Abstract
Transposable elements (TEs) are ubiquitous and mobile repeated sequences. They are major determinants of host fitness. Here, we characterized the TE content of the spotted wing fly Drosophila suzukii. Using a recently improved genome assembly, we reconstructed TE sequences de novo and found that TEs occupy 47% of the genome and are mostly located in gene-poor regions. The majority of TE insertions segregate at low frequencies, indicating a recent and probably ongoing TE activity. To explore TE dynamics in the context of biological invasions, we studied the variation of TE abundance in genomic data from 16 invasive and six native populations of D. suzukii. We found a large increase of the TE load in invasive populations correlated with a reduced Watterson estimate of genetic diversity a proxy of effective population size. We did not find any correlation between TE contents and bioclimatic variables, indicating a minor effect of environmentally induced TE activity. A genome-wide association study revealed that ca. 2,000 genomic regions are associated with TE abundance. We did not find, however, any evidence in such regions of an enrichment for genes known to interact with TE activity (e.g., transcription factor encoding genes or genes of the piRNA pathway). Finally, the study of TE insertion frequencies revealed 15 putatively adaptive TE insertions, six of them being likely associated with the recent invasion history of the species.
Keywords: Drosophila suzukii, transposable elements, biological invasion, populations, adaptation, PoolSeq
Introduction
Transposable elements (TEs) are selfish genetic elements. Despite being mostly neutral or deleterious, they persist and proliferate in populations by replicating within genomes (Doolittle and Sapienza 1980; Orgel and Crick 1980; Charlesworth B and Charlesworth D 1983). The interest for those sequences considerably rose in the 2000s, with the discovery of some TE insertions having a functional, and potentially adaptive, effect on their host (Mi et al. 2000; Daborn et al. 2002; Niu et al. 2019). The parallel completion of the first sequencing projects confirmed TE ubiquity and largely contributed to the growing interest for such sequences (C. elegans Sequencing Consortium 1998; Lander et al. 2001; Schnable et al. 2009).
The nature and intensity of TE deleterious effects may vary with their genomic localization (Mérel et al. 2020). First, TEs close to genes can alter their function. Second, TEs in highly recombining regions are more likely to promote ectopic recombination, that is, recombination between more-or-less identical sequences inserted at different locations in the genome. Third, recessive deleterious TEs are more likely to impact fitness when located on a chromosome in a hemizygous state (e.g., the X chromosome in males in an XY sex determination system). The strength of selection acting against TEs hence depends on the genomic region and may result in a local variation of TE density. In agreement with such expectations, TE density was found to be negatively correlated with gene density and local recombination rate in several species (Boissinot et al. 2001; Bartolomé et al. 2002). On the other hand, studies focusing on the D. melanogaster genome did not reveal a systematic lower TE content on the X-chromosome, which is hemizygous in males (Kofler et al. 2012; Cridland et al. 2013).
TE insertion frequencies reflect both TE activity and the selection acting upon them. Low-frequency TE insertions are likely to be recent, or strongly selected against, or both. Conversely, high-frequency TE insertions are likely to be old and only weakly subjected to purifying selection. As mentioned previously, TEs that are in the vicinity of genes and/or located in highly recombining regions are expected to be selected against. Accordingly, TE insertion frequencies were found to be negatively correlated with recombination rate and distance to the nearest gene in D. melanogaster (Kofler et al. 2012). In Drosophila, the overall distribution of TE frequencies seems compatible with an active repeatome (Kofler, Nolte, et al. 2015; Hill 2019). For example, 80% of the insertions have a frequency lower than 0.2 in D. melanogaster and its close relative D. simulans (Kofler, Nolte, et al. 2015).
Between population variation of TE content has been reported in various intraspecific studies. So far, the factors underlying such differences remain unclear. The effective population size (Ne) may play a prominent role in modulating TE contents. Considering that TEs are mostly deleterious, and that small Ne leads to a less-efficient purifying selection, small Ne should be associated with high TE content (Lynch and Conery 2003). In support of this hypothesis, Lynch and Conery (2003) found a significant correlation between genome size and estimates of the scaled mutation rate θ across populations representative of various species. Considering that the genome size is positively correlated with TE content, and estimates of θ are supposed to be proportional to Ne (with µ the mutation rate), this indeed suggested a correlation between TE content and Ne. At the intraspecific level, if a higher TE content in some populations has sometimes been suggested to result from a reduction of their Ne (García Guerreiro et al. 2008; García Guerreiro and Fontdevila 2011; Talla et al. 2017), to our knowledge the above expected correlation has not been reproduced at this evolutionary scale. Variation in TE content may also rely on changes in TE activity in relation with the environment (Vieira et al. 1999; Stapley et al. 2015). In Drosophila, several laboratory experiments suggest that TE activity may respond to the environment (García Guerreiro 2012; Horváth et al. 2017), but in natura studies considering the whole repeatome remain rare and a possible confounding effect of the demographic history cannot be excluded (Lerat et al. 2019). Finally, the host genotype may explain the intraspecific variation of TE abundance. For instance, in Drosophila, several studies found different levels of activity among isogenic lines (Biémont et al. 1987; Pasyukova and Nuzhdin 1993; Díaz-González et al. 2011).
The study of intraspecific variations in TE content and the underlying determining factors is valuable as TEs may also be important for adaptation (Daborn et al. 2002; Van’t Hof et al. 2016; Niu et al. 2019). Although some TE insertions exhibit a strong signal of positive selection and have been thoroughly validated experimentally, only a few studies aimed at identifying putatively adaptive insertions at a genome-wide level (González et al. 2008; Li et al. 2018; Rishishwar et al. 2018; Rech et al. 2019). In addition, most of these studies deal with D. melanogaster (González et al. 2008; González et al. 2010; Blumenstiel et al. 2014; Rech et al. 2019).The most comprehensive of these studies analyzed genomic data on 60 worldwide natural D. melanogaster populations and reported 57–300 putatively adaptive insertions (depending on the degree of evidence considered) among the ∼800 polymorphic insertions identified in the reference genome (Rech et al. 2019). Considering that approximately twice as many nonreference TE insertions as reference insertions may segregate in a single population (Kofler et al. 2012), quite a high number of TE-induced adaptations is therefore expected. However, it remains unclear how important TEs are as substrates of adaptation considering the paucity of studies and their focus on reference genome insertions.
Invasive species provide a unique opportunity to study the combined effect of in natura Ne variations and environmental variations both on TE abundance and TE adaptive potential. Invasive populations often go through demographic bottlenecks allowing them to test for an effect of Ne on TE abundance (Estoup et al. 2016). Individuals from invasive populations also encounter new environmental conditions, allowing to test for an effect of bioclimatic variables on TE abundance. Because of the need for colonizing individuals to adapt to new environmental conditions, biological invasions are often used to study rapid contemporary adaptation (Lavergne and Molofsky 2007; Rollins et al. 2015). Yet, the particular role of TEs in the rapid adaptation of invasive species remains speculative. In particular, TEs have been proposed to explain, at least in part, the paradox of invasive species, that is, the successful adaptation to a new environment despite a reduced genetic diversity caused by small founder population sizes (Stapley et al. 2015; Estoup et al. 2016; Marin et al. 2020). In response to environmental changes, TE sequences may be recruited and affect the expression of nearby genes. Furthermore, if a higher activity of TE is induced in response to environmental changes, the insertions could thus result in genetic variation and potentially beneficial alleles.
In this article, we focused on the spotted wing fly D. suzukii, a close relative of D. melanogaster, displaying one of the highest TE content among Drosophila (Sessegolo et al. 2016). Drosophila suzukii is native from Asia and has invaded independently the American and European continents where it was introduced probably in the late 2000s (Fraimout et al. 2017). Using the recently released high-quality genome assembly Dsuz-WT3_v2.0 based on Long PacBio Reads (Paris et al. 2020), we constructed a de novo TE database and found that TEs represented 47% of the genome. We further assessed TE insertion frequencies and TE abundance in 22 worldwide populations representative of the native area (n = 6) and of the two main invaded areas in Europe (n = 8) and America (n = 8). The study of TE frequencies showed that the repeatome is highly active in D. suzukii: 75% of insertions segregated at a frequency of <0.25. We found that the TE content was significantly higher in invasive populations and was correlated with a reduction of Ne. Finally, controlling for population structure, a genome scan conducted on polymorphic TE insertions identified 15 putatively adaptive TE insertions.
Results
A Highly Repetitive Reference Genome
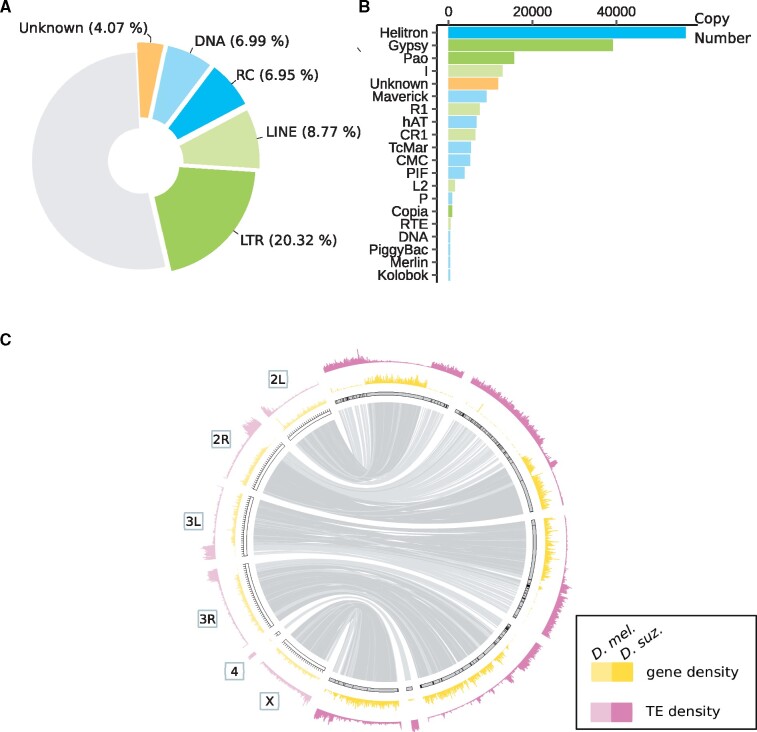
We found that the high-quality D. suzukii assembly Dsuz-WT3_v2.0 of Paris et al. (2020) is characterized by a high TE content. Overall, 47.07% of the reference assembly is annotated as repeated sequences(fig. 1A). In terms of genomic occupancy, long terminal repeat (LTR) is the predominant TE order with more than 20% of the sequence assembly corresponding to these elements, then LINEs (8.77%), DNA elements (6.99%), and rolling circles (RC: 6.95%). About 4.07% of the assembly is occupied by unknown repeated sequences. At a lower hierarchical level, the three most represented superfamilies are Gypsy, Helitron, and Pao, corresponding to 13.65%, 6.95%, and 6.44% of the assembly, respectively (supplementary table S1, Supplementary Material online). The average percentage of genomic occupancy per superfamily is 1.88%. Regarding TE copy numbers, the top three superfamilies are Helitron, Gypsy, and Pao (56,493, 39,189, and 15,555 copies, respectively) (fig. 1B). The average number of copies per superfamily is 4,963.
