Abstract
BACKGROUND
Pancreatic cancer (PC) is one of the deadliest malignancies with an alarming mortality rate. Despite significant advancement in diagnostics and therapeutics, early diagnosis remains elusive causing poor prognosis, marred by mutations and epigenetic modifications in key genes which contribute to disease progression.
AIM
To evaluate the various biological tumor markers collectively for early diagnosis which could act as prognostic biomarkers and helps in future therapeutics of PC in Kashmir valley.
METHODS
A total of 50 confirmed PC cases were included in the study to evaluate the levels of carbohydrate antigen 19-9 (CA 19-9), tissue polypeptide specific antigen (TPS), carcinoembryonic antigen (CEA), vascular endothelial growth factor-A (VEGF-A), and epidermal growth factor receptor (EGFR). Mutational analysis was performed to evaluate the mutations in Kirsten rat sarcoma (KRAS), Breast cancer type 2 (BRCA-2), and deleted in pancreatic cancer-4 (DPC-4) genes. However, epigenetic modifications (methylation of CpG islands) were performed in the promoter regions of cyclin-dependent kinase inhibitor 2A (p16; CDKN2A), MutL homolog 1 (hMLH1), and Ras association domain-containing protein 1(RASSF1A) genes.
RESULTS
We found significantly elevated levels of biological markers CA 19-9 (P ≤ 0.05), TPS (P ≤ 0.05), CEA (P ≤ 0.001), and VEGF (P ≤ 0.001). Molecular genetic analysis revealed that KRAS gene mutation is predominant in codon 12 (16 subjects, P ≤ 0.05), and 13 (12 subjects, P ≤ 0.05). However, we did not find a mutation in DPC-4 (1203G > T) and BRCA-2 (617delT) genes. Furthermore, epigenetic modification revealed that CpG methylation in 21 (P ≤ 0.05) and 4 subjects in the promoter regions of the p16 and hMLH1 gene, respectively.
CONCLUSION
In conclusion, CA 19-9, TPS, CEA, and VEGF levels were significantly elevated and collectively have potential as diagnostic and prognostic markers in PC. Global data of mutation in the KRAS gene commonly in codon 12 and rare in codon 13 could augment the predisposition towards PC. Additionally, methylation of the p16 gene could also modulate transcription of genes thereby increasing the predisposition and susceptibility towards PC.
Keywords: Pancreatic cancer, Genetic mutations, Epigenetic modifications, Biomarkers, Risk factors, Diagnostics
Core Tip: This study demonstrates that the collective evaluation of genetic mutations, epigenetic modifications in key genes and elevated levels of serum carbohydrate antigen 19-9, tissue polypeptide specific antigen, carcinoembryonic antigen, and vascular endothelial growth factor-A could be used as predictive biomarkers for diagnostics and prognostics in pancreatic cancer patients of the ethnic Kashmiri population. This could be useful to track the disease status of pancreatic cancer patients who are on a different regimen of chemotherapeutic interventions. To validate these results in the ethnic Kashmiri population, future studies need comprehensive, cohort, and replicative studies with large sample size.
INTRODUCTION
Pancreatic cancer (PC) is one of the deadliest malignancies among several solid malignancies. It is the 15th leading cancer in the world with an overall estimated incidence of 277000 new cases which is being diagnosed every year[1]. In the United States, PC is the fourth leading cause of death with a 5-year survival rate of less than 5%[2]. PC is mostly found in elderly people and has been reported to be associated with several risk factors[3]. The predominant risk factors include age, cigarette smoking, a high-fat diet, decreased serum levels of folate, diabetes mellitus, obesity, and chronic pancreatitis[4,5]. The familial history of pancreatitis increases the probability of developing PC by around 40%[6]. PC has the lowest prognosis among several solid-type tumors, mainly because almost 80% of PC patients are diagnosed when the disease is in the advanced or metastatic stage[7]. Owing to the lack of specific biological biomarkers used in clinical practice for the detection of PC and its nonspecific symptomology at the initial stage of the malignancy, the early diagnosis is extremely critical to detect and analyze disease progression[8]. Therefore, it is vital to identify specific biomarkers that play a key role in early diagnosis thereby improving the management and therapeutic outcome in PC.
Tumor biomarker(s) are the substances that can be examined in body fluids (blood, urine, and other fluids), synthesized and excreted by malignant cells within the tumor tissue besides exceeding the normal level potentiating its use for cancer diagnosis and/or prognosis[9]. Thus, ideally, the tumor markers should have high sensitivity and specificity, however, none of the tumor biomarkers have attained such precision[10]. Recent reports suggest that commonly used biomarkers for various malignancies include carbohydrate antigen 19-9 (CA 19-9), tissue polypeptide specific antigen (TPS), carcinoembryonic antigen (CEA), vascular endothelial growth factor-A (VEGF-A), and epidermal growth factor receptor (EGFR)[11]. The CA 19-9 and CEA are high molecular weight glycoproteins attached to the surface of tumor cells predominantly used in the diagnosis and prognosis of gut-associated cancers. However, marred by low sensitivity and specificity, they far from qualify for the diagnosis of other cancers[12]. The group of intermediate filament proteins to which TPS belongs is mainly used to measure cytokeratin 18 and 19 and expected to reflect the tumor progression. A few studies have examined TPS expression in PC; however, the findings are contentious. Although, individually TPS expression in PC may not provide significant information about the disease progression; in concert with other tumor biological markers it is worthwhile to evaluate its role for early diagnosis, prognosis, and to predict metastatic growth of PC[13]. A predominant dimeric, heparin-associated glycoprotein, VEGF-A has powerful pro-angiogenic and mitogenic activity. Elevated expression of VEGF-A enhances vascular permeability of endothelial cells and is reported to be involved in PC-associated angiogenesis[14], thus potentiates as a predictive biomarker. EGFR is a transmembrane protein that regulates cell growth and development. Mutation or elevated expression of EGFR is a key event in the pathogenesis of various malignancies such as glioblastoma, lung and oral carcinomas. There are reports of EGFR-mediated signaling associated with EGFR mutation in PC patients[15]. Consequently, these reports suggest that evaluation of serum EGFR levels in PC can be a promising putative biomarker for early diagnosis and prognosis to monitor the disease status post-therapeutic interventions. Although, individually the tumor biomarkers could aid in diagnostic and prognostic evaluation to a certain level, however, collectively, they can be more beneficial to track tumor progression and could be more useful to monitor disease status. Therefore, the current study aims to evaluate the collective role of various tumor biomarkers in PC patients for their potential role in early diagnosis and application in prognosis to examine post-treatment disease status in the ethnic Kashmiri population.
Genetic mutations play a pivotal role in tumor progression and genetic markers are critically important for the detection of malignant changes in PC[16]. Approximately, 97% of PC patients have alterations in genes that either follow the germline inheritance mode of transmission or occur sporadically[17]. These mutations could be either oncogenic (gain of function) or diminish tumor suppressor activity (loss of function). Gain-of-function in Kirsten rat sarcoma (KRAS) a proto-oncogene that encodes guanosine triphosphatase (GTPase), is one of the prominent mediators in signal transduction pathways that are implicated in neoplastic transformation and inflammation[18]. Approximately 95% of all cancers including PC are reported to harbor a KRAS gene mutation which is a key event in early tumorigenesis. The major KRAS activating gene mutations reportedly occur at codon 12 and less commonly at codon 13 and codon 61. Therefore, evaluation of genetic mutational analysis at the hotspot regions of the KRAS gene could help in early diagnosis and prognosis in PC. Breast cancer type 2 (BRCA-2), a tumor suppressor gene is associated with the maintenance of the genome by enhancing homologous recombination of a double-stranded break. Around 80% of BRCA-2 mutations are either frameshift or nonsense mutations that result in the formation of premature stop codons to encode non-functional BRCA-2 protein[19]. Almost 7.3% of PC patients have a hereditary mutation in the BRCA-2 gene which increases the risk of developing PC by approximately 20-fold[20] implicating a critical role of BRCA-2 in the early diagnosis of PC. Another vital gene, ‘deleted in pancreatic cancer-4’ (DPC-4) also known as SMAD family member 4, mothers against decapentaplegic homolog 4 (SMAD-4) is a tumor suppressor gene involved in the regulation of gene transcription. DPC-4 protein a downstream target of transforming growth factor-beta (TGF-β) pathway plays a critical role in the activation of TGF-β signaling thereby promotes neoplastic growth. It is reported that 30% of PC cases develop due to homozygous mutations in the DPC-4 gene[21,22]. Thus, mutational analysis of the DPC-4 gene could be a promising factor for the early diagnosis and prognosis of PC.
