Abstract
We have evaluated a new serological confirmatory test (INNO-LIA HTLV I/II Ab [INNO-LIA]) for human T-cell leukemia virus (HTLV) using a large collection of samples from Brazilian blood donors (São Paulo region) and compared the results with those obtained by Western blotting (WB) tests (WB2.3 and WB2.4). Blood donations were initially screened by enzyme-linked immunosorbent assays (ELISAs) based on viral lysates, and repeatedly reactive samples were further tested by WB2.3. When available, samples were also tested by PCR, two additional ELISAs based on recombinant antigens (recombinant ELISAs), a new-generation WB assay (WB2.4), and the INNO-LIA. Of the 18,169 samples tested, 292 (1.61%) were repeatedly reactive in the ELISAs (viral lysate based) and were further tested by WB2.3; 97 were positive (19 that were typed as HTLV type I [HTLV-I], 12 that were typed as HTLV type II [HTLV-II], and 66 that were nontypeable), 17 were negative, and 178 had indeterminate results. Of the samples with indeterminate results, 172 were tested by INNO-LIA, which could resolve 153 samples as negative. Regarding the positive samples, WB2.3 and INNO-LIA produced concordant results for all HTLV-I-positive samples, whereas for HTLV-II they agreed for 10 of 12 samples; the 2 samples with discordant results were considered to be positive for HTLV-II by WB with WB2.3 but negative for HTLV-II by INNO-LIA and the two recombinant ELISAs. Furthermore, of the 66 nontypeable samples, 60 underwent testing by INNO-LIA; 54 turned out to be negative by the latter test as well as by recombinant ELISAs. In conclusion, the new serological confirmatory assay for HTLV (INNO-LIA HTLV I/II Ab) resolved the results for the majority of the indeterminate and positive-untypeable samples frequently observed by WB assays.
The spread of the human T-cell lymphotropic viruses (HTLV; type I [HTLV-I] and type II [HTLV-II]) from known regions of endemicity to other parts of the globe has led public health authorities in many countries to institute routine screening procedures for these retroviruses. This is the case in Brazil, where nationwide blood bank screening for HTLV became mandatory in 1993. The prevalence of HTLV-I and HTLV-II in Brazil varies regionally (1, 6); prevalence rates may reach as high as 1.35% among blood donors and 35.2% among intravenous drug users in the northeast of the country, and HTLV-II infections are frequently found among certain Amerindian tribes (3, 5).
A major problem with the mass screening of blood for HTLV antibodies has been the unacceptably high rate of false reactivity associated with commercial HTLV enzyme-linked immunosorbent assay (ELISA) kits (9). The rate of discarded units subsequent to HTLV screening reached levels of 2.5% when testing first began in 1991. Unfortunately, confirmatory tests have hitherto offered only limited help in solving this problem. Indeed, the Western blotting (WB) confirmatory technology often gives rise to complex reactivity patterns, frequently rendering results inconclusive due to the presence of nonspecific bands. This makes counseling for ELISA-reactive blood donors even more complex and often requires the collection of a second sample for repetition of ELISA and WB.
Recently, a new HTLV confirmatory assay (INNO-LIA HTLV I/II Ab [INNO-LIA]; Innogenetics, Ghent, Belgium) appeared to be useful in resolving the results for samples with indeterminate results by WB for well-defined HTLV sera (13). This prompted us to compare the new test with a commercial WB assay with a large number of blood donations reactive for HTLV by ELISA screened routinely at our blood bank. As newer-generation ELISA and WB kits became available, we also subjected the repeatedly reactive sera to further investigations.
MATERIALS AND METHODS
Samples.
At the Fundação Pró-Sangue Hemocentro de São Paulo, a total of 18,169 blood donors were screened by ELISA in June 1995 for HTLV infection. Of these, 3,158 were tested with the Hemobio HTLV I/II kit (Embrabio, São Paulo, Brazil) and 15,011 were tested with the Vironostika HTLV-I kit (Organon Teknika, Boxtel, The Netherlands); both kits are based on HTLV lysates. All initially reactive samples were further tested by ELISA in duplicate. Samples that reacted repeatedly (n = 292) were additionally characterized by WB (HTLV blot 2.3 [WB2.3]; Diagnostic Biotechnology, Singapore). The available samples (n = 279) were then tested by the new INNO-LIA and by two new-generation HTLV ELISAs (see below). Whenever cells from a donation unit were available, further testing by an in-house PCR was performed (n = 230).
