Abstract
A blood culture from a 65-year-old febrile man undergoing hemodialysis revealed, 5 days after inoculation, an unusual gram-negative fusiform rod with darting motility. During another episode of fever 21 days later, this Campylobacter-like organism was again recovered from three blood cultures and subcultured under an H2-enriched microaerobic atmosphere. The organism was catalase negative and oxidase positive and hydrolyzed urea rapidly. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of whole-cell proteins was indistinguishable from that of “Flexispira rappini” LMG 8738 described by Archer et al. in 1988 (J. R. Archer, S. Romero, A. E. Ritchier, M. E. Hamacher, B. M. Steiner, J. H. Bryner, and R. F. Schell, J. Clin. Microbiol. 26:101–105, 1988). The analysis of the 16S ribosomal DNA sequence revealed a similarity of 99.3% between the two strains. The patient recovered completely after a 4-week course of meropenem therapy. This is the first reported case of a recurrent “F. rappini” bacteremia in an adult patient, which confirms that this organism may be an invasive pathogen in immunocompromised patients, like other newly described Helicobacter species.
“Flexispira rappini” is a provisional name for a fusiform bacterium with periplasmic fibers and bipolar tufts of sheathed flagella (2, 3). Phylogenetically, it belongs to the genus Helicobacter (16, 24) and shares its unusual morphological features with several other Helicobacter species, including Helicobacter bilis (8) and Helicobacter trogontum (14). Strains with the “F. rappini” morphology have been isolated from various animal sources, including aborted sheep fetuses (13), intestinal mucosae of laboratory mice (19), and stools of puppies (1). It has also been rarely recovered from human clinical specimens. The initial report was of isolates from stool samples of two men with mild chronic diarrhea (18). Very recently, the first case of “F. rappini” bacteremia, in a child with pneumonia (22), was described. Other Helicobacter species, like Helicobacter cinaedi and Helicobacter fennelliae, have been infrequently recovered from the blood of adult patients, mostly homosexual patients with human immunodeficiency virus (HIV) infection who presented with cellulitis (12, 20, 23, 25). In the present report, we describe the first case of “F. rappini” bacteremia in an adult patient. As the organism has never been validly described, the name has no standing in nomenclature and there is no official type strain for comparison. However, strain LMG 8738 (ATCC 43879), originally isolated in 1988 by Archer et al. from human diarrheic feces (1, 18), has been used as a reference culture in a variety of taxonomic and clinical studies and was used as a reference in the present study as well.
CASE REPORT
A 65-year-old HIV-seronegative patient was admitted in end-stage renal failure requiring hemodialysis a week before the septic problem described below. He had a history of chronic pancreatitis due to alcoholism and secondary diabetes mellitus requiring insulin. A long-term tobacco smoking habit resulted in severe generalized arteriopathy for which he had undergone replacement of aorto-bi-iliac bifurcation and right foot amputation. He had a previous history of repetitive episodes of infected skin lesions of the lower limbs that had been classified as chronic erysipelas and treated with flucloxacillin. Almost 2 months before the first septic episode, the patient suffered from a cellulitis of the forearm secondary to a cat scratch, which was treated with a 15-day course of clindamycin with slow improvement. A significant loss of renal function led us to initiate hemodialysis through a Hickman catheter. Three days later (day 0 of the first septic episode), he presented for his regular dialysis session with chills and fever of 38.5°C. He had no complaints of diarrhea or abdominal pain and had not recently traveled abroad. Physical examination showed no hepatosplenomegaly or