Abstract
Sensory systems are attractive evolutionary models to address how organisms adapt to local environments that can cause ecological speciation. However, tests of these evolutionary models have focused on visual, auditory, and olfactory senses. Here, we show local adaptation of bitter taste receptor genes in two neighboring populations of a wild mammal—the blind mole rat Spalax galili—that show ecological speciation in divergent soil environments. We found that basalt-type bitter receptors showed higher response intensity and sensitivity compared with chalk-type ones using both genetic and cell-based functional analyses. Such functional changes could help animals adapted to basalt soil select plants with less bitterness from diverse local foods, whereas a weaker reception to bitter taste may allow consumption of a greater range of plants for animals inhabiting chalk soil with a scarcity of food supply. Our study shows divergent selection on food resources through local adaptation of bitter receptors, and suggests that taste plays an important yet underappreciated role in speciation.
Keywords: local adaptation, bitter taste, ecological speciation, functional assay
Introduction
Understanding the link between speciation and natural selection has been a central topic in the field of evolutionary biology since Darwin (Darwin 1859). Ecological speciation is a model of speciation that occurs as a by-product of natural selection (Rundle and Nosil 2005; Schluter 2009; Schluter and Conte 2009), whereby divergent selection drives local adaptation to ecologically different environments, eventually leading to reproductive isolation between populations (Mayr 1947; Schluter 2001). Local environments can differ in a variety of biotic and abiotic factors, including food resources, climate, temperature, habitat, and interaction with other species such as predators and pollinators (Rundle and Nosil 2005; Sobel et al. 2010). Sensory systems are attractive evolutionary models to address how natural selection shapes local adaptation that can cause ecological speciation, because they could connect external environmental signals with internal physiological responses. However, although visual, auditory, and olfactory senses were suggested to be involved in numerous examples of ecological speciation (Kingston and Rossiter 2004; Seehausen et al. 2008; Keesey et al. 2020), the role of other sensory modalities, for example taste, has been underappreciated in speciation.
Two neighboring populations of a wild mammal, the blind mole rat Spalax galili, in northern Israel living in two contrasting soil environments—chalk and basalt—have been identified as a unique model of ecological sympatric speciation (Hadid et al. 2013; Li et al. 2015, 2016; Lövy et al. 2015, 2017) (fig. 1). One of the most significant differences between these soils is food resource. In general, basalt soil offers higher diversity and abundance of food resources than chalk soil (Lövy et al. 2015, 2017) (fig. 1). This is likely caused by a higher abundance of Sarcopoterium spinosum bushes in chalk soil (fig. 1), which effectively outcompete other herbaceous vegetation including plants with various underground storage organs such as geophytes—the staple food of mole rats (Mohammad and Alseekh 2013). It is thus suggested that mole rats living in chalk soil are exposed to more stressful conditions in terms of food supply. Therefore, food could be one of the important agents of divergent selection that subsequently promotes local adaptation to the ancestral chalk and derived basalt soil environments (Weinstein et al. 2006; Hadid et al. 2013; Li et al. 2015). In an earlier study (Li et al. 2015), we detected genetic divergence of several olfactory and taste receptors between the two neighboring populations, indicating that sensory systems may be involved in ecological speciation of the blind mole rat, although functional divergence of these receptors between populations remains unknown.
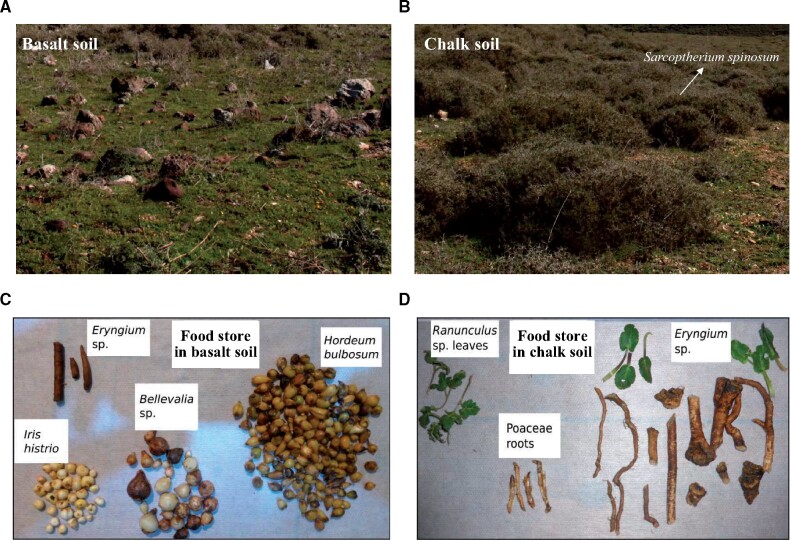
Fig. 1.
Vegetation characteristics and food stores in basalt and chalk soils. Distinct patterns of plant diversity and food stores were observed in basalt soil (A and C) and chalk soil (B and D). The chalk soil is covered by the highly abundant Sarcopoterium spinosum bushes. Pictures of the studied microsites were provided by Matěj Lövy.
In this study, we focused on the bitter taste receptors (Tas2rs) that can detect potentially toxic compounds in environments, and thus determine food selection. We hypothesized that functional divergence of Tas2rs may drive local adaptation to different food resources. Indeed, functional diversity of TAS2R1, TAS2R4, and TAS2R16 was detected in primates with different diets; such diversity could help different primate species to adapt to the food items they eat (Imai et al. 2012; Tsutsui et al. 2016). Moreover, functional differentiation of Tas2r20 receptor among giant panda populations was found to be associated with different contents of quercitrin in bamboo leaves (Hu et al. 2020). Local adaptation to different food resources may eventually lead to habitat-based assortative mating, which is considered to be a strong driving force in ecological speciation (Berner and Thibert-Plante 2015; Lövy et al. 2017; Kopp et al. 2018).
Here, we examine all Tas2r genes that are putatively functional in the two divergent soil populations of S. galili, including sequences that were previously reported as well as those that are newly generated in this study. We performed population genetic analysis, including FST measurement and haplotype phasing, to examine whether divergent selection plays a major role in the evolution of these putatively functional Tas2rs between the two populations. More importantly, we used cell-based functional assays of six differentiated Tas2rs to test for functional divergence between these two populations.