Besides genetic mutations, recent evidence suggests the epigenetic modifications such as DNA methylation plays a critical role in the pathogenesis of PC. In the recent past, reports suggest that methylation at the promoter regions of key tumor suppressor genes induces gene silencing and contributes to the development and progression of tumorigenesis[23]. Various tumor suppressor genes were inactivated by epigenetic modifications. Cyclin-dependent kinase inhibitor 2A (p16; CDKN2A), a tumor suppressor gene that encodes a member of cyclin-dependent kinase inhibitor which arrests the G1-S phase of the cell cycle to prevent tumor cell progression. Loss of the p16 gene is reported in 70% of cancers and around 10%-15% of the loss was due to promoter methylation[24]. Thus, screening of epigenetic modifications at the promoter region of the p16 gene could help in the early diagnosis of PC. MutL homolog 1 (hMLH1) a tumor suppressor gene that belongs to the mismatched repair gene family and prevents DNA damage by radiations and other associated mechanisms. hMLH1 is also reported to be inactivated epigenetically by promoter methylation which leads to DNA damage. The accumulation of mismatched and damaged DNA promotes tumor cell progression[25]. Ras association domain-containing protein 1(RASSF1A) is another tumor suppressor gene inactivated by promoter methylation. It is a component of RAS/PI3K/AKT and RAS/MAPK pathways. Recent reports suggest that epigenetic modifications in the RASSF1A promoter region promote tumor progression in various cancers including kidney, breast, lung, prostate, and thyroid[26]. Recent evidence suggests that 64% of pancreatic adenocarcinoma patients have RASSF1A hypermethylation at the promoter region, indicating that analysis of hypermethylation of RASSF1A at promoter region could be a promising approach for early diagnosis of PC. To summarize, these studies suggest that genetic mutations, epigenetic modulations, and elevated levels of serum biological markers play a critical role in the early diagnosis, therapeutics, and prognosis of various cancer. Therefore, keeping in consideration the documented role of tumor biomarkers, genetic mutations, and DNA methylation of tumor suppressor/protooncogenes in various malignancies including PC. The current study aimed to evaluate the serum levels of various biological tumor markers, genetic mutations, and epigenetic modifications of some key regulatory genes in PC. This would prove immensely helpful in the early diagnosis of PC, which helps in the identification of high-risk PC and may enable their development as biomarkers for future diagnostics, therapeutic interventions, and prognostics in the ethnic Kashmiri population.
MATERIALS AND METHODS
In this study, a total of 50 patients with pancreatic carcinoma and 50 healthy controls were included.
Inclusion criteria
Only the patients with histologically confirmed pancreatic carcinoma were included in this study. Written consent was taken at the very beginning from all the patients and healthy controls that were included in the study.
Exclusion criteria
The patients with a history of other malignancies and those who were not willing to comply with pre-requisite protocol were excluded from the study.
Physical examination and lifestyle habits
The study was designed and approved by the institutional review board of the Sher-i-Kashmir Institute of Medical Sciences (SKIMS), and informed consent was obtained from all participants. A comprehensive physical/clinical examination was performed in the Department of Medical Oncology, SKIMS, and the patients were evaluated for Jaundice (by examining features like yellowing of eyes and skin), pruritis (by examining features like redness, bumps, spots or blisters, dry/cracked skin and leathery/scaly skin), muscle wasting (by evaluating features like weakness or numbness in the limbs, loss of muscle coordination, tingling or weakness of the extremities, impaired balance while walking, fatigue and a general illness, facial weakness, progressive weakness, gradual memory loss and liver enlargement (were examined by features like abdominal pain, nausea/vomiting, fatigue, whitening of eyes and yellowing of the skin).
Besides the physical examination, lifestyle activities of the PC patients were also recorded which included smoking status, salt tea consumption, spicy and non-spicy food intake, dried vegetable consumption, mutton, and beef consumption, fish consumption, oil intake, urine habits, bowel habits, and daily physical activity were also recorded.
Laboratory findings
The basic clinical laboratory findings were performed by using automated analyzers. The laboratory findings are liver function test (like aspartate transaminase-AST, alanine transaminase-ALT, bilirubin, and alkaline phosphatase-ALP), diabetic status (hyperglycemia) by measuring glucose levels, and anemia by measuring red blood cell (RBC) count.
Diagnostic imaging
For any other malignancy PC patients were initially screened by using multiphase multidetector computed tomography (CT) scan, magnetic resonance imaging (MRI), ultrasonography (USG), endoscopic ultrasound (EUS), and chest X-ray (CXR).
Sample collection
A total of 5 mL blood sample was collected in clot activator and Ethylenediaminetetraacetic acid (EDTA) vial from PC patients. The serum was separated from the clot activator vials using centrifugation and was stored at -80 °C for further analysis. EDTA vials contain blood was stored at -20 °C for DNA extraction. A tissue chunk (12-50 μm thick tumor tissue section) was obtained from the PC patient by endoscopy using USG-guided probes for mutational analysis and epigenetic modifications.
Tumor biomarkers
Tumor markers including CA19-9, TPS, CEA, VEGF-A, and EGFR were estimated in the serum obtained from blood collected from the PC patients. Measurement of CA 19-9, CEA, VEGF-A, and EGFR levels in serum were performed by using a modular E-170 analyzer. However, TPS levels in the serum were measured by using an Immulite instrument.
Genetic mutation analysis
Genomic DNA was extracted by the phenol-chloroform method from mononuclear cells. Hypaque density gradient centrifugation was performed to extract leucocytes from blood and tissue samples obtained from PC patients. The quantity and quality control analysis of genomic DNA was performed by carrying out UV spectrophotometer (Eppendorf Biospectrometer®, Hamburg Germany) analysis and Gel electrophoresis, respectively. However, polymerase chain reaction (PCR) was carried out with a different set of primers for KRAS, DPC-4, and BRACA-2 genes under different PCR conditions. The PCR products obtained were subjected to Restriction Fragment Length Polymorphism (RFLP) using restriction enzymes BstN1 and BglI for mutational analysis of KRAS codon 12 and 13, respectively. GGA→TGA in exon 8, codon 358 of DPC-4 gene was analyzed by using MnlI restriction enzyme. 6174delT of BRCA-2 was analyzed by using allele-specific PCR technique and polyacrylamide gel electrophoresis (PAGE) was carried out to study any change in the BRCA-2 gene.
Epigenetic analysis
The epigenetic analysis was performed by examining the methylation status of the promoter and exon regions of genes including p16, RASSF1A, and hMLH1. The methylation status of p16, RASSF1A, and hMLH1 genes was determined by methylation-specific (MSP) PCR. Briefly, DNA extracted from tissue samples was first subjected to bisulfite conversion using EZ direct methylation kit. The bisulfite-converted DNA was then subjected to PCR using methylated and unmethylated primers specific for the respective genes. The results were analyzed on 2% agarose gel.
Statistical analysis
Numerical data collected from experiments for statistical analysis were performed by using non-parametrical statistical analysis tools which are the Kruskal-Wallis test and Mann-Whitney U test.
RESULTS
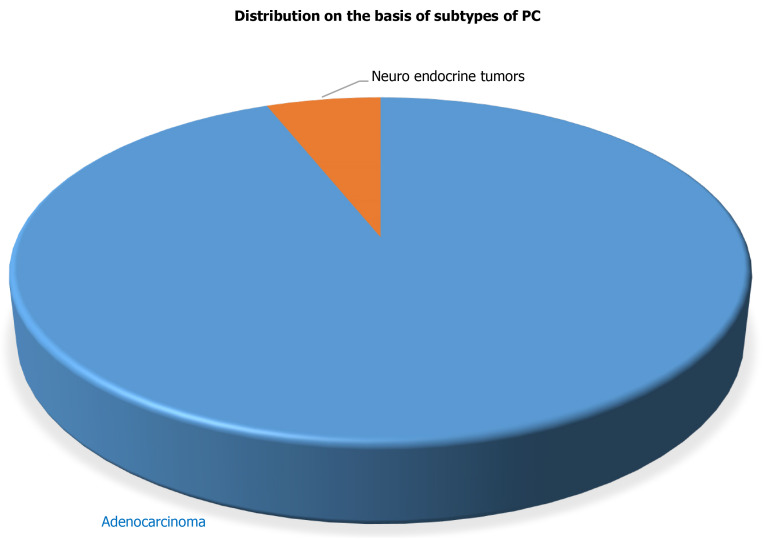
The current study included 50 PC patients with a mean age of 47.82 years at the time of diagnosis for the evaluation of various tumor biological markers for the PC diagnosis. Radio-diagnostics such as USG and CT, confirmed that all 50 patients had PC. Further, histopathological analysis supported the radio-diagnostic results and revealed that out of 50 confirmed PC patients, 47 PC patients had characterized to have adenocarcinoma whereas the remaining 3 PC patients have neuroendocrine carcinoma in the pancreas as shown in Figure 1. The other demographic parameters and daily activities of all confirmed 50 PC patients are presented in Table 1.
