Abstract
The number of olfactory receptor genes (ORs), which are responsible for detecting diverse odor molecules varies extensively among mammals as a result of frequent gene gains and losses that contribute to olfactory specialization. However, how OR expansions/contractions in fish are influenced by habitat and feeding habit and which OR subfamilies are important in each ecological niche is unknown. Here, we report a major OR expansion in a freshwater herbivorous fish, Megalobrama amblycephala, using a highly contiguous, chromosome-level assembly. We evaluate the possible contribution of OR expansion to habitat and feeding specialization by comparing the OR repertoire in 28 phylogenetically and ecologically diverse teleosts. In total, we analyzed > 4,000 ORs including 3,253 intact, 122 truncated, and 913 pseudogenes. The number of intact ORs is highly variable ranging from 20 to 279. We estimate that the most recent common ancestor of Osteichthyes had 62 intact ORs, which declined in most lineages except the freshwater Otophysa clade that has a substantial expansion in subfamily β and ε ORs. Across teleosts, we found a strong association between duplications of β and ε ORs and freshwater habitat. Nearly, all ORs were expressed in the olfactory epithelium (OE) in three tested fish species. Specifically, all the expanded β and ε ORs were highly expressed in OE of M. amblycephala. Together, we provide molecular and functional evidence for how OR repertoires in fish have undergone gain and loss with respect to ecological factors and highlight the role of β and ε OR in freshwater adaptation.
Keywords: Cyprinid fish, comparative genomics, olfactory receptor, evolutionary dynamics, ecological adaptation
Introduction
Olfaction is an important physiological function in animals because of its role in foraging, mate selection and avoiding predators or poisonous agents (Hara 1975; Su et al. 2009; Bazáes et al. 2013; Hughes et al. 2018). The vertebrate olfactory system is able to detect and discriminate various odor molecules in the environment using the multigene family of olfactory receptor genes (ORs). Vertebrate ORs belong to the family of G-protein-coupled receptors that are composed of seven α-helical transmembrane (TM) regions with conserved motifs (Mombaerts 1999). OR genes are predominantly expressed in sensory neurons of the main olfactory epithelium (OE) in nasal cavities both in mammals (Vassar et al. 1993; van der Linden et al. 2018) and fish (Churcher et al. 2015; Cong et al. 2019). The diversity and large number of OR genes facilitate the discrimination of a diverse range of environmental odor particles and are thought to be critical for adaptation to local environmental conditions.
The number of intact OR genes varies extensively among species of placental mammals ranging from ∼300 in orangutan to ∼2,000 in African elephants, and most species have a substantial number of OR pseudogenes (Niimura et al. 2014). The dramatic differences in OR repertoire and gene numbers among vertebrates result from frequent gene gains and losses through duplication and pseudogenization during evolution (Vandewege et al. 2016; Hughes et al. 2018; Niimura et al. 2018). As a consequence, such dynamic evolution of OR repertoire likely facilitates adaptation to different ecological niches (e.g., feeding ecology and habitat) (Hayden et al. 2010, 2014; Vandewege et al. 2016; Hughes et al. 2018). In mammals, aquatic and terrestrial species differ in total number of OR genes per gene family (Hayden et al. 2010, 2014) and the OR repertoire has not only expanded but also contracted in association with changes to local environmental conditions (Hughes et al. 2018). Together, these results suggest that OR gene expansion has played an important role in ecological adaptation in mammals, but comparatively less is known about the role of water-soluble OR in ecological adaptation in fish.
OR genes in vertebrates are classified into nine subfamilies, α, β, γ, δ, ε, ζ, η, θ, and κ (Niimura and Nei 2005; Niimura 2009). Most mammalian OR genes belonging to subfamily α and λ, known as “mammalian-like” genes, are expressed in air-filled medial diverticulum and responsible for detecting airborne odorants, whereas fish express subfamily δ, ε, ζ, and η, referred to as “fish-like” genes, are expressed in water-filled lateral diverticulum and associated with detecting water-soluble odorants. These water-soluble odorants mainly include amino acids, bile acids, gonadal steroids, and prostaglandins, which are nonvolatile (Hara and Zhang 1996; Cong et al. 2019). Because subfamily β OR genes were present both in aquatic and terrestrial vertebrates, they are recognized as both water soluble and airborne odorants (Niimura 2009).
Fish represent one of the largest vertebrate groups with at least 20,000 known species, colonizing tropical, temperate, and polar waters as well as virtually all fresh-water environments. Like other vertebrates, the fish olfactory system is critical for behavior related to feeding, reproduction, predator avoidance and odorant-oriented migration (Laberge and Hara 2001; Hamdani and Døving 2007). The recent availability of a wide taxonomic breadth of fish genome sequences offers an opportunity to explore the evolution of OR repertoires in this group. To date, most previous fish OR studies only focused on the gene identification, whereas the evolutionary dynamics of fish OR gene families and their role in adaptation to different ecological niches are completely unknown.
We tested the hypothesis that the evolution of fish OR gene repertoire has been influenced by habitat and feeding habit and identified which OR gene subfamilies are important for each ecological niche. We generated a high-quality reference genome assembly of a commercially important herbivorous fish, the blunt snout bream (Megalobrama amblycephala), to explore the OR gene family repertoire across 28 phylogenetic and ecologically diverse fish species.
Results
Genome Sequencing Assembly and Annotation
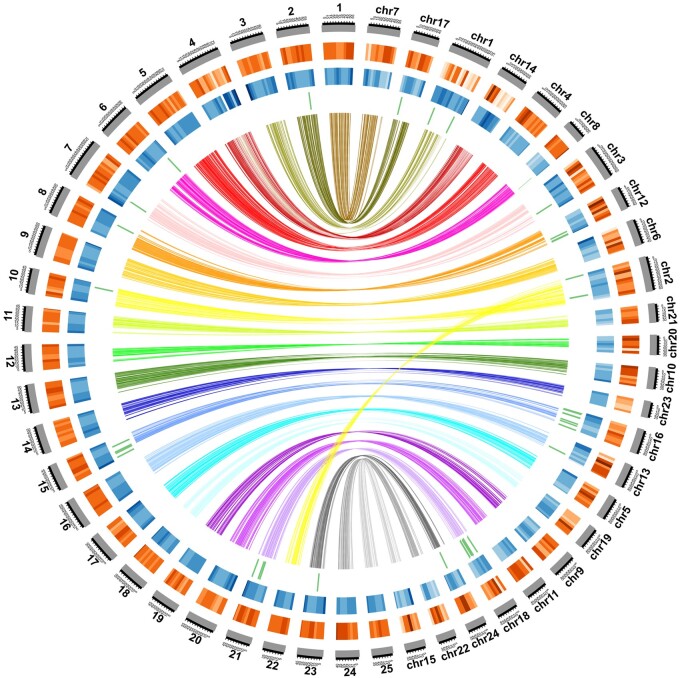
We produced a high quality reference genome using a combination of PacBio long reads (supplementary table 1, Supplementary Material online) and Illumina mate pair and paired end sequencing. The assembled genome includes 1,522 contigs and 868 scaffolds with a total length of 1.11 Gb (supplementary table 2, Supplementary Material online). BUSCO analysis identified 96.1% complete and 1.3% partial genes from the 4,584-gene Actinopterygii reference data set (supplementary table 3, Supplementary Material online). We used our previously published high-density linkage map (Liu et al. 2017) to order scaffolds and generate a chromosome-level assembly. Of 868 scaffolds in the assembly, 650 anchored onto 24 chromosomes with a total length of 1,078 Mb, representing 97.2% of the assembled genome sequences (table 1; supplementary table 4, Supplementary Material online). Although M. amblycephala shared a most recent common ancestor (MRCA) with Danio rerio ∼54 Ma (Liu et al. 2017), the genome of M. amblycephala retained strong collinearity with that of D. rerio (fig. 1). No large interchromosomal translocation was found between the 25 chromosomes of D. rerio and the 24 chromosomes of M. amblycephala. One notable difference is that M. amblycephala Chr2 corresponds to D. rerio Chr10 and 22 (fig. 1). We used Maker (Holt and Yandell 2011) to annotate the novel reference genome and quantify OR genes copies in M. amblycephala. In doing so, we discovered a notable expansion of OR genes that are mainly located on Chr16 and 18 (fig. 1). To evaluate whether this expansion was associated with ecological factors, we performed comparative analysis across other fish species with variation in habitat.
