSPL (Cys155, Cys239, Cys242, and Cys248) are located at similar positions to those of AfPhrB (Cys350, Cys438, Cys441, and Cys454), implying that they may also coordinate a [4Fe–4S] cluster as AfPhrB (fig. 3A). In the second model, the FAD-binding pocket of RfSPL is highly similar to that of SeCPDI (fig. 3C). Arg174 and Asp202 of RfSPL form a salt bridge on the re-face of FAD. Ala207 and Gly211 are located on the si-face of FAD. And, Asn208 is located at the side proximal to the N5 of FAD. Two tryptophans, Trp212 and Trp189 of RfSPL are superimposed with the proximal and intermediate residues of the tryptophan triad of class I CPD photolyases (Trp382 and Trp359 in EcCPDI, and Trp390 and Trp367 in SeCPDI), which may act as the electron transfer chain during photoreduction. The homolog of the distal residue of the Trp triad (Trp306 in EcCPDI, and Trp314 in SeCPDI) was not found in RfSPL. The third tryptophan may not be necessary for photoreduction of RfSPL because Trp189 may be already located at the protein surface. The substrate-binding pocket of RfSPL is formed by five residues, Glu112, Trp115, Asn171, Met175, and Trp214, which may accommodate a CPD lesion as well as that of SeCPDI (fig. 3D).
Fig. 3.
The modeled structures of RfSPL were shown in (A) cyan and (B) green cartoon representations which were constructed using the crystal structure of Agrobacterium fabrum FeS-BCP (AfPhrB, PDB ID: 4DJA, magenta ribbon) and that of Synechococcus elongatus class I CPD photolyase (SeCPDI, PDB ID: 1TEZ, blue ribbon) as the template, respectively. (C) and (D) were the detailed views of the putative FAD- and substrate-binding pockets of RfSPL modeled on the crystal structure of SeCPDI. The lower parts of (C) and (D) were the WebLogo presentations of the FAD- and substrate-binding sites of 221 class I/III CPD photolyases and those of 191 SPLs, respectively. The numbers under the abscissae indicated the residue numbers of EcCPDI/SeCPDI and RfSPL, respectively. The FAD, DMRL, and 8-HDF cofactors, the iron–sulfur cluster in the templates, the putative CPD substrate, and the key residues in the templates and the modeled structures were shown in the stick representations.
In the enzyme–substrate complex structure of SeCPDI, Arg350, Asn407, Gln411, Lys414, Gln461, and α6-helix of SeCPDI interact with the backbone of the substrate DNA and strengthen the binding of the substrate (Mees et al. 2004). In the second RfSPL model, Gln172, Arg228, Gln230, and Lys233 of RfSPL are located at similar positions to Arg350, Asn407, Gln411, and Lys414 of SeCPDI, which may also interact with the backbone of the substrate DNA (supplementary fig. S4, Supplementary Material online). The homologous residue of Gln461 in SeCPDI was not found in the model of RfSPL due to the failure of the modeling of the C-terminal structure. And, it seems that the RfSPL model do not have the homologous structure of the α6-helix of SeCPDI, although a possibility that an equivalent structure forming by the wrapped C-terminal in RfSPL cannot be ruled out. To elucidate the exact enzyme–substrate interaction of RfSPL, further crystallographic studies are needed.
RfSPL Is a CPD Photolyase with a FAD Cofactor and an Iron–Sulfur Cluster
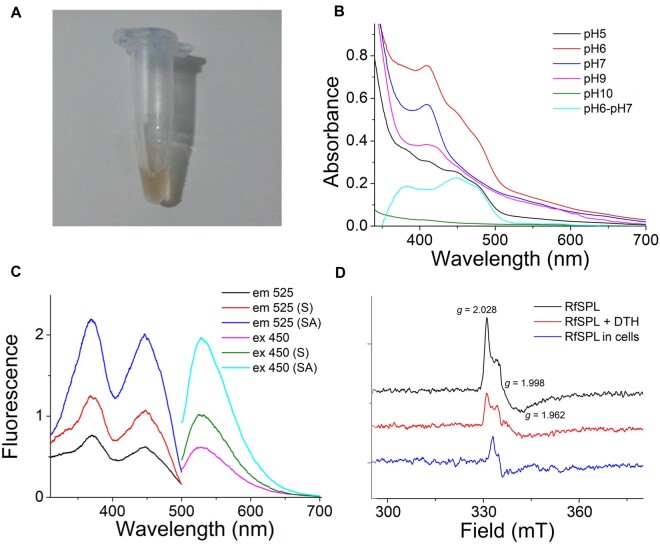
To verify the properties of SPLs inferred from bioinformatic analyses, we cloned and overexpressed one representative of them—RfSPL in E. coli. This protein was purified at different pH values using the affinity chromatography. The concentrated sample purified at pH 6.0 showed a brownish yellow color (fig. 4A). SDS–PAGE analysis of the concentrated sample gave a band with an apparent mobility corresponding to a molecular mass of 52 kDa, which is slightly higher than the calculated molecular mass of 46 kDa (supplementary fig. S5A, Supplementary Material online). The MALDI-TOF/TOF mass spectrometry gave a molecular mass of 47 kDa of the major species (supplementary fig. S5B, Supplementary Material online). There was an absorption peak at 410 nm of the RfSPL protein purified at pH 6.0, which is characteristic of the presence of an iron–sulfur cluster. Furthermore, an absorption shoulder around 450 nm was observed (fig. 4B, red line). The fluorescence spectroscopy of the RfSPL protein purified at pH 6.0 was analyzed. One emission peak at 525 nm, and two excitation peaks at 370 and 450 nm were observed. In contrast, the fluorescence spectra of a FeS-BCP from Vibrio cholerae exhibited an emission peak at 494 nm, and an excitation peaks at 420 nm, which were attributed to the antenna cofactor DMRL in the protein (Dikbas et al. 2019). Our results indicated that RfSPL might only contain a flavin cofactor without an antenna cofactor (fig. 4C, black and magenta lines). After boiling and centrifuging, the fluorescence of the sample supernatant increased ∼2 folds (fig. 4C, red and olive lines). When the pH value of the sample supernatant was lowered to 2.0, the fluorescence was further increased ∼2 folds (fig. 4C, blue and cyan lines). Thin layer chromatography confirmed that the released cofactor in the supernatant of the sample was FAD (supplementary fig. S6, Supplementary Material online). We also determined the contents of iron and acid-labile sulfide in RfSPL protein purified at pH 6.0 (supplementary figs. S7 and S8, Supplementary Material online). The results showed that one mol of the protein contained ∼0.15 mol of iron and ∼0.17 mol of acid-labile sulfide. The low occupancy of iron and acid-labile sulfide of the samples might be due to the decomposition of the iron–sulfur cluster under aerobic conditions. Nevertheless, the FAD occupancy of the samples was also very low, one mol of the protein contained only ∼0.043 mol of FAD. The ratio of iron: acid-labile sulfide: FAD was ∼3.5:4:1. Assuming that the holo-form RfSPL protein contains one FAD cofactor and one iron–sulfur cluster, the iron–sulfur cluster of RfSPL might be in the [4Fe–4S] or [3Fe–4S] form.