Results
Tas2r Identification and Gene Sequencing
Tas2rs are genes without introns, and have an average length of ∼900 nucleotides. Using a method previously described (Jiao et al. 2018), we identified 32 intact Tas2rs from the genome sequence of the blind mole rat. We then constructed a gene tree using IQ-TREE (Nguyen et al. 2015), based on the alignment of the 32 Tas2rs identified here and 288 intact rodent Tas2rs obtained from a previous study (Hayakawa et al. 2014). We found that 20 of the 32 Tas2rs have orthologous genes in other rodents, whereas the other 12 are species-specific (supplementary fig. S1, Supplementary Material online). In this study, we successfully amplified and sequenced the complete regions of 12 Tas2rs in 16 individuals from the basalt environment and 13 from the chalk environment. Combined with the other 20 Tas2rs previously sequenced in the same individuals (Li et al. 2015), our data set includes the sequences of all 32 intact Tas2rs in S. galili.
Genetic Differentiation of Tas2rs in the Two Populations
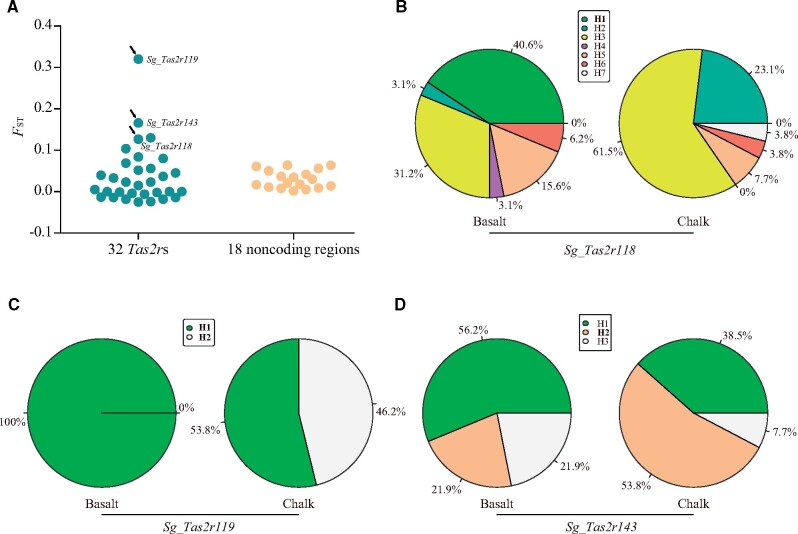
To examine the population genetic differentiation of these Tas2rs, pairwise FST values between the two populations were measured for each Tas2r gene using DnaSP (Librado and Rozas 2009). We found that ten Tas2rs are significantly differentiated between the chalk and basalt populations (P < 0.05, after false discovery rate [FDR] adjustment; supplementary table S1, Supplementary Material online). To further test whether Tas2rs genes evolved under natural selection, we utilized 18 randomly selected noncoding regions from the genome, which were previously sequenced in the same 29 individuals (Li et al. 2015). Only one of these regions showed significant genetic differentiation between the two populations (Li et al. 2015). Distinct distributions of FST values for Tas2r genes and noncoding regions were clearly observed (fig. 2A). Furthermore, the frequency of significantly differentiated loci is statistically higher in Tas2rs (10/32 = 31.25%) than in noncoding regions (1/18 = 5.56%) (P = 0.041, two-tailed Fisher’s exact test), suggesting that natural selection, rather than neutral processes, may have shaped the evolution of Tas2rs between the two populations.
Fig. 2.
Population genetic analysis of Spalax Tas2r genes. (A) The distribution of Fst value for 32 Tas2r genes and 18 noncoding regions. The arrow indicates that these Tas2rs exhibited functional divergence between the two populations. (B–D) Protein haplotype distribution of Sg_Tas2r118 (B), Sg_Tas2r119 (C), and Sg_Tas2r143 (D). For each gene, different protein haplotypes are marked with various colors. The percentages of each haplotype in each population are also shown. Significantly differentiated protein haplotypes were shown in bold.
Using the PHASE program (Stephens and Donnelly 2003), we inferred the nucleotide haplotypes for each Tas2r gene. A total of 214 nucleotide haplotypes were identified among the 32 Tas2rs (supplementary table S2, Supplementary Material online). In order to detect the haplotype differences that may be associated with functional differences, we further obtained the protein haplotypes for each Tas2r gene and compared the frequencies of each protein haplotype in the basalt population with those in the chalk population using Fisher’s exact test (supplementary tables S3 and S4, Supplementary Material online). Our results showed that there are six Tas2r genes (Sg_Tas2r118, Sg_Tas2r119, Sg_Tas2r134, Sg_Tas2r137, Sg_Tas2r143, and Sg_Tas2r582a) that have at least one significantly differentiated protein haplotype (fig. 2B–D and supplementary fig. S2, Supplementary Material online). For example, Sg_Tas2r118 has seven protein haplotypes (supplementary fig. S3, Supplementary Material online), of which six were found in the basalt population and five were found in the chalk population (fig. 2B). The frequency of protein haplotype Sg_Tas2r118 (H1) is significantly higher in the basalt population (13/32 = 40.6%) than in the chalk population (0/26 = 0%) (P = 0.001, after FDR adjustment, two-tailed Fisher’s exact test, fig. 2B and supplementary table S4, Supplementary Material online). Sg_Tas2r119 has two protein haplotypes; Sg_Tas2r119 (H1) is shared between the two populations, whereas Sg_Tas2r119 (H2) is unique to the chalk population, and occurs at a high frequency (12/26 = 46.2%) (fig. 2C and supplementary table S4, Supplementary Material online). Sg_Tas2r143 has three protein haplotypes, all of which are shared between the two populations (fig. 2D). However, the frequency of the protein haplotype Sg_Tas2r143 (H2) is statistically lower in the basalt population (7/32 = 21.9%) than in the chalk population (14/26 = 53.8%) (P = 0.045, after FDR adjustment, two-tailed Fisher’s exact test, fig. 2D and supplementary table S4, Supplementary Material online). Similar results can also be found in the other three differentiated Tas2r genes, including Sg_Tas2r134, Sg_Tas2r137, and Sg_Tas2r582a (supplementary fig. S2, Supplementary Material online). In addition, these six above-mentioned Tas2rs were also significantly differentiated between the two populations in the FST analysis (supplementary table S1, Supplementary Material online). This finding strongly suggests that these six Tas2r genes are under divergent selection and may exhibit functional divergence between the two populations.