Figure 1.
Distribution of samples (cases) based on subtypes of pancreatic cancer. PC: Pancreatic cancer.
Table 1.
Characteristics and clinical presentations of cases in the present study, n (%)
|
Patient characteristics
|
Cases, n = 50
|
P
value
|
| Age in yr | ||
| ≤ 50 | 28 (56.0) | 0.0377 |
| > 50 | 22 (44.0) | |
| Gender | ||
| Male | 29 (58.0) | 0.031 |
| Female | 21 (42.0) | |
| Family history | ||
| Smoker | ||
| Yes | 22 (44.0) | 0.034 |
| No | 28 (56.0) | |
| Lifestyle | ||
| Active | 44 (88.0) | 0.001 |
| Sedentary | 06 (12.0) | |
| Residence | ||
| Rural | 40 (80.0) | 0.01 |
| Urban | 10 (20.0) | |
| Dietary habits | ||
| Salt tea | ||
| Yes | 47 (94.0) | 0.01 |
| No | 03 (06.0) | |
| Spicy food | ||
| Yes | 28 (56.0) | 0.043 |
| No | 22 (44.0) | |
| Appetite | ||
| Yes | 25 (50.0) | 0.05 |
| No | 25 (50.0) | |
| Vegetables | ||
| Yes | 48 (100.0) | 0.001 |
| No | 02 (00.0) | |
| Non-veg. | ||
| Yes | 47 (100.0) | 0.001 |
| No | 03 (00.0) | |
| Edible oil | ||
| Saturated | 43 (86.0) | 0.01 |
| Unsaturated | 07 (17.0) | |
| Urine habits | ||
| Normal | 28 (56.0) | 0.05 |
| Disturbed | 22 (44.0) | |
| Bowel habits | ||
| Normal | 30 (60.0) | 0.05 |
| Disturbed | 20 (40.0) |
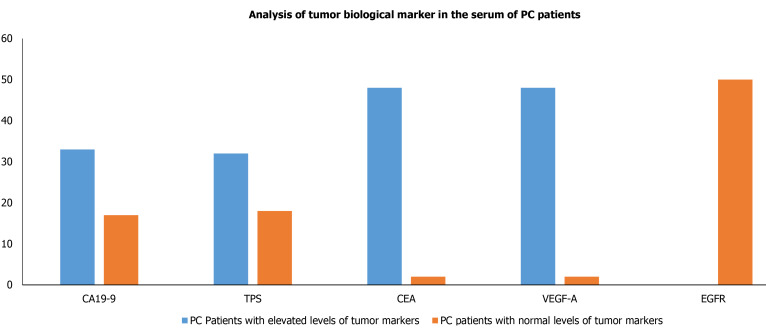
Owing to have relative ease in blood collection and non-invasive, it is preferred to evaluate the biological tumor markers in serum. Therefore, we also intended to evaluate the biological tumor markers which included CA 19-9, TPS, CEA, VEGF-A, and EGFR levels in the blood collected from PC patients. Our results demonstrated that the levels of serum biological tumor markers CA 19-9, CEA, VEGF-A, TPS, EGFR of PC patients were significantly raised in 33 (66%), 32 (64%), 48 (96%), 48 (96%) and 0 out of 50 PC patients, respectively (Table 2) and Figure 2.
Table 2.
Analysis of tumor biological marker in the serum of pancreatic cancer patients
|
Tumor marker
|
Normal level
|
PC patients with elevated levels of tumor markers
|
PC patients with normal levels of tumor markers
|
P
value for PC patients with elevated levels vs normal levels
|
| CA19-9 | < 37 U/mL | 33 | 17 | 0.05 |
| TPS | < 80 U/L | 32 | 18 | 0.05 |
| CEA | < 5 ng/mL | 48 | 02 | 0.003 |
| VEGF-A | 31.2-2000 pg/mL | 48 | 02 | 0.003 |
| EGFR | 62.5-4000 pg/mL | 0 | 50 | 0.001 |
PC: Pancreatic cancer; CA19-9: Carbohydrate antigen 19-9; TPS: Tissue polypeptide specific antigen; CEA: Carcinoembryonic antigen; VEGF-A: Vascular endothelial growth factor-A; EGFR: Epidermal growth factor receptor.
Figure 2.
Analysis of tumor biological marker (carbohydrate antigen 19-9, tissue polypeptide specific antigen, carcinoembryonic antigen, vascular endothelial growth factor-A, and epidermal growth factor receptor) in the serum of pancreatic cancer patients. PC: Pancreatic cancer; CA19-9: Carbohydrate antigen 19-9; TPS: Tissue polypeptide specific antigen; CEA: Carcinoembryonic antigen; VEGF-A: Vascular endothelial growth factor-A; EGFR: Epidermal growth factor receptor.
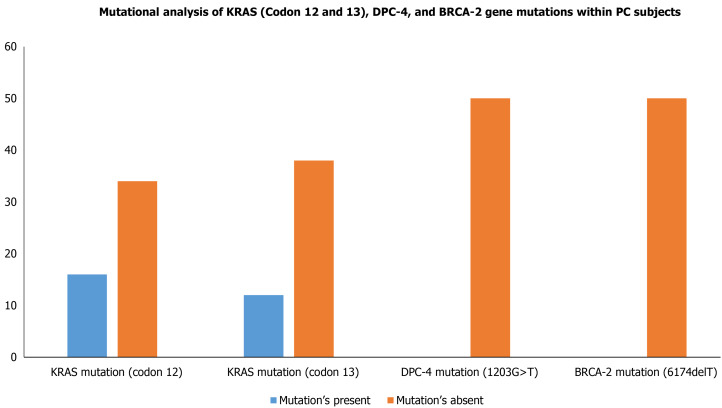
Although the PC progression is a heterogeneous and complex process that includes cell proliferation of intraepithelial and dysplasia to form a mass of cells, followed by an invasion of cells to neighboring tissues. Subsequently, one of the important driving factors of PC progression is genetic mutations of protooncogenes (gain-of-function) and tumor suppressor genes (loss-of-function). Among genetic mutations, KRAS mutation is the key point mutation followed by deletion mutation in tumor suppressor genes BRCA-2, DPC-4, and p16 in PC. To evaluate whether the PC patients in our study harbor these mutations, we sought to perform mutational analysis of KRAS hotspot codons (codon 12 and codon 13), DPC-4 (1203G>T), and BRCA-2 mutation (6174delT) in our PC subjects. Our mutational analysis results revealed that out of 50 PC patients, 16 and 12 PC patients had KRAS mutation at codons 12 and 13, respectively. However, we could not find mutation(s) at codons 12 and 13 of the KRAS gene in the remaining 34 and 38 PC patients, respectively Figure 3. The representative agarose gel picture of the amplification product of codons 12 and 13 of the KRAS gene and their RFLP pattern is shown in Figures 4 and 5. Subsequently, the mutational analysis of tumor suppressor gene DPC-4 (1203G>T) and BRCA-2 (6174delT) were also evaluated in all PC subjects. Interestingly, we did not find any mutations in DPC-4 and BRCA-2 mutation at (1203G>T) (6174delT) sites, respectively Figure 3. The representative agarose gel picture of DPC-4 (1203G>T) amplification and RFLP pattern is shown in Figure 6, respectively, and that of amplification and RFLP pattern of BRCA-2 (6174delT) is shown in Figure 7. The results obtained from the genetic mutation analysis are summarized in Table 3.