Table 1.
Summary statistics for different assemblies of the M. amblycephala genome
| V1.0 | V2.0 | |
|---|---|---|
| Total genome size (Mb) | 1,116 | 1,109 |
| N50 length of scaffold (kb) | 840 | 3,154 |
| N50 length of contig (kb) | 49 | 2,397 |
| Total GC content (%) | 37.30 | 37.64 |
| Complete BUSCOs (%) | 81.4 | 96.1 |
| Fragmented BUSCOs (%) | 9.1 | 1.3 |
| Protein-coding genes number | 23,696 | 30,357 |
| Average gene length (bp) | 15,797 | 16,093 |
| Number of markers in genetic map | 5,317 | 13,653 |
| Scaffolds anchored on linkage groups (LGs) | 1,434 | 650 |
| Length of scaffolds anchored on LGs (Mb) | 779.54 (70.0%) | 1,078 (97.2%) |
V1.0 is the version published in Liu et al. (2017); V2.0 is the final hybrid assembly in this study.
Fig. 1.
Whole genome alignment between M. amblycephala (right) and D. rerio (left). From inside to outside: green bars, OR genes; blue bars, GC content within a 50-kb sliding window; orange bars, gene distribution on each chromosome; gray, the genetic linkage map.
Identification of or Repertoires Across Fish Genomes
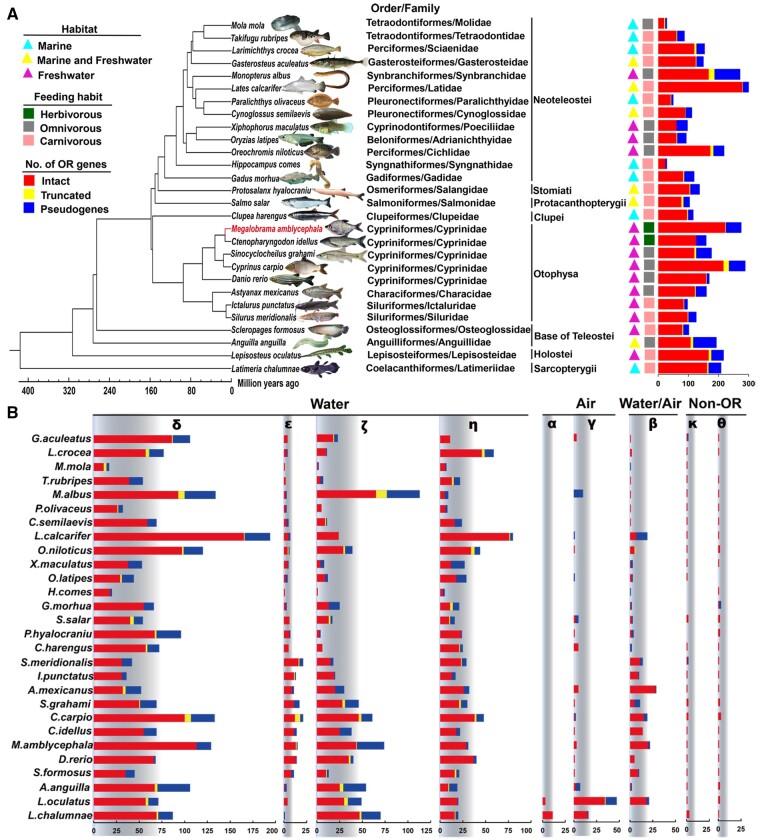
We identified the olfactory genes in the genome assemblies of M. amblycephala and 27 other fish species for which deep-coverage genome sequences are available (supplementary table 5, Supplementary Material online). These 28 species span 19 different fish orders and include seven other Otophysa species in addition to M. amblycephala (fig. 2A). Following an extensive homology search and manual curation, we identified 4,288 OR genes and classified them into three categories, intact genes (putatively functional genes, n = 3,253), truncated genes (n = 122), and pseudogenes (n = 913, fig. 2A). The proportion of pseudogenes among fish species was highly variable and ranged from 5.3% in D. rerio to 37.8% in Xiphophorus maculatus. We also found extensive variation in the size of OR repertoires and the number of intact genes ranged from 20 in Mola to 279 in Lates calcarifer while the reference, M. amblycephala, contained 223 intact OR genes (fig. 2A). The number of OR genes varies in a lineage-specific manner, for example, as in Tetraodontiformes, M. mola and Takifugu rubripes which have low numbers of OR (20 and 61, respectively) whereas in Cypriniformes we found more than 120 OR genes in each species.
Fig. 2.
Phylogenetic tree and number of OR genes in 28 fish species. (A) Phylogenetic tree depicting the evolutionary relationships of 28 fish species. Genome of M. amblycephala sequenced in this study is highlighted in red. Habitat and feeding habits are depicted for each species. Bars represent the total OR numbers. (B) The number of OR genes from each subfamily across 28 fish species. Red, yellow and blue bars represent intact genes, truncated genes and pseudogenes, respectively. “Water,” “Air,” and “Water/Air” represent the detection of water-soluble, airborne and both water and airborne odorants, respectively.
OR Gene Family Phylogeny and Classification
To examine the evolutionary relationships and classify the subfamilies of OR genes, we constructed a Maximum Likelihood (ML) tree using 3,253 amino acid sequences from 28 fish genomes (supplementary fig. 1, Supplementary Material online). Here, we restricted our analysis to only include intact genes because most pseudogenes contained deletions and truncated genes were much shorter than the intact genes. The OR genes clearly separated into nine a priori subfamilies α, β, γ, δ, ε, ζ, η, θ, and κ (Niimura 2009) and the major clades of the tree were supported by high bootstrap values (more than 70% in 1,000 replicates). The δ subfamily is the largest subfamily accounting for more than 50% of the total number of identified ORs followed by subfamily ζ and η. OR genes generally form tandem arrays that are highly conserved across distantly related species (supplementary fig. 2, Supplementary Material online).
The number of genes belonging to OR subfamilies is highly variable among fish species (fig. 2B;supplementary table 6, Supplementary Material online). OR genes belonging to subfamily α are almost completely absent in fish with the exception of 3 and 11 copies in Lepisosteus oculatus and Latimeria chalumnae. Interestingly, the airborne odorants subfamily γ of OR genes, is present in low numbers with one to five copies in some teleost fish (Gasterosteus aculeatus, Oreochromis niloticus, Salmo salar, Protosalanx hyalocranius, Clupea harengus, Astyanax mexicanus, Scleropages formosus, Anguilla, and five Cyprinid fishes) but with 34 copies in L. oculatus and 15 in L. chalumnae (fig. 2B;supplementary table 6, Supplementary Material online). Subfamily δ, ε, ζ, η, and β OR genes are abundant in all tested fish (supplementary table 6, Supplementary Material online). Particularly notable expansions include β and ε OR genes, which have expanded in eight Otophysa fish species with 5 − 28 and 9 − 16 copies, respectively (fig. 2B;supplementary table 6, Supplementary Material online).