Functional Divergence of Tas2rs in the Two Populations
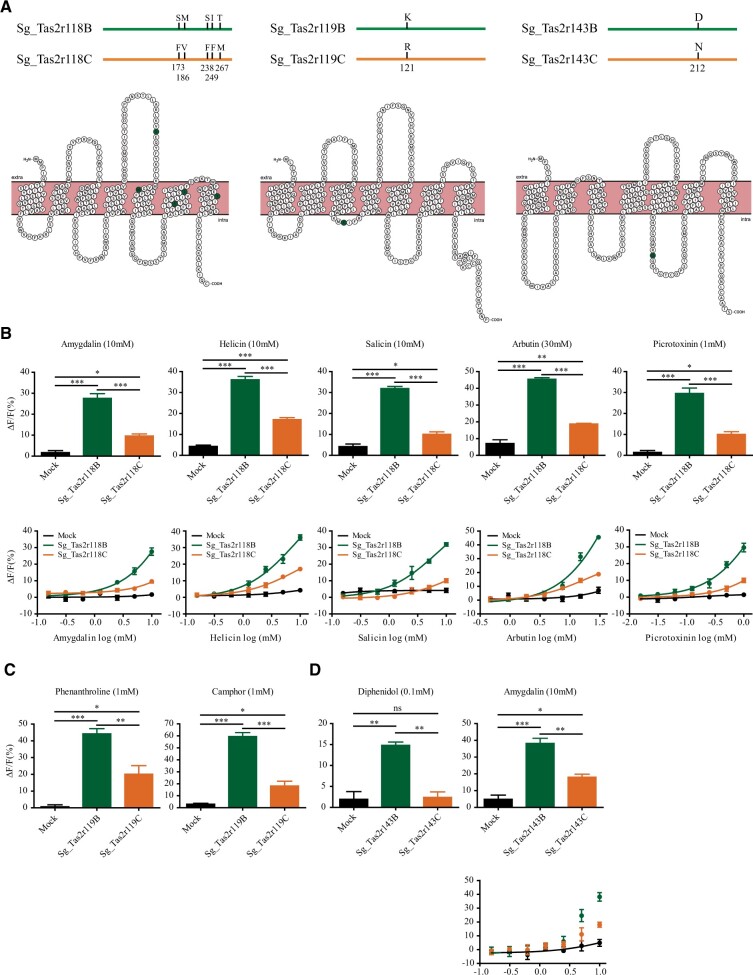
Of the six Tas2rs that were significantly differentiated between the two populations based on the FST and protein haplotype analysis (fig. 2 and supplementary fig. S2, Supplementary Material online), significantly differentiated haplotypes of four Tas2rs (Sg_Tas2r119, Sg_Tas2r134, Sg_Tas2r137, and Sg_Tas2r143) have only one amino acid change, and that of the remaining two Tas2rs (Sg_Tas2r118 and Sg_Tas2r582a) have multiple amino acid changes (fig. 3A and supplementary fig. S2, Supplementary Material online). It is worth noting that these amino acid changes are not fixed in either population because the two populations separated very recently (Li et al. 2015). Based on the frequencies of protein haplotypes in these Tas2rs (fig. 2B–D and supplementary fig. S2, Supplementary Material online), we selected two protein haplotypes for each Tas2r, one from the basalt population and the other from the chalk population, for cell-based functional assays. For example, for Sg_Tas2r118, Sg_Tas2r118 (H1), and Sg_Tas2r118 (H3) were selected, hereafter renamed as Sg_Tas2r118B (B denotes Basalt) and Sg_Tas2r118C (C denotes Chalk) (fig. 3A). A similar renaming was applied to each selected protein haplotype for the other five Tas2rs, where each haplotype is appended with a B or C for basalt or chalk, respectively (fig. 3A and supplementary fig. S2, Supplementary Material online). We examined the responses of the two selected protein haplotypes of each of the six Tas2rs to 20 bitter compounds with distinct chemical structures (supplementary table S5, Supplementary Material online). Our screening results showed that although no functional divergence was found between the two selected protein haplotypes of Sg_Tas2r134, Sg_Tas2r137, and Sg_Tas2r582a (supplementary table S6, Supplementary Material online), the two protein haplotypes of the remaining three Tas2rs (Sg_Tas2r118, Sg_Tas2r119, and Sg_Tas2r143) exhibited clear difference on the responses to two to five bitter compounds (fig. 3). We confirmed that the functional divergence was not due to the difference of Tas2r haplotype expression levels in HEK293 cells by immunofluorescence assay (supplementary fig. S4, Supplementary Material online). The bitter receptor Sg_Tas2r118 can be activated by five natural bitter compounds, including four beta-glucopyranosides (amygdalin, helicin, salicin, and arbutin) and one sesquiterpenoid (picrotoxinin) (fig. 3B). Interestingly, we found that the responsiveness of the protein haplotype Sg_Tas2r118B to each of the five compounds is significantly higher than that of Sg_Tas2r118C (one-way ANOVA, Tukey test for multiple comparisons) (fig. 3B). Furthermore, we obtained the dose–response curves for all five identified compounds (fig. 3B). Our results showed that Sg_Tas2r118B appears to be more sensitive than Sg_Tas2r118C to each of the five compounds, although the EC50 value cannot be accurately measured in this study owing to the toxicity of these bitter compounds at high concentration in vitro. We also found that Sg_Tas2r119 recognized two compounds, including one natural (camphor) and one synthetic (phenanthroline) (fig. 3C). Moreover, Sg_Tas2r143 recognized three bitter compounds, including two natural (amygdalin and salicin) and one synthetic (diphenidol) (fig. 3D). Similarly, the responsiveness of the protein haplotype Sg_Tas2r119B to each ligand is significantly higher than that of the protein haplotype Sg_Tas2r119C (one-way ANOVA, Tukey test for multiple comparisons) (fig. 3C). Based on the dose–response curves, Sg_Tas2r119B appears to be more sensitive than Sg_Tas2r119C to the two agonists (fig. 3C). These trends hold true for Sg_Tas2r143 (fig. 3D). Thus, our findings clearly show that the basalt-type haplotype for each of the three Tas2rs has a greater response intensity and sensitivity than the chalk-type haplotype.
Fig. 3.
Functional divergence between basalt-type and chalk-type protein haplotypes of three Tas2r genes. (A) Basalt-type and chalk-type protein haplotypes of Sg_Tas2r118, Sg_Tas2r119, and Sg_Tas2r143. The amino acid difference between basalt-predominant and chalk-predominant protein haplotypes is indicated. Snake plot of each Tas2r was generated based on the sequence of basalt-type protein haplotype. (B) Functional divergence between Sg_Tas2r118B and Sg_Tas2r118C to four beta-glucopyranosides (amygdalin, helicin, salicin, and arbutin) and one sesquiterpenoid (picrotoxinin). (C) Functional divergence between Sg_Tas2r119B and Sg_Tas2r119C to phenanthroline and camphor. (D) Functional divergence between Sg_Tas2r143B and Sg_Tas2r143C to diphenidol, amygdalin, and salicin. All bar graphs and dose-dependent curves were generated with GRAPHPAD PRISM 5. Analysis of variance with Tukey’s multiple comparisons test was used for statistical analysis (*P < 0.05, **P < 0.01, ***P < 0.001).