Fig. 4.
RfSPL is an iron–sulfur cluster containing flavoprotein. (A) The concentrated RfSPL sample purified at pH 6.0 exhibited a brownish yellow color. (B) The UV-vis absorption spectra of the RfSPL samples purified at various pH values: pH 5.0, black line; pH 6.0, red line; pH 7.0, blue line; pH 9.0, magenta line; pH 10.0, olive line. The differential spectrum of the sample purified at pH 6.0 and that purified at 7.0 was represented in a cyan line. The result of the RfSPL sample purified at pH 8.0 was not shown which was similar to that of the sample purified at pH 7.0. (C) The fluorescence spectra of the RfSPL sample purified at pH 6.0, the sample supernatant after boiling and centrifuging (indicated by (S)), and the sample supernatant when the pH value of which was lower to 2.0 (indicated by (SA)). For the excitation spectra (black, red, and blue lines), the emission wavelength was set to 525 nm. For the emission spectra (magenta, olive, and cyan lines), the excitation wavelength was set to 450 nm. (D) The EPR spectra of RfSPL. The black and red lines were the signals of the RfSPL sample purified at pH 6.0 before and after treatment with 3 mM sodium dithionite (SDT) for 10 min. The blue line was the signal of overexpressed RfSPL protein in Escherichia coli cells.
The RfSPL samples purified at the pH between 7.0 and 9.0 also had the absorption peaks at ∼410 nm, but lacked the absorption shoulder around 450 nm (fig. 4B, blue and magenta lines). The differential spectrum of the sample purified at pH 6.0 and that purified at 7.0 was represented in a cyan line in figure 4B, which supported that the sample purified at pH 6.0 bound FAD. Chemical analyses showed that the samples purified at the pH between 7.0 and 9.0 also contained iron and acid-labile sulfide. But the FAD cofactor of these samples was scarce, which might be released during purification at this pH range. The RfSPL sample purified at pH 5.0 did not have the absorption peak at ∼410 nm, indicating that the iron–sulfur cluster was decomposed at this pH value. However, the sample had absorption around 450 nm, implying that it retained the FAD cofactor (fig. 4B, black line). The absorption between 300 and 700 nm of the RfSPL sample purified at pH 10.0 was weak, indicating that both the FAD cofactor and the iron–sulfur cluster had been released (fig. 4B, olive line). The RfSPL samples purified at pH 5.0 or pH 10.0 were very unstable, which tended to aggregate shortly after purification.
To get more information about the iron–sulfur cluster of RfSPL, electron paramagnetic resonance (EPR) spectroscopy was employed. The E. coli expressed RfSPL protein purified at pH 6.0 exhibited an intense signal with g values of 2.028, 1.998, and 1.962 at 10 K (fig. 4D, black line). This signal was undetectable above 40 K. After a brief treatment with sodium dithionite (SDT), the signal of the sample was decreased (fig. 4D, red line). These results implied that the protein contained an oxidized iron–sulfur cluster, such as [4Fe–4S]3+ or [3Fe–4S]+. Compared with the reported EPR spectra, it was found that the EPR spectrum of E. coli expressed RfSPL is more like those of [3Fe–4S]+ (Beinert and Thomson 1983; Breton et al. 1995). The signal of the RfSPL sample purified at pH 7.0 was very weak, the reason of which is unclear. The whole E. coli cell sample with overexpressed RfSPL protein was also investigated. A broad signal between 300 and 370 mT was observed (supplementary fig. S9, Supplementary Material online). After substituting the signal of the uninduced whole-cell sample from that of the overexpressed whole-cell sample, a sharp signal around the g value of ∼2 was obtained (fig. 4D, blue line). It resembles that of the purified protein, but much narrower. The different environments of the protein might be responsible for the difference of the signals from the purified protein and the whole-cell sample.
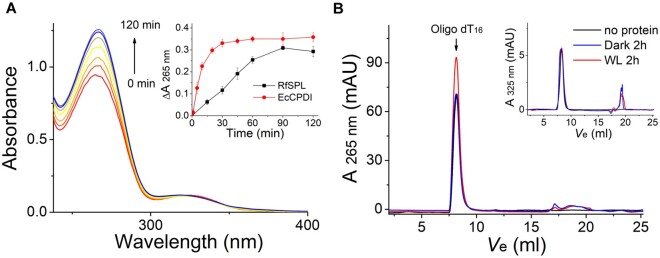
Then we investigated the photorepair activity of E. coli expressed RfSPL in vitro. The fresh protein purified at pH 6.0 was mixed with UV-irradiated oligo(dT)16 and DTT, and illuminated with white light (irradiance of ∼800 W m−2). It was observed that the absorption at 265 nm was gradually increased, but the absorption at 325 nm was almost unchanged during illumination (fig. 5A). However, the aggregation of the protein, and the decomposition of the iron–sulfur cluster in the protein might interfere the absorption measurement. To rule out these interferences, the protein in the assay mixture was removed by boiling and centrifugation after illumination, and the supernatant was analyzed by size exclusion chromatography. The results showed that the absorption at 265 nm of the oligo(dT)16 substrate peak was unambiguously increased after 2 h illumination compared with the control kept in the dark and that illuminated in the absence of the RfSPL protein (fig. 5B). The absorption at 325 nm of the substrate peak was barely changed after illumination (fig. 5B, the inset). The restoration of the absorption at 265 nm was indicative of the repairing of CPDs (which has little absorption at 265 nm) to pyrimidines (ε265 = 8,500 M−1 cm−1) (Thiagarajan et al. 2010). The 6-4 photoproduct has an absorption peak at 325 nm (ε325 = 6,000 M−1 cm−1) (Yamamoto et al. 2013). There was no absorbance decrease at 325 nm, indicating that the 6-4 photoproducts were not repaired. In summary, we tentatively conclude that RfSPL is a CPD photolyase with a FAD cofactor and an iron–sulfur cluster.
Fig. 5.
RfSPL is a CPD photolyase. The assay mixture contained 0.1 mg/ml Escherichia coli expressed RfSPL protein (with ∼0.1 µM binding FAD), 10 µM UV-oligo(dT)16, and 1 mM DTT in Protein buffer at pH 6.0. The mixture was illuminated under a white-light LED (irradiance of ∼800 W m−2). (A) The time-dependent absorption spectra of the assay mixture. In the inset, the change of absorbance at 265 nm versus time was shown. An activity assay result of EcCPDI under similar conditions was also shown to give a reference. (B) The assay mixture was boiling and centrifugation after illumination for 2 h, and the supernatant was analyzed by size exclusion chromatography (red line, WL 2 h). The elution profile of a control that illuminated in the absence of the RfSPL protein was shown in a black line (no protein), and that of a control kept in the dark for 2 h was shown in a blue line (dark 2 h). The elution peak of the oligo(dT)16 substrate was indicated by an arrow. The restoration of absorption at 265 nm was indicative of the conversion of CPDs to pyrimidines. The elution profiles detected at 325 nm were shown in the inset. The absorption at 325 nm of the substrate peaks was almost unchanged before and after illumination.