lymphadenopathy. Blood tests showed a C-reactive protein level of 7.8 mg/dl and a leukocyte count of 13,900/μl. Three blood culture sets were rapidly drawn through the recently placed Hickman catheter, because the patient refused hospitalization. He was treated empirically with a single dose of vancomycin (2 g) and amikacin (3 mg/kg) intravenously and improved over a period of days with resolution of fever. After 5 days, one aerobic blood culture vial was detected as positive and showed unusual spindle-shaped gram-negative rods on Gram staining (Fig. 1), with a motility suggesting a Campylobacter-like organism (CLO) by phase-contrast microscopy (strain H1353). The patient appeared well until day 21, when he was admitted to the hospital because of recurrent fever (38.8°C), dyspnea, and a productive cough. Empirical antibiotic therapy with oral co-trimoxazole was begun because a bronchopneumonic superinfection was suspected. Endobronchic fibroscopy was not performed because of the patient’s refusal. Laboratory tests showed an increased C-reactive protein level of 18.6 mg/dl and a leukocyte count of 16,800/μl. Two aerobic blood culture vials taken on admission were positive after 5 days of incubation for the same CLO, which was then identified as “F. rappini.” Investigations proposed for detection of any septic source (echocardiography and gallium scintigraphy) were refused by the patient. On the basis of the blood culture results and the absence of a clinical response to empirical therapy, treatment with co-trimoxazole was discontinued and replaced with meropenem (500 mg/day) on day 25. Clinical improvement was rapidly noted, the fever subsided, and laboratory test results returned to normal values. Considering a possible endovascular infection, meropenem was continued for a total of 4 weeks, along with the recommended management of Campylobacter fetus bacteremia (9, 20). The patient recovered completely, with no recurrence of bacteremia during a 9-month follow-up.
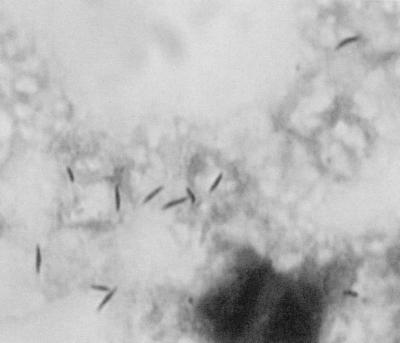
FIG. 1.
Photomicrograph of a Gram-stained smear of the blood culture broth, showing fusiform “F. rappini” rods. Magnification, ×1,000.
MATERIALS AND METHODS
Blood culture protocol.
Our standing protocol for routine blood cultures with adults recommends aseptic collection of two or three samples of 20 ml of venous blood by peripheral venipuncture. Samples are directly inoculated into Bactec Plus Aerobic/F* and Anaerobic/F* blood culture vials. These vials are monitored with the Bactec 9240 system (Becton Dickinson Europe, Meylan, France) and incubated for 5 days at 37°C. Only vials having positive growth are examined with a Gram-stained smear of the vial’s broth and subcultured onto Columbia agar with 5% sheep blood incubated aerobically (aerobic vials) and onto Schaedler agar with 5% sheep blood incubated anaerobically (anaerobic vials). Additional media or atmospheres are used when required for subculture of fastidious organisms, based on the Gram stain morphology.
Growth conditions and microscopic examination.
Gram staining and phase-contrast microscopy of a wet mount preparation were performed by standard methods. The broths were subcultured onto Columbia agar supplemented with 5% sheep blood and, exceptionally, onto Columbia agar supplemented with 5% fresh horse blood. These different plates were incubated in the following atmospheres: aerobic, anaerobic, 5% CO2 enriched, and microaerobic. Three types of microaerobic atmosphere were tried: first, by using a candle jar; second, by using an anaerobic jar with a CampyPak Plus (Becton Dickinson) microaerobic atmosphere-generating envelope; and third, by generating a 3% H2-enriched microaerobic atmosphere in an anaerobic jar with the Anoxomat system (Mart Microbiology BV, Lichtenvoorde, The Netherlands). These cultures were incubated at different temperatures (25, 37, and 42°C). The atmosphere in the jars was made moist by adding a wet towel in the bottom.
Biochemical characterization.