The screening assays (two ELISAs) were performed in duplicate, with results expressed as a mean value of the ratio (sample signal/cutoff).
Additional screening assays.
When sufficient repeatedly reactive sera were available, additional testing by the HTLV ELISA from Murex Diagnostics (Dartford, United Kingdom) (n = 273) and by Ortho Diagnostic System (Raritan, N.J.) (n = 256) was performed. Both kits are newer-generation assays that use recombinant proteins and selected synthetic peptides as HTLV antigens. Due to the more limited availability of serum, the additional assays were performed only once and the results are expressed as a ratio. A sample was considered reactive if the ratio was equal to or greater than 1. A ratio of 0.8 was considered “grey-zone reactivity,” which is generally considered to be an indication for repeating the test.
Confirmatory assays.
Originally, WB2.3 was used to characterize all repeatedly reactive samples. The interpretation of the results obtained by this WB was done according to the manufacturer’s criteria. Some samples with a positive nontypeable WB pattern were also submitted to further testing with the HTLV WB version 2.4 (WB2.4) from Diagnostic Biotechnology (n = 57). This new generation of the WB assay uses a truncated HTLV-I recombinant gp21 (rgp21) protein which resolves most of the samples falsely reactive for rgp21 with the previous version of the WB assay, WB2.3.
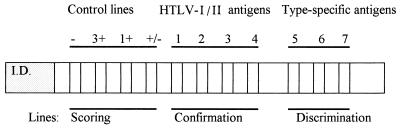
Samples were also tested by INNO-LIA (n = 279), and the interpretation criteria were supplied by the manufacturer. The INNO-LIA kit uses recombinant antigens and synthetic peptides derived from both HTLV-I and HTLV-II protein sequences. In addition to these HTLV antigens, control lines are used for semiquantitative evaluation of the results, as well as for sample addition and reagent controls. The layout of the strips is shown in Fig. 1.
FIG. 1.
Layout of INNO-LIA strips. The antigen lines are compared to the scoring lines to provide a relative intensity for each line. If a sample is confirmed to be positive, according to the criteria presented in Materials and Methods and Fig. 2, HTLV type determination can be obtained by comparing the relative intensities of the antigen lines in the discrimination area. Line 1, antigen gag p19; line 2, gag p24; line 3, env gp46; line 4, env gp21; line 5, gag p19-1; line 6, env gp46-1; line 7, env gp46-2.
The assay procedure can be summarized as follows: serum or plasma samples were diluted 1:100 and were incubated at room temperature (25°C) overnight; this was followed by three washing steps with washing buffer before the addition of an alkaline phosphatase anti-human immunoglobulin conjugate. The three washing steps were performed again, followed by the addition of a chromogen. Color development was then stopped with an appropriate stop solution. Following the visual interpretation protocol, after color development, each line was compared to the control lines, and the intensities were scored as follows: 0 (−), no response or an intensity less than that of the cutoff line; 0.5 (±), intensity equal to that of the cutoff line; 1 (+), intensity between those of the cutoff line and the 1+ control line; 2 (++), intensity between those of the 1+ control line and the 3+ control line; 3 (+++), intensity equal to that of the 3+ control line; 4 (++++), intensity greater than that of the 3+ control line.