Figure 3.
Mutational analysis of KRAS (Codon 12 and 13), DPC-4, and BRCA-2 gene mutations within pancreatic cancer cases. KRAS: Kirsten rat sarcoma; DPC-4: Deleted in pancreatic cancer-4; BRCA-2: Breast cancer type 2.
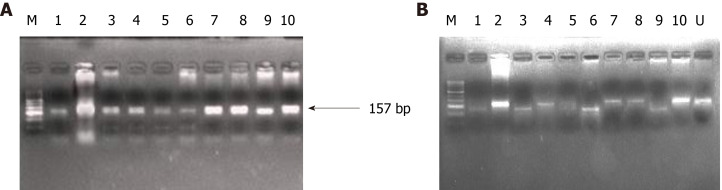
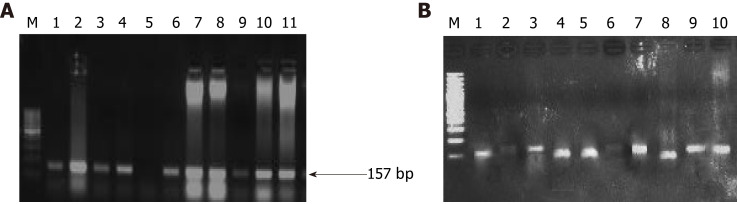
Figure 4.
Representative agarose gel picture of polymerase chain reaction amplification (A) and restriction fragment length polymorphism using BstN1 (B) for KRAS codon 12. The arrow represents the 157 bp amplicon and M denotes the DNA marker (50 bp). Lane M represents a DNA marker (50 bp). Lanes 2, 4, 7, 8, and 10 represent the mutant band (undigested) of 157 bp. Lane 1, 3, 6, and 9 represent the wild band (digested) of 128 bp. U represents the undigested band used as mutant control.
Figure 5.
Representative gel picture of polymerase chain reaction amplification (A) and restriction fragment length polymorphism using BglI (B) for K-RAS codon 13. The arrow represents the 157 bp amplicon and M denotes the DNA marker (100 bp). Lane M represents a DNA marker (100 bp). Lanes 2, 3, 6, 7, 8, 9, 10, and 11 represent the mutant band (undigested) of 157 bp. Lanes 1, 4, 5, and 8 represent a wild band (digested) of 125 bp.
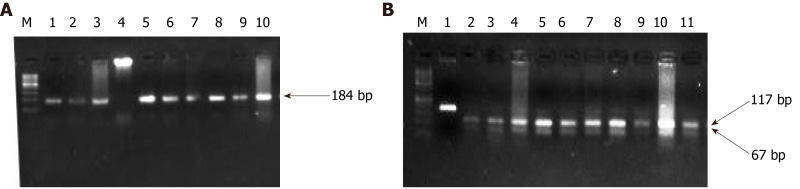
Figure 6.
Representative gel picture of polymerase chain reaction amplification and restriction fragment length polymorphism using MnlI of DPC-4. The arrow represents the 184bp amplicon and M denotes the DNA marker (100 bp). Lane M represents a DNA marker (50 bp). 117 bp and 67 bp represent the wild bands (digested). U represents the mutant control band of 184 bp (undigested).
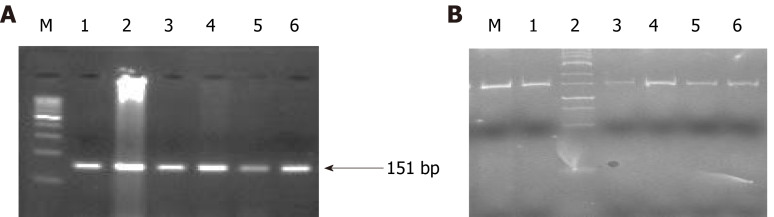
Figure 7.
Representative agarose gel picture of AS-polymerase chain reaction amplification (A) and gel picture representing polyacrylamide gel electrophoresis (20% gel) (B) of BRCA-2. All the lanes show 151 bp amplicon which is the wild band and M denotes the DNA marker (100 bp). M is the marker lane (25 bp). Here also, the only wild band (151 bp) is observed in all the lanes.
Table 3.
Mutational analysis of KRAS (Codon 12 and 13), DPC-4, and BRCA-2 gene mutations within pancreatic cancer subjects
|
Key genes in PC patients evaluated for mutational analysis
|
Mutation’s present
|
Mutation’s absent
|
P
value for mutations present vs absent
|
| KRAS mutation (codon 12) | 16 | 34 | 0.05 |
| KRAS mutation (codon 13) | 12 | 38 | 0.05 |
| DPC-4 mutation (1203G>T) | 0 | 50 | 0.001 |
| BRCA-2 mutation (6174delT) | 0 | 50 | 0.001 |
KRAS: Kirsten rat sarcoma; DPC-4: Deleted in pancreatic cancer-4; BRCA-2: Breast cancer type 2.
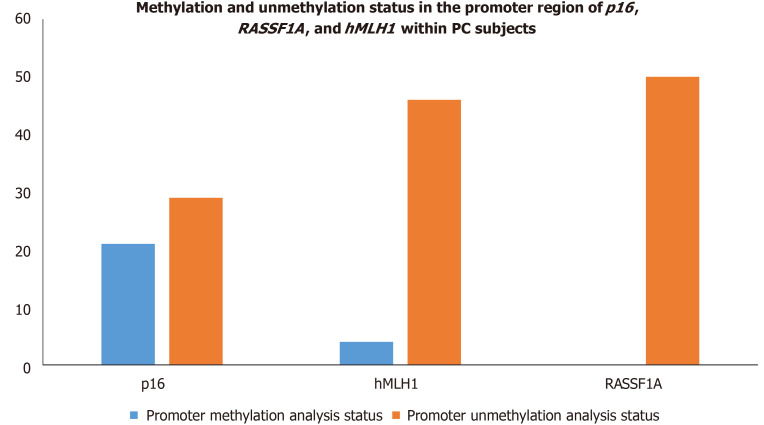
Epigenetic alterations have been documented to play a crucial role in PC progression. The p16, RASSF1A, and hMLH1 are key tumor suppressor genes regulating mismatch repair to minimize DNA damage and are frequently inactivated by epigenetic modification in various malignancies. Therefore, we intended to investigate the epigenetic modification (methylation of CpG islands) of p16, RASSF1A, and hMLH1 genes by determining the methylation in their respective promoter regions in all PC subjects. Our epigenetic modification results demonstrated that 21 out of 50 PC subjects were found methylated in the CpG islands of the promoter region of the p16 gene while the remaining 29 were unmethylated. However, the CpG islands in the promoter region of RASSF1A were found to be unmethylated in all 50 PC patients. Additionally, we observed that 4 out of 50 PC patients showed methylation patterns in the promoter region of the hMLH1 gene, whereas the remaining 46 PC patients had the hMLH1 gene unmethylated in their promoter regions. The representative agarose gel pic of MS-PCR for hMLH1 and RASSF1A is shown in Figure 8. The methylation and unmethylation status as observed in the present study for p16, hMLH1, and RASSF1A are summarized in Table 4 and Figure 9.
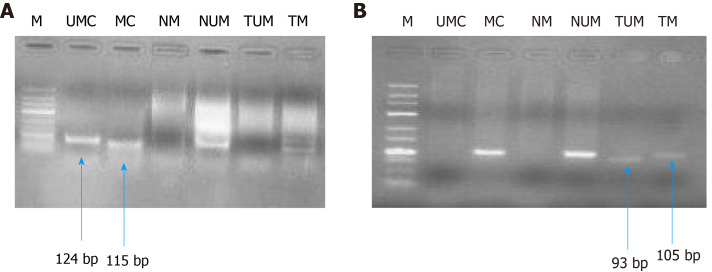
Figure 8.
Gel picture representing MS-polymerase chain reaction for hMLH1 (A, 100 bp) and RASSF1A (B, 25 bp). M represents DNA marker; UMC represents unmethylated control; MC represents methylated control; NM represents normal methylated; NUM represents normal unmethylated; TUM represents tumor unmethylated; TM represents tumor methylation.
Table 4.