Catalase production was tested by placing a small amount of growth from the plates on a glass slide into a drop of 10% hydrogen peroxide. Oxidase activity was tested on a Bactident Oxidase strip (Diagnostica Merck, Darmstadt, Germany). Hydrolysis of urea was detected with urea-indole medium (bioMérieux, Marcy l’Etoile, France). Presence of alkaline phosphatase and leucine aminopeptidase and hydrolysis of indoxyl acetate and hippurate were all detected with diagnostic tablets from Rosco Diagnostica (Taastrup, Denmark). Reduction of nitrate was tested with nitrate broth and reagents from Difco Laboratories (Detroit, Mich.). Glucose fermentation was tested by using the method of Hugh and Leifson. A triple sugar iron (TSI) slant was inoculated and examined for the detection of H2S production. Tests for tolerance to nalidixic acid and cephalothin were performed with 30-μg disks (bioDiscs; bioMérieux) on BBL Mueller-Hinton agar with 5% sheep blood (Becton Dickinson).
Antimicrobial susceptibility testing.
A 0.5 McFarland suspension of the isolate was inoculated in parallel onto Mueller-Hinton agar with 5% sheep blood and Mueller-Hinton agar supplemented with 5% fresh horse blood, incubated at 37°C under hydrogen-enriched microaerobic conditions for 48 h, and tested by a disk diffusion method (Neo-Sensitabs; Rosco Diagnostica). The susceptibility of the isolate to penicillin G, ampicillin, cefazolin, ceftriaxone, meropenem, erythromycin, clindamycin, clarithromycin, doxycycline, gentamicin, amikacin, ciprofloxacin, nitrofurantoin, co-trimoxazole, and metronidazole was evaluated. The susceptibility test results were interpreted according to the National Committee for Clinical Laboratory Standards breakpoints based on the critical inhibition zone size diameters proposed by the test manufacturer for testing rapidly growing aerobic bacteria (18a), except for metronidazole and clarithromycin, for which tentative guidelines proposed for Helicobacter pylori testing were applied.
SDS-PAGE analysis of whole-cell proteins.
Strain H1353 was grown on Mueller-Hinton agar with 5% horse blood at 37°C in a microaerobic atmosphere for 72 h. Protein samples were prepared, separated by polyacrylamide gel electrophoresis (PAGE), digitized, and subjected to comparative numerical analysis with the GelCompar system version 4.0 (Applied Maths, Kortrijk, Belgium) as described previously (17). Briefly, discontinuous gels (1.5 mm thick) were run overnight at a constant current (6 mA per gel) and temperature in a vertical slab apparatus. The separation gel is 12.6 cm long and contains 12% total acrylamide (the monomer solution contains 30% total acrylamide with 2.67% cross-linking in 0.375 M Tris-HCl [pH 8.8] and 0.1% sodium dodecyl sulfate [SDS]); the stacking gel is 12 mm long and contains 5% total acrylamide (the monomer solution again contains 30% total acrylamide with 2.67% cross-linking in 0.125 M Tris-HCl [pH 6.8] and 0.1% SDS). Protein bands are stained with Coomassie blue R-250 in 50% (vol/vol) methanol–10% (vol/vol) acetic acid. These conditions allow separation of proteins and peptides in the molecular weight range of 14,000 to 116,000. The profile was recorded and stored on a personal computer. The similarity between the whole-cell protein profiles of strain H1353 and of a collection of about 250 strains representing all presently named and several unnamed Helicobacter taxa was calculated and expressed by the Pearson product moment correlation coefficient converted for convenience to a percent value.
DNA preparation.
DNA was extracted as described by Niemann et al. (15).
16S rDNA sequencing.
Part of the ribosomal DNA (rDNA) operon, comprising nearly the complete 16S DNA, was amplified by PCR. The forward primer was AGA GTT TGA TCC TGG CTC AG, corresponding to positions 8 to 27 in the Escherichia coli 16S rRNA numbering system. The reverse primer was AAG GAG GTG ATC CAG CCG CA, complementary to positions 1541 to 1522 in the E. coli 16S rRNA numbering system. PCR-amplified 16S rDNAs were purified by using a QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany). Sequence analysis was performed with an Applied Biosystems 377 DNA sequencer by the protocols of the manufacturer (Perkin-Elmer, Applied Biosystems Div., Foster City, Calif.), using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (with AmpliTaq DNA polymerase, Fs). The sequencing primers were those given by Coenye et al. (4). Sequence assembly was performed by using the program AutoAssembler (Perkin-Elmer, Applied Biosystems Div.), and phylogenetic analysis was performed by using the GeneCompar 2.0 software package (Applied Maths).