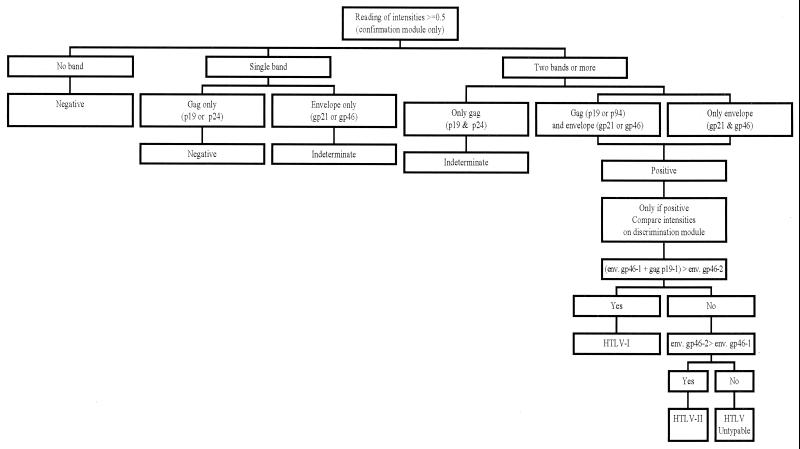
A sample was classified as positive if it reacted with at least one envelope antigen (gp21 or gp46) and one gag antigen (p19 or p24). Alternatively, two reactive envelope antigens (gp21 and gp46) also indicated a positive sample. When an isolated band or no reaction appeared, the sample was considered negative. When two gag antigens were reactive, the sample was considered indeterminate. Discrimination between HTLV-I and HTLV-II was indicated by the corresponding specific antigens present on the same strip (Fig. 2).
FIG. 2.
Criteria for interpreting the results obtained with the INNO-LIA strips; reactivity to none, one, or more of the specified antigens is interpreted as negative, indeterminate, or positive, as assigned. The positive samples are discriminated as HTLV-I or HTLV-II depending on the intensity of the color of the strips; e.g., if the env gp46-1 plus gag p19-1 bands are more intense than the env gp46-2 band, the sample is typed as HTLV-I.
Finally, blood units from donors repeatedly reactive for HTLV were used to obtain peripheral blood mononuclear cells for PCR. For erythrocyte lysis, 10 ml of blood was mixed with 10 ml of 0.4% saponin and 0.5% NaCl, and the mixture was centrifuged at 2,000 × g for 10 min. The peripheral blood mononuclear cell pellet was washed twice with 0.9% NaCl and resuspended in 100 μl of lysis buffer (10 mM Tris-buffer [pH 8.3], 0.5% Nonidet P-40, 0.5% Tween 20) and digested with 40 μg of proteinase K at 65°C for 2 h. The DNA was purified by phenol-chloroform extraction. All samples were subjected to PCR amplification for two HTLV genes: pol and tax. If a discordant result was obtained, PCR was repeated in duplicate for both sets of primers; for the pol gene, 1 μg of DNA was amplified in one round with primers SK110 and SK111 (7) in a reaction mixture containing 2.0 mM MgCl2 and 0.2 mM each primer. The PCR product was detected by liquid hybridization with probe SK112 for HTLV-I and probe SK188 for HTLV-II; for detection of the tax gene, DNA was amplified by a nested PCR with SK43 and SK44 in the first round and TAX1 (5′-GTGTTTGGCGATTGTGTACA-3′) and TAX2 (5′-CCATCGATGGGGTCCCA-3′) in the second round in a reaction mixture containing 2.5 mM MgCl2 and 0.2 mM each primer. The PCR product was analyzed upon electrophoresis on a 1% agarose gel.
RESULTS
Screening.
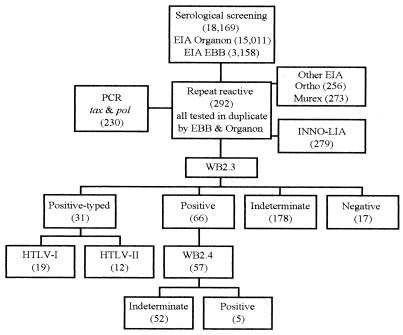
The screening of 18,169 blood donations for HTLV by viral lysate-based ELISAs resulted in 292 (1.6%) samples repeatedly reactive for HTLV-I or HTLV-II. Subsequent confirmatory testing by analysis with WB2.3 indicated the following: 31 serum samples (10.6%) were typed as HTLV-I or HTLV-II, 66 serum samples (22.6%) were HTLV positive but nontypeable, 178 serum samples (60.9%) were indeterminate, with various patterns, and 17 serum samples (5.9%) were negative (Table 1; Fig. 3). If only the results of WB2.3 were taken into consideration, the prevalence of HTLV antibodies was 0.53% (97 of 18,169). However, of the 66 HTLV-positive but nontypeable samples, 57 could be retested with a later version of the WB kit (WB2.4) with a truncated rgp21 (GD21), which is less prone to nonspecific reactivity (8). By testing with WB2.4, only 5 of 57 samples remained positive and were still nontypeable. The results for the remaining 52 samples became indeterminate by losing their reactivity to the rgp21 antigen originally present in WB2.3.