Gains and Losses of OR Genes in Fish during Evolution
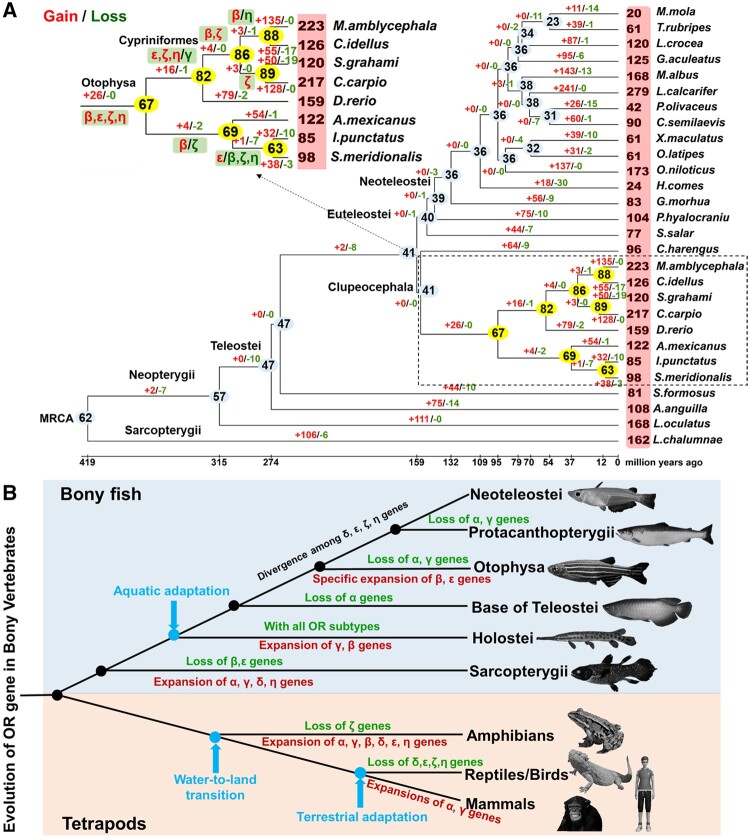
To investigate the evolutionary dynamics in the number of OR genes in teleost, we estimated the OR repertoire size in ancestral species and calculated the numbers of gene gains and losses for each branch during the evolution and speciation of the 28 fish species. The results showed that gains and losses of OR genes have occurred frequently in each taxonomic lineage (fig. 3A). Two species with similar numbers of OR genes may have very different OR gene repertoires. For example, X. maculatus and Oryzias latipes both have 61 OR genes at present, whereas their common ancestor was estimated to have 32 (fig. 3A). Similarly, in Silurus meridionalis and Ictalurus punctatus, each species has ∼90 ORs, but only ∼60 is shared between them. The MRCA of Osteichthyes was estimated to have had 62 intact ancestral OR genes with all subfamilies represented. Several major gain and loss events happened, including the loss of 10 OR genes in the Teleostei lineage and 26 gained in the Otophysa clade when compared with their MRCA, Neopterygii and Clupeocephala lineage, respectively. These 26 gained OR genes belong to subfamily β, ε, ζ, and η (fig. 3A).
Fig. 3.
Evolutionary dynamics of OR genes in fish and tetrapod. (A) Changes in the number of OR genes during the evolution of 28 fish species inferred from intact genes. Numbers in the red rectangular box indicates the number of intact OR genes in each fish species. The estimated number in light blue and yellow oval represents the number of OR genes at an ancestral node. Estimated numbers of gene gains and gene losses in each branch are also shown with red plus and green minus signs. (B) Schematic illustration of the evolution of OR gene families in bony vertebrates.
In mammals and birds, the number of subfamily α and γ subfamilies of OR genes are preferentially expanded (fig. 3B), whereas subfamily δ, ε, ζ, and η were completely lost (Niimura 2009). Amphibians retain nearly all OR subfamilies except ζ which are sensitive to both water-soluble and airborne odorants (Niimura 2009), consistent with their aquatic and terrestrial lifecycles. The L. oculatus genome also contains all subfamilies of OR genes.
Phylogenetic Generalized Least-Squares Regression Analysis
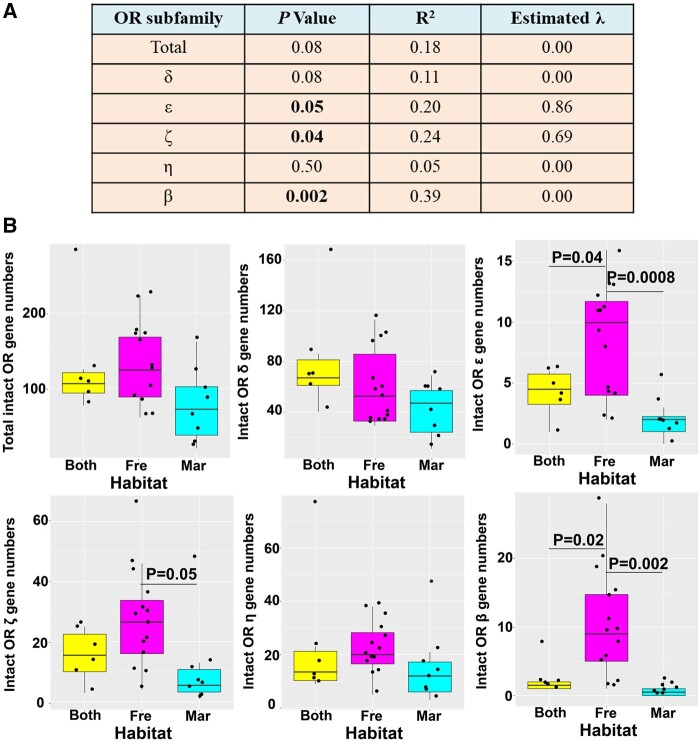
To investigate whether OR repertoire size is related to the ecological factors, we performed a phylogenetic generalized least squares (PGLS) regression analyses. We used two ecological niche factors as predictor variables for this analysis: Including habitat (marine, freshwater [FW] and both marine and FW) and feeding habit (carnivorous, omnivorous, and herbivorous) (supplementary note 1, Supplementary Material online). Although the habitat was not a significant predictor of the total number of intact OR genes, we found a strong effect of habitat on the number of subfamily β (Pagel’s λ = 0, P = 0.002), ε (Pagel’s λ = 0.86, P = 0.05), and ζ (Pagel’s λ = 0.69, P = 0.04) OR genes (fig. 4A). However, we did not find a significant association between the number of OR repertoires and feeding habits (supplementary fig. 3A, Supplementary Material online).
Fig. 4.
OR genes subfamilies were compared among fish with different habitats. (A) Phylogenetic generalized least squares (PGLS) regression analysis for number of intact OR genes in fish species (n = 28) versus ecological niches (habitats). (B) Box plot of intact OR gene numbers among groups of divergent ecological niches. One-way ANOVA analysis and Tukey HSD test were conducted to determine statistical difference among groups. Mar, marine; Fre, freshwater; Both, both in marine and freshwater.