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession number for the 16S rDNA sequence of strain H1353 is AF118017.
RESULTS
The strain was recovered from a total of four aerobic blood culture vials, one inoculated with blood on day 1, two inoculated on day 21 and one inoculated on day 22, all after 5 days of incubation. The anaerobic vials remained negative. They were nevertheless subcultured at the end of the incubation period onto Columbia agar supplemented with 5% sheep blood in aerobic, anaerobic, and microaerobic atmospheres.
Growth requirements and microscopic examination.
The organism was a slender, fusiform gram-negative rod with occasional incurvation (Fig. 1) and showing a darting motility. Growth was obtained at 37 and 42°C, only under the H2-enriched microaerobic conditions (Table 1). After incubation for 48 h, greyish, glistening, flat colonies spreading to confluence over the entire agar surface were observed on 5% sheep blood agar and 5% fresh horse blood agar.
TABLE 1.
Comparative characteristics of “F. rappini” and selected Helicobacter speciesa
| Test | H. pylori | H. cinaedi | H. fennelliae | H. trogontum | H. bilis |
F. rappini
|
||
|---|---|---|---|---|---|---|---|---|
| Reference 1 | Reference 18 | This report | ||||||
| Oxidase | + | + | + | + | + | + | + | + |
| Catalase | + | + | + | + | + | − | − | − |
| Urease | + | − | − | + | + | + | − | + |
| Hippurate hydrolysis | − | − | − | − | − | − | − | − |
| Nitrate reduction | − | + | − | − | + | − | − | − |
| Microaerobic growth at (°C): | ||||||||
| 25 | − | − | − | − | − | − | − | − |
| 37 | + | + | + | + | + | + | + | + |
| 42 | − | − | − | + | + | + | + | + |
| Susceptibility to: | ||||||||
| Nalidixic acid | R | S | S | R | R | R | S | R |
| Cephalothin | S | I | S | R | R | R | R | R |
| Indoxyl acetate hydrolysis | − | − | + | ND | − | ND | ND | − |
| H2S production (TSI) | − | ND | ND | + | ND | − | ND | No growth |
Phenotypic characterization.
The results of phenotypic characterization are shown in Table 1 in comparison with previously reported data on “F. rappini” and related Helicobacter species. Hydrolysis of urea was detected in 1 h. The isolate was positive for oxidase, alkaline phosphatase, and leucine aminopeptidase and negative for production of catalase, reduction of nitrate, indoxyl acetate, hippurate hydrolysis, and glucose fermentation. No growth was observed on the TSI slant. The bacterium was considered to be resistant to nalidixic acid and cephalothin because no zone of growth inhibition was observed.
Susceptibility to antibiotics.
The strain was considered to be susceptible to the tested drug when the inhibition zone was greater than 30 mm, as found with ceftriaxone, meropenem, erythromycin, clindamycin, clarithromycin, doxycycline, gentamicin, amikacin, ciprofloxacin, nitrofurantoin, and metronidazole. The strain was considered to be resistant to penicillin G and cefazolin because no zone of growth inhibition was observed. Susceptibility to ampicillin and co-trimoxazole appeared to be decreased (inhibition zone diameters of 28 and 22 mm, respectively).
SDS-PAGE protein profile.
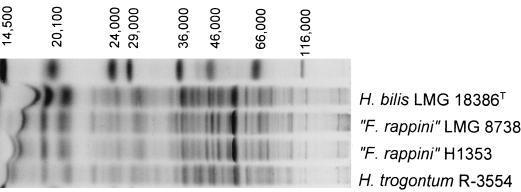
Comparison of the whole-cell protein pattern of strain H1353 with the database revealed that the profile of this strain resembled those of the H. bilis and H. trogontum reference strains but was virtually indistinguishable from that of “F. rappini” LMG 8738, as shown in Fig. 2. The similarity to reference strains of other Helicobacter species was low (data not shown).