Identification of Genetic Changes Responsible for Functional Divergence of Sg_Tas2r118 between the Two Populations
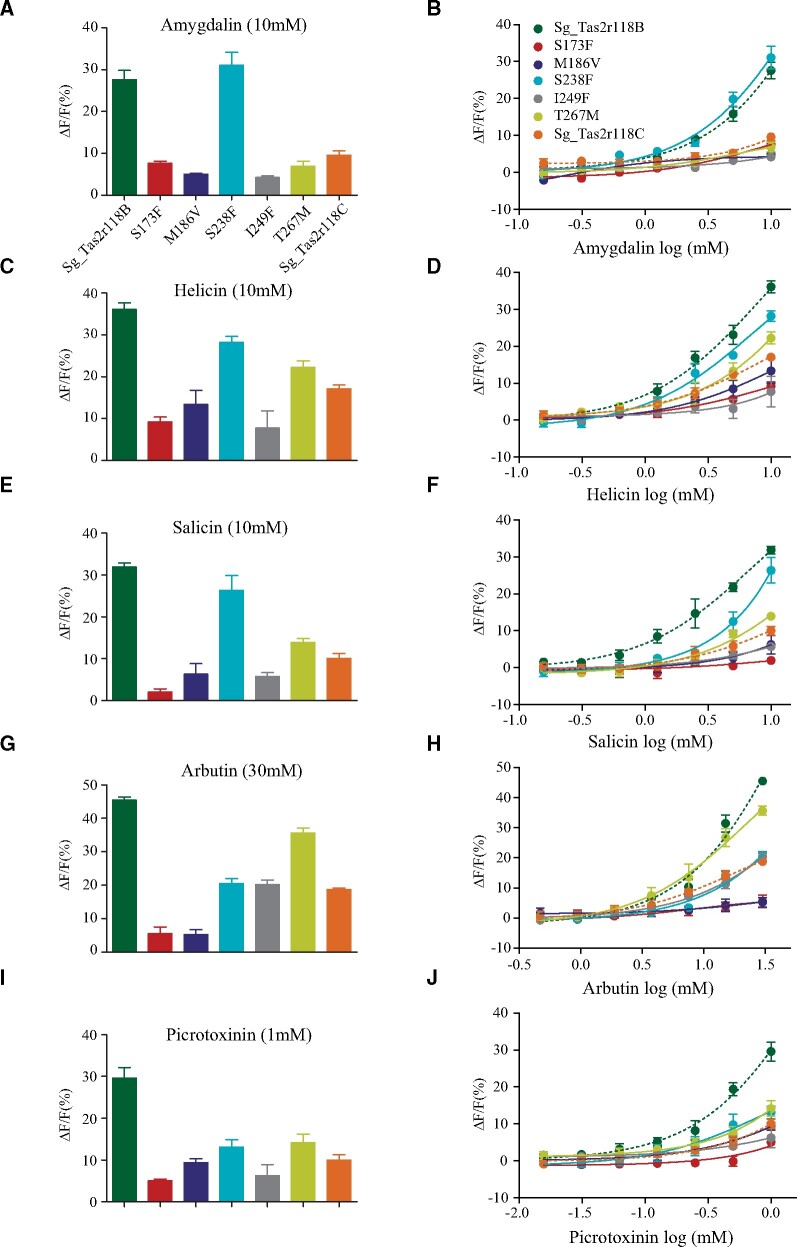
Given that there is only one amino acid difference between the two selected protein haplotypes of Sg_Tas2r119 and Sg_Tas2r143 (fig. 3A), we reasoned that K121R and D212N are exclusively responsible for the functional divergence of Sg_Tas2r119 and Sg_Tas2r143, respectively (fig. 3C and D). However, the specific residues that determine functional divergence of Sg_Tas2r118 between the two populations are still unclear, because five amino acids are different between the protein haplotypes Sg_Tas2r118B and Sg_Tas2r118C (fig. 3A). To assess the functional consequences of these five sites, we performed site-directed mutagenesis to generate five mutant receptors (S173F, M186V, S238F, I249F, and T267M) by replacing each of the five amino acids of protein haplotype Sg_Tas2r118B with that of Sg_Tas2r118C. For example, we mutated the amino acid S173 of Sg_Tas2r118B to F (phenylalanine) to generate the mutant receptor S173F. HEK293 cells expressing these mutant receptors along with Gα16-gust44 were then assayed for their responses to the five identified agonists. We assessed the expression levels of five Tas2r mutants in HEK293 cells by immunofluorescence assay and found similar expression levels (14.8–20.7%) for different Tas2r mutants (supplementary fig. S4, Supplementary Material online). The responsiveness of these mutant receptors to agonists was either similar or significantly reduced when compared with that of Sg_Tas2r118B (fig. 4 and supplementary table S7, Supplementary Material online). For example, compared with the background Sg_Tas2r118B, the mutant receptor S238F showed a similar response to amygdalin (fig. 4A and supplementary table S7, Supplementary Material online). The other four mutant receptors showed significantly reduced responses, similar to the response of Sg_Tas2r118C to amygdalin (fig. 4A and supplementary table S7, Supplementary Material online). The dose–response curves further confirm that both Sg_Tas2r118B and S238F showed similar levels of sensitivity, whereas Sg_Tas2r118C and the other four mutant receptors showed similar levels of sensitivity (fig. 4B). These findings suggest that the difference in response of Sg_Tas2r118B and Sg_Tas2r118C to amygdalin is likely caused by the other four amino acid replacements, rather than S238F. Such general pattern was also observed in the other three beta-glucopyranosides (fig. 4), indicative of the obvious effects of those four amino acid replacements (S173F, M186V, I249F, and T278M) on the recognition of beta-glucopyranosides. Nonetheless, we noticed that Sg_Tas2r118B and its mutant receptor S238F showed distinct levels of sensitivity to salicin (fig. 4F), suggesting that the replacement S238F may also be involved in salicin binding and contribute to functional divergence between Sg_Tas2r118B and Sg_Tas2r118C to salicin. As for the nonglucopyranoside agonist picrotoxinin, all five mutant receptors showed significantly reduced responses when compared with Sg_Tas2r118B (fig. 4I and supplementary table S7, Supplementary Material online), or similar responses when compared with Sg_Tas2r118C (fig. 4I and supplementary table S7, Supplementary Material online). The dose–response curves showed that Sg_Tas2r118B is the most sensitive among all assayed receptors, and that five mutant receptors and Sg_Tas2r118C exhibit similar degrees of sensitivity (fig. 4J), indicating that all five amino acid replacements are likely to result in functional divergence between Sg_Tas2r118B and Sg_Tas2r118C to picrotoxinin.
Fig. 4.