To further explore the difference of OR repertoires among specified niches groups, one-way ANOVA analysis was performed (fig. 4B). The results indicated that the numbers of subfamily β (P = 0.002), ε (P = 0.0008), and ζ (P = 0.05) OR genes in FW species were significantly higher than that in marine fish species. Additionally, the number of subfamily β (P = 0.02) and ε (P = 0.04) genes in FW fish were also significantly higher than in fish living in both FW and marine water. For feeding habits, only the number of β OR genes in the herbivorous species was significantly higher than in carnivorous species (P = 0.05) (supplementary fig. 3B, Supplementary Material online).
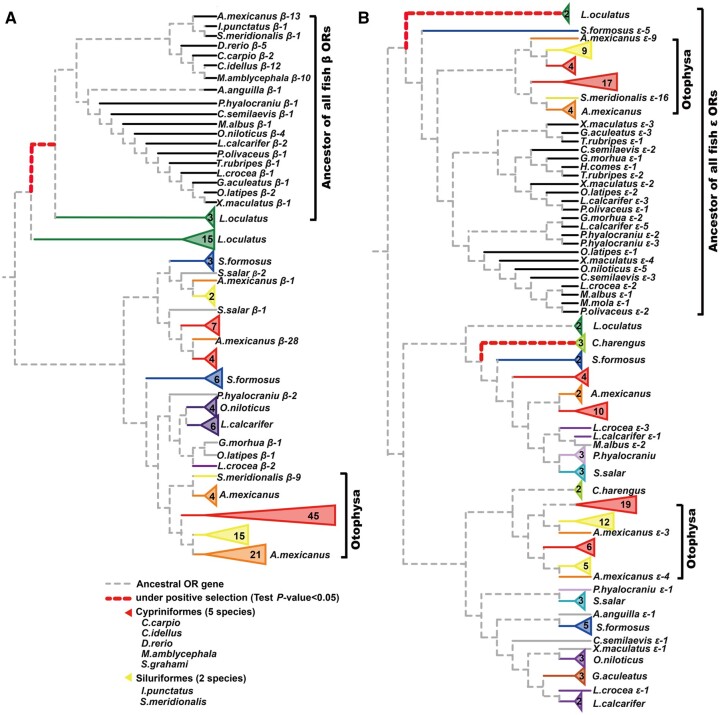
Molecular Evolution of Subfamily β and ε
Our phylogenetic analysis of 164 β and 161 ε OR genes in fish uncovered lineage-specific expansions in M. amblycephala and the other fish species of the Otophysa lineage (fig. 5; supplementary figs. 4 and 5, Supplementary Material online). The majority of fish species included in this study have one or two copies of β OR genes, whereas all Otophysa species show an expansion of the β subfamily (more than 10 on average). Specifically, multiple highly supported clades of β OR genes suggest recurrent and lineage-specific expansions for Otophysa β ORs (fig. 5A). Similarly, genes in subfamily ε also independently duplicated multiple times in the Otophysa lineage (fig. 5B). To test whether these duplicated copies are under selection, we used an adaptive branch-site random effects likelihood (aBSREL) model to explore the presence/absence of selection by measuring rates of nonsynonymous to synonymous (dN/dS = ω) substitutions in genes. We found that four and three branches show strong support being under significant positive selection (test P-value < 0.05) in β and ε ORs trees, respectively (fig. 5A;supplementary table 7, notes 2 and 3, Supplementary Material online). Of note, ORs of the subfamily β show evidence of positive selection in the ancestor of fish species included in this study, but not in branches that would represent lineage-specific expansions (fig. 5).
Fig. 5.
Molecular evolution of subfamily β (A) and ε (B) OR genes in 28 fish species. Fish from the same order are labeled with same color. The red bold dashed lines in the branches of the trees have P < 0.05 (corrected for multiple testing) considered to have experienced diversifying positive selection.
Expression Patterns of OR Genes
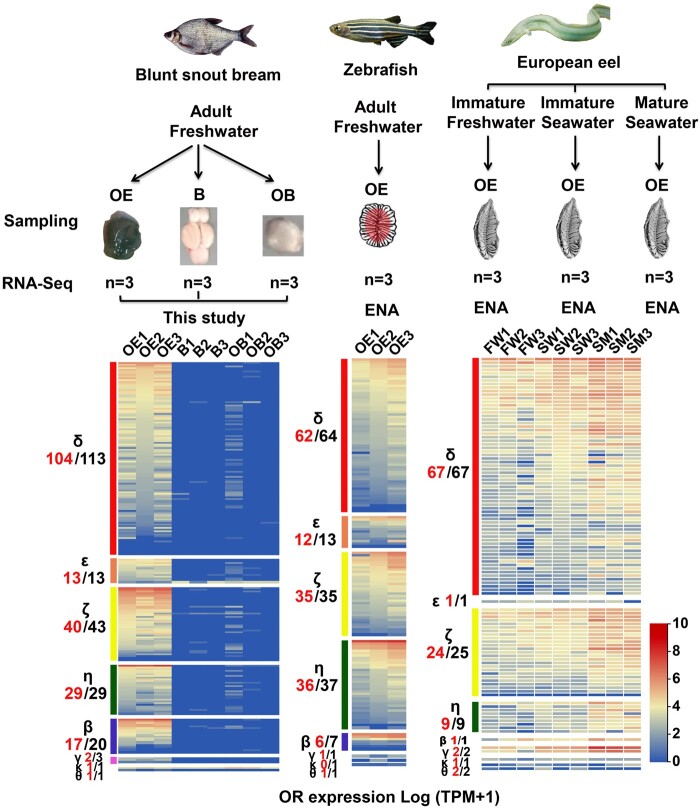
To determine which of the OR genes are being used, we performed RNA-seq on the OE, whole brain (B), and olfactory bulb (OB) of adult M. amblycephala and analyzed previously reported OE transcriptome data from adult D. rerio and of different life stages of A. anguilla (fig. 6). Our results demonstrate that the majority of candidate OR genes are expressed in the OE (fig. 6). In M. amblycephala, OR genes were not expressed in brain and OB, whereas 207 of 223 intact ORs were expressed in OE. In D. rerio, 153 of 159 intact ORs were expressed in OE. In A. anguilla, all the OR genes were expressed in OE except one ζ subfamily. Importantly, the expression level of the majority ORs of A. anguilla were higher in sexually mature male (SM) than in sexually immature FW and seawater (SW) OE. These results suggest that most intact ORs were expressed and putatively functional in the OE of these species.
Fig. 6.
Gene expression profiles of ORs in M. amblycephala, D. rerio and A. anguilla. A total of nine sequencing libraries, including three olfactory epithelium (OE), three whole brain (B) and three olfactory bulb (OB) of M. amblycephala were sequenced. Three RNA-seq raw data of OE from adult D. rerio and nine OE from A. anguilla were also analyzed. RNA expression levels are represented on a log2(X + 1) scale of normalized TPM (0, not expressed; 10, highly expressed). OR genes in each subfamily are displayed in descending order of their expression values. Numbers labeled in red represent the number of expressed OR genes while numbers in black are the total number of intact genes in each subfamily.