Methylation and unmethylation status in the promoter region of p16, RASSF1A, and hMLH1within pancreatic cancer subjects
|
Genes
|
Promoter methylation analysis status
|
Promoter unmethylation analysis status
|
P
value for promoter methylation vs ummethylation
|
| p16 | 21 | 29 | 0.05 |
| RASSF1A | 0 | 50 | 0.001 |
| hMLH1 | 4 | 46 | 0.165 |
Figure 9.
Methylation and unmethylation status in the promoter region of p16,RASSF1A, and hMLH1 within pancreatic cancer subjects. PC: Pancreatic cancer.
DISCUSSION
Despite new therapeutic approaches to improve the outcome of PC patients by the introduction of molecular target approaches and combinatorial therapy, there is an unmet need to find the prospective biomarkers for early diagnosis of PC[27]. Therefore, the aim of the current study was to collective evaluation of tumor biological markers, mutational status, and epigenetic modulations in PC patients of the ethnic Kashmiri population for early diagnosis. Our findings revealed the elevated levels of serum biomarkers CA 19-9, TPS, CEA, and VEGF-A, in the blood samples of PC patients, however, EGFR levels were found to be in the normal range. The mutational analysis demonstrated that the KRAS gene mutation which is the major driver in PC progression was found in codons 12 (16 subjects) and 13 (12 subjects). Furthermore, DNA of CpG islands of 21 subjects was found significantly methylated in the promoter regions of the p16 gene. Collectively, these results suggest that in combination with mutational analysis and epigenetic modulations (CpG methylation), the biological tumor markers evaluated in PC subjects could be valuable for early diagnostics and could strongly predict the PC prognostics. Additionally, these types of studies could further strengthen the validation of biological tumor markers and have a promising perspective for the predisposition and susceptibility towards PC.
A biological tumor marker is an entity in the body that gives information about a diagnosis, prognosis, and therapeutic modalities for a particular disease. The preferred entity to be eligible as a biomarker should be available in body fluids and non-invasive[28]. One of the important tumor biomarkers used in various malignancies is a high molecular weight glycoprotein CA 19-9. Biochemically, a carbohydrate antigen, CA19-9 is mainly expressed by the cells of the pancreaticobiliary system. Previous studies suggest that CA 19-9 levels were elevated in gut-associated malignancies such as gastric, bile duct, colorectal, and ovarian cancers. Owing to its relatively higher sensitivity and specificity among other biomarkers in PC patients, CA 19-9 is an important and valuable biomarker in the diagnostics of PC[29]. Although reports suggest a significant progress in overall survival and reduction in CA19-9 levels in PC, however, a recent study by Hess et al[30] did not support these findings. O’Brien et al[31] reported that CA 19-9 levels were raised in PC patients and may act as a better biomarker for the early diagnosis of PC. Besides, the levels of CA 19-9 were found directly associated with tumor size, tumor burden, and stage of tumorigenesis in PC, the pre-and post-operative levels of CA 19-9 in PC patients could be used as a prognosticator. Consistent with these findings, our results revealed that out of 50 PC patients 33 had significantly elevated levels of CA 19-9 in their blood samples, which indicates that more studies with a large cohort size are needed in the future to validate CA 19-9 as a better early diagnostic biomarker in PC.
Another valuable biomarker used in the diagnosis of various malignancies is TPS. It is essentially an antigen that binds to the epitope of soluble cytokeratin 18 fragments. The striking feature of TPS is to differentiate between PC and chronic pancreatitis and it is a better marker than CA 19-9 for differentiating PC and pancreatitis[32]. Previous studies suggested that serum TPS levels have a better correlation with gastric, colorectal, and pancreatic cancer than CA 19-9, CA 195, or CEA biomarkers[33,34]. Consistent with the previous studies, our results revealed that 48 out of 50 confirmed PC patients had significantly elevated levels of TPS, which suggests that elevated levels of serum TPS are better correlated with PC than CA 19-9 and could act as a better diagnostic and prognostic biomarker in PC. CEA, a glycoprotein, first identified in 1965, is present normally in the fetal pancreas, gastrointestinal tract, and liver. In the adolescent stage, it is found in lesser quantity in endodermal tissue and colon. CEA was used as a diagnostic marker of PC decades before and is now replaced by markers that have greater sensitivity for the detection of PC[35]. Elevated serum levels of CEA have been documented in more than 60% of cases of PC. Consistent with previous findings, our results demonstrated that 64% of patients (32 out of 50 confirmed PC) had elevated levels of serum CEA. However, if used with other biomarkers for early diagnostics, CEA could be adding up great value to early diagnostics of PC[36]. High expression of VEGF-A is associated with tumor size and progression. Overexpression of VEGF-A has been reported in head and neck, non-small cell lung, ovarian, endometrial, osteosarcoma, bladder, B cell lymphoma, ocular adnexal lymphoma, papillary renal cell carcinoma, and pancreatic cancers. VEGF-A is reported to have an 80 gene loci whose alterations are reported in hepatocarcinoma, lung, pancreatic, and endometrial cancers[36]. A study conducted by Seo et al[37] demonstrated that 93% of ductal pancreatic adenocarcinomas showed high expression of VEGF-A protein. In the recent past, around 77% of VEGF-A expression was observed in PC tissues whereas only 15% of VEGF-A expression was found in the normal range[38]. Consistent with these findings, our results showed that out of 50 confirmed PC subjects, 48 cases had elevated levels of VEGF-A expression, which indicates that VEGF-A plays a critical role and had a strong causal association with PC progression, thus could act as a valuable tumor biomarker in combination with other biomarkers for early diagnosis of PC.
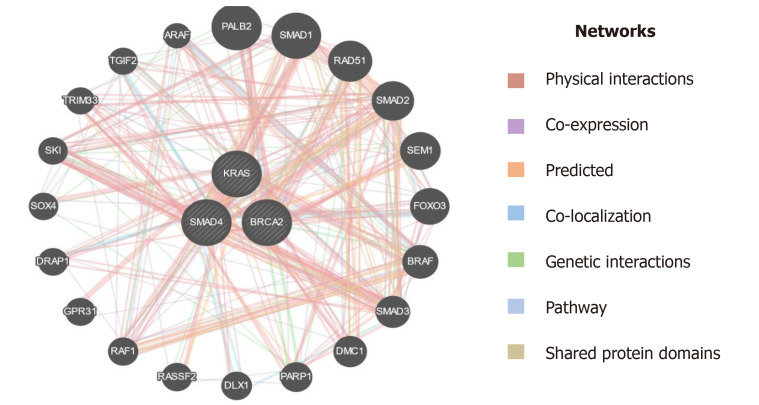
Besides, the currently available biomarkers for PC diagnostics, it is worthy to introduce genetic markers to develop more sophisticated tools for early detection of PC. PC is a disease that harbors somatic as well as hereditary mutations. Approximately, 5%-10% of the familial PC is caused by a mutation in a myriad of genes and surges the predisposition to PC by several-fold[24]. Previous studies reported that several genes showed a strong causal association with PC progression, among these most important are KRAS, DPC-4, and BRCA-2 which in turn are associated with different other genes using different interactions like physical interaction, genetic interaction, shared protein interaction, etc. as depicted in Figure 10[39]. The most common mutation reported in PC is KRAS mutation and is the earliest recognizable event in its pathogenesis. Studies have reported that mutations in the KRAS gene are mainly limited to codon 12 and rarely on codon 13[40,41]. The pathological mutation in KRAS encodes constitutive Ras protein which belongs to GTP binding protein family. The constitute Ras protein facilitates the oncogenic signaling pathway which leads to inflammation, deregulated cellular growth, cell motility, and remodeling of the cytoskeletal elements. KRAS gene mutations are known to be driver mutations that occur sporadically. It accounts for 30% of early neoplasms of the pancreas and nearly 100% in pancreatic adenocarcinomas. Besides PC, a mutation in the KRAS gene is adequate to promote lung cancer, colon cancer, breast cancer, and other cancers as well[42,43]. Recent clinical data suggest that KRAS mutations act as significant prognostic biomarker to predict therapeutic intervention for PC management. Kim et al[44] demonstrated that out of 136 PC patients, 70 PC patients harbored point mutation in codon 12 of the KRAS gene, and these patients have shown dismal response to gemcitabine-based chemotherapy compared to those who had wild type allele for KRAS gene[44]. Another study revealed that out of 173 PC patients, 121 were found to harbor point mutations in codon 12 of the KRAS gene, and among them are nonresponders to erlotinib. However, patients with wild-type alleles displayed a promising overall survival rate[45]. Consistent with these studies, our mutational analysis results revealed that 16 PC patients had KRAS point mutation at codon 12, interestingly we observe point mutation in the KRAS gene at codon 13. These findings suggest that further studies are needed to validate the high frequency of point mutation in codon 13 of the KRAS gene. BRCA-1 and BRCA-2 germline mutations substantially increase the lifetime risk of breast cancer tumorigenesis. Recent reports suggest that the mutations in these genes also have a strong causal association with other cancers including PC[46]. The primary role of the BRCA-2 gene is the maintenance of the genome by enhancing homologous recombination of a double-stranded break. Approximately, 80% of BRCA-2 mutations are either frameshift or non-sense mutations which result in the formation of premature stop codons to encode non-functional BRCA-2 protein. BRCA-2 mutations have been found in 7.3% of familial PC patients which indicates an increased risk of cancers by about 20-fold[47]. DPC-4 (SMAD-4) is a tumor suppressor gene intricated in the regulatory mechanisms of gene transcription. Approximately, 30% of PC cases have been reported to harbor homozygous mutations in the DPC-4 gene[21]. The mutated DPC-4 gene-encoded hyperactivated Smad-4 protein leads to the activation of TGF-β pathways thereby promote cell proliferation and tumor growth. Mutations in the DPC-4 gene have been reported in approximately 50% of PCs and serves as a leading cause of protein inactivation[48]. Inconsistent with previous studies, our mutational analysis of BRCA-2 and DPC-4 genes revealed a zero frequency of DPC-4 1203 G>T and BRCA-2 6174 deletion in PC patients. Collectively, these findings suggest that a larger sample size is needed to validate our results in the ethnic Kashmiri population.