TABLE 1.
PCR analysis of 230 samples tested by WB with WB2.3
| WB2.3 result | No. of samples with the following PCR result:
|
|||||
|---|---|---|---|---|---|---|
| Negative | tax only | HTLV-I positive | HTLV-II positive | Not tested | Total | |
| Negative | 12 | 0 | 0 | 0 | 5 | 17 |
| Indeterminate | 143 | 3 | 0 | 0 | 32 | 178 |
| Positive | 65 | 1 | 0 | 0 | 0 | 66 |
| HTLV-I positive | 2 | 0 | 13 | 0 | 4 | 19 |
| HTLV-II positive | 2 | 0 | 0 | 8 | 2 | 12 |
| Total | 224 | 4 | 13 | 8 | 43 | 292 |
FIG. 3.
Blood donor samples (the numbers of samples are given in parentheses) were tested by the various assays, as specified (see descriptions in Materials and Methods); the numbers of samples with the indicated results by WB with WB2.3 and WB2.4 (positive, indeterminate, or negative) are also given in parentheses. EBB, Embrabio; EIA, ELISA.
PCR.
Amplification of the pol and tax genes was performed for 230 of the 292 ELISA-reactive samples. The results are summarized in Table 1 (see also Fig. 3). Of the 31 samples typed with WB2.3 (19 HTLV-I and 12 HTLV-II), 25 were analyzed by PCR; only 21 (84%) were positive (both tax and pol genes were amplified). The 143 WB-indeterminate samples were also tested by PCR, and none was amplified with both sets of primers; 3 samples were positive only by the tax PCR. Another tax-positive pol-negative sample was found among the 66 samples nontypeable but positive by WB. These samples were not considered to be positive. Unfortunately, a second fresh sample could not be obtained from these donors to confirm the PCR results in terms of sensitivity. The remaining positive but nontypeable or indeterminate samples were all negative by PCR.
INNO-LIA versus WB2.3.
Of the 292 samples tested with WB2.3, 279 could be further investigated by INNO-LIA (Table 2; Fig. 3); 54 of 60 WB2.3-positive but nontypeable samples and 153 of 172 WB2.3-indeterminate samples were resolved as negative by INNO-LIA. Both WB and INNO-LIA had the same results for 19 HTLV-I-positive and 10 of 12 HTLV-II-positive samples. The two remaining HTLV-II-positive samples were negative by both recombinant ELISAs and were indeterminate by WB with WB2.4.
TABLE 2.
INNO-LIA results compared to WB2.3 results
| WB2.3 result | No. of samples with the following INNO-LIA result:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| HTLV-I positive | HTLV-II positive | Positive | Indeterminate | Negative | Total tested | Not tested | Total | |
| HTLV-I positive | 19 | 0 | 0 | 0 | 0 | 19 | 0 | 19 |
| HTLV-II positive | 0 | 10 | 0 | 0 | 2 | 12 | 0 | 12 |
| Positive | 1 | 0 | 4 | 1 | 54 | 60 | 6 | 66 |
| Indeterminate | 6 | 2 | 8 | 3 | 153 | 172 | 6 | 178 |
| Negative | 0 | 0 | 0 | 2 | 14 | 16 | 1 | 17 |
| Total | 26 | 12 | 12 | 6 | 223 | 279 | 13 | 292 |
INNO-LIA testing resulted in the detection of seven additional HTLV-I-positive samples that were either positive (n = 1) or indeterminate (n = 6) by WB with WB2.3. Two samples were HTLV-II positive by INNO-LIA, while they were indeterminate by WB with WB2.3 and WB2.4. Finally, 12 samples were positive but nontypeable by INNO-LIA; of these, 4 were nontypeable but positive and 8 were indeterminate by WB with WB2.3. The reactivity patterns of the 21 samples with discrepant results are presented in Table 3; of these, 13 samples (samples 255, 1270, 314, 256, 274, 297, 419, 426, 432, 460, 1091, 1100, and 1175) were positive by more than one ELISA, with 6 of them also being positive by one of the WBs. Two samples that were borderline by the two ELISAs were either positive by the WB (sample 1164) or PCR positive only for the tax gene (sample 404).