Fig. 1.
Main features of the TE content in the Drosphilla suzukii reference genome. (A) TE genomic occupancy. Piechart illustrating genomic sequence occupancy of each TE order (in percentages of the assembly). (B) TE copy numbers. Bar plot representing TE copy numbers for the 20 TE superfamilies displaying the highest copy numbers. (C) Distribution of TEs and genes. TE density and gene density are shown for windows of 200 kb. Lighter shades correspond to D. melanogaster and darker shades correspond to D. suzukii. The maximum value of gene density is 54 for D. suzukii and 102 for D. melanogaster. The maximum number of TE fragments is 713 for D. suzukii and 442 for D. melanogaster. Syntenic relationships between D. melanogaster and D. suzukii assemblies are shown inside using light links for regions of low gene density (<7 genes per 200 kb in D. suzukii assembly) and dark links for regions of high gene density (≥7 genes/200 kb). Contigs are surrounded by black strokes. Ticks on D. melanogaster assembly are separated by 1 Mb.
Syntenic relationships with D. melanogaster genome have been established for 212 of the 546 contigs of D. suzukii assembly. A total of 241 Mb of the 268 Mb assembly have a clearly identified counterpart in the D. melanogaster genome (fig. 1C and supplementary table S2, Supplementary Material online). Considering the observed bimodal distribution of gene density, we partitioned the D. suzukii assembly into gene-rich regions (≥ 7 genes/200 kb; 121.8 Mb) and gene-poor regions (<7 genes/200 kb; 108 Mb) (fig 1C and supplementary fig. S1, Supplementary Material online). TE fragment density also follows a bimodal distribution: 127.4 Mb corresponds to TE-rich regions (≥165 TE fragments/200 kb) and 102.4 Mb to TE-poor regions (<165 TE fragments/200 kb) (fig. 1C and supplementary fig. S2, Supplementary Material online). Note that in the D. melanogaster assembly the bimodal character of both gene and TE fragments density is less clear (supplementary figs. S3–S4, Supplementary Material online). In the D. suzukii assembly, gene-poor regions are enriched with TEs, and gene-rich regions are depleted of TEs (χ2 = 786.47, df = 1, P value <2.2 × 10−16). We did not find any difference in mean TE density between autosomal and X-linked contigs (, , W = 78,900, P value = 0.11). This conclusion holds when comparing autosomal and X-linked contigs as defined in Paris et al. (2020) using a female-to-male read mapping coverage ratio (, , W = 79,088, P value = 0.38). However, when considering only gene-rich regions, the mean TE density was far higher for X-linked contigs (,, W = 47,394, P value <2.2 × 10−16). Once again, this conclusion holds when using autosomal and X-linked contigs as defined by Paris et al. (2020) (, , W = 47,557, P value < 2.2 × 10−16).
An Active Repeatome in the Watsonville Reference Population
The female used to establish the WT3 isofemale strain corresponding to the genome assembly was collected in Watsonville (CA, USA) (Paris et al. 2020). To thoroughly evaluate TE activity in this reference population, we assessed TE insertion frequencies in a PoolSeq sample of 50 D. suzukii individuals from Watsonville. Because TEs are mostly deleterious, rare TE insertions are likely to be recent insertions, not yet eliminated by selection, whereas fixed TE insertions are presumably old insertions weakly submitted to selection. It is worth stressing that, for the study of TE frequencies and abundances, we first used simulated PoolSeq data to validate our pipelines and to evaluate their performance and their sensibility to parameters such as sequencing coverage or number of individuals (see Appendix A in supplementary material, Supplementary Material online, for details).
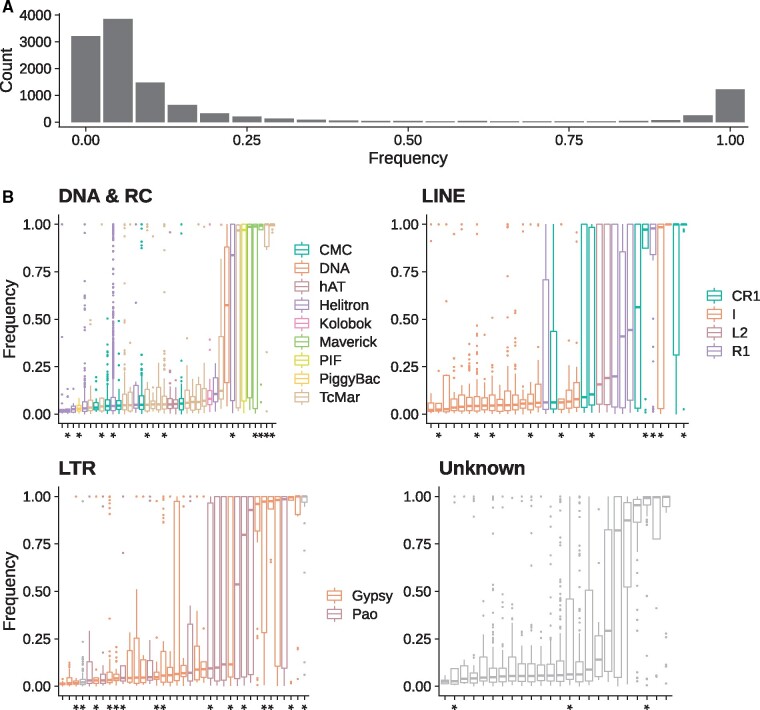
A total of 11,751 insertions were recovered in the reference population. The frequency distribution is approximately L-shaped (fig. 2A) with a majority of insertions segregating at low frequency (N = 9,599, f < 0.25) and 1,639 insertions are found at high frequency in the reference population (f ≥ 0.75). Only a minority of insertions are of intermediate frequency (N = 513, 0.25 ≤ f < 0.75). Among the 654 families/pseudofamilies found in the whole data set (see Materials and Methods), 472 were present in the reference population, 99 belonged to the DNA order, 96 to the LINE order, 182 to the LTR order, 45 to the RC, and 50 were Unknown. Only 133 TE families/pseudofamilies have more than 10 insertions: 30 DNA families/pseudofamilies, 34 LINEs, 37 LTR, 8 RC, and 24 Unknown. The vast majority of these families presented a median frequency lower than 0.25 (N = 97) (fig. 2B). Only six families displayed a median frequency between 0.25 and 0.75. Finally, 30 families had a median frequency superior or equal to 0.75. We did not find evidence that the number of TE families in these categories differed between TE orders (supplementary table S3, Supplementary Material online; χ2 = 3.54, df = 8, P value = 0.90). However, the mean frequency was slightly different (, , , , , Kruskal–Wallis χ2 = 4940.80, df = 4, P value <2.2 × 10−16). TE insertion frequencies were not evenly distributed along with the assembly: mean TE insertion frequency was considerably lower in gene-rich windows. (, , W = 19,556, P value <2.2e−16; supplementary fig. S5, Supplementary Material online).
Fig. 2.
TE activity in the Drosophila suzukii reference population from Watsonville (USA). (A) Frequency distributions of TE insertions. (B) Population frequencies for each TE family. Pseudofamilies are indicated by a star on the x-axis. Only families/pseudofamilies with more than ten insertions in the reference population are shown. DNA and RCs have been grouped for graphical reasons.