Preliminary Study on the Physiological Properties and Functions of RfSPL
Although the RfSPL protein expressed in E. coli provided a lot of information about this protein, the status of RfSPL in vivo might be different. Therefore, we carried out the study on the physiological properties and functions of RfSPL. For convenience, the model stain R. fascians ATCC 12974 (=NBRC 12155) was used. At first, transcription of the Rfspl gene in the stationary phase culture of the strain was measured by real-time RT–PCR. It was found that the mRNA level of Rfspl was about one-sixth of that of the Rfphr gene, the later encoding a class I CPD photolyase in R. fascians (supplementary fig. S10A, Supplementary Material online). This result ruled out the possibility that Rfspl is a nontranscriptable pseudogene. Then two R. fascians strains were constructed. In one strain named R. fascians oSPL, RfSPL was overexpressed by transforming the plasmid pRARfSPL, which is a derivate of the Rhodococcus–E. coli shuttle vector pRESQ (van der Geize et al. 2002) that containing a His-tagged Rfspl gene under the control of a strong Rhodococcus promoter Pami (Jiao et al. 2018). The other strain was named R. fascians dSPL, in which the Rfspl gene was knock out by double cross-over homologous recombination (Schafer et al. 1994). The real-time RT–PCR experiments showed that the mRNA level of Rfspl in R. fascians oSPL was increased about 12 folds compared with the wild-type strain; whereas the Rfspl mRNA expression in R. fascians dSPL was not detectable (supplementary fig. S10A, Supplementary Material online). The photoreactivation kinetics of the wild-type strain, R. fascians oSPL, and R. fascians dSPL was determined (supplementary fig. S10B, Supplementary Material online). It was found that photoreactivation efficiency of R. fascians oSPL was significantly higher than the other two strains. The photoreactivation efficiency of R. fascians dSPL was slightly lower than that of the wild-type strain, but the difference was not significant. This indicated that RfSPL plays a minor role in photoreactivation in vivo.
The RfSPL protein homologously expressed in the R. fascians oSPL strain was purified via immobilized metal ion affinity chromatography and gel filtration. Similar to the RfSPL protein expressed in E. coli, the homologously expressed RfSPL samples gave a main band of ∼52 kDa, both in SDS–PAGE and Western-blotting analyses (supplementary fig. S10C and D, Supplementary Material online). Chemical and fluorescence spectroscopy analyses confirmed that homologously expressed RfSPL contained iron, acid-labile sulfide, and FAD; the mole ratio of which was ∼3.2:4.2:1. The FAD occupancy of the homologously expressed samples was higher than those expressed in E. coli (∼0.2 vs. ∼0.043). The specific activity of homologously expressed RfSPL to UV-irradiated oligo(dT)16 was also about 5-fold higher than that of E. coli expressed RfSPL. The absorption spectrum of homologously expressed RfSPL exhibited peaks at 415 nm, 447 nm, 471 nm, and a shoulder at 506 nm (supplementary fig. S10E, Supplementary Material online, BL 0 min). The absorption peak at 471 nm and shoulder at 506 nm might be attributed to some pigments of R. fascians that bound to the protein nonspecifically, but which could hardly be removed by dialysis or ultrafiltration. Prolong illumination of the sample supplemented with 10 mM DTT under blue light caused absorption decrease around 450 nm; and further decrease of absorption was observed after adding 5 mM SDT (supplementary fig. S10E, Supplementary Material online). This suggested that the FAD cofactor in homologously expressed RfSPL could be photo- and chemically reduced. In one EPR experiment, homologously expressed RfSPL exhibited a broad signal near zero field (supplementary fig. S10F, Supplementary Material online), which was probably due to a [3Fe–4S]0 cluster in the S = 2 state (Hagen et al. 1985; Breton et al. 1995). But in another experiment, this signal was not detected. Further experiments are needed to confirm the natural state of the iron–sulfur cluster in RfSPL.
Discussion
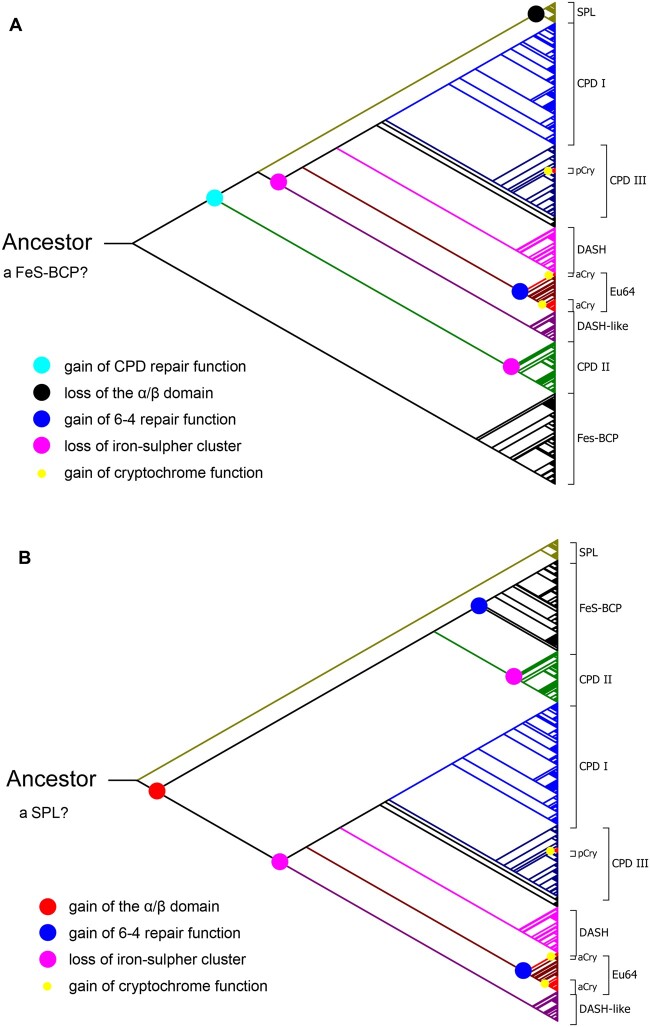
It is widely accepted that photolyases are ancient DNA repair enzymes, which have already been evolved far before the oxygen accumulation in atmosphere and the formation of the ozone layer (Eisen and Hanawalt 1999; Lucas-Lledo and Lynch 2009; Vechtomova et al. 2020). Photolyases might be more crucial for repairing UV-inducing DNA lesions in the primordial organisms, since the intense solar UV irradiation could reach the surface of the early Earth without the attenuation by the ozone layer (Cockell 1998; Eisen and Hanawalt 1999). However, the evolutionary scenario of photolyases was not fully elucidated. It was proposed that a prokaryotic 6-4 photolyase with an iron–sulfur cluster was the first common ancestor of photolyases (Zhang et al. 2013). But CPDs are the major UV-induced DNA lesions. Therefore, it is unlikely that the 6-4 photorepair occurred earlier than CPD photorepair in the evolution of photolyases. Nevertheless, if the ancestor of photolyases was a 6-4 photolyase, it would make the evolutionary pathway of photolyases more complex that functional changes of 6-4 photorepair and CPD photorepair would occur several times (fig. 6A).