FIG. 2.
Electrophoretic protein profiles of H. bilis, H. trogontum, and “F. rappini” reference strains and of strain H1353. The molecular weight markers used (top lane) are indicated from left to right: lysozyme, 14,500; trypsin inhibitor, 20,100; trypsinogen, 24,000; carbonic anhydrase, 29,000; glyceraldehyde-3-phosphate dehydrogenase, 36,000; egg albumin, 45,000; bovine albumin, 66,000; and beta-galactosidase, 116,000.
16S rDNA sequencing.
The 16S rDNA sequence of strain H1353 was determined and was compared to other sequences in the sequence databases (6a). The closest match (99.6%) found was to the 16S rRNA gene of a putative “F. rappini” strain described by Tee et al. (22) (GenBank accession no. AF034135); a similarity level of 99.3% to the “F. rappini” reference strain LMG 8738 (GenBank accession no. M88137) was found.
DISCUSSION
Since the discovery of H. pylori, the genus Helicobacter has been expanding rapidly (7). Except for H. pylori, which is adapted to the human gastric mucosa, Helicobacter species are zoonotic pathogens that are occasionally found in human clinical specimens. Newly described Helicobacter species have been isolated from the stomachs, the livers, or the intestinal tracts of various animals, including rodents, dogs (6, 11), cats (11), chickens, and monkeys. The majority are proven or suspected gastrointestinal or hepatic pathogens for animals only. Some of these organisms are reported to be potentially pathogenic for humans with immunodeficiency (12, 20, 23, 25).
In the present study, a strain with the “F. rappini” morphology was isolated from a blood culture of a 65-year-old HIV-seronegative patient with end-stage renal failure. Analysis of the nearly complete 16S rRNA gene (about 1,450 bp) revealed over 99% sequence similarity to 16S rRNA gene sequences of several “F. rappini” strains; a 98.0% sequence similarity to the H. cinaedi type strain (GenBank accession no. M88150) was the highest similarity to the corresponding genes of related Helicobacter species. The high similarities to the “F. rappini” strains suggest that strain H1353 belongs to the same species. However, Stackebrandt and Goebel (21) demonstrated that these high levels of 16S rRNA gene sequence similarity do not guarantee species identity, as was prematurely assumed by Tee and coworkers (22); this problem was illustrated for members of the genus Helicobacter by Jalava et al. (10), who indicated that strains of multiple Helicobacter species shared over 99% of their 16S rRNA gene sequences.
We used comparative whole-organism protein electrophoresis for species-level identification of strain H1353. This method has been used successfully to identify a wide variety of Helicobacter strains and was shown to correlate with a level of DNA-DNA hybridization which is generally considered the standard for species discrimination (5, 11, 23, 25). The whole-cell protein pattern of strain H1353 had some degree of similarity with those of H. bilis and H. trogontum reference strains but was indistinguishable from that of “F. rappini” LMG 8738. Although the taxonomic status of “F. rappini” has not been finally settled, this degree of whole-cell protein pattern similarity unambiguously indicates that strains H1353 and LMG 8738 likely belong to the same taxon. In addition, all biochemical data determined in the present study correspond with the data reported by Archer et al. (1) for strain LMG 8738. The biochemical and physiological characteristics of “F. rappini” and selected Helicobacter species are shown in Table 1.
“F. rappini” is considered a natural inhabitant of the intestinal mucosa of mice (19). The source of infection and portal of entry in the present case are unclear. The patient owned two cats, one puppy, and several rabbits. He presented with cellulitis due to a scratch made by a cat owned by one of his neighbors. Any of these pets could have been a source of contamination with “F. rappini”. For practical reasons, a search for the microorganism from these animal reservoirs was not attempted. Contact with a puppy was reported in the two other cases of human infections (18, 22). Transmission could have occured by animal contact with the patient’s chronic skin lesions of the lower limbs, the cat scratch associated with secondary cellulitis of the forearm, or inhalation followed by pneumopathy, as suggested for a child with pneumonia (22). Our patient was immunocompromised (due to diabetes mellitus and end-stage renal failure), and severe degenerative arteriopathy was a predisposing condition for a possible endovascular infection, as observed in bacteremia caused by C. fetus (9, 20) or nontyphoid Salmonella.