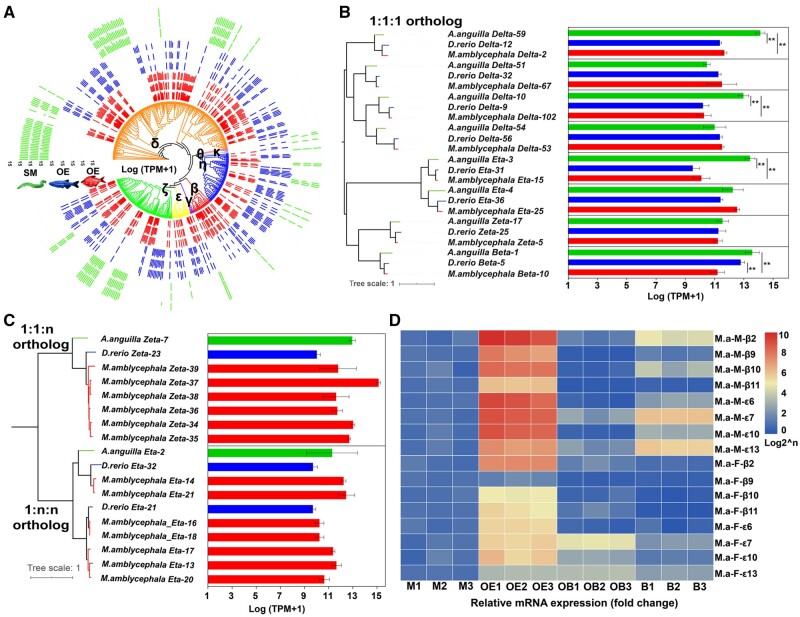
To evaluate whether there are differences in OR expression levels among the three species, we plotted phylogenetic trees generated from all intact OR sequences overlaid with the normalized values corresponding to their expression (fig. 7A). Half of eight identified 1:1:1 OR orthologs were expressed at significantly higher levels (P < 0.01) in A. anguilla than in M. amblycephala OE (fig. 7B). Among the 1:1:n and 1:n:n orthologs, nearly all the duplicated ORs are highly expressed (fig. 7C). To further validate the transcriptome results and verify the functional significance of subfamily β and ε OR genes, we selected eight representative β and ε ORs in four tissues (muscle, OE, OB, and brain) of adult male and female M. amblycephala using real-time quantitative PCR (qRT-PCR) (fig. 7D). All the tested β and ε OR genes were highly expressed in OE of male and female M. amblycephala (fig. 7D), whereas these were not expressed in muscle tissue, OB and brain except β2 ε7, ε13 with slight expression in male M. amblycephala brain. The expression trend of these ORs revealed using qRT-PCR analysis was consistent with that detected in the OE transcriptome analysis.
Fig. 7.
Comparison of OR genes expression patterns among different fish species. (A) Unrooted phylogenetic tree and normalized expression levels for all intact OR genes among M. amblycephala, D. rerio and A. anguilla. Bars indicate the relative expression level represented on a log2(X + 1) scale of normalized TPM. (B) and (C) Phylogenetic tree (left) and expression levels (right) of 1:1:1 and 1: n: n OR orthologs in A. anguilla, D. rerio and M. amblycephala. Bars showed the RNA-seq expression values (mean ± SD, n = 3) for the OR genes in fish OE. One-way ANOVA analysis and Tukey HSD test were conducted to determine statistical difference among species (**P < 0.01). (D) The relative expression levels of representative β and ε OR genes detected by qRT-PCR. Expression in olfactory epithelium (OE), olfactory bulb (OB) and brain tissues (B) were normalized to the expression in muscle (M). M.a-M, male M. amblycephala; M.a-F, female M. amblycephala.
Discussion
Sensory systems play a crucial role in the aquatic lifestyle of fish in feeding, migration, spawning, and defense (Hara 1975). Gene expansion in ORs is thought to have played an important role in the transition from aquatic to terrestrial living in mammals (Niimura 2009; Hayden et al. 2010), but the role of ORs expansion among aquatic species has not been previously studied using comparative evolutionary analysis. Using a high-quality genome assembly of M. amblycephala and data from 27 taxonomically diverse fish species, we found that the numbers of intact OR genes have changed extensively during fish evolution. We find evidence for an expansion of OR genes in FW species, in particular as regards members of the β and ε subfamilies, which most likely reflects ecological adaptation.
Genomic surveys have revealed that ORs represent the largest gene family in mammals (Zhang and Firestein 2002) and some studies indicate that OR repertoires vary widely among vertebrates even between closely related taxa (Matsui et al. 2010; Niimura et al. 2014, 2018). The variation in the number of intact OR genes is >10-fold among the 28 teleosts examined in this study, ranging from 20 in M. mola to 279 in L. calcarifer (supplementary table 6, Supplementary Material online). This result is consistent with previous analyses of the total number of OR genes from the whole genome data of T. rubripes (44 ORs), Tetraodon nigroviridis (42 ORs) (Alioto and Ngai 2005), D. rerio (167 ORs), Hippocampus comes (26 ORs) (Lin et al. 2016), and Larimichthys crocea (112 ORs) (Ao et al. 2015). The MRCA between fish and tetrapods had at least nine subfamilies of OR genes (Niimura and Nei 2005) and at least six (δ, ε, ζ, η, β, and γ) out of the seven classified subfamilies of intact OR genes are retained in fish (fig. 3; supplementary table 6, Supplementary Material online). The pattern is opposite in mammals, where only two subfamilies (α and γ) are retained, but their copy numbers are greatly expanded (Niimura 2009).
Most fish possess a well-developed olfactory system and have a relatively large OB and OE. But why do the number of intact OR genes vary so much among fish species? We predicted that OR genes might vary according to different habitats and lifestyles. Gene duplications followed by subfunctionalization is one possible mechanism for genetic adaptation (Chang and Duda 2012). Comparative studies of OR gene families have demonstrated birth and death of genes as a model of OR evolution involving tandem duplications and chromosomal rearrangements (Niimura and Nei 2003; Nei and Rooney 2005; Hughes et al. 2018; McKenzie and Kronauer 2018). Analysis of three cyprinid fishes showed that fish ORs are found in several genomic clusters (supplementary fig. 2, Supplementary Material online). Within gene clusters, different subfamilies of OR genes are largely contiguous and located close to each other and with the same transcriptional orientation, suggesting tandem duplication as a mechanism of gene expansion, consistent with a previous fish study (Alioto and Ngai 2005).
Studies on reptiles, birds and mammals have suggested that expansion and contraction of OR genes are related to major changes in habitat and lifestyle (Niimura 2009; Hayden et al. 2010, 2014; Khan et al. 2015; Vandewege et al. 2016; Hughes et al. 2018). For example, previous study explored OR diversity in 2 reptiles and 48 birds, which indicated species and lineage-specific variation in subfamily of OR genes associated with aquatic and terrestrial adaptations (Khan et al. 2015). Similarly, a study on OR genes in sauropsida suggests that adaption related to life history evolution have shaped the unique OR repertoires in different members of this subfamily (Vandewege et al. 2016). Our taxonomically diverse sampling allowed us to assess the relationship between habitat and feeding behavior in relation to OR gene expansion in fish. Transitions to different aquatic environments may be facilitated by the olfactory sensory organs and OR genes if olfaction is critical to survival. For example, elimination of visual function resulted in increased olfactory capabilities in the blind cave fish, A. mexicanus (Blin et al. 2018). In the present study, we observed a substantial expansion of subfamily β OR genes in A. mexicanus genome, which may compensate to some extent their loss of visual information (supplementary table 6, Supplementary Material online).
Our phylogenetically controlled environmental association analysis (fig. 4) indicated that although the total number of intact OR genes was unaffected by environmental factors, there is a strong association between the number of subfamily β, ε, and ζ OR genes and FW habitat. Most teleosts possess one or two β OR genes, whereas fish in the Otophysa lineage possess more than 10 on average (fig. 5A;supplementary table 6, Supplementary Material online). Why might β OR genes have specifically expanded in FW Otophysa species? A possible explanation for this observation is that subfamily β OR genes are important for detecting both water soluble and airborne odorants (Niimura 2009). The large number of β and ε OR genes in the FW Otophysa lineage may allow them to detect and discriminate a wide range of odorant stimuli that are important for survival in FW. Previous research has suggested that expansion or contraction of gene families can be a random process driven by drift and/or by natural selection. Earlier studies highlighted that positive selection shaped the diversification and variation of the OR gene family in ants (Engsontia et al. 2015), bees (Brand and Ramírez 2017), and fish (Alioto and Ngai 2005; Hussain et al. 2009). However, in this study, no signatures of positive selection were found in the β and ε ORs showing copy number expansion (fig. 5).