Identification of key residues that were responsible for the functional divergence of Sg_Tas2r118B and Sg_Tas2r118C. Five mutant receptors were generated using site-directed mutagenesis, and their responses to five agonists were measured. (A, C, E, G, and I) Quantitative analysis of responses of mutant receptors to amygdalin (10 mM), helicin (10 mM), salicin (10 mM), arbutin (30 mM), and picrotoxinin (1 mM), respectively. See the statistical comparisons in supplementary table S7, Supplementary Material online. (B, D, F, H, and J) Dose-dependent responses of mutant receptors to amygdalin, helicin, salicin, arbutin, and picrotoxinin, respectively. Sg_Tas2r118B and Sg_Tas2r118C (dashed lines) are also included for comparison. Different receptors are indicated by various colors.
We also performed structure modeling and molecular docking for each haplotype of the three Tas2rs to validate the effects of amino acid variants on protein–ligand interaction (supplementary fig. S5, Supplementary Material online). Among those variants examined in this study, only one amino acid, F249 of Sg_Tas2r118C, was predicted to be directly involved in the interaction with helicin, salicin, and arbutin (supplementary fig. S5, Supplementary Material online). Therefore, our structure–function analysis confirms an impact of the replacement I249F of Sg_Tas2r118 on ligand recognition, and suggests that other amino acid variants may also be involved in other aspects of receptor activation, such as signal transduction and changes in the stability of protein conformation (Singh et al. 2011; Thomas et al. 2017).
Discussion
In this study, we performed population genetic analysis and functional characterization of Tas2rs between the two neighboring populations of blind mole rats inhabiting divergent soil conditions. We found that ten of 32 Tas2rs show significant differentiation between the two populations based on the FST measurements. Of these differentiated Tas2rs, six were also identified by protein haplotype analysis, indicating that natural selection shapes their genetic divergence. Cell-based functional assays of six Tas2rs further confirmed the functional divergence between the basalt-predominated protein haplotype and the chalk-predominated protein haplotype.
The genetic divergence of bitter taste receptors between the two different soil populations has been reported previously by sequencing 20 Tas2rs (Li et al. 2015). Here, we sequenced the remaining 12 Tas2rs in the same individuals, generating a full data set that includes all putatively functional Tas2rs. Ten of the 32 Tas2rs were identified as significantly differentiated loci, eight whose orthologs have been detected in other rodent species and two that are species-specific. A similar pattern was also observed in two other chemosensory gene families: olfactory receptor genes and vomeronasal receptor genes (Li et al. 2015; Jiao et al. 2019). This is consistent with rapid evolution of chemosensory receptors that directly respond to new environments (Niimura and Nei 2006; McBride 2007; Brand et al. 2015). However, the significant population differentiation indicated by high FST values could have resulted from the different frequencies of either nonsynonymous or synonymous variations between populations. Although the former are responsible for functional variations, the latter are less important in protein function because they do not lead to amino acid changes. We therefore compared the frequencies of protein haplotypes between the two populations. The protein haplotype was inferred by merging the nucleotide haplotypes with synonymous variations. We identified six Tas2rs with at least one significantly differentiated protein haplotype. Interestingly, these six Tas2rs can also be identified by FST analysis, suggesting that population differentiation of these Tas2rs were mainly determined by different frequencies of nonsynonymous variations. In contrast, different frequencies of synonymous variations in the other four Tas2rs that led to significant population differentiation were identified by FST analysis but not the protein haplotype analysis. Such remarkable genetic differentiations of bitter taste receptors were also observed in subspecies of chimpanzees (Sugawara et al. 2011; Hayakawa et al. 2012).
Our functional assays demonstrated that functional divergence appears to be present between the basalt-type and chalk-type protein haplotypes of three Tas2rs (Sg_Tas2r118, Sg_Tas2r119, and Sg_Tas2r143). In general, the basalt-type protein haplotype of each gene is more sensitive than the chalk-type protein haplotype to agonists. It appears that K121R leads to functional divergence of Sg_Tas2r119, owing to only one amino acid change between the two protein haplotypes, whereas D212N accounts for functional divergence of Sg_Tas2r143. By assaying the mutant receptors of Sg_Tas2r118, we found that all five amino acid replacements can affect ligand sensitivity, with the exception of S238F, which is less important in affecting amygdalin binding. These findings suggest that these five amino acids are involved in the recognition of both types of chemical compounds with distinct structures: beta-glucopyranosides (amygdalin, helicin, salicin, and arbutin) and sesquiterpenoids (picrotoxinin). Interestingly, mutations in three of these five amino acid positions in human TAS2R16 (ortholog to Sg_Tas2r118) have been associated with interaction with salicin, thus paralleling our study (Thomas et al. 2017).
Blind mole rats are herbivorous; their diet consists of plants with underground storage organs, such as bulbs, corms, and roots. The bitter taste meditated by Tas2rs has been considered to play an important role for the survival of animals, especially for herbivores and insectivores (Li and Zhang 2014; Wang and Zhao 2015). However, we observed a significantly lower intensity and sensitivity of bitter taste receptors to bitter compounds in the ancestral chalk population compared with the derived basalt population (Weinstein et al. 2006; Li et al. 2015). We used codeml in PAML 4 (Yang 2007) to infer the ancestral states of three functionally divergent Tas2rs at the ancestral node of the two populations (supplementary fig. S6, Supplementary Material online). Our results showed that the basalt-type haplotype is derived, suggesting that positive selection may promote the functional divergence of Tas2rs. These findings indicate that the bitter taste is dull in mole rats inhabiting the ancestral chalk soil environment, and then becomes sensitive in individuals adapted to basalt soils. Moreover, most of the bitter compounds used in this study are common in plants. For example, beta-glucopyranosides and camphor can be found in two plant species (Ranunculus asiaticus and Eryngium creticum) growing in both soils (Toki et al. 1996; Lövy et al. 2015; Kikowska et al. 2016); the latter is the most frequent plant in the chalk soil at the studied microsite (Lövy et al. 2015). In other words, these bitter compounds or other compounds with similar chemical structures occur in the daily food of mole rats living in the two soils. Under such selective pressures, Tas2rs of the mole rats are likely locally adapted.