Our strain appeared to be more susceptible to antibiotics than the strain described by Archer et al. (1). However, it should be stressed that culture conditions and interpretive criteria are not standardized for this type of microorganism. The results of antimicrobial susceptibility testing were consistent with the clinical failure of co-trimoxazole treatment and with cure obtained by using meropenem therapy. Erythromycin may also be a good therapeutic choice, as previously reported (22).
Our results suggest that uncultured CLOs may be grown on standard media if optimal culture conditions are provided, particularly the special microaerophilic atmosphere with hydrogen enrichment and high relative humidity. Because of the very characteristic darting motility of these organisms, phase-contrast microscopy of a wet mount preparation of positive blood cultures is very helpful for selecting the conditions for incubation of subcultures on solid medium.
The identification of CLOs is difficult, and confirmation is unavailable because they have fastidious growth requirements and because determination of the conventional phenotypic characters is not well standardized and provides poor discrimination between species. The comparison of SDS-PAGE profiles of whole-cell proteins by numerical analysis combined with the use of minimal phenotypic tests enables clear and rapid identification of these organisms (9, 17). This method compares favorably with the more costly technique of 16S rRNA sequence analysis. However, access to the latter method and to the reference sequence databases is more widely available than access to the former method, which is performed only by specialized laboratories.
ACKNOWLEDGMENTS
We thank K. Vandemeulebroecke for technical assistance and Isabelle Salmon and Raf De Ryck for assistance with the preparation of figures.
REFERENCES
- 1.Archer J R, Romero S, Ritchie A E, Hamacher M E, Steiner B M, Bryner J H, Schell R F. Characterization of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:101–105. doi: 10.1128/jcm.26.1.101-105.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryner J H, Littleton J, Gates C, Kirkbride C A, Ritchie A E. Microbe 86, abstract book of the XIV International Congress of Microbiology, Manchester, England. 1986. Flexispira rappini gen. nov., sp. nov., a Gram-negative rod from mammalian fetus and feces; pp. 11–18. [Google Scholar]
- 3.Bryner J H. Proceedings of the Fourth International Workshop on Campylobacter Infections, Gôteborg, Sweden. 1987. Flexispira rappini gen. nov., sp. nov., a motile urease-producing rod similar to Campylobacter pyloridis, abstr. 256. [Google Scholar]
- 4.Coenye, T., E. Falsen, M. Vancanneyt, B. Hoste, J. R. W. Govan, K. Kersters, and P. Vandamme. Identification of Alcaligenes faecalis-like isolates from the environment and human clinical samples; description of Ralstonia gilardii sp. nov. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 5.Costas M, On S L W, Owen R J, Lopez-Urquijo B, Lastovica A J. Differentiation of Helicobacter species by numerical analysis of their one-dimensional electrophoretic protein patterns. Syst Appl Microbiol. 1993;16:396–404. [Google Scholar]
- 6.Eaton K A, Dewhirst F E, Paster B J, Tzellas N, Coleman B E, Paola J, Sherding R. Prevalence and varieties of Helicobacter species in dogs from random sources and pet dogs: animal and public health implications. J Clin Microbiol. 1996;34:3165–3170. doi: 10.1128/jcm.34.12.3165-3170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.European Bioinformatics Institute. 6 January 1999, posting date. Sequence databases. [Online.] http://www2.ebi.ac.uk/fasta3/. [3 March 1999, last date accessed.]