Fig. 6.
Two possible evolutionary pathways of CPF. The first common ancestor of CPF might be (A) a 6-4 photolyase with an iron–sulfur cluster like a FeS-BCP, or (B) a CPD photolyase with a single α-helical domain and an iron–sulfur cluster like a SPL. Proposed major evolutionary events are indicated by colored dots. SPL, short photolyase-likes; CPD I, class I CPD photolyases; CPD III, class III CPD photolyases; pCry, plant cryptochromes; DASH, DASH proteins; Eu64, eukaryotic 6-4 photolyases; aCry, animal cryptochromes; DASH-like, DASH-like proteins; CPD II, class II CPD photolyases; FeS-BCP, FeS-BCP proteins.
In this work, we identified a novel class of photolyases named SPLs. SPLs contain only an α-helical domain which may bind a FAD cofactor and a CPD substrate, but lack the α/β domain of normal photolyases. Therefore, SPLs are shorter than other normal photolyases. It was demonstrated that a representative SPL from R. fascians—RfSPL, had an iron–sulfur cluster and CPD repair activity. The phylogenetic analysis showed that all SPLs constituted one monophyletic group, which were located between the group of class II CPD photolyases and a large group which is consisted of class I/III CPD photolyases, DASH-like proteins, DASH proteins, and eukaryotic 6-4 photolyases. The phylogenetic tree obtained by this analysis was unrooted, with an apparent root on the branch near the group of FeS-BCPs (fig. 1). If the root was manually placed on the branch near the group of SPLs, a more convincing evolutionary pathway of CPF could be obtained (fig. 6B). The first common ancestor of CPF might be a CPD photolyase with a single α-helical domain and an iron–sulfur cluster like a SPL. The other members of CPF acquire the α/β domain, and then the antenna cofactor-binding ability by gene fusion during evolution. FeS-BCPs and eukaryotic 6-4 photolyases gained 6-4 photorepair activity through two independent functional change events. The iron–sulfur cluster is retained only in SPLs and FeS-BCPs. The other members of CPF lost the iron–sulfur cluster during evolution to adapt to the aerobic environment. The plant and animal cryptochromes were evolved from class III CPD photolyases and eukaryotic 6-4 photolyases through later and independent functional change events, respectively (fig. 6B).
The function of the iron–sulfur cluster in SPLs and FeS-BCPs is not fully understood. It was proposed that the iron–sulfur cluster in A. fabrum FeS-BCP (AfPhrB) was an integral component of the protein fold (Zhang et al. 2013; Graf et al. 2015). This may also be true for that in SPLs. The RfSPL protein purified at pH 5.0 was very unstable, which might be partially due to the decomposition of the iron–sulfur cluster at this pH. Recently, it was reported that two tyrosines (Tyr424 and Tyr430) between the substrate-binding site and the iron–sulfur cluster in AfPhrB might serve as electron transmitters between DNA and the iron–sulfur cluster, which were demonstrated to be crucial for the substrate binding and repair activity (Graf et al. 2015; Zhang et al. 2017). In SPLs, there is a conserved tryptophan/tyrosine (Trp176 in RfSPL) located at the similar position, which may perform the same function as the two tyrosines in FeS-BCPs do. In recent years, increasing numbers of DNA repair and processing enzymes were found to contain the iron–sulfur clusters, such as DNA glycosylases Endonuclease III (Kuo et al. 1992) and MutY (Fromme et al. 2004), DNA helicases XPD and FancJ (Rudolf et al. 2006), nuclease AddAB (Yeeles et al. 2009), spore photoproduct lyases (Buis et al. 2006), DNA polymerases (Netz et al. 2011), and the large subunit of primase PriL (which also has high structural similarity to FeS-BCPs) (Sauguet et al. 2010; Zhang et al. 2013). Beside the structural role, the iron–sulfur clusters in the DNA repair and processing enzymes may also participate the DNA-mediated charge transfer for DNA damage detection and long-range communication between these enzymes (Fuss et al. 2015). This function may degenerate in the other photolyases due to the UV-induced DNA lesions becoming rare under the protection of the ozone layer.
The occupancy of FAD and iron–sulfur cluster was very low in E. coli expressed RfSPL. This might be due to the lack of essential components in E. coli for assembling of the RfSPL protein. Homologous expression of RfSPL using the R. fascians oSPL strain increased the occupancy of FAD and iron–sulfur cluster, and the specific activity of the protein. However, there are still some problems to be solved. First, the yields of the protein were not high by homologous expression, because the promoter Pami may not be strong enough. Second, the growth of R. fascians is slow; and the cells are hydrophobic, which often aggregate during culturing. Third, R. fascians contains pigments which may bind to the proteins nonspecifically and hamper identifying the spectra of the proteins.
In EPR experiments, E. coli expressed RfSPL exhibited signals like those of [3Fe–4S]+, and a signal like those of [3Fe–4S]0 was observed in a homologously expressed RfSPL sample. However, it was possible that the [3Fe–4S]+ or [3Fe–4S]0 cluster in the RfSPL protein samples was an artefact from degradation of the [4Fe–4S] cluster under aerobic conditions. Based on sequence alignment, it was found that there are 12 residues between the third and the fourth coordinating cysteines in AfPhrB, whereas there are only five residues between those in RfSPL (fig. 2). The iron–sulfur cluster in RfSPL might be more exposed, which caused it to be more susceptible to oxidation than that in AfPhrB. Although R. fascians was stated as an aerobic organism, it was reported that it was able to grow under anaerobic conditions (Cnovas et al. 1997). RfSPL may be functional when the organism is growing under anaerobic conditions. Otherwise, functional RfSPL may be located at a part of the cell where is relatively anaerobic even under aerobic conditions. Further work is planned to explore the functions of RfSPL in vivo under different conditions. It is also planned to use anaerobic conditions to express and purify the SPL proteins, and to discover more stable SPLs for investigation.