Why do the mole rats from different soils possess distinct capacity to detect bitter substances? We speculate that the basalt soil may have a larger abundance of food sources but may also have a higher proportion of toxic plants, such as the predominant plant Ornithogalum lanceolatum (comprising 28% of all underground storage organs of plants in the basalt soil) (Lövy et al. 2015). The bulbs of Ornithogalum species are poisonous because they contain a variety of cardiotoxic cardenolides (Burrows and Tyrl 2012). Thus, animals in the basalt soil tend to have more food to eat but have evolved a keen sense of bitter taste to deal with the higher chance of encountering toxins. By contrast, the proportion of Ornithogalum lanceolatum among all underground storage organs of plants in the chalk soil is very low (∼2%) (Lövy et al. 2015). As a result, chalk soil may offer fewer types of food sources that might be less toxic. This scenario suggests that adaptation of bitter taste could have effectively hindered migration between the two populations: basalt animals avoid migration to chalk soil because of the scarcity of food supply, whereas chalk animals avoid migration to basalt soil as they would be toxified due to their dull bitter taste. We propose a potential tradeoff between the two populations: more food and more toxin versus less food and less toxin. Given the role of taste adaptation in hindering migration between the two populations, we speculate that local adaptation of taste receptors should be a cause of incipient speciation. Although it is a matter of speculation, it would be interesting to test this scenario in the future. In addition, our study reinforces the recent discovery that bitter taste receptors of a small rodent were locally adapted to a desert condition where food resources are limited (Tigano et al. 2020).
Taken together, we found that functional divergence of bitter taste receptors may have played a major role in food selection during the process of ecological speciation of S. galili. Divergent selection could arise from the difference in food recourses between the two soils, which then drives local adaptation of bitter taste receptors. The keen sense of bitter taste provides a physiological basis for the mole rats living in the basalt soil to obtain food with less toxic compounds, whereas the dull bitter taste helps mole rats in the chalk soil to compromise the bitter compounds in their foods. Local adaptation of bitter taste can lead to food choice and habitat preference, such that mole rats preferring to reside in the soil to which they are adapted will have higher fitness (Rundle and Nosil 2005). The subsequent barriers to gene flow and prezygotic isolation would evolve between the two populations due to habitat-based assortative mating (Lövy et al. 2017, 2020), which may lead to premating reproductive isolation.
Limitations
There are two limitations to this work. First, our collection of bitter compounds (ten natural and ten synthetic) used in the cell-based assay is quite small. Although the ten natural compounds include several bitter substances commonly used for chemical defense in plants, they cannot reflect the real composition of bitter substances present in the daily food of the mole rat. Thus, some of Tas2r functional divergence we detected here may not be relevant to the difference of dietary ecology between the two soils. Second, our study only focused on genetic and functional analysis of bitter taste receptors, proposing a hypothesis that taste may play an important role in ecological speciation. We are not able to conduct physiological and behavioral experiments on mole rats. It would be interesting to extract bitter compounds from plants growing in the two soils, and test these compounds in cell-based assays and behavioral experiments in the future. These measurements could provide direct evidence linking the bitter taste sensitivity to food choice by the mole rats.
Conclusions
This study used population genetic analysis and functional experiments to provide clear evidence of local adaptation of bitter taste receptor genes in two populations of S. galili inhabiting contrasting soil environments. We found that basalt-type bitter receptors showed higher response intensity and sensitivity compared with chalk-type receptors. Hence, it is likely that the new and rich habitat of basalt, which includes many geophytes with bitter bulbs and corms, selected for higher bitterness, leading to local adaptation of bitter taste. Our study shows divergent selection on food resources through local adaptation of bitter receptors, and suggests a role of taste that has been underappreciated in speciation.
Materials and Methods
Identification and Nomenclature of Tas2r Genes
We downloaded the genome sequence of the blind mole rat Spalax galili (GenBank assembly: GCF_000622305.1) (Fang et al. 2014) and performed TBlastN searches (Altschul et al. 1990) to identify Tas2r genes using protein sequences of known rodent Tas2rs as queries. A total of 32 Tas2r genes were found to possess an intact open reading frame longer than 270 amino acids in length and were considered putatively functional. The nucleotide sequences of these intact Tas2rs are provided in supplementary data S1, Supplementary Material online. We followed the nomenclature of Tas2rs proposed in Euarchontoglires (Hayakawa et al. 2014). Specifically, we first constructed a gene tree (supplementary fig. S1, Supplementary Material online) using IQ-TREE with 1,000 bootstrap replicates (Nguyen et al. 2015) based on the alignment of nucleotide sequences of the 32 Tas2rs identified here, along with all intact rodent Tas2rs identified in a previous study (Hayakawa et al. 2014). We next used “Sg_Tas2rx” to denote the bitter taste receptor genes in S. galili (102≤x ≤ 146 in orthologs between mouse and S. galili; 200 < x < 300 in the ancestral orthologs of rodents that are not defined in mouse; 500 < x < 600 in rodent-specific orthologs).
DNA Sequencing
We sequenced all 32 Tas2rs from 29 individuals of S. galili, including 16 sampled from the basalt soil and 13 sampled from the chalk soil. Of these 32 Tas2rs, 20 have been reported in our previous study (Li et al. 2015). Genomic DNAs were isolated from the muscle tissues that were stored at −20 °C, using a DNeasy Blood and Tissue Kit (Qiagen). Primers were designed based on the genome sequence of the blind mole rat (supplementary table S8, Supplementary Material online). The details of polymerase chain reactions and subsequent sequencing were described previously (Li et al. 2015; Jiao et al. 2019).
Population Genetic Analysis
Fixation index (FST) was used to measure the genetic differentiation between the basalt and chalk populations for each Tas2r gene (Weir and Cockerham 1984), and the nearest-neighbor statistical test (Hudson 2000) was conducted to evaluate the significance of this genetic differentiation. Using PHASE v2.1 software, the protein haplotypes for each Tas2r gene were inferred with a Bayesian statistical method (Stephens and Donnelly 2003).
Bitter Compounds
Our bitter compound library includes ten naturally occurring and ten synthetic compounds (supplementary table S5, Supplementary Material online). All examined compounds were purchased from Sigma–Aldrich, except for diphenidol hydrochloride (Reagent World). These compounds were dissolved in Dulbecco’s phosphate-buffered saline (DPBS) or dimethyl sulphoxide (DMSO) and then diluted with DPBS to prepare the tested solutions. Given the high toxicity of DMSO to the transfected cells, the final DMSO concentration in the tested solutions must be less than 0.1%. The highest concentrations of bitter compounds were obtained from a previous study (Meyerhof et al. 2010).
Preparation of Spalax Tas2r Constructs and Site-Directed Mutants
The complete coding sequences of three Spalax Tas2r genes were amplified from genomic DNA, and then subcloned into pEAK10 vector, with the first 45 amino acid residues of rat somatostatin receptor 3 as the signal peptide at the N-terminal. Point mutations in Tas2r118 were introduced by site-directed mutagenesis, using the QuikChange method (Agilent Technologies, La Jolla, CA). All constructs were verified by Sanger sequencing.