Figure 10.
Interaction of KRAS, SMAD4 (DPC4), and BRCA2 with other genes based on various parameters. KRAS: Kirsten rat sarcoma; DPC-4: Deleted in pancreatic cancer-4; BRCA-2: Breast cancer type 2.
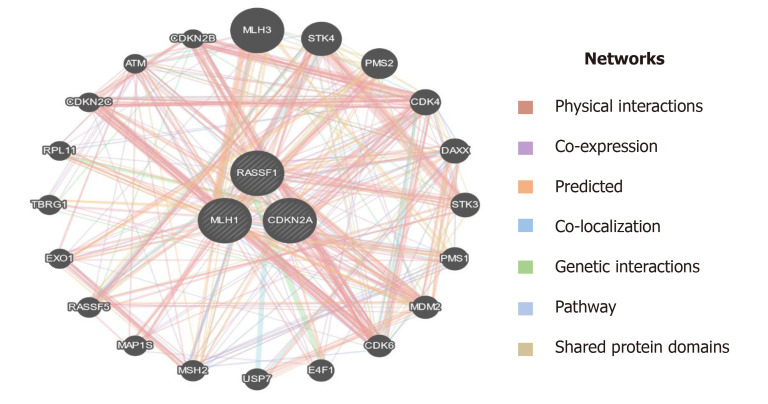
Epigenetic modulations play a critical role in tumorigenesis. Change in DNA methylation of tumor suppressor genes has indispensable importance in therapeutics and could serve as biomarkers for diagnostics and prognostics in various cancers[49]. p16 is a tumor suppressor gene that encodes cyclin-dependent kinase inhibitors to arrest the cellular growth of malignant cells. Besides point mutations and homozygous deletions in the p16 gene, recent evidence suggests that methylation of CpG islands in the promoter regions of p16 stimulates transcriptional silencing of the p16 gene and contributes to PC progression. In the recent past, hypermethylation in the promoter region of p16 is significantly raised in chronic pancreatitis compared to normal; suggesting that hypermethylation in the promoter region of the p16 gene might deregulate cell cycle kinetics and could promote PC progression. Further, reports demonstrated that p16 hypermethylation in chronic pancreatitis might increase the risk of PC development many-fold[48]. Moore et al[50] demonstrated the role of p16 promoter hypermethylation and associated molecular pathways involved in exocrine and endocrine development of PC. Further, studies suggest that the reduction in overall survival rate associated with p16 alterations signifies the fact that p16 could act as an important diagnostic and prognostic biomarker in resected ductal PC patients[51]. RASSF1A is another tumor suppressor and an important component of RAS/PI3K/AKT and RAS/RAF/MEK/ERK pathways that have been epigenetically inactivated in various sporadic human malignancies. A higher frequency of promoter methylation status of RASSF1A has been implicated in several cancers. The highest frequency of RASSF1A promoter hypermethylation was reported in prostate cancer (99%), followed by lung cancer (95%) and breast cancer (88%)[52]. Recent reports suggest that a low frequency of hMLH1 hypermethylation was detected in PC. The loss of the hMLH1 gene which encodes for Mut L protein homology 1 is common in various cancers. Further, whole-genome sequencing revealed that somatic hMLH1 mutations are rare in cancers with an observed frequency of < 1%[53]. In the present study, we carried out epigenetic modifications (CpG methylation) of promoter regions of p16, RASSF1A, and hMLH1 genes. Our results demonstrate that significant hypermethylation (CpG islands) was reported in the promoter regions of the p16 gene in PC patients. However, we observed an extremely low frequency of methylation in the promoter region of the hMLH1 gene in PC patients. Interestingly, the CpG methylation in the promoter region of the RASSF1A gene was completely absent in PC patients. Additionally, in-silico analysis suggest that plethora of genes are associated through various interactions with the key genes (hMLH1, RASSF1A, and CDKN2A) as in Figure 11, which could play a key role in the progression of PC[39]. Together, these results suggest that a cohort and comprehensive study with larger sample size is needed to document our findings in the ethnic Kashmiri population.
Figure 11.
Interaction of MLH1, RASSF1, and CDKN2A (p16) with other important genes which have a role in the progression of pancreatic cancer.
CONCLUSION
In conclusion, the present study strongly suggests that the elevated levels of serum CA 19-9, TPS, CEA, and VEGF-A can be used as predictive biomarkers in PC patients of the ethnic Kashmiri population and may act as prognostic biomarkers to benefit the patients who are on a different regimen of chemotherapeutic interventions. Further, mutational analysis data suggest that besides harboring point mutation in codon 12 of KRAS gene, the PC patients of the current study significantly harbored codon 13-point mutation as well, which is very rarely reported in the previous studies. This may act as a genetic risk predictor in the development of PC. Additionally, considerable hypermethylation (CpG islands) in the promoter region of the p16 gene in the current study may lead to silencing of the p16 gene and could also increase the predisposition towards PC. However, we could not find the association of DPC-4G>T and BRCA-2 6174 deletion mutations and hypermethylation of CpG islands in the promoter region of RASSF1A and hMLH1 gene towards the risk of PC. To validate these results in the Kashmiri population the future studies need to be comprehensive and with larger sample sizes.
ARTICLE HIGHLIGHTS
Research background
Pancreatic cancer (PC) is one of the deadliest malignancies with an alarming mortality rate. Despite significant advancement in diagnostics and therapeutics, early diagnosis remains elusive causing poor prognosis, marred by mutations and epigenetic modifications in key genes which contribute to disease progression.
Research motivation
To explore the various biological tumor markers collectively and mutational analysis of key regulatory genes for early diagnosis and prognosis of PC.
Research objectives
To evaluate various biological tumor markers collectively in PC and their association with genetic mutation and epigenetic modification of key regulatory genes that could act as early diagnostic and prognostic biomarkers and will help in future therapeutics of PC in Kashmir valley.
Research methods
The current study includes 50 confirmed PC cases to evaluate the levels of carbohydrate antigen 19-9 (CA 19-9), tissue polypeptide specific antigen (TPS), carcinoembryonic antigen (CEA), vascular endothelial growth factor-A (VEGF-A), and epidermal growth factor receptor (EGFR) by enzyme-linked immunosorbent assay (ELISA) method. Mutational analysis of key genes Kirsten rat sarcoma (KRAS), Breast cancer type 2 (BRCA-2), and deleted in pancreatic cancer-4 (DPC-4) genes was performed to evaluate the mutations at hotspot regions. Furthermore, epigenetic modifications were performed in the promoter regions of cyclin-dependent kinase inhibitor 2A (p16; CDKN2A), MutL homolog 1 (hMLH1), and Ras association domain-containing protein 1 (RASSF1A) genes.