Although we demonstrated the RfSPL has CPD repair activity, its activity is relatively weak. And, it was observed that there was only a slight reduction in photoreactivation efficiency in the Rfspl knock out R. fascians strain dSPL. Nevertheless, it was observed that almost all organisms that contain SPLs have normal photolyases. For instance, there are three CPF genes in R. fascians: one is the Rfspl gene; and the other two genes encode a class I CPD photolyase (RfCPDI) and a prokaryotic 6-4 photolyase (RfFeS-BCP), respectively (supplementary table S3, Supplementary Material online). Preliminary study also showed that RfCPDI plays a major role in photoreactivation in R. fascians; and to a lesser extent, does RfFeS-BCP (data not shown). Therefore, it cannot rule out that SPLs in the contemporary organisms are “remnants” of ancient proteins that only have residual repair activity at present time. And, in most instances, their repair function can be replaced by normal photolyases. Otherwise, they may perform some other functions under certain conditions. Intriguingly, it was found that most SPLs (except for some members of the subgroup C) contain C-terminal extensions in various lengths. This feature was reminiscent of the plant and animal cryptochromes, which also have C-terminal extensions with different functions. Whether SPLs act as bacterial cryptochromes that regulating expression of other genes deserves further research.
During the submission of this paper, a distinct class of photolyases named NewPHL was described by Lars-Oliver Essen group (Emmerich et al. 2020), which is actually the same class as SPL. Two NewPHL(SPL) members from Dinoroseobacter shibae and Methylobacterium mesophilicum were characterized. It was shown that these two proteins are specific for CPD lesions within single-stranded DNA, and lack any bound antenna cofactors. It was not surprising that no iron–sulfur cluster was detected in these two proteins, because both of them belong to the subgroup A of SPL, no member of which was found to have the four conserved cysteines. Nevertheless, we agree with their opinion that NewPHL(SPL) may represent the ancestor of CPF. Studies on SPLs may shed new light not only on the evolution of CPF, but also on the evolution of the life on the Earth.
Materials and Methods
Phylogenetic Analyses
To assemble a comprehensive set of CPF proteins, TBlastN searches were performed on the web site of the National Centre for Biotechnology Information (NCBI, http://blast.ncbi.nlm.nih.gov) using three query protein sequences with known crystal structures: Escherichia coli class I CPD photolyase (EcCPDI, PDB ID: 1DNP) (Park et al. 1995), Methanosarcina mazei class II CPD photolyase (MmCPDII, PDB ID: 2XRY) (Kiontke et al. 2011) and A. fabrum FeS-BCP (AfPhrB, PDB ID: 4DJA) (Zhang et al. 2013). For prokaryotes, the RefSeq Genome Database (refseq_genomes) of certain species was searched. For eukaryotes, the Reference RNA sequences (refseq_rna) of certain species were searched. The “Organism” option was set to a certain taxonomic level (normally to a class or an order). If there were too many results (>100) retrieved in one search, the lower taxonomic levels were selected and searches were performed again. If there were a few tens or less of results in one search, several representative sequences were selected from them. Some poorly conserved sequences with the alignment scores <40 were excluded. Finally, a total of 574 sequences of 271 organisms from all three kingdoms were retrieved. The NCBI RefSeq ID of the sequences and the corresponding organism names were listed in supplementary table S1, Supplementary Material online. These sequences were subjected to a multiple sequence alignment using CLUSTAL W (Thompson et al. 1994) or MUSCLE (Edgar 2004). In some trials, a number of sequences of the large subunits of the archaeal/eukaryotic primases (PriLs) were included in the alignments, which have structural similarity with photolyases (Sauguet et al. 2010; Zhang et al. 2013). Some alignments were manually adjusted in the putative FAD- and substrate-binding regions, based on the 3 D alignment results of the known crystal structures of EcCPDI, MmCPDII, and AfPhrB, which were obtained by using PyMOL (Schrödinger, LLC). Then phylogenetic analyses were performed by MEGA 7.0 (Kumar et al. 2016), using the maximum likelihood method based on the Le_Gascuel_2008 with Freq. (+F) model (Le and Gascuel 2008) and 500 bootstrap iterations.
To explore the category of SPLs, a SPL sequence of R. fascians (WP_032397095.1) was used as the query protein sequence, and TBlastN searches were performed as above. A total of 191 SPL sequences of 187 eubacteria were retrieved. The RefSeq ID of the SPL sequences and the corresponding organism names were listed in supplementary table S2, Supplementary Material online. Phylogenetic analyses were performed as above with these SPL sequences.
Primary Structure Analysis and Tertiary Structure Modeling
To obtain structural information of SPLs, the aligned sequences of five representative SPLs: R. fascians SPL (RfSPL), S. elongatus (formerly known as Anacystis nidulans) SPL (SeSPL), Cyclobacterium marinum SPL (CmSPL), Ruegeria lacuscaerulensis SPL (RlSPL), and Curvibacter gracilis SPL (CgSPL), together with those of EcCPDI and AfPhrB were isolated and inspected. Based on the structural information of EcCPDI and AfPhrB, the putative FAD-binding, substrate-binding, and iron–sulfur cluster coordination sites were assigned to these SPL sequences. The aligned result was rendered by using ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/) (Robert and Gouet 2014). To construct homology tertiary structural models of RfSPL, the sequence of RfSPL was analyzed with SWISS-MODEL (https://swissmodel.expasy.org) (Waterhouse et al. 2018). Template search were performed. Two top-ranked structures, AfPhrB (PDB ID: 4DJA) and S. elongatus class I CPD photolyase (SeCPDI, PDB ID: 1TEZ) (Mees et al. 2004), were selected as the templates to build the models. The FAD- and substrate-binding sites of 221 class I/III CPD photolyases and those of 191 SPLs were represented using WebLogo (Crooks et al. 2004), respectively.
Bacterial Strains, Gene Cloning, and Manipulation
Rhodococcus fascians strain MCCC 1A03174 was purchased from the Marine Culture Collection of China (MCCC), which has a high similarity to the model strain R. fascians NBRC 12155.
The Rfspl gene of this strain was amplified by PCR. Sequencing analysis showed that the Rfspl gene of R. fascians MCCC 1A03174 has 96% identities to the homologous genomic region of R. fascians NBRC 12155. The nucleotide sequence of the Rfspl gene of R. fascians MCCC 1A03174 has been deposited to GenBank with the accession number MK474076. The Rfspl(MK474076) gene was cloned into pET22b between the restriction sites of NdeI and NotI to obtain the plasmid pETRfSPL22.
Rhodococcus fascians strain ATCC 12974 (=NBRC 12155) was purchased from Mingzhoubio (Zhejiang, China). To construct an overexpression plasmid, a synthetic strong Rhodococcus promoter Pami (Jiao et al. 2018) with the start codon and a N-terminal His-tag coding sequence was inserted into the Rhodococcus–E. coli shuttle vector pRESQ (van der Geize et al. 2002), to obtain pRA1. The Rfspl gene of R. fascians ATCC 12974 was amplified by PCR, and cloned into pRA1 to construct pRARfSPL. The pRARfSPL plasmid was electro-transformed into R. fascians ATCC 12974 to obtain the overexpression strain R. fascians oSPL.