TABLE 3.
Detailed analysis of reactivity patterns of samples with discrepant results by screening and confirmatory assays
| Sample no. | ELISA ratioab
|
WB2.3 reactivity pattern | WB resultb
|
INNO-LIA reactivity pattern | INNO-LIA result | PCR resultc | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| First ELISA
|
Second ELISA
|
|||||||||
| Org | Emb | Mur | Orth | WB2.3 | WB2.4 | |||||
| 255 | 1.50 | 1.97 | 0.27 | 1.07 | p19, gp21, p24, p26, p28, p53 | Ind | NT | p19, p24, gp46, gp46-1 | HTLV-1 | Neg |
| 348 | 0.61 | 0.28 | 0.26 | 0.51 | p24 | Ind | NT | p19, p24, gp46, gp46-1 | HTLV-I | Neg |
| 404 | 0.85 | 0.81 | 0.34 | 0.40 | p19, gp21, p26, p28 | Ind | NT | p19, gp46, gp46-1 | HTLV-I | tax |
| 445 | 3.06 | 0.04 | 0.29 | 0.28 | p24, p28 | Ind | NT | p24, pg46, gp46-1 | HTLV-I | Neg |
| 461 | 1.55 | 0.16 | 0.27 | 0.96 | rgp21, p24 | Pos | Pos | p24, gp46, gp46-1 | HTLV-I | Neg |
| 1157 | 0.76 | 1.26 | 0.72 | 0.73 | p19, gp21, p26, p28, p53 | Ind | Ind | p19, gp46, gp46-1 | HTLV-I | Neg |
| 1270 | 1.05 | 2.18 | 0.28 | 0.75 | p19, gp21, p26, p28, p32, p36 | Ind | NT | p19, gp46, p19-1, gp46-1 | HTLV-I | Neg |
| 314 | 1.43 | 1.64 | 1.53 | NT | p19, gp21, p28 | Ind | NT | p19, gp46, gp46-2 | HTLV-II | tax |
| 343 | 1.95 | 0.89 | 0.31 | 0.26 | p24 | Ind | Ind | p24, gp46, gp46-2 | HTLV-I | Neg |
| 256 | 2.87 | 2.01 | 0.29 | 0.06 | p24, p53 | Ind | Ind | p19, p24, gp21 | Pos | Neg |
| 274 | 1.42 | 1.55 | 0.34 | 0.88 | p19, gp21, p26, p28, p32, p36, p53 | Ind | NT | p19, gp46 | Pos | Neg |
| 297 | 2.45 | 1.28 | 1.00 | 0.71 | rgp21, p24 | Pos | Ind | p24, gp46, gp21 | Pos | Neg |
| 419 | 0.85 | 1.59 | 0.26 | 1.22 | p19, gp21, p26, p28, p32, p36, p53 | Ind | Ind | p19, gp46 | Pos | NT |
| 426 | 2.03 | 3.17 | 0.30 | 0.64 | p19, gp21, p26, p28, p32, p36, p53 | Ind | Ind | p19, gp46 | Pos | Neg |
| 432 | 2.98 | 1.99 | 0.31 | 1.97 | p19, gp21, p26, p28, p32, p36, gp46, p53 | Ind | Ind | p19, gp21 | Pos | Neg |
| 441 | 1.22 | 0.55 | 0.28 | 0.57 | p19, gp21, p28 | Ind | NT | p19, p24, gp46 | Pos | Neg |
| 460 | 1.58 | 0.36 | 10.24 | 0.28 | rgp21, p19, gp21 | Pos | Pos | p19, gp21 | Pos | Neg |
| 1091 | 1.23 | 1.64 | 0.24 | 0.47 | rgp21, p19, gp21 | Pos | Ind | p19, gp21 | Pos | Neg |
| 1100 | 1.66 | 2.39 | 0.32 | 0.44 | rgp21, p19, gp21, p26, p28, p36 | Pos | Ind | p19, gp46 | Pos | Neg |
| 1164 | 0.49 | 0.87 | 0.89 | 0.53 | p19, gp21, p24, p26, p28, p53 | Ind | Pos | p19, gp21 | Pos | Neg |
| 1175 | 1.71 | 2.34 | 0.27 | 0.21 | p19, gp21 | Ind | Pos | p19, gp46 | Pos | Neg |
ELISA ratios (signal/cutoff) were calculated as described by each manufacturer. For the kits from Organon (Org) and Embrabio (Emb), the results are average ratios of two determinations. For the kits from Murex (Mur) and Ortho (Orth), the results are ratios of a single determination.