Research results
Besides significant elevation in levels of tumor markers CA 19-9 (P ≤ 0.05), TPS (P ≤ 0.05), CEA (P ≤ 0.001), and VEGF (P ≤ 0.001), our mutational analysis observed that KRAS gene mutation is predominant in codon 12 (16 subjects, P ≤ 0.05), and 13 (12 subjects, P ≤ 0.05). Additionally, epigenetic modification analysis suggests that CpG methylation was observed in 21 (P ≤ 0.05) and 4 subjects in the promoter regions of the p16 and hMLH1 gene, respectively.
Research conclusions
The study revealed the significant elevation of serum biological markers in PC patients and the causal association of hotspot mutations and epigenetic modification of key with PC pathogenesis thus indicates the potential of biological markers, mutational status, and epigenetic modifications of key genes collectively for predisposition, susceptibility as well as diagnostics and prognostics of PC.
Research perspectives
The study strongly suggests that the elevated levels of serum CA 19-9, TPS, CEA, and VEGF-A can be used as predictive biomarkers in PC subjects. Additionally, mutational analysis epigenetic modifications in the promoter region of key genes may act as prognostic biomarkers to benefit the patients who are on a different regimen of chemotherapeutic interventions. Further to validate these results, future studies need comprehensive, cohort, and explicative studies with large sample sizes.
ACKNOWLEDGEMENTS
The authors extend their appreciation to the State Department of Science and Technology, Jammu and Kashmir, India.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Institutional Ethics committee (IEC), Sher-i-Kashmir Institute of Medical Sciences, Srinagar, Jammu and Kashmir, India
Conflict-of-interest statement: The authors declare no conflicts of interest.
STROBE statement: The authors have read the STROBE Statement, and the manuscript was prepared and revised according to the STROBE Statement.
Manuscript source: Unsolicited manuscript
Peer-review started: April 7, 2021
First decision: May 27, 2021
Article in press: July 13, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shafqat S S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Xing YX
Contributor Information
Bilal Rah, Advanced Centre for Human Genetics, Sher-i-Kashmir Institute of Medical Sciences, Srinagar 190011, Jammu and Kashmir, India.
Manzoor Ahmad Banday, Department of Medical Oncology, Sher-i-Kashmir Institute of Medical Sciences, Srinagar 190011, Jammu and Kashmir, India.
Gh Rasool Bhat, Advanced Centre for Human Genetics, Sher-i-Kashmir Institute of Medical Sciences, Srinagar 190011, Jammu and Kashmir, India.
Omar J Shah, Department of Surgical Gastroenterology, Sher-i-Kashmir Institute of Medical Sciences, Srinagar 190011, Jammu and Kashmir, India.
Humira Jeelani, Advanced Centre for Human Genetics, Sher-i-Kashmir Institute of Medical Sciences, Srinagar 190011, Jammu and Kashmir, India.
Fizalah Kawoosa, Department of Immunology and Molecular Medicine, Sher-i-Kashmir Institute of Medical Science, Srinagar 190011, Jammu and Kashmir, India.
Tahira Yousuf, Advanced Centre for Human Genetics, Sher-i-Kashmir Institute of Medical Sciences, Srinagar 190011, Jammu and Kashmir, India.
Dil Afroze, Advanced Centre for Human Genetics, Sher-i-Kashmir Institute of Medical Sciences, Srinagar 190011, Jammu and Kashmir, India. afrozedil@gmail.com.
Data sharing statement
No additional data are available.
References
- 1.Hadden M, Mittal A, Samra J, Zreiqat H, Sahni S, Ramaswamy Y. Mechanically stressed cancer microenvironment: Role in pancreatic cancer progression. BiochimBiophysActa Rev Cancer . 2020;1874:188418. doi: 10.1016/j.bbcan.2020.188418. [DOI] [PubMed] [Google Scholar]
- 2.Saad AM, Turk T, Al-Husseini MJ, Abdel-Rahman O. Trends in pancreatic adenocarcinoma incidence and mortality in the United States in the last four decades; a SEER-based study. BMC Cancer . 2018;18:688. doi: 10.1186/s12885-018-4610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawla P, Sunkara T, Gaduputi V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J Oncol . 2019;10:10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Midha S, Chawla S, Garg PK. Modifiable and non-modifiable risk factors for pancreatic cancer: A review. Cancer Lett . 2016;381:269–277. doi: 10.1016/j.canlet.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Avgerinos KI, Spyrou N, Mantzoros CS, Dalamaga M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism . 2019;92:121–135. doi: 10.1016/j.metabol.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Chen S, Brune KA, Hruban RH, Parmigiani G, Klein AP. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J ClinOncol . 2007;25:1417–1422. doi: 10.1200/JCO.2006.09.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev ClinOncol . 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 8.McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol . 2018;24:4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marrugo-Ramírez J, Mir M, Samitier J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int J MolSci . 2018;19 doi: 10.3390/ijms19102877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Mohan A, Guleria R. Biomarkers in cancer screening, research and detection: present and future: a review. Biomarkers . 2006;11:385–405. doi: 10.1080/13547500600775011. [DOI] [PubMed] [Google Scholar]
- 11.Brase JC, Wuttig D, Kuner R, Sültmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer . 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rückert F, Pilarsky C, Grützmann R. Serum tumor markers in pancreatic cancer-recent discoveries. Cancers (Basel) . 2010;2:1107–1124. doi: 10.3390/cancers2021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barak V, Goike H, Panaretakis KW, Einarsson R. Clinical utility of cytokeratins as tumor markers. ClinBiochem . 2004;37:529–540. doi: 10.1016/j.clinbiochem.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 14.van der Bilt AR, Terwisscha van ScheltingaAG, Timmer-Bosscha H, Schröder CP, Pot L, Kosterink JG, van der Zee AG, Lub-de Hooge MN, de Jong S, de Vries EG, Reyners AK. Measurement of tumor VEGF-A levels with 89Zr-bevacizumab PET as an early biomarker for the antiangiogenic effect of everolimus treatment in an ovarian cancer xenograft model. Clin Cancer Res . 2012;18:6306–6314. doi: 10.1158/1078-0432.CCR-12-0406. [DOI] [PubMed] [Google Scholar]
- 15.Lassman AB, Roberts-Rapp L, Sokolova I, Song M, Pestova E, Kular R, Mullen C, Zha Z, Lu X, Gomez E, Bhathena A, Maag D, Kumthekar P, Gan HK, Scott AM, Guseva M, Holen KD, Ansell PJ, van den Bent MJ. Comparison of Biomarker Assays for EGFR: Implications for Precision Medicine in Patients with Glioblastoma. Clin Cancer Res . 2019;25:3259–3265. doi: 10.1158/1078-0432.CCR-18-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut . 2012;61:1085–1094. doi: 10.1136/gut.2010.236026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducreux M, Cuhna AS, CaramellaC , Hollebecque A, Burtin P, Goéré D, Seufferlein T, Haustermans K, Van Laethem JL, Conroy T, Arnold D ESMO Guidelines Committee. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol . 2015;26 Suppl 5:v56–v68. doi: 10.1093/annonc/mdv295. [DOI] [PubMed] [Google Scholar]
- 18.Matikas A, Mistriotis D, Georgoulias V, Kotsakis A. Targeting KRAS mutated non-small cell lung cancer: A history of failures and a future of hope for a diverse entity. Crit Rev OncolHematol . 2017;110:1–12. doi: 10.1016/j.critrevonc.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Blackford A, Parmigiani G, Kensler TW, Wolfgang C, Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Eshleman JR, Goggins M, Jaffee EM, Iacobuzio-Donahue CA, Maitra A, Klein A, Cameron JL, Olino K, Schulick R, Winter J, Vogelstein B, Velculescu VE, Kinzler KW, Hruban RH. Genetic mutations associated with cigarette smoking in pancreatic cancer. Cancer Res . 2009;69:3681–3688. doi: 10.1158/0008-5472.CAN-09-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reznik R, Hendifar AE, Tuli R. Genetic determinants and potential therapeutic targets for pancreatic adenocarcinoma. Front Physiol . 2014;5:87. doi: 10.3389/fphys.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 Mutations in Pancreatic Cancer. Cancers (Basel) . 2017;9 doi: 10.3390/cancers9050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev GastroenterolHepatol . 2020;17:153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 23.Mishra NK, Guda C. Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer. Oncotarget . 2017;8:28990–29012. doi: 10.18632/oncotarget.15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol . 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith L, Anti S, White N, Saldanha SN. The Genetics and Epigenetics of Colorectal Cancer Health Disparity. In: Epigenetic Mechanisms in Cancer. Elsevier, 2018: 87-115. [Google Scholar]