Demography as Driver of TE Contents in D. suzukii Populations
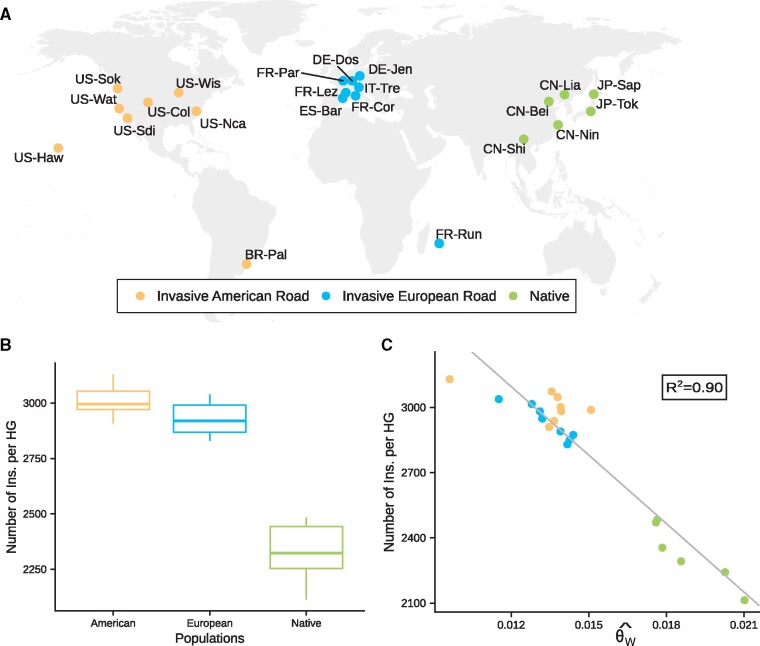
Our estimation of TE abundance in the 22 genotyped D. suzukii populations (fig. 3A) indicates substantial variation across populations, with significantly more TEs in invasive than in native populations and a strong correlation with the Watterson estimate of genetic diversity obtained from single nucleotide polymorphisms (SNPs) corresponding to a proxy of effective population size (fig. 3B and C). The mean number of insertions per haploid genome (HG) and per population was 2,793, ranging from 2,113 in the Chinese population CN-Nin to 3,129 in the Hawaiian population (US-Haw). There was a significant effect of the continent on the mean number of families/pseudofamilies per population: American and European populations had more families/pseudofamilies than native populations (, , , Kruskal–Wallis χ2 = 10.505, df = 2, P value = 0.0052). American and European populations also had more insertions per HG than native populations (, , , Kruskal–Wallis χ2 = 14.4, df = 2, P value = 7.3 × 10−4). We found a negative linear correlation between the total number of insertions per HG and per population and the Watterson estimate of genetic diversity obtained from SNPs , a proxy of effective population size (t = −13.42, df = 20, P value = 1.8 × 10−11, fig. 3C). The variation of explains a large proportion of the variance in the total number of insertions per HG across the populations (R2 = 0.90). The correlation remains significant when considering only native populations (t = −5.22, df = 4, P value = 6.4 × 10−3), or only invasive populations (t = −3.06, df = 14, P value = 8.6 × 10−3), or only European populations (t = −5.46, df = 6, P value = 1.6 × 10−3), but not when considering only American populations (t = −1.89, df = 6, P value = 0.11). The correlation between the number of insertions per HG per population and was also assessed individually for the 83 TE families/pseudofamilies showing an amplitude of variation equal or higher than three copies per HG. After a Benjamini–Hochberg correction for multiple testing, we found a significant correlation for 63 TE families (P-adjusted < 0.05, with R2 ranging from 21% to 90% when significant).
Fig. 3.
TE dynamics in native and invasive Drosophila suzukii populations. (A) Geographic location and historical status of the 22 D. suzukii population samples genotyped using a pool-sequencing methodology (Fraimout et al. 2017). (B) TE content in D. suzukii populations, as the numbers of insertions per HG. (C) Correlation between TE content and Watterson’s θ in D. suzukii population samples.
To verify that the observed variations of reflect variations of population effective sizes such as those encountered by D. suzukii populations during their spread out of Asia, we performed a simulation work (see supplementary methods, Supplementary Material online, for details). We followed the evolution of in a population whose initial size has been chosen to reflect the size of native populations. After a burn-in period of 7.5 N generations, allowing this population to reach an equilibrium, a bottleneck is simulated. An example where population size is divided by 200 and then multiplied by 1.9 every generation is shown in supplementary fig. S6, Supplementary Material online. We observe that 100 generations after the bottleneck, that is, roughly the time separating the invasion from the sampling, is lower than before the bottleneck and has reached values compatible with those observed in our samples of invasive populations. We tested several combinations of bottleneck intensities and population growth rates and found a negative correlation between and the bottleneck intensity (t = 8.2475, df = 73, P value = 4.83 × 10−12; supplementary fig. S7, Supplementary Material online).
The comparison of TE insertion frequency distributions between continents indicates that invasive populations are characterized by more insertions of low–intermediate frequencies (0 < f < 0.5) and nearly fixed insertions (0.90 < f < 1) (supplementary fig. S8, Supplementary Material online). This pattern is compatible with an overall relaxation of selection, leading to an increase in TE insertion frequencies. The excess of TE insertions of very low frequencies (0 < f < 0.1) could also be the signature of greater TE activity, as a result of environmentally induced changes in activity or acquisition of TEs by horizontal transfer, for example. To test if TE families horizontally transferred during the invasion could contribute to the higher number of TE copies in invasive HGs, putatively horizontally transferred families/pseudofamilies were identified as families/pseudofamilies that are absent in all native populations. For these families/pseudofamilies, we evaluated the number of insertions per HG number as well as the TE insertion frequency spectrum (supplementary fig. S9, Supplementary Material online). We found that these families/pseudofamilies correspond to a very low number of TE insertions, overall or for insertions of very low frequencies, and cannot explain the differences in the amount observed between native and invasive populations.
Note that, to allow unbiased comparison between samples, we performed a read subsampling in the TE calling pipeline used to produce TE abundance estimates, but also the frequency spectrum presented in this paragraph. Because this subsampling may affect the ability to detect TE insertions, especially insertions segregating at low frequencies, we investigated a potential effect of this technical correction on our results. We found the frequency spectrum to be overall similar with and without subsampling. However, low-frequency insertions (0 < f < 0.1) were more abundant in native populations than invasive ones without subsampling (supplementary fig. S10, Supplementary Material online), whereas the opposite was true with subsampling. This result suggests that the subsampling may lead to a lack of detection of some insertions segregating at a very low frequency in native populations. However, even without subsampling, the number of insertions per HG is greater in invasive populations (, , , Kruskal–Wallis χ2 = 15.564, df = 2, P value = 0.00041). Furthermore, the correlation between overall TE content and in D. suzukii populations remains true (t = −-5.79, df = 20, P value = 1.41 × 10−5, R2 = 63%).
Environmental and Genotypic Effects on TE Abundance
Because did not explain all the observed variation in TE abundance among the 22 sampled populations, that is, 20 out of the 83 investigated families/pseudofamilies do not display any significant correlation between variations of abundance and the estimations of and some others show only a weak correlation (e.g., R2 = 21%), we tested the effect of two other factors: the environmentally induced changes in TE activity and the genetically determined changes.
To test for an effect of environmentally induced changes in TE activity, we used partial Mantel tests. We tested the correlation between 19 bioclimatic variables and TE family abundance, for the 83 TE families showing amplitude of variation superior or equal to three copies per HG, correcting for population structure. After correction for multiple testing, we did not find any significant correlation (Benjamini–Hochberg correction for multiple testing, P-adjusted < 0.05).
To evaluate the effect of genetic variation on TE abundance we performed a genome-wide scan for association using methods controlling for population structure. To that end, we relied on the 13,530,656 bi-allelic variants (mostly SNPs) previously described on the same data set (Olazcuaga et al. 2020). We searched for an association between allelic frequencies and the population abundance of the 83 TE families/pseudofamilies mentioned above using the BayPass software. Globally, we found 1,950 genomic regions showing evidence of association with population abundance of at least one TE family. Each region included one or several significant SNP/InDel separated by less than 1 kb. On average, each region was associated with the number of insertions per HG of 2.51 families (min = 1, max = 44) and contained 1.37 SNPs/InDels (min = 1, max = 49). Four hundred and fifty-two (23.44%) regions overlapped with repeated sequences as annotated in the reference genome, which is more than expected by drawing SNP-/InDel-associated regions randomly (, q0.025=7.60%, q0.975=11.16%; supplementary fig. S11A, Supplementary Material online). Only seven of these regions contain a TE of the same family/pseudofamily as the TE abundance they were associated with. Regarding genes, 737 (37.80%) regions were associated with at least one gene, which is less than expected under random expectations (, quantile0.025=62.40%, quantile0.975=70.76%; supplementary fig. S11B, Supplementary Material online). Due to their known role in the activity of TEs, we further searched for enrichment in genes encoding transcription factors and piRNA pathway effectors among the genes located within our candidate regions. We did not observe any significant enrichment in genes encoding transcription factors (observed: 4.05%, expected: , quantile0.025 = 4.80%, quantile0.975 = 8.40%; supplementary fig. S11C, Supplementary Material online) nor in genes involved in the piRNA pathway (observed: 0.051%, expected: , q0.025 = 0%, q0.975=0.60%; supplementary fig. S11D, Supplementary Material online). Among the top ten regions, corresponding to the regions associated with the highest number of TE families/pseudofamilies, three appeared to be nongenic, four could not be attributed to D. melanogaster genome and three were associated with the mitochondrial genome.
A Small Number of Putatively Adaptive TE Insertions
We investigated the presence of putatively adaptive insertions using a genome scan combining three methods controlling for population structure implemented in BayPass (Olazcuaga et al. 2020). First, we assessed overall differentiation (based on the XtX statistics, a global differentiation statistics). Second, we focused on insertions that may have facilitated the recent invasion. To do so, we studied allele frequency differences between two groups of populations (based on the C2 statistics, which contrasts allelic frequencies between user-defined groups of populations): American invasive versus native populations (), European invasive versus native populations (), all invasive versus native populations (). Third, we carried out genome-wide association with each of the 19 bioclimatic variables (based on the BF).
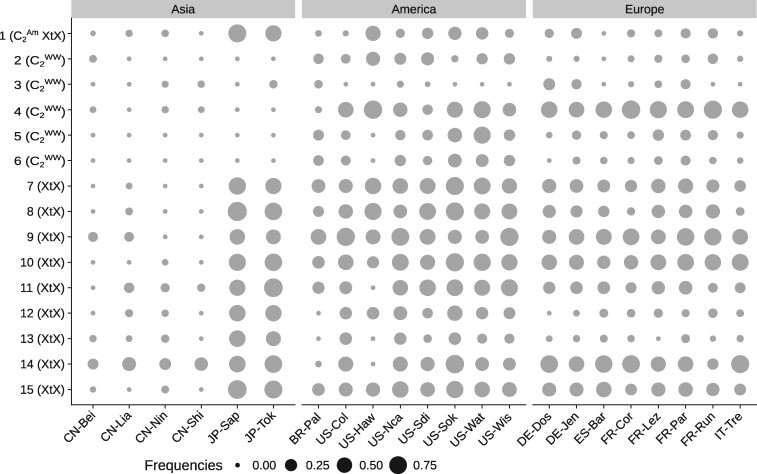
The genome scan was conducted on 7,004 polymorphic TE insertions (minor allelic frequency (MAF) > 0.025, 5,944 autosomal insertions, and 1,060 X-linked insertions treated separately). We identified a total of 15 putatively adaptive insertions (12 located on autosomal and three on X-linked contigs) (table 1 and fig. 4). Nine of these insertions were outliers when considering the global differentiation statistics XtX. Note that their frequencies were distinct between native Chinese (low frequencies) and native Japanese populations (high frequencies). One insertion was an outlier for both the XtX and statistics. Finally, the last five insertions were outliers for the statistics. No significant association was found between TE insertion frequencies and the 19 bioclimatic variables investigated.