- 7.Fox J G. The expanding genus of Helicobacter: pathogenic and zoonotic potential. Semin Gastrointest Dis. 1997;8:124–141. [PubMed] [Google Scholar]
- 8.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe R A, Clarke T, Wilcox M H, Vandamme P, Spencer R C. Campylobacter fetus subspecies fetus septicaemia: SDS-PAGE as an aid to speciation. J Infect. 1995;31:229–232. doi: 10.1016/s0163-4453(95)80032-8. [DOI] [PubMed] [Google Scholar]
- 10.Jalava K, Kaartinen M, Utriainen M, Happonen I, Hänninen M L. Helicobacter salomonis sp. nov., a canine gastric Helicobacter sp. related to Helicobacter felis and Helicobacter bizzozeronii. Int J Syst Bacteriol. 1997;48:975–982. doi: 10.1099/00207713-47-4-975. [DOI] [PubMed] [Google Scholar]
- 11.Jalava K, On S L W, Vandamme P, Happonen I, Sukura A, Hänninen M L. Isolation and identification of Helicobacter spp. from canine and feline gastric mucosa. Appl Environ Microbiol. 1998;64:3998–4006. doi: 10.1128/aem.64.10.3998-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiehlbauch J A, Tauxe R V, Baker C N, Wachsmuth I K. Helicobacter cinaedi-associated bacteremia and cellulitis in immunocompromised patients. Ann Intern Med. 1994;121:90–93. doi: 10.7326/0003-4819-121-2-199407150-00002. [DOI] [PubMed] [Google Scholar]
- 13.Kirkbride C A, Gates C E, Collins J E, Ritchie M S. Ovine abortion associated with an anaerobic bacterium. J Am Vet Med Assoc. 1985;186:789–791. [PubMed] [Google Scholar]
- 14.Mendes E N, Queiroz D M M, Dewhirst F E, Paster B J, Moura S B, Fox J G. Helicobacter trogontum sp. nov. isolated from the rat intestine. Int J Syst Bacteriol. 1996;46:916–921. doi: 10.1099/00207713-46-4-916. [DOI] [PubMed] [Google Scholar]
- 15.Niemann S, Puehler A, Tichy H V, Simon R, Selbitschka W. Evaluation of the resolving power of three different DNA fingerprinting methods to discriminate among isolates of a natural Rhizobium meliloti population. J Appl Microbiol. 1997;82:477–484. doi: 10.1046/j.1365-2672.1997.00141.x. [DOI] [PubMed] [Google Scholar]
- 16.Paster B J, Lee A, Fox J G, Dewhirst F E, Tordoff L A, Fraser G J, O’Rourke J L, Taylor N S, Ferrero R. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991;41:31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- 17.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organism protein fingerprints. In: Goodfellow M, O’Donnell A G, editors. Modern microbial methods. Chemical methods in prokaryotic systematics. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1994. pp. 493–521. [Google Scholar]
- 18.Romero S, Archer J R, Hamacher M E, Bologna S M, Schell R F. Case report of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:142–143. doi: 10.1128/jcm.26.1.142-143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Rosco Diagnostica. Neo-Sensitabs susceptibility testing user’s guide. Taastrup, Denmark: Rosco Diagnostica; 1997. [Google Scholar]
- 19.Schauer D B, Ghori N, Falkow S. Isolation and characterization of “Flexispira rappini” from laboratory mice. J Clin Microbiol. 1993;31:2709–2714. doi: 10.1128/jcm.31.10.2709-2714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skirrow M B, Jones M D, Sutcliffe E, Benjamin J. Campylobacter bacteraemia in England and Wales, 1981–91. Epidemiol Infect. 1993;110:567–573. doi: 10.1017/s0950268800050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 22.Tee W, Leder K, Karroum E, Dyall-Smith M. “Flexispira rappini” bacteremia in a child with pneumonia. J Clin Microbiol. 1998;36:1679–1682. doi: 10.1128/jcm.36.6.1679-1682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandamme P, Falsen E, Pot B, Kersters K, De Ley J. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J Clin Microbiol. 1990;28:1016–1020. doi: 10.1128/jcm.28.5.1016-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991;41:88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- 25.Van der Ven A J A M, Kullberg B J, Vandamme P, Meis J F G M. Helicobacter cinaedi bacteremia associated with localized pain but not with cellulitis. Clin Infect Dis. 1996;22:710–711. doi: 10.1093/clinids/22.4.710. [DOI] [PubMed] [Google Scholar]