Calculating total mRNA abundance of each OR permitted us to assess which receptors are functionally expressed in the OE transcriptomes of M. amblycephala and D. rerio (FW fish), as well as A. anguilla (both living in FW and SW). We present evidence that almost all the intact OR genes belonging to different subfamilies are expressed and putatively functional in OE of these species (fig. 6). Similar OR expression patterns have also been observed in the mouse (Ibarra-Soria et al. 2014) and human (Olender et al. 2016). Although the number of intact ORs (108) in A. anguilla is lower than in D. rerio (159 intact ORs) and in M. amblycephala (223 intact ORs), the expression levels of four out of eight 1:1:1 OR orthologs in SW stage A. anguilla were significantly higher than that in FW M. amblycephala (fig. 7B). Previous work describing differential expression of OR genes in OE of wild Atlantic salmon (Johnstone et al. 2011) and European eel (Churcher et al. 2015) suggests that the regulation of these genes is associated with different physiological states and responses to environmental cues.
In summary, we present the first phylogenetic comparative analysis of OR genes expansion and contraction in fish. We show that expansions of subfamily β and ε OR genes have occurred in the FW Otophysa lineage and that β and ε OR genes expansion is consistently associated with FW habitats in other species. Gene expression analyses indicated that nearly all the intact OR genes including expanded subfamily β and ε OR genes in FW fish are expressed in olfactory organs, strongly supporting their functional importance, which have potentially facilitated FW adaptation.
Materials and Methods
Genome Sequencing and Assembly
Genomic DNA was isolated from whole blood of a double haploid fish as described in our previous study (Liu et al. 2017). The genomic DNA was fragmented to 20 kb and sequenced with the Pacific Biosystems RSII platform. The genome assembly was performed using a combination of sequencing technologies: PacBio RS II reads, Illumina paired-end reads (PE), and Illumina mate-pair reads (MP). Briefly, high-quality Illumina PE reads were separately assembled into Illumina contigs using Platanus (v1.2.4) (Kajitani et al. 2014). Next, low-quality PacBio subreads with a read length shorter than 1,000 bp or a quality score lower than 0.8 were filtered out. The remaining PacBio subreads were error-corrected with MECAT (v 1.0) (Xiao et al. 2017). Then, the error-corrected PacBio reads and Illumina contigs were combined to perform a hybrid assembly using the DBG2OLC (Ye et al. 2016) pipeline. Illumina reads (65× coverage) were mapped to the contigs using BWA-aln. This alignment was then used to correct the assembly with Pilon 1.22 (Walker et al. 2014). A total of 69.15 Gb of Illumina MP data (approximately 61×), with an insert size varying between 2 and 20 kb, was used to scaffold the assembly with SSPACE (v3.0) (Boetzer et al. 2011). Then, the gaps were filled with GapFiller (v1.10) (Nadalin et al. 2012) and PBjelly (v1.2) (English et al. 2012). To further estimate the completeness of the assembly, Core Eukaryotic Genes Mapping Approach was performed using benchmarking universal single-copy orthologs (BUSCO) (SimAo et al. 2015).
Chromosome-Level Assembly and Synteny Analyses between M. amblycephala and D. rerio
A RAD genetic linkage map of M. amblycephala (Liu et al. 2017) composed of 14,648 SNP markers was used to organize and orientate the scaffolds into chromosome-sized sequences. The RAD tags corresponding to these 14,648 SNP markers were mapped onto the genome using BWA-aln. Then, we placed and oriented the scaffolds and contigs relative to each other with the physical and genetic positions of the mapped markers. To identify syntenic blocks, the protein sequences from M. amblycephala and D. rerio were searched against each other using BLASTp (E < 1e−5). The results were subjected to MCScan (–a, –e : 1e–5, –u : 1, –s : 5) to determine syntenic blocks.
Genome Annotation
We used a combination of homology-based, de novo and RNA-seq annotation methods to predict genes in the assembled genome. For the sequence similarity-based prediction, protein sequences from D. rerio, G. aculeatus, L. crocea, L. calcarifer, L. oculatus, O. niloticus, O. latipes, and S. grahami were mapped to M. amblycephala genome using genBlastA v1.0.1 (She et al. 2009) with an E-value threshold of 1e−5. Subsequently, homologous genome sequences were aligned against the matched proteins to define gene models using GeneWise (v2.2.0) (Birney et al. 2004). For the de novo gene predictions, AUGUSTUS (v2.5.5) (Stanke et al. 2006) was used to identify candidate protein-encoding genes in the masked genome with self-trained model parameters. Then unigene sequences from nine transcriptoms sequenced this study were used to search transcripts region using BLAT v. 36 and PASA (Haas et al. 2003). Finally, the homology-based, de novo-derived and transcript gene sets were merged to generate a high confidence gene set using Maker 2.31.10 (Holt and Yandell 2011). To assign functions to the gene models, we performed BLASTP, with an E-value threshold of ≤10−5, using the NR, KOG, SwissProt, and TrEMBL databases (Uniprot release 2017-09). The gene motifs and domains were determined using InterProScan (version 5.16) (Zdobnov and Apweiler 2001) against public protein databases. Functional annotation was carried out using Blast2GO (Conesa et al. 2005).
Phylogenetic Tree Construction and Divergence Time Estimation
We first extracted 22,708 orthogroups conserved among 28 species (C. harengus, C. idellus, C. semilaevis, C. carpio, D. rerio, G. morhua, G. aculeatus, H. comes, I. punctatus, L. crocea, L. calcarifer, L. chalumnae, L. oculatus, M. amblycephala, M. mola, M. albus, O. niloticus, O. latipes, P. olivaceus, P. hyalocraniu, S. salar, S. meridionalis, S. grahami, T. rubripes, X. maculatus, A. anguilla, A. mexicanus, and S. formosus) using OrthoFinder v2.3.11 (Emms and Kelly 2019). Of these, 41 single copy gene families were detected by the following steps. Firstly, we did pairwise alignment among protein sequences through Diamond v0.9.24.125 (Buchfink et al. 2014). Secondly, we use MCL software to cluster the pairwise comparison of protein sequences, do multiply alignment with Mafft (Katoh and Standley 2013) and construct phylogeny tree of each orthogroups through FastTree2 (Price et al. 2010). At last, we set the root by Species Tree Root Inference from Duplication (STRIDE) (Emms and Kelly 2017). Divergence times were calculated by the PAML mcmctree program (Reis and Yang 2011). The parameters were set as the following: the approximate likelihood calculation method, the correlated molecular clock, REV substitution model. And three fossil calibration time used to calculate divergence times of the other branch were: L. chalumnae versus D. rerio 416 − 422 Ma; D. rerio versus G. aculeatus: 149.85 − 165.2 Ma, O. latipes vs G. aculeatus: 96.9 − 150.9 Ma.