Table 1.
Description of the 15 Putatively Adaptive TE Insertions.
| Insertion | Statistics | Gene Vicinity | Outlier SNP Nearby | A/X | TE Order |
|---|---|---|---|---|---|
| 1 | XtX | ASPP | F | A | RC |
| 2 | dia | F | A | RC | |
| 3 | — | T | A | DNA | |
| 4 | NA | F | X | RC | |
| 5 | inaE | F | X | RC | |
| 6 | — | F | X | DNA | |
| 7 | XtX | Mical | F | A | DNA |
| 8 | XtX | CG30015 | F | A | RC |
| 9 | XtX | — | F | A | RC |
| 10 | XtX | CR31386 | F | A | Unknown |
| 11 | XtX | — | F | A | RC |
| 12 | XtX | Dop1R2 | F | A | Unknown |
| 13 | XtX | jing | F | A | RC |
| 14 | XtX | CG14282 | F | A | Unknown |
| 15 | XtX | GATAe | F | A | RC |
Note.—Each insertion is an outlier when considering one or a combination of the global differentiation statistics (XtX) and statistics contrasting allelic frequencies between native populations and populations of the invasive American road () or populations of the invasive European road () or all invasive populations (). The fourth column indicates whether an SNP potentially evolving under positive selection had been detected less than 5 kb away in Olazcuaga et al. (2020) (F = false, T = true). The fifth column indicates whether the insertion is located on an autosomal (A) or X-linked contig (X).
Fig. 4.
Frequencies of each of the 15 putatively adaptive insertions in the 22 Drosophila suzukii populations. Insertion number is indicated on the left together with the associated BayPass statistics. XtX corresponds to a global differentiation statistic, C2 to a statistic contrasting allelic frequencies between native populations and populations of the invasive American road () or populations of the invasive European road () or all invasive populations ().
One of the 15 putatively adaptive insertions was close (i.e., 399 bp away) to an SNP/InDel that had previously been identified in a region potentially associated with D. suzukii invasive success (table 1) (Olazcuaga et al. 2020). Note that in their study Olazcuaga et al. (2020) analyzed 14 millions SNPs/InDels in the same populations as those studied here. They found 204 markers significantly associated with the invasive status. Because they found a lack of clustering of candidate markers, suggesting a small extent of linkage disequilibrium across the D. suzukii populations, we did not consider the presence of an outlier SNP potentially evolving under positive selection nearby as a necessary condition for an insertion to be classified as putatively adaptive.
Out of the 15 putatively adaptive insertions, for one insertion, we did not find any homologous regions in D. melanogaster, four others were in genomic regions without any genes, and the ten remaining were associated with genes.
We further investigated signatures of selection around candidate insertions by estimating local Tajima’s D statistics in the SNP/InDel data set. Low values of Tajima’s D indicate an excess of rare mutations, one possible signature of a selective sweep due to positive selection. To test whether each of our candidate insertions was associated with selective sweeps, we computed the linear correlation between its frequency and local Tajima’s D values (supplementary fig. S12, Supplementary Material online). Five statistically significant correlations were found corresponding to the insertions nos. 4, 9, 10, 12, and 15 (Pearson’s product–moment correlation, P < 0.05). Only a single insertion was associated with an extreme local Tajima’s D (insertion no. 15; Tajima’s D < quantile0.05), and only for a single population. The visualization of Tajima’s D at a larger scale (i.e., 10 kb upstream–10 kb downstream the insertion) confirms the lack of strong effect of the investigated insertions on Tajima’s D (supplementary fig. S13, Supplementary Material online). It is worth noting that, if the effect of our candidate TE insertion on Tajima’s D is globally low, a close investigation of Tajima’s D suggests that, at least in some cases, it is the absence rather than the presence of the insertion that may be adaptive. As a matter of fact, although the correlation involving an extreme local Tajima’s D was negative, the four other significant correlations between local Tajima’s D and insertion frequency were positive.
Discussion
For most species, the repeatome is still a poorly known genomic compartment and much remains to be understood regarding its variability, dynamics, functional, and fitness impacts. This is all the more important given that TEs appear to be ubiquitous, prompt to invade new genomes (Kofler, Hill, et al. 2015), and they may drastically impact the host phenotype (Nikitin and Woodruff 1995; Daborn et al. 2002; Van’t Hof et al. 2016). Here we capitalized on a recently generated long-read genome assembly and a large set of populational PoolSeq data (Olazcuaga et al. 2020; Paris et al. 2020) to thoroughly characterize the TE content of the nonmodel invasive species D. suzukii.
An Abundant, Unevenly Distributed, and Active Repeatome
The observed 47% of TEs in the genome of D. suzukii confirmed that this species ranks among the Drosophilid with the highest TE content (Kim et al. 2020). Our estimate is somewhat higher than those reported in previous studies in D. suzukii (Chiu et al. 2013; Ometto et al. 2013; Paris et al. 2020). Considering that the assembly of repeats is often impossible using short paired-end (PE) reads (Rius et al. 2016), it is not surprising that we recovered more TEs in a long reads genomic assembly than previous studies investigating TE contents using PE reads assemblies (Chiu et al. 2013; Ometto et al. 2013). In addition, we here performed a de novo reconstruction of TE sequences, which allowed us to identify more TE families/pseudofamilies, compared with the previous research work based on sequence homology on the same assembly (35%) (Paris et al. 2020). Overall, de novo reconstruction of TE sequences from long-read assemblies, such as the 15 Drosophila species assemblies recently generated using nanopore sequencing (Miller et al. 2018), should greatly improve our knowledge of TE diversity in Drosophila.
In agreement with the gene disruption hypothesis and observations in a variety of species (Bartolomé et al. 2002; Medstrand et al. 2002; Wright et al. 2003), we observed a depletion of TE copies in gene-rich regions of the D. suzukii genome. Although it is likely that TEs are strongly selected against in these regions due to their negative effect on gene function or expression (Lee and Karpen 2017; Mérel et al. 2020), it is also possible that TE copies are depleted in these regions because they promote ectopic recombination. In agreement with the latter hypothesis, gene-rich regions are also known to display a high recombination rate in D. melanogaster (Adams et al. 2000). The generation of a genomic map of recombination rates in D. suzukii would be needed to disentangle the respective effects of ectopic recombination and gene disruption.
At the chromosomal scale, we did not find a lower density of TEs on the X chromosome compared with autosomes. This pattern indicates that, if X-linked recessive insertions are more efficiently selected against than autosomal insertions, the effect on TE abundance is either low or balanced by another process. When comparing only gene-rich regions, we even found a higher density of TEs on the X chromosome than on autosomes. Three nonmutually exclusive explanations can be invoked: 1) there may be a higher insertion rate on the X chromosome, similar to what was previously found in D. melanogaster (Adrion et al. 2017); 2) the recombination rate may be lower on the X chromosome, which would lead to a stronger Muller’s ratchet; and 3) the strength of selection may be reduced by a smaller effective population size for the X chromosome.
Similar to what has been found in D. melanogaster and D. simulans (Kofler, Nolte, et al. 2015) and to what is probably common among Drosophila species (Hill 2019), the pattern of TE insertion frequencies in D. suzukii is compatible with an active repeatome. We found differences in the mean insertion frequency between TE orders, which suggests differences in activity but could also result from variation in the strength of purifying selection acting against the different orders (Petrov et al. 2003; Lee and Karpen 2017). Considering the trap model of TE dynamics (i.e., a model in which newly invading TEs are quickly inactivated by host defense, Zanni et al. 2013; Kofler et al. 2018), an active repeatome suggests a recurrent turnover of TEs, potentially due to horizontal transfer events. Investigating TE activity in D. melanogaster and D. simulans, Kofler and colleagues (Kofler, Nolte, et al. 2015) suggested that such a turnover is influenced by the colonization history of those species. They propose that the high activity of DNA transposons in D. simulans results from horizontal transfer events from D. melanogaster during D. simulans worldwide colonization. In agreement, we detected more families/pseudofamilies in invasive populations of D. suzukii than in the native ones, suggesting that new TE families may have been acquired during the recent colonization of new areas. However, we found a low number of insertions for putatively horizontally transferred TEs, that is, that were absent in all native populations.
Demography, Rather than Environment or Genotype, Drives TE Content
In agreement with the Lynch and Conery hypothesis (Lynch and Conery 2003), we found that the TE content in D. suzukii is negatively correlated with the Watterson estimate of genetic diversity that may be viewed as a proxy of the effective population size Ne. The negative correlation between and TE content was significant when considering only European invasive populations, invasive populations as a whole, or only native populations, but was not significant when considering only American invasive populations. Although a few studies suggest an increase of TE content following colonization (Nardon et al. 2005; García Guerreiro et al. 2008; García Guerreiro and Fontdevila 2011; Talla et al. 2017), to our knowledge, it is the first time that a correlation between TE content and Ne is found at the intraspecific level. Although several factors may affect Ne, the variation observed is likely to result from demographic processes. Indeed, both European and American invasive populations have encountered bottlenecks (Fraimout et al. 2017). In agreement with this idea, the invasive population from Hawaii, which experienced the strongest bottleneck (Fraimout et al. 2017), showed the smallest values. It is interesting to note that the negative correlation between and TE content remains significant when considering only native populations suggesting that other demographic event than bottleneck may also be involved (e.g., different stable effective population sizes and gene flow patterns).