To knock out the Rfspl gene, the left and right flanking sequences of Rfspl with the length ∼1,000 bp (HL and HR), and a kanamycin resistant gene (kan) from pET28a were amplified. The fragments of HL, kan, and HR were ligated and inserted into the suicide plasmid pK18mobsacB (Schafer et al. 1994) to construct pKdRfSPL. The pKdRfSPL plasmid was transferred by intergeneric conjugation from the E. coli strain S17-1 to R. fascians ATCC 12974. Transconjugants were selected on nutrient agar (NA) plates with 25 μg ml−1 kanamycin and 50 μg ml−1 nalidixic acid. A single colony was grown in nonselective medium at 25 °C for 24 h, and then plated onto NA plates containing 10% sucrose, incubated at 25 °C for ∼3 days. The colonies were verified by PCR and sequencing. A strain in which the Rfspl gene had been successfully knocked out was named R. fascians dSPL.
Photoreactivation Experiments
Photoreactivation of wild-type R. fascians ATCC 12974, R. fascians oSPL, and R. fascians dSPL was investigated using a protocol described previously (Xu et al. 2015) with some modifications. The R. fascians strains were cultured in the cultivation medium (containing 20 g glucose, 1 g yeast extract, 7 g tryptone, 0.38 g K2HPO4, 0.5 g KH2PO4 and 0.5 g MgSO4·7H2O per liter) at 25 °C for ∼3 days to early stationary phase. The cells were harvested, washed, and diluted with saline to a proper scale (normally ∼1:1,000). The cell suspension was irradiated under 254 nm UVC light to give a dose of ∼100 J m−2. Then cell suspension was illuminated under white light (irradiance of ∼200 W m−2). The illuminated samples were withdrawn at intervals, and seriously diluted. Aliquots (5 μl) were dropped on to NA plates, and colonies were counted after incubation at 25 °C for ∼3 days. All experiments were performed under a red LED lamp at 23 °C.
Real-Time RT–PCR Analysis
The genomic DNA-free total RNA samples of R. fascians strains were prepared by using the AP method (Xu et al. 2019) with an additional lysozyme lysis step, and then used as templates for reverse transcription (RT). Real-time PCR analysis was performed on an ABI StepOne Real-time PCR system (Applied Biosystems) with Forget-Me-Not qPCR Master Mix (Biotium). The amplification conditions were as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s. The mRNA levels of Rfspl in the wild-type strain R. fascians ATCC 12974, R. fascians oSPL, and R. fascians dSPL were measured using Rfphr (encoding a class I CPD photolyase) or RfsecA (encoding a protein translocation ATPase) as the reference. The two reference genes gave similar results.
Protein Expression and Purification
The pETRfSPL22 plasmid was transformed into competent E. coli Rosetta(DE3) cells. The transformed cells were grown at 37 °C in LB medium with 100 μg ml−1 ampicillin until A600 reached 1.0–2.0. Then the cells were induced with 0.5 mM IPTG and cultured at 20 °C for 20 h. The induced cells were collected by centrifugation, resuspended in Start buffer (containing a certain buffer reagent of 50 mM, 200 mM NaCl, 50 mM imidazole, and 10% glycerol) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMFS), 20 μM Bestatin, 3 μM Pepstatin A, and 3 μM E-64, then disrupted by French Press. The supernatant of the lysate was loaded onto a Ni2+-chelating Sepharose column. The column was washed with the Start buffer, and then eluted with Elution Buffer (containing a certain buffer reagent of 50 mM, 200 mM NaCl, 500 mM imidazole, and 10% glycerol). The eluted protein was concentrated, and the buffer was quickly changed to Protein buffer (containing a certain buffer reagent of 50 mM, 200 mM NaCl, and 10% glycerol) using a Sephadex G25 column. The buffer reagents at various pH values were tested: sodium acetate, pH 5.0; Mes, pH 6.0; Tris, pH 7.0 and 8.0; Glycine, pH 9.0 and 10.0. The protein concentration was determined by the Micro-Biuret method (Goa 1953) after precipitating the protein using deoxycholate and trichloroacetic acid. The molecular weight of the protein was analyzed by using AB Sciex 5800 MALDI-TOF/TOF mass spectrometry.
To purify the homologously expressed RfSPL protein, R. fascians oSPL was cultured in the cultivation medium at 25 °C for ∼3 days. The cells were harvested by centrifugation. The purification procedure was the same as above using the Mes, pH 6.0 buffers. The protein samples were analyzed by SDS–PAGE and Western blotting using an anti-6×His-tag mouse monoclonal antibody (BBI, D191001).
Optical Spectroscopic Analysis of RfSPL
The UV-vis absorption spectra of the protein samples were recorded on a UV-1800 spectrophotometer equipped with a TCC-240A temperature controller (Shimadzu). The scanning range was from 200 to 700 nm. During recording of the spectra of the protein samples, the temperature of the sample was held at 18 ± 0.5 °C.
The fluorescence spectra were recorded on an F-4600 fluorescence spectrophotometer (Hitachi). To determine the emission spectra, the excitation wavelength was set to 450 nm, the slit widths were 2.5 nm, and the scan range was from 500 to 700 nm. To determine the excitation spectra, the emission wavelength was set to 525 nm, the slit widths were 2.5 nm, and the scan range was from 200 to 500 nm.
Determination of Iron and Acid-Labile Sulfide in RfSPL
The iron content of RfSPL was determined by using a protocol modified from a previous report (Lovenberg et al. 1963). Briefly, protein solution of 500 μl was mixed with 25 μl concentrated HCl, and heated for 10 min to 80 °C. After 10 min of centrifugation at 12,000×g, 500 μl supernatant was mixed with 250 μl of 0.2 M potassium biphthalate and 500 μl of 0.3% (w/v) 1,10-phenanthroline (phen). The absorbance spectrum was measured from 350 to 750 nm. A standard curve of the absorbance at 511 nm was made using ferrous ammonium sulfate in the range of 0–25 μM.
Determination of acid-labile sulfide in RfSPL was performed by an adaptation of a published method (Gong and Carmeli 2003). Briefly, a protein sample of 400 μl was mixed with 200 μl of Zn acetate (1% w/v) and 10 μl of NaOH (12% w/v). The tube was closed and vortexed for 1 min. After incubation at room temperature for 20 min, 100 μl of N,N′-dimethyl-p-phenylenediamine (DMPD, 1% w/v in 5 M HCl) was rapidly added and mixed by gentle swirling, then 20 μl of FeCl3 (23 mM in 1.2 M HCl) was quickly added. The tube was closed immediately and vortexed vigorously for 1 min. The mixture was incubated at room temperature for 20 min in the dark. The precipitate was removed by centrifugation at 12,000×g for 10 min. The supernatant was carefully pipetted out and the absorbance spectrum was measured from 400 to 800 nm. The absorbance values at 670 or 745 nm were recorded. Standard curves were made using fresh sodium sulfide in 10 mM NaOH in the range of 0–20 μM.