Identification of OR Genes from Fish Genome Sequences
To identify OR genes from 28 complete fishes genome (supplementary table 5, Supplementary Material online), we followed a bioinformatics pipeline similar to previously described methods (Niimura and Nei 2007; Niimura 2009) with some modifications. Briefly, we used a first-round of TBLASTN searches (Altschul et al. 1997) with a cutoff E-value of 1e−5 against each fish genome sequence using a set of known functional OR genes sequences from D. rerio, O. latipes, O. niloticus, L. chalumnae, L. oculatus, L. vexillifer, and T. rubripes as queries. We then predicted the structure of the OR genes using the blast-hit sequence with GeneWise (Birney et al. 2004), extending in both 3’ and 5’ directions along the genome sequences. Hits shorter than 200 bp were discarded. All best-hits from the genome sequence were extracted. We classified OR genes with interrupting stop codons or frame-shifts were as pseudogenes. We classified a truncated gene as those with a partially intact sequence encoding a part of an OR and were validated by alignment to functional genes using the program MUSCLE (Edgar 2004). The longest coding sequences from the start (ATG) codon to a stop codon with an uninterrupted open reading frame and seven transmembrane domains were considered as intact OR genes. To classify OR genes into different subfamilies, we used BLASTP to intact ORs into putative subfamilies based on the classification of zebrafish and pufferfish OR genes (Niimura and Nei 2005). A phylogenetic tree was constructed using the ML method in MEGA 5.10 (Tamura et al. 2011) with 1,000 replicates to verify and correct the putative BLASTP-based assignments. Finally, we aligned all identified intact OR genes found in this study using MUSCLE. The alignment was manually corrected and used to construct a phylogenetic tree by FastTree2 (Price et al. 2010). The phylogenetic tree was displayed and labeled using Interactive Tree Of Life (iTOL) v4 (Letunic and Bork 2019). All the identified intact and truncated OR nucleic and translated protein sequences in this study are shown in supplementary notes 4 and 5, Supplementary Material online.
Collinear Analysis of OR Genes Among M. amblycephala, D. rerio, and C. idellus
Syntenic blocks between M. amblycephala and D. rerio, and between M. amblycephala and C. idellus were firstly identified using BLASTp and MCScan following methods described above. Then, the OR genes from these three fish were positioned back on their genome to determine which genes belong to which syntenic region. Finally, variant sites among OR genes located in syntenic region were identified by multiple sequence alignment using MUSCLE.
Estimation of OR Genes Gain and Loss Events
We used CAFE′ (De Bie et al. 2006) to reconstruct the OR repertoires and calculate copy numbers for ancestral OR genes in each lineage using all the intact ORs identified from fish genomes. Firstly, a data file containing the sizes of all the OR gene subfamilies were prepared. Divergence times for each node in the CAFE′ analyses were estimated from the phylogenetic tree (fig. 2A). The CAFE′ method employs a random birth and death model to estimate gene gains and losses in each lineage. The global parameter λ described both the gene birth (λ) and death (μ = −λ) rate across all branches in the tree for all gene subfamilies was estimated using ML. A conditional P-value was calculated for each gene family, and families with conditional P-values of <0.05 were considered to have a notable gain or loss.
Molecular Evolution and Selection Analyses of Subfamily β and ε OR Genes
All intact subfamily β and ε OR genes from 28 fish genomes (β, n = 164 and ε, n = 161) were aligned to prepare the alignments. The OR genes trees were constructed using the ML method in MEGA 5.10. We used the aBSREL approach (Smith et al. 2015) in order to test for evidence of episodic positive selection implemented in Datamonkey (Weaver et al. 2018). In aBSREL, a likelihood ratio test was performed to compare the null model (ω = 1) against the alternative, where the branch was undergoing some form of selection (ω ≠ 1). We used a threshold of P < 0.05 (after correction for multiple testing) to infer positive selection at the marked branches.
PGLS Regression Analysis
We use phylogenetic generalized least-squares regressions (PGLSs) to establish the relationships between the numbers of OR genes and ecological factors (feeding habit and habitat) of each fish species while controlling for phylogenetic effects. Feeding habit was categorized as carnivorous, omnivorous or herbivorous and habitat was categorized as marine, FW , or both marine and FW (supplementary note 1, Supplementary Material online). The PGLS analyses were performed using the R packages “caper” (Orme 2013). The input phylogenetic tree was from our present study (fig. 2A). Here, we used Pagel’s λ (Pagel 1999) with the value ranging from 0 (phylogenetic independence) to 1 (phylogenetic dependence) to estimate the degree of phylogenetic signal of each trait.
RNA-Seq Analysis
The experimental procedures were approved by the Animal Care and Use Committee of Huazhong Agricultural University. The OE, OB, and whole brain (B) were collected from the adult male M. amblycephala and immediately frozen in liquid nitrogen. All samples were prepared in triplicate. Total RNA was isolated from each sample with RNAiso Plus (TaKaRa, Dalian, China). A total of nine RNA-seq libraries were constructed and sequenced in BGI by BGISEQ-500 platform (Shenzhen, China). For comparison, the raw data from OE of adult D. rerio and different life stages, FW, SW, and SM of A. anguilla were downloaded for further analyses. The detailed sampling information is shown in supplementary table 8, Supplementary Material online.
All raw data were assessed using fastp v0.20.0 (Chen et al. 2018). After removing the adapters and poly N or low-quality sequences (Q < 15), the remainder were termed as clean reads. High quality clean reads from each sample were separately aligned to M. amblycephala (this study), D. rerio (GRCz11.99, Ensembl) and A. anguilla (GCA_000695075.1, NCBI) genome using Hitsat2 v2.0.4 (Pertea et al. 2016). Estimated mapped read counts and transcript lengths were used to calculate transcripts per million (TPM) using RSEM (v1.2.25) with the default settings (Li and Dewey 2011). To reduce the variation among the three fish species, the DESeq2 package (Love et al. 2014) was also used to estimate the size factors and dispersion and to generate a normalized counts matrix using the 4,404 single-copy orthologs. TPM values were subsequently transformed to log2 (TPM + 1).
qRT-PCR Analysis
A total of 8 representative β and ε OR genes in four tissues (muscle, OE, OB, and B) of adult male and female M. amblycephala were detected by using qRT-PCR. The OE, OB, B, and muscle tissues were collected from the adult male (n = 9) and female M. amblycephala (n = 9) and immediately frozen in liquid nitrogen. Tissues from three individuals were pooled to extract RNA and three independent biological replicates for male and female were separately prepared. cDNA was synthesized from 1 μg of total RNA using a reverse transcriptase kit from TaKaRa Biochemicals (TaKaRa, Dalian, China). Primers were designed using Primer 5.0 software (supplementary table 9, Supplementary Material online). β-actin served as an internal normalization control for qRT-PCR analysis. PCR reactions contain 1 μl cDNA, 1 μl forward and reverse primers, 10 μl SYBR Green PCR Master Mix (TaKaRa, Dalian, China). A qRT-PCR was performed using a Roter-gene Q (Qiagen, Hilden, Germany) with one cycle of predenaturation at 95 °C for 45 s, followed by 40 cycles of amplification at 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 30 s.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31772901), the Fundamental Research Funds for the Central Universities (2662020SCQD002), the China Agriculture Research System of MOF and MARA (No. CARS-45-08), and the financial support from China Scholarship Council.
Author Contributions
H.L., L.A., and W.W. conceived the project. H.L. analyzed the data and wrote the paper. W.W. initiated and provided scientific input. L.A. and E.D.E. modified the manuscript and discussed the data. C.C. did the collinear analysis and OR genes gain and loss estimation. M.L. and N.L. conducted the biological experiments. Genome assembly and annotation were performed by Y.H. H.Z. identified the OR gene families. The samples for RAD-Seq were prepared by Z.G. All authors read and approved the final manuscript and declare that they have no competing interests.