Functional Assays of Spalax Tas2rs
Human embryonic kidney 293 (HEK293) cells (PEAKrapid) were obtained from ATCC (CRL-2828) and cultured in Opti-MEM medium with 6% fetal bovine serum. The cells were seeded in 96-well plates coated with poly-l-lysine, at a density of 40,000–50,000 per well. After 20–22 h, cells were transiently transfected with a Tas2r construct (0.1 μg/well) along with a coupling chimeric G protein Gα16-gust44 (0.1 μg/well) using Lipofectamine 2000 (0.5 μl/well). Cells that were transfected with only Gα16-gust44 were used as negative controls (mock transfection). After transfection for one day, the cells were washed once with DPBS and loaded with the calcium-sensitive dye Fluo-4 AM for 1 h in the dark. After three washes with DPBS, the cells were assayed in a FlexStation III reader (Molecular Devices) to monitor the fluorescence changes every 2 s for 2 min. Calcium mobilization traces were recorded. Changes in fluorescence (ΔF) were quantified as the peak fluorescence minus baseline fluorescence. The percentage of ΔF relative to baseline (F) averaged from triplicate experiments was used to quantify the calcium mobilization. Calcium mobilization traces, bar graphs, and dose-dependent curves were generated with GraphPad Prism 5.
Immunofluorescence Assays
HEK293 (PEAKrapid) cells were seeded onto poly-lysine coated coverslips in 12-well plates and transfected with a Tas2r construct (1 µg/well for each construct), along with Gα16-gust44 (1 µg/well) by Lipofectamine (4 µl/well). After 24 h of transfection, cells were washed twice with warm phosphate-buffered saline (PBS) and placed at 4 °C for 2 h. The cell surface staining was performed by incubation with concanavalin A, Alexa Fluor 633 Conjugate (C21402, Thermo Fisher, 1:5) for 1 h, followed by three rinses with ice cold PBS buffer. The cells were then fixed by 4% paraformaldehyde in PBS for 20 min. Cells were washed three times with PBS and incubated with 0.1% Triton X-100 in PBS for 10 min. After washed with PBS, cells were incubated for 1 h in 10% fetal bovine serum in PBS to block unspecific binding. Next, an anti-HSV antibody (ab19355, Abcam, 1:500) in PBS supplemented with 10% fetal bovine serum was applied overnight at 4 °C. A donkey anti-rabbit secondary antibody conjugated with Alexa Fluor 488 (ANT024, Wuhan antgene Biotechology, 1:1,000) was then used for detection of the HSV tag. The nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were captured with confocal laser scanning microscopy (Leica TCS SP8). To evaluate the expression level of Tas2r in HEK293 cells, three independent areas were counted.
Structure Modeling and Molecular Docking
The 3D structures of bitter taste receptors were obtained by GPCR-I-TASSER (Zhang et al. 2015). The model with the highest C-score was used in subsequent docking analysis. Different bitter tastants were docked with the corresponding receptors using Autodock Vina (Trott and Olson 2010). The interactions in receptor–ligand complexes were characterized using PLIP (Salentin et al. 2015).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Yi Wang, Wei Hong, and Xin Zheng for technical assistance in the laboratory, and Yasuka Toda for providing the plasmid pEAK10. This work was supported by National Natural Science Foundation of China (31722051 and 31672272 to H.Z. and 32000385 to H.J.), Natural Science Foundation of Hubei Province (2019CFA075 to H.Z.), China Postdoctoral Science Foundation (2020M672407 to H.J.), China National Postdoctoral Program for Innovative Talents (BX20200255 to H.J.) and Hubei Provincial Postdoctoral Foundation (to H.J.), and by the National Institute on Deafness and Other Communication Disorders at the National Institutes of Health (R01DC010842 to P.J.). Additionally, E.N. thanks the Ancell-Teichert Research Foundation for Genetic and Molecular Evolution for financial support in the study of Spalax galili.
Author Contributions
H.Z. conceived and designed the research. H.J. performed the sequencing work and population genetic analysis. H.J., Q.W., Q.L., and W.S. conducted the functional assays. P.J. supervised the functional assays and commented the manuscript. B.-J.W. performed structure modeling and molecular docking. H.J. and Q.W. analyzed the data. M.L. provided the photographs of studied microsites and commented the manuscript. K.L. and E.N. provided the samples and edited the manuscript. H.J. and H.Z. wrote the manuscript with input from all authors.
Data Availability
All sequences that were newly generated in this article have been deposited in the GenBank (accession numbers: MN750256–MN750313 and MN815131–MN815768).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Berner D, Thibert-Plante X.. 2015. How mechanisms of habitat preference evolve and promote divergence with gene flow. J Evol Biol. 28(9):1641–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, Ramirez SR, Leese F, Quezada-Euan JG, Tollrian R, Eltz T.. 2015. Rapid evolution of chemosensory receptor genes in a pair of sibling species of orchid bees (Apidae: Euglossini). BMC Evol Biol. 15(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows GE, Tyrl RJ.. 2012. Toxic plants of North America. 8th ed. Chichester, UK: John Wiley & Sons. [Google Scholar]
- Darwin C.1859. On the origin of species by means of natural selection, or the preservation of favored races in the struggle for life. London: John Murray. [Google Scholar]
- Fang X, Nevo E, Han LJ, Levanon EY, Zhao J, Avivi A, Larkin D, Jiang XT, Feranchuk S, Zhu YB, et al. 2014. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat Commun. 5:3966. [DOI] [PubMed] [Google Scholar]
- Hadid Y, Tzur S, Pavlicek T, Sumbera R, Skliba J, Lovy M, Fragman-Sapir O, Beiles A, Arieli R, Raz S, et al. 2013. Possible incipient sympatric ecological speciation in blind mole rats (Spalax). Proc Natl Acad Sci U S A. 110(7):2587–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Sugawara T, Go Y, Udono T, Hirai H, Imai H.. 2012. Eco-geographical diversification of bitter taste receptor genes (TAS2Rs) among subspecies of chimpanzees (Pan troglodytes). PLoS One 7(8):e43277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Suzuki-Hashido N, Matsui A, Go Y.. 2014. Frequent expansions of the bitter taste receptor gene repertoire during evolution of mammals in the Euarchontoglires clade. Mol Biol Evol. 31(8):2018–2031. [DOI] [PubMed] [Google Scholar]