Electron Paramagnetic Resonance Spectroscopy
Electron paramagnetic resonance (EPR) spectra of RfSPL were obtained at X-band (9.396 GHz) and 10 K using an EMX plus 10/12 EPR Spectrometer (Bruker) equipped with an ESR910 Liquid Helium cryostat (Oxford Instruments). Microwave power employed was 2 mW. Modulation frequency and modulation amplitude were 100 kHz and 0.3 mT, respectively.
Activity Assays In Vitro
The repair activity of the RfSPL protein samples in vitro was analyzed as described (Mu et al. 2005; Xu et al. 2017) with some modifications. Oligo-thymidylate [oligo(dT)16] of 650 μM in water was irradiated under a 254-nm UVC lamp (20 W) at a distance of 5 cm for 2 h, to produce the substrate containing both CPDs (∼2.5 per molecule) and 6-4 photoproducts (∼1 per molecule). The assay volume was 600 µl, which contained 0.1 mg/ml protein (with ∼0.1 µM binding FAD), 10 µM UV-oligo(dT)16, and 1 mM DTT in Protein buffer at pH 6.0. The mixture was illuminated under a white-light LED (irradiance of ∼800 W m−2). Absorption changes at 265 and 325 nm were recorded to determine the CPD and 6-4 repair activity. Otherwise, the mixture was boiled and centrifuged. And, the supernatant was subjected to size exclusion chromatography by using a Superdex peptide column (GE Healthcare). The activity was evaluated from the increment of the absorption area of the substrate elution peak at 265 nm. The repair activity of EcCPDI was also determined to give a reference. The conditions were as above, except that the protein concentration was 0.1 µM, and Protein buffer at pH 7.0 was used.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Prof. Aixin Yan, Prof. Qinhua Song, Prof. Jiafu Chen, Prof. Xiuqing Yang, Dr Lu Yu, Dr Hongmin Liu, Dr Yongliang Jiang, and Prof. Congzhao Zhou for their helpful discussions and suggestions. We thank Lulu Tang, Xingrui Chen, Fei Liu, Dongyang Tian, Caiyun Zhang, and Yuting Li for performing some of the experiments. A portion of this work was performed on the Steady High Magnetic Field Facilities, High Magnetic Field Laboratory, Chinese Academy of Sciences. We thank Dr Wei Tong for the assistance with the EPR experiments. This work was supported by the National Natural Science Foundation of China (31971199); the Natural Science Foundation of Anhui Province (1808085MH272); and the Young Backbone Talents Fund Project of Wannan Medical College (2019). The nucleotide sequence of the RfSPL gene from R. fascians strain MCCC 1A03174 has been deposited to GenBank under accession number MK474076. The homology models of RfSPL constructed using the structures of AfPhrB (4DJA) and SeCPDI (1TEZ) as the templates were deposited to Model Archive (https://www.modelarchive.org) with the accession codes ma-gm2th and ma-6dkqz. The authors declare they have no competing interests.
References
- Aubert C, Vos MH, Mathis P, Eker AP, Brettel K.. 2000. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature 405(6786):586–590. [DOI] [PubMed] [Google Scholar]
- Beinert H, Thomson AJ.. 1983. Three-iron clusters in iron-sulfur proteins. Arch Biochem Biophys. 222(2):333–361. [DOI] [PubMed] [Google Scholar]
- Beukers R, Eker AP, Lohman PH.. 2008. 50 years thymine dimer. DNA Repair (Amst). 7(3):530–543. [DOI] [PubMed] [Google Scholar]
- Breton JL, Duff JL, Butt JN, Armstrong FA, George SJ, Petillot Y, Forest E, Schafer G, Thomson AJ.. 1995. Identification of the iron-sulfur clusters in a ferredoxin from the archaeon Sulfolobus acidocaldarius. Evidence for a reduced [3Fe-4S] cluster with pH-dependent electronic properties. Eur J Biochem. 233(3):937–946. [DOI] [PubMed] [Google Scholar]
- Buis JM, Cheek J, Kalliri E, Broderick JB.. 2006. Characterization of an active spore photoproduct lyase, a DNA repair enzyme in the radical S-adenosylmethionine superfamily. J Biol Chem. 281(36):25994–26003. [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M.. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 62:335–364. [DOI] [PubMed] [Google Scholar]
- Cnovas M, Garc’a-Cases L, Iborra JL.. 1997. Shifts in metabolism and morphology of Rhodococcus fascians when debittering synthetic citrus juices in the absence of aeration. Biotechnol Lett. 19(12):1181–1184. [Google Scholar]
- Cockell CS.1998. Biological effects of high ultraviolet radiation on early earth – a theoretical evaluation. J Theor Biol. 193(4):717–729. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikbas UM, Tardu M, Canturk A, Gul S, Ozcelik G, Baris I, Ozturk N, Kavakli IH.. 2019. Identification and characterization of a new class of (6-4) photolyase from Vibrio cholerae. Biochemistry 58(43):4352–4360. [DOI] [PubMed] [Google Scholar]
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Hanawalt PC.. 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 435(3):171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich HJ, Saft M, Schneider L, Kock D, Batschauer A, Essen LO.. 2020. A topologically distinct class of photolyases specific for UV lesions within single-stranded DNA. Nucleic Acids Res. 48(22):12845–12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL.. 2004. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427(6975):652–656. [DOI] [PubMed] [Google Scholar]
- Fuss JO, Tsai CL, Ishida JP, Tainer JA.. 2015. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim Biophys Acta. 1853(6):1253–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisselbrecht Y, Fruhwirth S, Schroeder C, Pierik AJ, Klug G, Essen LO.. 2012. CryB from Rhodobacter sphaeroides: a unique class of cryptochromes with new cofactors. EMBO Rep. 13(3):223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goa J.1953. A micro biuret method for protein determination; determination of total protein in cerebrospinal fluid. Scand J Clin Lab Invest. 5(3):218–222. [DOI] [PubMed] [Google Scholar]
- Gong XM, Carmeli C.. 2003. Determination of acid-labile sulfide in photosystem I in the presence of various detergents. Anal Biochem. 321(2):259–262. [DOI] [PubMed] [Google Scholar]
- Graf D, Wesslowski J, Ma H, Scheerer P, Krauß N, Oberpichler I, Zhang F, Lamparter T.. 2015. Key amino acids in the bacterial (6-4) photolyase PhrB from Agrobacterium fabrum. PLoS One 10(10):e0140955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen WR, Dunham WR, Johnson MK, Fee JA.. 1985. Quarter field resonance and integer-spin/half-spin interaction in the EPR of Thermus thermophilus ferredoxin. Possible new fingerprints for three iron clusters. Biochim Biophys Acta. 828(3):369–374. [DOI] [PubMed] [Google Scholar]
- Huber C, Wachtershauser G.. 1997. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science 276(5310):245–247. [DOI] [PubMed] [Google Scholar]
- Imlay JA.2006. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 59(4):1073–1082. [DOI] [PubMed] [Google Scholar]
- Jiao S, Yu H, Shen Z.. 2018. Core element characterization of Rhodococcus promoters and development of a promoter-RBS mini-pool with different activity levels for efficient gene expression. N Biotechnol. 44:41–49. [DOI] [PubMed] [Google Scholar]