- 26.Wang G, Zhao D, Spring DJ, DePinho RA. Genetics and biology of prostate cancer. Genes Dev . 2018;32:1105–1140. doi: 10.1101/gad.315739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herreros-Villanueva M, Bujanda L. Non-invasive biomarkers in pancreatic cancer diagnosis: what we need vs what we have. Ann Transl Med . 2016;4:134. doi: 10.21037/atm.2016.03.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Costa JJ, Goldsmith JC, Wilson JS, Bryan RT, Ward DG. A Systematic Review of the Diagnostic and Prognostic Value of Urinary Protein Biomarkers in Urothelial Bladder Cancer. Bladder Cancer . 2016;2:301–317. doi: 10.3233/BLC-160054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sattar Z, Ali S, Hussain I, Sattar F, Hussain S, Ahmad S. Diagnosis of pancreatic cancer. In: Theranostic Approach for Pancreatic Cancer. Elsevier, 2019: 51-68. [Google Scholar]
- 30.Hess V, Glimelius B, Grawe P, Dietrich D, Bodoky G, Ruhstaller T, Bajetta E, Saletti P, Figer A, Scheithauer W, Herrmann R. CA 19-9 tumour-marker response to chemotherapy in patients with advanced pancreatic cancer enrolled in a randomised controlled trial. Lancet Oncol . 2008;9:132–138. doi: 10.1016/S1470-2045(08)70001-9. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien DP, Sandanayake NS, Jenkinson C, Gentry-Maharaj A, Apostolidou S, Fourkala EO, Camuzeaux S, Blyuss O, Gunu R, Dawnay A, Zaikin A, Smith RC, Jacobs IJ, Menon U, Costello E, Pereira SP, Timms JF. Serum CA19-9 is significantly upregulated up to 2 years before diagnosis with pancreatic cancer: implications for early disease detection. Clin Cancer Res . 2015;21:622–631. doi: 10.1158/1078-0432.CCR-14-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slesak B, Harlozinska-Szmyrka A, Knast W, Sedlaczek P, van Dalen A, Einarsson R. Tissue polypeptide specific antigen (TPS), a marker for differentiation between pancreatic carcinoma and chronic pancreatitis. A comparative study with CA 19-9. Cancer . 2000;89:83–88. doi: 10.1002/1097-0142(20000701)89:1<83::aid-cncr12>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 33.Kornek G, Schenk T, Raderer M, Djavarnmad M, Scheithauer W. Tissue polypeptide-specific antigen (TPS) in monitoring palliative treatment response of patients with gastrointestinal tumours. Br J Cancer . 1995;71:182–185. doi: 10.1038/bjc.1995.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jelski W, Mroczko B. Biochemical Markers of Colorectal Cancer - Present and Future. Cancer Manag Res . 2020;12:4789–4797. doi: 10.2147/CMAR.S253369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammarstrom S, Shively JE, Paxton RJ, Beatty BG, Larsson A, Ghosh R, Bormer O, Buchegger F, Mach JP, Burtin P. Antigenic sites in carcinoembryonic antigen. Cancer Res . 1989;49:4852–4858. [PubMed] [Google Scholar]
- 36.Wu FT, Stefanini MO, Mac Gabhann F, Kontos CD, Annex BH, Popel AS. A systems biology perspective on sVEGFR1: its biological function, pathogenic role and therapeutic use. J Cell Mol Med . 2010;14:528–552. doi: 10.1111/j.1582-4934.2009.00941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo Y, Baba H, Fukuda T, Takashima M, Sugimachi K. High expression of vascular endothelial growth factor is associated with liver metastasis and a poor prognosis for patients with ductal pancreatic adenocarcinoma. Cancer . 2000;88:2239–2245. doi: 10.1002/(sici)1097-0142(20000515)88:10<2239::aid-cncr6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 38.Stacker SA, Achen MG. The VEGF signaling pathway in cancer: the road ahead. Chin J Cancer . 2013;32:297–302. doi: 10.5732/cjc.012.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res . 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim ST, Lim DH, Jang KT, Lim T, Lee J, Choi YL, Jang HL, Yi JH, Baek KK, Park SH, Park YS, Lim HY, Kang WK, Park JO. Impact of KRAS mutations on clinical outcomes in pancreatic cancer patients treated with first-line gemcitabine-based chemotherapy. Mol Cancer Ther . 2011;10:1993–1999. doi: 10.1158/1535-7163.MCT-11-0269. [DOI] [PubMed] [Google Scholar]
- 41.Ghimessy A, Radeczky P, Laszlo V, Hegedus B, Renyi-Vamos F, Fillinger J, Klepetko W, Lang C, Dome B, Megyesfalvi Z. Current therapy of KRAS-mutant lung cancer. Cancer Metastasis Rev . 2020;39:1159–1177. doi: 10.1007/s10555-020-09903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev . 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 43.Kodaz H, Kostek O, Hacioglu MB, Erdogan B, Kodaz CE, Hacibekiroglu I, Turkmen E, Uzunoglu S, Cicin I. Frequency of RAS mutations (KRAS, NRAS, HRAS) in human solid cancer. Breast Cancer . 2017:7. [Google Scholar]
- 44.Kim D, Xue JY, Lito P. Targeting KRAS(G12C): From Inhibitory Mechanism to Modulation of Antitumor Effects in Patients. Cell . 2020;183:850–859. doi: 10.1016/j.cell.2020.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boeck S, Ormanns S, Haas M, Bächmann S, Laubender RP, Siveke JT, Jung A, Kirchner T, Heinemann V. Translational research in pancreatic cancer: KRAS and beyond. Pancreas . 2014;43:150–152. doi: 10.1097/MPA.0b013e31829629f6. [DOI] [PubMed] [Google Scholar]
- 46.Wong W, Raufi AG, Safyan RA, Bates SE, Manji GA. BRCA Mutations in Pancreas Cancer: Spectrum, Current Management, Challenges and Future Prospects. Cancer Manag Res . 2020;12:2731–2742. doi: 10.2147/CMAR.S211151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, Kern SE. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res . 1996;56:5360–5364. [PubMed] [Google Scholar]
- 48.Maitra A, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Best Pract Res ClinGastroenterol . 2006;20:211–226. doi: 10.1016/j.bpg.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 49.van Kampen JG, Marijnissen-van Zanten MA, Simmer F, van der Graaf WT, Ligtenberg MJ, Nagtegaal ID. Epigenetic targeting in pancreatic cancer. Cancer Treat Rev . 2014;40:656–664. doi: 10.1016/j.ctrv.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Moore PS, Orlandini S, Zamboni G, Capelli P, Rigaud G, Falconi M, Bassi C, Lemoine NR, Scarpa A. Pancreatic tumours: molecular pathways implicated in ductal cancer are involved in ampullary but not in exocrine nonductal or endocrine tumorigenesis. Br J Cancer . 2001;84:253–262. doi: 10.1054/bjoc.2000.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerdes B, Ramaswamy A, Ziegler A, Lang SA, Kersting M, Baumann R, Wild A, Moll R, Rothmund M, Bartsch DK. p16INK4a is a prognostic marker in resected ductal pancreatic cancer: an analysis of p16INK4a, p53, MDM2, an Rb. Ann Surg . 2002;235:51–59. doi: 10.1097/00000658-200201000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, Boehm BO, Pfeifer GP, Hoang-Vu C. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene . 2003;22:3806–3812. doi: 10.1038/sj.onc.1206582. [DOI] [PubMed] [Google Scholar]
- 53.Lawrence YR, Moughan J, Magliocco AM, Klimowicz AC, Regine WF, Mowat RB, DiPetrillo TA, Small W Jr, Simko JP, Golan T, Winter KA, Guha C, Crane CH, Dicker AP. Expression of the DNA repair gene MLH1 correlates with survival in patients who have resected pancreatic cancer and have received adjuvant chemoradiation: NRG Oncology RTOG Study 9704. Cancer . 2018;124:491–498. doi: 10.1002/cncr.31058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.