Data Availability
The data underlying this article (raw data of whole-genome sequencing, genome assembly and RNA-seq data) have been deposited at NCBI BioProject database under Bioproject ID PRJNA343584.
References
- Alioto TS, Ngai J.. 2005. The odorant receptor repertoire of teleost fish. BMC Genomics 6:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao J, Mu Y, Xiang LX, Fan D, Feng M, Zhang S, Shi Q, Zhu L-Y, Li T, Ding Y, et al. 2015. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 11:e1005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazáes A, Olivares J, Schmachtenberg O.. 2013. Properties, projections, and tuning of teleost olfactory receptor neurons. J Chem Ecol. 39:451–464. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and Genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin M, Tine E, Meister L, Elipot Y, Bibliowicz J, Espinasa L, Rétaux S.. 2018. Developmental evolution and developmental plasticity of the olfactory epithelium and olfactory skills in Mexican cavefish. Dev Biol. 441:242–251. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Brand P, Ramírez SR.. 2017. The evolutionary dynamics of the odorant receptor gene family in corbiculate bees. Genome Biol Evol. 9:2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2014. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12:59–60. [DOI] [PubMed] [Google Scholar]
- Chang D, Duda TF Jr. 2012. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol. 29:2019–2029. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher AM, Hubbard PC, Marques JP, Canário AVM, Huertas M.. 2015. Deep sequencing of the olfactory epithelium reveals specific chemosensory receptors are expressed at sexual maturity in the European eel Anguilla. Mol Ecol. 24:822–834. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Cong X, Zheng Q, Ren W, Chéron JB, Fiorucci S, Wen T, Zhang C, Yu H, Golebiowski J, Yu Y.. 2019. Zebrafish olfactory receptors ORAs differentially detect bile acids and bile salts. J Biol Chem. 294:6762–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW.. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 22:1269–1271. [DOI] [PubMed] [Google Scholar]
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2017. STRIDE: species tree root inference from gene duplication events. Mol Biol Evol. 34:3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific biosciences RS long-read sequencing technology. PLoS One 7:e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Robertson HM, Satasook C.. 2015. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Res Notes. 8:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdani EH, Døving KB.. 2007. The functional organization of the fish olfactory system. Prog Neurobiol. 82:80–86. [DOI] [PubMed] [Google Scholar]
- Hara TJ, Zhang C.. 1996. Spatial projections to the olfactory bulb of functionally distinct and randomly distributed primary neurons in salmonid fishes. Neurosci Res. 26(1):65–74. [DOI] [PubMed] [Google Scholar]
- Hara TJ.1975. Olfaction in fish. Prog Neurobiol. 5:271–335. [DOI] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC.. 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 20:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Goodbla A, Murphy WJ, Dávalos LM, Teeling EC.. 2014. A cluster of olfactory receptor genes linked to frugivory in bats. Mol Biol Evol. 31:917–927. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GM, Boston ES, Finarelli JA, Murphy WJ, Higgins DG, Teeling EC.. 2018. The birth and death of olfactory receptor gene families in Mammalian niche adaptation. Mol Biol Evol. 35:1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A, Saraiva LR, Korsching SI.. 2009. Positive Darwinian selection and the birth of an olfactory receptor clade in teleosts. Proc Natl Acad Sci U S A. 106:4313–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Soria X, Levitin MO, Saraiva LR, Logan DW.. 2014. The olfactory transcriptomes of mice. PLoS Genet. 10:e1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone KA, Lubieniecki KP, Koop BF, Davidson WS.. 2011. Expression of olfactory receptors in different life stages and life histories of wild Atlantic salmon (Salmo salar). Mol Ecol. 20(19):4059–4069. [DOI] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, Yang Z, Maldonado E, Li C, Zhang G, Gilbert MTP, Jarvis ED, O’brien SJ, Johnson WE, Antunes A.. 2015. Olfactory receptor subgenomes linked with broad ecological adaptations in sauropsida. Mol Biol Evol. 32:2832–2843. [DOI] [PubMed] [Google Scholar]
- Laberge F, Hara TJ.. 2001. Neurobiology of fish olfaction: a review. Brain Res Rev. 36:46–59. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Fan S, Zhang Y, Xu M, Zhang H, Yang Y, Lee AP, Woltering JM, Ravi V, Gunter HM, et al. 2016. The seahorse genome and the evolution of its specialized morphology. Nature 540:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen C, Gao Z, Min J, Gu Y, Jian J, Jiang X, Cai H, Ebersberger I, Xu M, et al. 2017. The draft genome of blunt snout bream (Megalobrama amblycephala) reveals the development of intermuscular bone and adaptation to herbivorous diet. Gigascience 6:gix039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Go Y, Niimura Y.. 2010. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol Biol Evol. 27:1192–1200. [DOI] [PubMed] [Google Scholar]
- McKenzie SK, Kronauer DJ.. 2018. The genomic architecture and molecular evolution of ant odorant receptors. Genome Res. 28:1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P.1999. Seven transmembrane proteins as odorant and chemosensory receptors. Science 286:707–711. [DOI] [PubMed] [Google Scholar]
- Nadalin F, Vezzi F, Policriti A.. 2012. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics 13:S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP.. 2005. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 39:121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K.. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K.. 2018. Acceleration of olfactory receptor gene loss in primate evolution: possible link to anatomical change in sensory systems and dietary transition. Mol Biol Evol. 35:1437–1450. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. 2003. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci U S A. 100:12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. 2005. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A. 102:6039–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One 2(8):e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y.2009. On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol Evol. 1:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender T, Keydar I, Pinto JM, Tatarskyy P, Alkelai A, Chien MS, Fishilevich S, Restrepo D, Matsunami H, Gilad Y, et al. 2016. The human olfactory transcriptome. BMC Genomics 17(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D, Freckleton R, Thomas G, Petzoldt T.. 2013. The caper package: comparative analysis of phylogenetics and evolution in R. R Package Version 5:1–36. [Google Scholar]
- Pagel M.1999. Inferring the historical patterns of biological evolution. Nature 401(6756):877–884. [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL.. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis MD, Yang Z.. 2011. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 28:2161–2172. [DOI] [PubMed] [Google Scholar]
- She R, Chu JSC, Wang K, Pei J, Chen N.. 2009. genBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 19:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO online supplementary information: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL.. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B.. 2006. AUGUSTUS: a b initio prediction of alternative transcripts. Nucleic Acids Res. 34:435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CY, Menuz K, Carlson JR.. 2009. Olfactory perception: receptors, cells, and circuits. Cell 139:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden C, Jakob S, Gupta P, Dulac C, Santoro SW.. 2018. Sex separation induces differences in the olfactory sensory receptor repertoires of male and female mice. Nat Commun. 9:5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewege MW, Mangum SF, Gabaldón T, Castoe TA, Ray DA, Hoffmann FG.. 2016. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol Evol. 8:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R.. 1993. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74:309–318. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL.. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol. 35:773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao CL, Chen Y, Xie SQ, Chen KN, Wang Y, Han Y, Luo F, Xie Z.. 2017. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods. 14:1072–1074. [DOI] [PubMed] [Google Scholar]
- Ye C, Hill CM, Wu S, Ruan J, Ma ZS.. 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep. 6:31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R.. 2001. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S.. 2002. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 5:124–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article (raw data of whole-genome sequencing, genome assembly and RNA-seq data) have been deposited at NCBI BioProject database under Bioproject ID PRJNA343584.