Our analysis is controlled for sequencing bias, that is, coverage and insert size, and we are confident in the biological significance of the correlation observed here. However, it is worth stressing that our data set of TE insertions corresponds to a small fraction of the repeatome. Indeed, the mean number of scored insertions per HG per population in this analysis is markedly below the number of TE copies recovered in the reference genome (2,793 vs. 173,720). To qualify this statement, note that a part of the large difference between these values is likely to be explained by an overestimation of TE copy numbers in the reference genome. For example, we found almost 40,000 copies of the Gypsy element. Considering a mean length of 6,000 pb per copy, 240 Mb of the assembly would be occupied by Gypsy elements, whereas we found only 47% of TEs in the 270 Mb assembly. It is likely that some copies are deleted or degraded copies, or that some copies interrupted by secondary insertions are counted several times. We believe that this is due to an impossibility to properly call TE insertions when TEs are too close or even nested (Vendrell-Mir et al. 2019). It is thus possible that the negative correlation that we found here exists only for some part of the genome. Especially, it is likely that regions of low TE density, where most TE insertions are polymorphic, display the strongest answer to a reduction of selection efficacy. This is simply because polymorphic insertions can increase in frequency whereas fixed insertions cannot. One could also argue that the efficiency of selection is a function of the product between Ne and s (with s the selection coefficient). Therefore, the effects of a reduction of Ne should be especially marked in regions where selection against TEs is strong, such as TE-poor/gene-rich regions.
It appears from our simulation work that the 100 generations which passed between the colonization and the sampling of flies are not sufficient for to reach equilibrium and thus is not necessarily reflecting the strength of selection. Nevertheless, our simulation work suggests that our estimations of reflect the intensity of the recent bottleneck experienced by these populations (Fraimout et al. 2017), and thus a reduction of the strength of selection in the recent past. We are therefore convinced that reflects the intensity of selection, at the time of sampling for native populations, or a few generations earlier for invasive populations. We think that it is a plausible explanation to the observed correlation between TE content and .
In addition, if our analysis pinpoints toward demography as the main driver of TE content, one limitation remains to be mentioned. Our estimation of the impact of the strength of selection on the number of insertions per HG may be overestimated. Insertions of very low frequency are difficult to detect and expected to be more abundant in populations, where the strength of selection is strong and/or that were submitted to a weaker founder effect, that is, populations of high . We may thus underestimate TE content in these populations.
Beyond variations in selection intensity, changes in TE activity could also contribute to TE content variation in D. suzukii populations. It may explain, for example, that for 20 families, TE content is not correlated with . This could result from a higher activity of TEs in invasive populations that leads to a higher number of TEs of low population frequency in invasive populations. Alternatively, we may envision a role of environmental changes on TE activity. We found no significant effect on TE abundance for all the 19 environment variables tested. This might be surprising at first sight given the large number of studies showing an association between TE activity and external factors, such as temperature or viral infection (García Guerreiro 2012; Ryan et al. 2016; Horváth et al. 2017; Roy et al. 2020). Several factors may explain this discrepancy. First, it is important to notice that, in Drosophila, most of these studies rely on lab experiments, some of them exploring environmental conditions unlikely in natura (see García Guerreiro 2012 for a review). To our knowledge, none of these studies established a link between TE activity and the natural environment without any possible confounding effect from population structure and demographic features. Second, as often in Drosophila, most of such research works were carried out on the same particular species, D. melanogaster, so that so far we do not know much about interspecific variability. Third, although partial Mantel tests allowed revealing 15 significant correlations between TE abundance and environmental variables in A. thaliana populations (Quadrana et al. 2016), we consider our results as conservative, especially regarding the long discussion about the statistical performance of partial Mantel tests (Diniz-Filho et al. 2013). More sophisticated statistical methods may be needed to tackle such relationships in more detail.
Considering that several studies on Drosophila suggest a genotype effect on TE activity (Biémont et al. 1987; Pasyukova and Nuzhdin 1993; Díaz-González et al. 2011; Adrion et al. 2017), we performed a Genome-Wide Association Study (GWAS) on TE abundance to assess this effect in natural populations and identify the genomic regions involved. Overall, we found ca. 2,000 genomic regions associated with TE abundance. These regions were not enriched in transcription factor genes nor genes of the piRNA pathway. It may be because those regions are involved in tolerance to TEs rather than in the control of their activity. For example, in D. melanogaster, the bruno gene has been shown to control tolerance to P-element transposition, and one may expect more tolerant genotypes to accumulate more TEs (Kelleher et al. 2018). As far as we know, no such GWAS study has been carried out in Drosophila populations. Our results are somewhat similar to those found in A. thaliana, in which although a strong causal link between one transcription factor and the abundance of two TE families was found, no enrichment for any particular function was observed (Quadrana et al. 2016). Because our candidate regions are enriched for TE overlapping regions, two different types of loci may play a particular role in the modulation of TE activity in D. suzukii populations. The first would be TE regulatory sequences. The second would be piRNA clusters, that is, the TE cemeteries generating piRNAs, small RNAs that silence TEs by means of sequence complementarity. Additional analyses such as the sequencing of piRNAs to locate piRNA clusters, or annotation of TE structures should help us shed light on this issue.
A Potential Adaptive Role for a Limited Number of TEs
Similar to studies investigating TE adaptive potential in D. melanogaster populations (González et al. 2008, 2010; Rech et al. 2019), we found several putatively adaptive TE insertions in our D. suzukii data set. Overall, we found 15 insertions, six of which may have eased the worldwide invasion of D. suzukii. It is important to note that we are probably missing some insertions, and thus likely underestimating the number of adaptive insertions sites.
Overall, we did not capture a strong signal of a selective sweep near the candidate adaptive TE insertions. This may be due to overall large effective population sizes as suggested in Olazcuaga et al. (2020), but also to the fact that Tajima’s D is unlikely to detect soft selective sweep, that is, adaptation from standing variation or multiple successive beneficial mutations (Pennings and Hermisson 2006). An appealing perspective would be to sequence candidate regions in individual strains and use a haplotype-based analysis. For example, the recently introduced Comparative Haplotype Identity (xMD) statistics (Lange and Pool 2016; Villanueva-Cañas et al. 2017) has been shown to perform well for soft sweeps. If the effect of our candidate TE insertion on Tajima’s D is globally low, it highlighted the possibility that the absence rather than the presence of the insertion may be adaptive, at least for some of our candidate insertions. More specifically, for four insertions a positive correlation was found between local Tajima’s D and insertion frequency. However, the only extreme local Tajima’s D was found in the population where the putatively adaptive insertion is at its highest frequency, indicating that it is probably the insertion itself rather than the absence that might be adaptive.
One added value to our analysis based on GWAS is that the same type of analysis has been carried out using SNPs/InDel (Olazcuaga et al. 2020). The authors of this study found 204 markers strongly associated with invasion success distributed over the whole genome. If we compare this number with our six TE insertions, it seems unlikely that TEs solely may explain the genetic paradox of invasive species (Stapley et al. 2015). It is worth noting that the level of variation remains high in invasive D. suzukii populations (Fraimout et al. 2017). Hence, it would be interesting to carry out similar analyses in invasive species that experienced a more intense depletion of genetic variation during invasion (Prentis et al. 2009; Zhang et al. 2010; Roux et al. 2011) to assess whether TEs are more likely to be adaptive in invasive populations with low levels of genetic diversity.
At first sight our finding of 15 putatively adaptive polymorphic insertions in worldwide populations of D. suzukii contrasts with the 41 to 300 putatively adaptive polymorphic insertions found in worldwide populations of D. melanogaster (Rech et al. 2019). The difference is even more glaring considering that we analyzed 7,004 polymorphic insertions, against ∼800 in (Rech et al. 2019). This suggests a largely higher rate of TE-induced adaptations during D. melanogaster invasion and this despite the much larger, still active and diverse repeatome of D. suzukii. It may be due to historical differences between the two species (Stephan and Li 2007; Fraimout et al. 2017) or intrinsic differences with respect to the repeatome contents. However, differences in methodology are likely to play an important role in preventing the comparison. For example, our analysis relies essentially on the search of overly differentiated TEs across populations with a correction for population structure (Gautier 2015; Olazcuaga et al. 2020), whereas in the analysis used for D. melanogaster, there is no direct methodological control for population structure. Applying our statistical methodologies to the D. melanogaster data set (Kapun et al. 2020), which also consist in PoolSeq data, would help determine if methodology differences can explain the observed discrepancy.
Our study of TE-induced adaptation strongly calls for validation of candidate insertions. Allele-specific expression assays would allow evaluating if these insertions affect nearby gene expression (Gonzalez et al. 2009). This would consist of testing a difference of nearby gene expression between the two alleles of an F1 hybrid between strains with and without the insertion. Although such test should control for genotype effect, compared with a simple test of differential expression between strains, it does not preclude an effect of an SNP/InDel close to the insertion. Using a CRISPR/Cas9 methodology would also allow (in)validate that the TE(s) of interest is the causative agent of gene expression change and would allow direct testing for a phenotypic effect.
Conclusion
Our study illustrates the value of an approach combining a long read-based genome assembly, a de novo reconstruction of TE sequences, and PoolSeq population data, to characterize the repeatome of a nonmodel species. Our set of analyses especially highlighted that the particularly large D. suzukii repeatome is probably active and shaped by purifying selection, similar to that of D. melanogaster’s. Additional data, such as local recombination rate, would also help us shed light on the nature of selection acting on TEs. The analysis of TE abundance variations in invasive and native populations suggests that a reduction of purifying selection intensity, in response to demographic processes, can significantly increase TE content. Our study also indicates that positive selection may act on TE insertions in response to selective factors that remain to be determined. Experimental validation will allow us to (in)validate a functional impact of our putatively adaptive insertions. Overall, the natural extent of the trends we uncovered here should be explored in more detail, for instance through the application of similar methods to other (invasive) species that would allow us to evaluate the impact of a stronger bottleneck on both TE content increase and TE adaptive potential.