- Kiontke S, Geisselbrecht Y, Pokorny R, Carell T, Batschauer A, Essen LO.. 2011. Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J. 30(21):4437–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Martin W.. 2005. On the origin of genomes and cells within inorganic compartments. Trends Genet. 21(12):647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritsky MS, Telegina TA, Vechtomova YL, Buglak AA.. 2012. Why flavins are not competitors of chlorophyll in the evolution of biological converters of solar energy. Int J Mol Sci. 14(1):575–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritsky MS, Telegina TA, Vechtomova YL, Kolesnikov MP, Lyudnikova TA, Golub OA.. 2010. Excited flavin and pterin coenzyme molecules in evolution. Biochemistry (Mosc). 75(10):1200–1216. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CF, McRee DE, Fisher CL, O’Handley SF, Cunningham RP, Tainer JA.. 1992. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science 258(5081):434–440. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25(7):1307–1320. [DOI] [PubMed] [Google Scholar]
- Lopez L, Fasano C, Perrella G, Facella P.. 2021. Cryptochromes and the circadian clock: the story of a very complex relationship in a spinning world. Genes (Basel). 12(5):672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovenberg W, Buchanan BB, Rabinowitz JC.. 1963. Studies on the chemical nature of clostridial ferredoxin. J Biol Chem. 238:3899–3913. [PubMed] [Google Scholar]
- Lucas-Lledo JI, Lynch M.. 2009. Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family. Mol Biol Evol. 26(5):1143–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons TW, Reinhard CT, Planavsky NJ.. 2014. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506(7488):307–315. [DOI] [PubMed] [Google Scholar]
- Mees A, Klar T, Gnau P, Hennecke U, Eker AP, Carell T, Essen LO.. 2004. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 306(5702):1789–1793. [DOI] [PubMed] [Google Scholar]
- Mu W, Zhang D, Xu L, Luo Z, Wang Y.. 2005. Activity assay of His-tagged E. coli DNA photolyase by RP-HPLC and SE-HPLC. J Biochem Biophys Methods. 63(2):111–124. [DOI] [PubMed] [Google Scholar]
- Muller M, Carell T.. 2009. Structural biology of DNA photolyases and cryptochromes. Curr Opin Struct Biol. 19(3):277–285. [DOI] [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ.. 2011. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 8(1):125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberpichler I, Pierik AJ, Wesslowski J, Pokorny R, Rosen R, Vugman M, Zhang F, Neubauer O, Ron EZ, Batschauer A, et al. 2011. A photolyase-like protein from Agrobacterium tumefaciens with an iron-sulfur cluster. PLoS One 6(10):e26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparin AI.1938. The origin of life. New York: MacMillan. [Google Scholar]
- Park HW, Kim ST, Sancar A, Deisenhofer J.. 1995. Crystal structure of DNA photolyase from Escherichia coli. Science. 268(5219):1866–1872. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P.. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42(Web Server issue):W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF.. 2006. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell. 23(6):801–808. [DOI] [PubMed] [Google Scholar]
- Russell MJ, Hall AJ.. 1997. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J Geol Soc Lond. 154(3):377–402. [DOI] [PubMed] [Google Scholar]
- Sancar A.2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 103(6):2203–2237. [DOI] [PubMed] [Google Scholar]
- Sauguet L, Klinge S, Perera RL, Maman JD, Pellegrini L.. 2010. Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS One 5(4):e10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A.. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145(1):69–73. [DOI] [PubMed] [Google Scholar]
- Scheerer P, Zhang F, Kalms J, von Stetten D, Krauß N, Oberpichler I, Lamparter T.. 2015. The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways. J Biol Chem. 290(18):11504–11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboada B, Ciria R, Martinez-Guerrero CE, Merino E.. 2012. ProOpDB: prokaryotic Operon DataBase. Nucleic Acids Res. 40(Database issue):D627–D631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan V, Villette S, Espagne A, Eker AP, Brettel K, Byrdin M.. 2010. DNA repair by photolyase: a novel substrate with low background absorption around 265 nm for transient absorption studies in the UV. Biochemistry 49(2):297–303. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ.. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L.. 2002. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol Microbiol. 45(4):1007–1018. [DOI] [PubMed] [Google Scholar]
- Vechtomova YL, Telegina TA, Kritsky MS.. 2020. Evolution of proteins of the DNA photolyase/cryptochrome family. Biochemistry (Mosc). 85(Suppl 1):S131–S153. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46(W1):W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.2005. Light-driven enzymatic catalysis of DNA repair: a review of recent biophysical studies on photolyase. Biochim Biophys Acta. 1707(1):1–23. [DOI] [PubMed] [Google Scholar]
- Xu L, Sun L, Guan G, Huang Q, Lv J, Yan L, Ling L, Zhang Y.. 2019. The effects of pH and salts on nucleic acid partitioning during phenol extraction. Nucleosides Nucleotides Nucleic Acids. 38(4):305–320. [DOI] [PubMed] [Google Scholar]
- Xu L, Tian C, Lu X, Ling L, Lv J, Wu M, Zhu G.. 2015. Photoreactivation of Escherichia coli is impaired at high growth temperatures. J Photochem Photobiol B Biol. 147:37–46. [DOI] [PubMed] [Google Scholar]
- Xu L, Wen B, Wang Y, Tian C, Wu M, Zhu G.. 2017. Residues at a single site differentiate animal cryptochromes from cyclobutane pyrimidine dimer photolyases by affecting the proteins’ preferences for reduced FAD. Chembiochem. 18(12):1129–1137. [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Martin R, Iwai S, Plaza P, Brettel K.. 2013. Repair of the (6-4) photoproduct by DNA photolyase requires two photons. Angew Chem Int Ed Engl. 52(29):7432–7436. [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Plaza P, Brettel K.. 2017. Repair of (6-4) lesions in DNA by (6-4) photolyase: 20 years of quest for the photoreaction mechanism. Photochem Photobiol. 93(1):51–66. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Cammack R, Dillingham MS.. 2009. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J Biol Chem. 284(12):7746–7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma H, Bowatte K, Kwiatkowski D, Mittmann E, Qasem H, Krauß N, Zeng X, Ren Z, Scheerer P, et al. 2017. Crystal structures of bacterial (6-4) photolyase mutants with impaired DNA repair activity. Photochem Photobiol. 93(1):304–314. [DOI] [PubMed] [Google Scholar]
- Zhang F, Scheerer P, Oberpichler I, Lamparter T, Krauss N.. 2013. Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proc Natl Acad Sci U S A. 110(18):7217–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.