- Hu X, Wang G, Shan L, Sun S, Hu Y, Wei F.. 2020. TAS2R20 variants confer dietary adaptation to high-quercitrin bamboo leaves in Qinling giant pandas. Ecol Evol. 10(12):5913–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR.2000. A new statistic for detecting genetic differentiation. Genetics 155(4):2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Suzuki N, Ishimaru Y, Sakurai T, Yin LJ, Pan WS, Abe K, Misaka T, Hirai H.. 2012. Functional diversity of bitter taste receptor TAS2R16 in primates. Biol Lett. 8(4):652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H, Hong W, Nevo E, Li K, Zhao H.. 2019. Convergent reduction of V1R genes in subterranean rodents. BMC Evol Biol. 19(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H, Wang Y, Zhang L, Jiang P, Zhao H.. 2018. Lineage-specific duplication and adaptive evolution of bitter taste receptor genes in bats. Mol Ecol. 27(22):4475–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesey IW, Grabe V, Knaden M, Hansson BS.. 2020. Divergent sensory investment mirrors potential speciation via niche partitioning across Drosophila. eLife 9:e57008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikowska M, Dworacka M, Kędziora I, Thiem B.. 2016. Eryngium creticum – ethnopharmacology, phytochemistry and pharmacological activity. A review. Rev Bras Farmacogn. 26(3):392–399. [Google Scholar]
- Kingston T, Rossiter SJ.. 2004. Harmonic-hopping in Wallacea’s bats. Nature 429(6992):654–657. [DOI] [PubMed] [Google Scholar]
- Kopp M, Servedio MR, Mendelson TC, Safran RJ, Rodriguez RL, Hauber ME, Scordato EC, Symes LB, Balakrishnan CN, Zonana DM, et al. 2018. Mechanisms of assortative mating in speciation with gene flow: connecting theory and empirical research. Am Nat. 191(1):1–20. [DOI] [PubMed] [Google Scholar]
- Li D, Zhang J.. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol Biol Evol. 31(2):303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Hong W, Jiao H, Wang GD, Rodriguez KA, Buffenstein R, Zhao Y, Nevo E, Zhao H.. 2015. Sympatric speciation revealed by genome-wide divergence in the blind mole rat Spalax. Proc Natl Acad Sci U S A. 112(38):11905–11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Wang L, Knisbacher BA, Xu QQ, Levanon EY, Wang HH, Frenkel-Morgenstern M, Tagore S, Fang XD, Bazak L, et al. 2016. Transcriptome, genetic editing, and microRNA divergence substantiate sympatric speciation of blind mole rat, Spalax. Proc Natl Acad Sci U S A. 113(27):7584–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J.. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. [DOI] [PubMed] [Google Scholar]
- Lövy M, Šklíba J, Hrouzková E, Dvořáková V, Nevo E, Šumbera R.. 2015. Habitat and burrow system characteristics of the blind mole rat Spalax galili in an area of supposed sympatric speciation. PLoS One 10(7):e0133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövy M, Šklíba J, Šumbera R, Nevo E.. 2017. Soil preference in blind mole rats in an area of supposed sympatric speciation: do they choose the fertile or the familiar? J Zool. 303(4):291–300. [Google Scholar]
- Lövy M, Šumbera R, Heth G, Nevo E.. 2020. Presumed ecological speciation in blind mole rats: does soil type influence mate preferences? Ethol Ecol Evol. 32(1):46–59. [Google Scholar]
- Mayr E.1947. Ecological factors in speciation. Evolution 1(4):263–288. [Google Scholar]
- McBride CS.2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci U S A. 104(12):4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M.. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 35(2):157–170. [DOI] [PubMed] [Google Scholar]
- Mohammad AG, Alseekh SH.. 2013. The effect of Sarcopoterium spinosum on soil and vegetation characteristics. CATENA 100:10–14. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. 2006. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 51(6):505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle HD, Nosil P.. 2005. Ecological speciation. Ecol Lett. 8(3):336–352. [Google Scholar]
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M.. 2015. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 43(W1):W443–W447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D.2001. Ecology and the origin of species. Trends Ecol Evol. 16(7):372–380. [DOI] [PubMed] [Google Scholar]
- Schluter D.2009. Evidence for ecological speciation and its alternative. Science 323(5915):737–741. [DOI] [PubMed] [Google Scholar]
- Schluter D, Conte GL.. 2009. Genetics and ecological speciation. Proc Natl Acad Sci U S A. 106(Suppl 1):9955–9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HDJ, Miyagi R, van der Sluijs I, Schneider MV, Maan ME, Tachida H, Imai H, et al. 2008. Speciation through sensory drive in cichlid fish. Nature 455(7213):620–626. [DOI] [PubMed] [Google Scholar]
- Singh N, Pydi SP, Upadhyaya J, Chelikani P.. 2011. Structural basis of activation of bitter taste receptor T2R1 and comparison with class A G-protein-coupled receptors (GPCRs). J Biol Chem. 286(41):36032–36041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JM, Chen GF, Watt LR, Schemske DW.. 2010. The biology of speciation. Evolution 64(2):295–315. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P.. 2003. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 73(5):1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara T, Go Y, Udono T, Morimura N, Tomonaga M, Hirai H, Imai H.. 2011. Diversification of bitter taste receptor gene family in western chimpanzees. Mol Biol Evol. 28(2):921–931. [DOI] [PubMed] [Google Scholar]
- Thomas A, Sulli C, Davidson E, Berdougo E, Phillips M, Puffer BA, Paes C, Doranz BJ, Rucker JB.. 2017. The bitter taste receptor TAS2R16 achieves high specificity and accommodates diverse glycoside ligands by using a two-faced binding pocket. Sci Rep. 7(1):7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano A, Colella JP, MacManes MD.. 2020. Comparative and population genomics approaches reveal the basis of adaptation to deserts in a small rodent. Mol Ecol. 29(7):1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki K, Takeuchi M, Saito N, Honda T.. 1996. Two malonylated anthocyanidin glycosides in Ranunculus asiaticus. Phytochemistry 42(4):1055–1057. [Google Scholar]
- Trott O, Olson AJ.. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 31(2):455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui K, Otoh M, Sakurai K, Suzuki-Hashido N, Hayakawa T, Misaka T, Ishimaru Y, Aureli F, Melin AD, Kawamura S, et al. 2016. Variation in ligand responses of the bitter taste receptors TAS2R1 and TAS2R4 among New World monkeys. BMC Evol Biol. 16(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhao H.. 2015. Birds generally carry a small repertoire of bitter taste receptor genes. Genome Biol Evol. 7(9):2705–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein Y, Navon O, Altherr R, Stein M.. 2006. The role of lithospheric mantle heterogeneity in the generation of Plio-Pleistocene alkali basaltic suites from NW Harrat Ash Shaam (Israel). J Petrol. 47(5):1017–1050. [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. [DOI] [PubMed] [Google Scholar]
- Yang Z.2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang JY, Jang R, Zhang Y.. 2015. GPCR-I-TASSER: a hybrid approach to G protein-coupled receptor structure modeling and the application to the human genome. Structure 23(8):1538–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences that were newly generated in this article have been deposited in the GenBank (accession numbers: MN750256–MN750313 and MN815131–MN815768).