Materials and Methods
Creation of a TE Database
A TE database was created by merging previously established consensus of Drosophila TE families and de novo reconstructed consensus of D. suzukii TE families. The previously established consensus were obtained by extracting all Drosophila consensus from Dfam and Repbase databases (release 2016–2018 for both) (Hubley et al. 2016; https://www.girinst.org/repbase/, last accessed December 01, 2019). De novo reconstruction was performed using an assembly of an American strain from Watsonville, sequenced using PacBio long reads technology, and the REPET package (v2.5) (Flutre et al. 2011; Paris et al. 2020). Obtained TE sequences were annotated by homology to previously established consensus of Drosophila TE families. Finally, all consensus were clustered in families using UClust (Edgar 2010). See supplementary methods, Supplementary Material online, for details on the method. The generated TE database is accessible at: https://github.com/vmerel/Dsu-TE.
Annotation of the Reference Genome
To recover TE fragments and TE genomic sequence occupancy, the reference genome assembly was masked using RepeatMasker and the above TE database (-gccalc, -s, -a, -cutoff 200, -no_is, -nolow, -norna, -u; v 1.332) (http://www.repeatmasker.org/, last accessed December 01, 2019). TE density was evaluated as the number of TE fragments completely within nonoverlapping genomic windows of 200 kb. TE copies were reconstructed from TE fragments using OneCodeToFindThemAll (Bailly-Bechet et al. 2014). Gene density was computed from a run of augustus (–species=fly, –strand=both, –genemodel=complete; v2.5.5) (Stanke et al. 2008) as the number of genes completely within nonoverlapping genomic windows of 200 kb. For comparison, the same pipeline was applied to D. melanogaster assembly (release 6). Promer was used to generate alignments between D. melanogaster and D. suzukii masked assemblies and establish syntenic relationships (MUMmer v3.23) (Kurtz et al. 2004). The promer output was filtered out using the delta-filter module to obtain a one-to-one mapping of reference to query (-q, -r). A file containing alignment coordinates for alignments of minimum length 100 bp, and in which overlapping alignments were merged, was generated with the show-coords module (-b, -L 100, -r). Because the abundance of repeated sequences and the use of masked assemblies may result in multiple small alignments, alignments separated by less than 20 kb were merged using a custom script. Note that only alignments involving the 2L, 2R, 3L, 3R, X, and four chromosomes of D. melanogaster were kept at this step, and if a D. suzukii contig aligned to several D. melanogaster chromosomes only the best pair was conserved (i.e., the pair producing the longest alignment). Graphical visualization of the results was produced using Circos (Krzywinski et al. 2009).
Fly Samples and Pool Sequencing
Pool-sequencing (PoolSeq) data originate from Olazcuaga et al. (2020), where the detailed associated protocol is described. Briefly, adult wild flies were sampled between 2013 and 2016 from 22 localities of both native and invasive areas (fig. 3A) (Fraimout et al. 2017). Six samples were collected in the native Asian area, more precisely in four Chinese and two Japanese localities. The remaining 16 samples were chosen to be representative of two separate invasion roads: the American invasion road and the European invasion road. The American invasion road is represented by one Hawaiian sample, one Brazilian sample, and six samples from the United States. The European invasion road corresponds to two German samples, four French samples (including one from La Réunion Island), one Italian sample, and one Spanish sample. For each population sample, DNA extraction was performed from the thoraxes of 50–100 flies and used to prepare PE libraries (insert size of ∼550 bp). PE sequencing was achieved using an HiSeq 2500 from Illumina to obtain 2 × 125 bp reads. Reads were trimmed using the trim-fastq.pl script in the PoPoolation package (–min-length 75, –quality-threshold 20; v1.2.2) (Kofler et al. 2011).
TE Frequencies and Abundances
To obtain TE insertion frequencies and abundances in PoolSeq samples a calling of TEs was done using PoPoolationTE2 (Kofler et al. 2016), the reference genome, and the newly constructed database. To make sure that no reads from TE sequences could map on the masked assembly, TE reads were simulated, and mapped on the masked assembly and aligned positions were also masked. Reads simulation was performed using the script create-reads-for-te-sequences.py (Kofler et al. 2016): reads of 125 bp reads, coverage of 1,024 X per TE sequence in the database. Because we do not expect a split read-based TE calling tool such as PoPoolationTE2 to accurately call for insertions shorter than the insert size, TE sequences shorter than 500 bp were removed before calling. Moreover, as PoPoolationTE2 filters out insertions with reads mapping on more than one family, families with cross-mapping were grouped in pseudofamilies. Two families were brought together if at least 1% of reads from one sequence of the first family were mapped on a sequence of the second family (read simulation: 125 bp reads, coverage of 100X per consensus). The classification of each pseudofamily corresponds to that of the families that compose it. In case of divergent classification between families of a pseudofamily, this pseudofamily was classified as unknown. One exception has been made for a pseudofamily regrouping ten families: one annotated as an I element and nine as Helitron; this pseudofamily was considered as Helitron. Concerning the TE calling, reads were mapped using bwa bwasw (v0.7.17) (Li and Durbin 2010) and PE information restored using the se2pe script provided with the PoPoolationTE2 package (v1.10.04) (Kofler et al. 2016). One unique pileup file was generated with all samples specifying a minimum mapping quality of 15. The remaining modules of PoPoolationTE2 were used as follows: identifySignatures: –mode joint, –signature-window minimumSampleMedian, –min-valley minimumSampleMedian, –min-count 2; updatestrand: –map-qual 15, –max-disagreement 0.5; frequency; filterSignatures –min-coverage 10, –max-otherte-count 2, –max-structvar-count 2; pairupSignatures –min-distance -200, –max-distance 300. The final output contained frequencies in the 22 populations for each called TE insertion. To allow an unbiased comparison across samples, the frequencies spectrum (supplementary figs. S8 and S9, Supplementary Material online) and estimates of TE abundance came from a slightly different pipeline. The following parameters were modified: –target-coverage 30; identifySignatures –mode separate. This modification makes it possible, in particular, to account for differences in coverage and insert sizes between samples. TE abundances, as the numbers of insertions per HG per population, were estimated in each sample by summing insertion frequencies. See Appendix A in supplementary material, Supplementary Material online, for the validation work on simulated data.
Evaluation of Population Genetics Statistics
We estimated Watterson’s theta () and Tajima’s D statistics in nonoverlapping 1,000 bp windows using PoPoolation (v1.2.2) (Kofler et al. 2011). Forward and reverse trimmed reads were mapped separately using bwa aln (-o 2 -d 12 -e 12 -n 0.01; v0.7.17) (Li and Durbin 2010). A PE alignment file was generated using bwa sampe. Reads were filtered for a minimum mapping quality of 20 and a pileup file generated with samtools (v1.7) (Li et al. 2009). Each pileup file was split into two files: one corresponding to autosomal contigs and another corresponding to X-linked contigs (autosomal and X-linked contigs as determined in Olazcuaga et al. 2020). PoPoolation was used as follows: –min-count 2 –min-coverage 8 –max-coverage 250 –min-qual 20. The pool-size argument was modified accordingly between autosomal and X-linked pileup.
Genome-Wide Association Study with TE Family Abundance
All genome scans were performed using BayPass (v2.2) (Gautier 2015; Olazcuaga et al. 2020), a package aiming at identifying markers evolving under selection and/or associated with population-specific covariates, taking into account the shared history of the populations. For each SNP/InDel previously called in these PoolSeq samples (Olazcuaga et al. 2020), we estimated 83 Bayes factors (BFs), reflecting their association with the number of insertions per HG of 83 families/pseudofamilies (based on a linear regression model). The 83 chosen TE families/pseudofamilies were those displaying an amplitude of variation of at least three insertions per HG across the complete data set. To improve computing time BayPass was run on data subsets. Data-concerning TE abundance was split into three subsets of 28, 28, and 27 families, respectively. For SNPs/InDel, we used the data subsets of Olazcuaga et al. (2020), for which the 11,564,472 autosomal variants are divided into 154 subsets and the 1,966,184 X-linked variants into 26 subsets. Note that the calling was done on an unmasked assembly. Because we used the importance sampling algorithm implemented in Baypass to assess BFs, and single-run estimations may be unstable, a total of three runs were performed for each combination of TE subsets–SNP/InDel subsets and the median of BFs computed (Gautier et al. 2018). Note that different pool size files were used for autosomal and X-linked variants to take into account differences in the number of autosomes and X chromosomes in each PoolSeq sample. An SNP/InDel was considered to be associated with the abundance of a family/pseudofamily if it met two criteria. First, in accordance with Jeffrey’s rule, a BF greater than 20 was required. Second, in order to ensure a low number of false positives, the value of BF also had to be greater than or equal to the maximum of a null distribution, that is, a distribution for “neutral” SNPs evolving independently from covariates. Two null distributions had previously been obtained for each covariate, one for autosomes (5,550,000 markers) and one for gonosomes (1,950,000 markers). For this purpose the function simulate.baypass(nsnps = 75,000, pi.maf = 0) was used with 74 autosomal and 26 gonosomal matrices.
SNP/InDel locations were used to define genomic regions associated with TE abundance. Variants were gathered if separated by less than 1 kb. For each region, we looked for overlapping TEs using the RepeatMasker annotation (gff file, see Annotation of the reference genome). We also investigated gene content. First, if the spanned genomic interval was less than 1 kb, the region was obtained by adding 500 bp on both sides. Second, we retrieved homologous regions in the D. melanogaster genome using BLAT against the D. melanogaster masked assembly downloaded from UCSC Genome Browser (http://hgdownload.soe.ucsc.edu/goldenPath/dm6/bigZips/, last accessed December 01, 2019; BLAT v.36x4, -t=dnax -q=dnax). We then checked for genes overlapping the best hit subject sequence using the UCSC Genome Browser gff annotation file. Note that if the best-hit score was lower than 100, we considered that no homologous region was retrieved. The number of transcription factor genes among the genes retrieved was obtained by comparing their IDs to those of the gene group Transcription factor on flybase (https://flybase.org/reports/FBgg0000745.html, last accessed January 01, 2020). Similarly, the number of genes involved in the piRNA pathway was obtained by comparing gene IDs to those listed in Ozata et al. (2019). To test whether the candidate regions were enriched in TEs, we generated random expectations by applying the above to 1,000 randomly selected SNPs 250 times. For computing time reasons, for genes, transcription factor genes, or genes involved in the piRNA pathway, we used 500 randomly selected SNPs 125 times.
Correlation between Climatic Variables and TE Family Abundance
Partial Mantel tests were used to test the correlation between bioclimatic variables and TE family abundance correcting for population structure (as in Quadrana et al. 2016). Following 19 bioclimatic variables from the worldclim data set (Fick and Hijmans 2017) were considered: annual mean temperature, mean diurnal range, isothermality, temperature seasonality, max temperature of the warmest month, minimum temperature of the coldest month, temperature annual range, mean temperature of wettest quarter, mean temperature of driest quarter, mean temperature of warmest quarter, mean temperature of coldest quarter, annual precipitation, precipitation of wettest month, precipitation of driest month, precipitation seasonality, precipitation of wettest quarter, precipitation of driest quarter, precipitation of warmest quarter, and precipitation of coldest quarter. The 83 families with an amplitude of variation of at least three insertions per HG between populations were considered. The population structuring of genetic diversity is summarized by the scaled covariance matrix of population allele frequencies (Ω) estimated with BayPass, one autosomal subset randomly chosen was used (the correlation of the posterior means of the estimated Ω elements across SNP subsamples had previously been verified, Olazcuaga et al. 2020). Partial Mantel tests were conducted using the R package ecodist (Goslee and Urban 2007). P values were further adjusted to account for multiple testing applying the Benjamini–Hochberg correction (Benjamini and Hochberg 1995).
Screening for Putatively Adaptive TE Insertions
A genome scan for putatively adaptive TE insertions was performed using BayPass (v2.2) with the output of the TE frequency pipeline (Gautier 2015; Olazcuaga et al. 2020). Insertions with MAF inferior to 0.025 were removed before the analysis. Autosomal and X-linked contigs were analyzed separately. Three statistics were computed to detect putatively adaptive TE insertions: XtX, C2, and the BF for Environmental Association Analysis. Briefly, XtX corresponds to a global differentiation statistics, C2 contrasts allelic frequencies between user-defined groups of populations, and BF measures the support of the association between a marker and a covariate (usually an environmental variable). Because BF was computed using the importance sampling algorithm, and single-run estimations may be unstable, BF was estimated as the median over five estimates obtained from independent runs of BayPass (Gautier et al. 2018). In accordance with Jeffrey’s rule, a BF superior to 20 deciban (dB) was considered as decisive evidence supporting an association (Jeffreys 1961). XtX and C2 estimates came from one single run and simulation were used to determine a significance threshold. The R function simulate.baypass() provided within the BayPass package was used to simulate read count data (nsnp = 10,000, pi.maf = 0). We used the physical coverage estimated from the pileup file using the module stat-coverage of PoPoolationTE2 (Kofler et al. 2016). BayPass was run on this simulated data set to estimate the null distribution of the XtX and the C2 statistics. An insertion was considered as overly differentiated (for XtX) or associated with the tested contrast (for C2) if the corresponding statistics exceeded the 99.9% quantile of the estimated null distribution. The populations whose frequencies were contrasted using the C2 were: populations of the invasive American road and the native ones (), populations of the invasive European road and the native ones (), invasive populations, and the native ones (). This choice was made according to the invasion roads inferred using microsatellite markers (Fraimout et al. 2017), the populations structure assessed with SNP/InDel markers called in these samples (Olazcuaga et al. 2020), and the population structure assessed here with TE markers (supplementary fig. S14, Supplementary Material online). For each putatively adaptive insertion, gene vicinity in a 1 kb region centered on the insertion was investigated as described in the paragraph “Genome-Wide Association Study with TE family abundance.” The presence of the insertion in a region of the selective sweep was assessed using Tajima’s D. For the 22 populations, we investigated if the Tajima’s D estimated in the 1 kb window containing this insertion was inferior to the quantile 0.05 of Tajima’s D distribution in this population. More precisely, to prevent for a difference between autosome and X chromosome, autosomal insertions were compared with the autosomal Tajima’s D distribution and X-linked insertions to the X chromosome Tajima’s D distribution (with autosomal and X-linked contigs as defined in Paris et al. 2020). We also checked if the insertion was close to SNPs/InDels previously identified as potentially adaptive during D. suzukii invasion (considering a maximum distance of 5 kb) (Olazcuaga et al. 2020).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the French National Research Agency (ANR-16-CE02-0015-01—SWING) and performed using the computing facilities of the CC LBBE/PRABI. We sincerely thank C. Mermet-Bouvier for technical help. We are also grateful to B. Prud’homme and F. Sabot for constructive discussion about this article.
Data Availability
The reference assembly used in this article is available on NCBI website (https://www.ncbi.nlm.nih.gov/genome/18317?genome_assembly_id=910956) (Paris et al. 2020). Population sequencing data are available online (SRA repository under the BioProject accession number PRJNA576997) (Olazcuaga et al. 2020). The generated TE database and scripts are available in a github (https://github.com/vmerel/Dsu-TE).
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287(5461):2185–2195. [DOI] [PubMed] [Google Scholar]
- Adrion JR, Song MJ, Schrider DR, Hahn MW, Schaack S.. 2017. Genome-wide estimates of transposable element insertion and deletion rates in Drosophila melanogaster. Genome Biol Evol. 9(5):1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M, Haudry A, Lerat E.. 2014. “One code to find them all”: a Perl tool to conveniently parse RepeatMasker output files. Mobile DNA 5(1):13. [Google Scholar]
- Bartolomé C, Maside X, Charlesworth B.. 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol. 19(6):926–937. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 57(1):289–300. [Google Scholar]
- Biémont C, Aouar A, Arnault C.. 1987. Genome reshuffling of the copia element in an inbred line of Drosophila melanogaster. Nature 329(6141):742–744. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Chen X, He M, Bergman CM.. 2014. An age of allele test of neutrality for transposable element insertions. Genetics 196(2):523–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV.. 2001. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 18(6):926–935. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282(5396):2012–2018. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 1983. The population dynamics of transposable elements. Genet Res. 42(1):1–27. [Google Scholar]
- Chiu JC, Jiang X, Zhao L, Hamm CA, Cridland JM, Saelao P, Hamby KA, Lee EK, Kwok RS, Zhang G, et al. 2013. Genome of Drosophila suzukii, the spotted wing Drosophila. G3 (Bethesda) 3(12):2257–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Macdonald SJ, Long AD, Thornton KR.. 2013. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol Biol Evol. 30(10):2311–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, Goff GL, Feil E, Jeffers S, Tijet N, Perry T, Heckel D, Batterham P, et al. 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297(5590):2253–2256. [DOI] [PubMed] [Google Scholar]
- Díaz-González J, Vázquez JF, Albornoz J, Domínguez A.. 2011. Long-term evolution of the roo transposable element copy number in mutation accumulation lines of Drosophila melanogaster. Genet Res (Camb). 93(3):181–187. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, Soares TN, Lima JS, Dobrovolski R, Landeiro VL, de Campos Telles MP, Rangel TF, Bini LM.. 2013. Mantel test in population genetics. Genet Mol Biol. 36(4):475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C.. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284(5757):601–603. [DOI] [PubMed] [Google Scholar]
- Edgar RC.2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Estoup A, Ravigné V, Hufbauer R, Vitalis R, Gautier M, Facon B.. 2016. Is there a genetic paradox of biological invasion? Annu Rev Ecol Evol Syst. 47(1):51–72. [Google Scholar]
- Fick SE, Hijmans RJ.. 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 37(12):4302–4315. [Google Scholar]
- Flutre T, Duprat E, Feuillet C, Quesneville H.. 2011. Considering transposable element diversification in de novo annotation approaches. PLoS One 6(1):e16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraimout A, Debat V, Fellous S, Hufbauer RA, Foucaud J, Pudlo P, Marin J-M, Price DK, Cattel J, Chen X, et al. 2017. Deciphering the routes of invasion of Drosophila suzukii by means of ABC random forest. Mol Biol Evol. 34(4):980–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Guerreiro MP, Chávez-Sandoval BE, Balanyà J, Serra L, Fontdevila A.. 2008. Distribution of the transposable elements bilbo and gypsy in original and colonizing populations of Drosophila subobscura. BMC Evol Biol. 8(1):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Guerreiro MP, Fontdevila A.. 2011. Osvaldo and Isis retrotransposons as markers of the Drosophila buzzatii colonisation in Australia. BMC Evol Biol. 11(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Guerreiro MPG.2012. What makes transposable elements move in the Drosophila genome? Heredity (Edinb) 108(5):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M.2015. Genome-wide scan for adaptive divergence and association with population-specific covariates. Genetics 201(4):1555–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Yamaguchi J, Foucaud J, Loiseau A, Ausset A, Facon B, Gschloessl B, Lagnel J, Loire E, Parrinello H, et al. 2018. The genomic basis of color pattern polymorphism in the Harlequin Ladybird. Curr Biol. 28(20):3296–3302.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Karasov TL, Messer PW, Petrov DA.. 2010. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PL0S Genet. 6(4):e1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA.. 2008. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 6(10):e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Macpherson JM, Petrov DA.. 2009. A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol Biol Evol. 26(9):1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslee SC, Urban DL.. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Soft. 22(7):1–19. [Google Scholar]
- Hill T.2019. Transposable element dynamics are consistent across the Drosophila phylogeny, despite drastically differing content. bioRxiv. 651059.doi:10.1101/651059. [Google Scholar]
- Horváth V, Merenciano M, González J.. 2017. Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends Genet. 33(11):832–841. [DOI] [PubMed] [Google Scholar]
- Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AFA, Wheeler TJ.. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res. 44(D1):D81–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys H.1961. Theory of probability, 3rd edition. Oxford: Oxford University Press. [Google Scholar]
- Kapun M, Barrón MG, Staubach F, Obbard DJ, Wiberg RAW, Vieira J, Goubert C, Rota-Stabelli O, Kankare M, Bogaerts-Márquez M, et al. 2020. Genomic analysis of European Drosophila melanogaster populations reveals longitudinal structure, continent-wide selection, and previously unknown DNA viruses. Mol Biol Evol. 37(9):2661–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Jaweria J, Akoma U, Ortega L, Tang W.. 2018. QTL mapping of natural variation reveals that the developmental regulator bruno reduces tolerance to P-element transposition in the Drosophila female germline. PLoS Biol. 16(10):e2006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Wang JR, Miller DE, Barmina O, Delaney E, Thompson A, Comeault AA, Peede D, D’Agostino ERR, Pelaez J, et al. 2020. Highly contiguous assemblies of 101 Drosophilid genomes. bioRxiv. doi:10.1101/2020.12.14.422775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Betancourt AJ, Schlötterer C.. 2012. Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster. PLoS Genet. 8(1):e1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Gómez-Sánchez D, Schlötterer C.. 2016. PoPoolationTE2: comparative population genomics of transposable elements using Pool-Seq. Mol Biol Evol. msw137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Hill T, Nolte V, Betancourt AJ, Schlötterer C.. 2015. The recent invasion of natural Drosophila simulans populations by the P-element. Proc Natl Acad Sci U S A. 112(21):6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Nolte V, Schlötterer C.. 2015. Tempo and mode of transposable element activity in Drosophila. PLoS Genet. 11(7):e1005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Orozco-terWengel P, Maio ND, Pandey RV, Nolte V, Futschik A, Kosiol C, Schlötterer C.. 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6(1):e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Senti K-A, Nolte V, Tobler R, Schlötterer C.. 2018. Molecular dissection of a natural transposable element invasion. Genome Res. 28(6):824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA.. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL.. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- Lange JD, Pool JE.. 2016. A haplotype method detects diverse scenarios of local adaptation from genomic sequence variation. Mol Ecol. 25(13):3081–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne S, Molofsky J.. 2007. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci U S A. 104(10):3883–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Karpen GH.. 2017. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. Elife 6:doi:10.7554/eLife.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Goubert C, Guirao‐Rico S, Merenciano M, Dufour A-B, Vieira C, González J.. 2019. Population-specific dynamics and selection patterns of transposable element insertions in European natural populations. Mol Ecol. 28(6):1506–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-W, Hou X-H, Chen J-F, Xu Y-C, Wu Q, González J, Guo Y-L.. 2018. Transposable elements contribute to the adaptation of Arabidopsis thaliana. Genome Biol Evol. 10(8):2140–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 302(5649):1401–1404. [DOI] [PubMed] [Google Scholar]
- Marin P, Genitoni J, Barloy D, Maury S, Gibert P, Ghalambor CK, Vieira C.. 2020. Biological invasion: the influence of the hidden side of the (epi)genome. Funct Ecol. 34(2):385–400. [Google Scholar]
- Medstrand P, van de Lagemaat LN, Mager DL.. 2002. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 12(10):1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérel V, Boulesteix M, Fablet M, Vieira C.. 2020. Transposable elements in Drosophila. Mob DNA. 11(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, . et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403(6771):785–789. [DOI] [PubMed] [Google Scholar]
- Miller DE, Staber C, Zeitlinger J, Hawley RS.. 2018. Highly contiguous genome assemblies of 15 Drosophila species generated using nanopore sequencing. G3 (Bethesda) 8(10):3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420(6915):520. [DOI] [PubMed] [Google Scholar]
- Nardon C, Deceliere G, Loevenbruck C, Weiss M, Vieira C, Biémont C.. 2005. Is genome size influenced by colonization of new environments in dipteran species? Mol Ecol. 14(3):869–878. [DOI] [PubMed] [Google Scholar]
- Nikitin AG, Woodruff RC.. 1995. Somatic movement of the mariner transposable element and lifespan of Drosophila species. Mut Res/DNAging. 338(1–6):43–49. [DOI] [PubMed] [Google Scholar]
- Niu X-M, Xu Y-C, Li Z-W, Bian Y-T, Hou X-H, Chen J-F, Zou Y-P, Jiang J, Wu Q, Ge S, et al. 2019. Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc Natl Acad Sci U S A. 116(14):6908–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olazcuaga L, Loiseau A, Parrinello H, Paris M, Fraimout A, Guedot C, Diepenbrock LM, Kenis M, Zhang J, Chen X, et al. 2020. A whole-genome scan for association with invasion success in the fruit fly Drosophila suzukii using contrasts of allele frequencies corrected for population structure. Mol Biol Evol. 37(8):2369–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto L, Cestaro A, Ramasamy S, Grassi A, Revadi S, Siozios S, Moretto M, Fontana P, Varotto C, Pisani D, et al. 2013. Linking genomics and ecology to investigate the complex evolution of an invasive Drosophila pest. Genome Biol Evol. 5(4):745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FHC.. 1980. Selfish DNA: the ultimate parasite. Nature 284(5757):604–607. [DOI] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD.. 2019. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 20(2):89–108. [DOI] [PubMed] [Google Scholar]
- Paris M, Boyer R, Jaenichen R, Wolf J, Karageorgi M, Green J, Cagnon M, Parinello H, Estoup A, Gautier M, et al. 2020. Near-chromosome level genome assembly of the fruit pest Drosophila suzukii using long-read sequencing. Sci Rep. 10(1):11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova EG, Nuzhdin SV.. 1993. Doc and copia instability in an isogenic Drosophila melanogaster stock. Mol Genet. 240(2):302–306. [DOI] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J.. 2006. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2(12):e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE.. 2003. Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol. 20(6):880–892. [DOI] [PubMed] [Google Scholar]
- Prentis P, Sigg D, Raghu S, Dhileepan K, Pavasovic A, Lowe A.. 2009. Understanding invasion history: genetic structure and diversity of two globally invasive plants and implications for their management. Div Distrib. 15(5):822–830. [Google Scholar]
- Quadrana L, Bortolini Silveira A, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V.. 2016. The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5:e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech GE, Bogaerts-Márquez M, Barrón MG, Merenciano M, Villanueva-Cañas JL, Horváth V, Fiston-Lavier A-S, Luyten I, Venkataram S, Quesneville H, et al. 2019. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. PLoS Genet. 15(2):e1007900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishishwar L, Wang L, Wang J, Yi SV, Lachance J, Jordan IK.. 2018. Evidence for positive selection on recent human transposable element insertions. Gene 675:69–79. [DOI] [PubMed] [Google Scholar]
- Rius N, Guillén Y, Delprat A, Kapusta A, Feschotte C, Ruiz A.. 2016. Exploration of the Drosophila buzzatii transposable element content suggests underestimation of repeats in Drosophila genomes. BMC Genomics 17(1):344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins LA, Richardson MF, Shine R.. 2015. A genetic perspective on rapid evolution in cane toads (Rhinella marina). Mol Ecol. 24(9):2264–2276. [DOI] [PubMed] [Google Scholar]
- Roux JJL, Brown GK, Byrne M, Ndlovu J, Richardson DM, Thompson GD, Wilson JRU.. 2011. Phylogeographic consequences of different introduction histories of invasive Australian Acacia species and Paraserianthes lophantha (Fabaceae) in South Africa. Div Distrib. 17(5):861–871. [Google Scholar]
- Roy M, Viginier B, Saint-Michel É, Arnaud F, Ratinier M, Fablet M.. 2020. Viral infection impacts transposable element transcript amounts in Drosophila. Proc Natl Acad Sci U S A. 117(22):12249–12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CP, Brownlie JC, Whyard S.. 2016. Hsp90 and physiological stress are linked to autonomous transposon mobility and heritable genetic change in nematodes. Genome Biol Evol. 8(12):3794–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115. [DOI] [PubMed] [Google Scholar]
- Sessegolo C, Burlet N, Haudry A.. 2016. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biol Lett. 12(8):20160407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Stapley J, Santure AW, Dennis SR.. 2015. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol. 24(9):2241–2252. [DOI] [PubMed] [Google Scholar]
- StephanW, , Li H.. 2007. The recent demographic and adaptive history of Drosophila melanogaster. Heredity 98(2):65–68. [DOI] [PubMed] [Google Scholar]
- Talla V, Suh A, Kalsoom F, Dinca V, Vila R, Friberg M, Wiklund C, Backström N.. 2017. Rapid increase in genome size as a consequence of transposable element hyperactivity in wood-white (Leptidea) butterflies. Genome Biol Evol. 9(10):2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van’t Hof AE, Campagne P, Rigden DJ, Yung CJ, Lingley J, Quail MA, Hall N, Darby AC, Saccheri IJ.. 2016. The industrial melanism mutation in British peppered moths is a transposable element. Nature 534(7605):102–105. [DOI] [PubMed] [Google Scholar]
- Vendrell-Mir P, Barteri F, Merenciano M, González J, Casacuberta JM, Castanera R.. 2019. A benchmark of transposon insertion detection tools using real data. Mob DNA 10(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira C, Lepetit D, Dumont S, Biémont C.. 1999. Wake up of transposable elements following Drosophila simulans worldwide colonization. Mol Biol Evol. 16(9):1251–1255. [DOI] [PubMed] [Google Scholar]
- Villanueva‐Cañas JL, Rech GE, Cara MAR, González J.. 2017. Beyond SNPs: how to detect selection on transposable element insertions. Methods Ecol Evol. 8(6):728–737. [Google Scholar]
- Wright SI, Agrawal N, Bureau TE.. 2003. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 13(8):1897–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanni V, Eymery A, Coiffet M, Zytnicki M, Luyten I, Quesneville H, Vaury C, Jensen S.. 2013. Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc Natl Acad Sci U S A. 110(49):19842–19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-Y, Zhang D-Y, Barrett S.. 2010. Genetic uniformity characterizes the invasive spread of water hyacinth (Eichhornia crassipes), a clonal aquatic plant. Mol Ecol. (9):191774–1786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The reference assembly used in this article is available on NCBI website (https://www.ncbi.nlm.nih.gov/genome/18317?genome_assembly_id=910956) (Paris et al. 2020). Population sequencing data are available online (SRA repository under the BioProject accession number PRJNA576997) (Olazcuaga et al. 2020). The generated TE database and scripts are available in a github (https://github.com/vmerel/Dsu-TE).