NT, not tested; Ind, indeterminate; Pos, positive.
Neg, negative; tax, positive only for tax PCR.
DISCUSSION
Screening for HTLV by viral lysate-based ELISAs in Brazil has hitherto led to a high rate of discarding of units of blood. The majority of the samples repeatedly reactive by ELISA indeed have indeterminate results upon complete investigation by supplemental tests such as WB or are negative as determined by INNO-LIA and PCR. Although the new version of the WB assay, WB2.4, has substantially decreased the high rate of false-positive results usually obtained by WB with WB2.3 (4, 8), a large number of indeterminate results are still observed due to reactivity with bands other than GD21.
All WB-indeterminate and nontypeable but positive samples were negative by PCR with both sets of primers (Table 1), in agreement with other studies that have shown a low percentage of HTLV infections among individuals whose samples have indeterminate results by WB and these reactivity patterns (10, 11). Four samples were positive only for tax primers. Unfortunately, a second sample could not be obtained from these donors. It is possible that they truly represent infected persons because HTLV infections have been found in samples with low levels of reactivity by serological tests (12). However, in the absence of a second sample to confirm the initial PCR results, one should be cautious about the overall interpretation of the serological test results for those samples.
In Table 3 we presented the reactivity patterns for 21 discrepant samples that tested positive by INNO-LIA. Those that tested negative by PCR and the two recombinant ELISAs (n = 12) were probably from noninfected individuals. However, since none of the PCR or recombinant ELISAs showed a fully optimized sensitivity, it is not possible to reach a final conclusion about these samples. Although nine samples were found to be negative by PCR, they were positive by at least one of the new-generation ELISAs and/or by one of the WB versions, indicating possible infection.
Nevertheless, if upon prospective study the samples with nontypeable but positive patterns by INNO-LIA represented false-reactive samples, more stringent criteria for Brazilian samples could be validated. For instance, in the confirmation module of the INNO-LIA, when the gp21 antigen was not reacting, we observed that truly positive samples reacted at least with the three other antigens (p19, p24, and gp46), while potentially false-reactive samples showed limited reactivities to only two of these antigens. This additional stringency criterion would resolve the results for most of the PCR-negative and INNO-LIA-positive samples. A prospective study by a standardized PCR method can help in further validation of stringency criteria in terms of both sensitivity and specificity. We believe that more stringent criteria might be necessary in tropical areas like Brazil, where a high frequency of nonspecific reactions to HTLV antigens is often reported (2). The screening for infections with virtually all HTLV variants may be compromised by omitting reactivities to the gag antigens. Thus, the samples with indeterminate or positive but nontypeable results by INNO-LIA represent a suitable and focused target in attempts to isolate new hypothetical HTLV strains.
In conclusion, the sensitivity of INNO-LIA was high in that it could detect all samples reactive by all assays. The two samples positive for HTLV-II by WB with WB2.3 but negative by INNO-LIA proved to be falsely typed on the basis of WB2.4 and recombinant ELISA results. INNO-LIA eliminates the majority of the samples with indeterminate results by WB (153 of 172) and positive but nontypeable samples (54 of 60). Furthermore, the typing capability is more accurate by INNO-LIA than by WB.
ACKNOWLEDGMENT
We thank Fred Shapiro for critical review of the manuscript.
REFERENCES
- 1.Ferreira Junior O C, Vaz R S, Carvalho M B, Guerra C, Fabron A L, Rosemblit J, Hamerschlak N. Human T-lymphotropic virus type I and type II infections and correlation with risk factors in blood donors from Sao Paulo, Brazil. Transfusion. 1995;35:258–263. doi: 10.1046/j.1537-2995.1995.35395184284.x. [DOI] [PubMed] [Google Scholar]
- 2.Garin B, Gosselin S, de The G, Gessain A. HTLV-I/II infection in a high viral endemic area of Zaire, Central Africa: comparative evaluation of serology, PCR, and significance of indeterminate Western blot pattern. J Med Virol. 1994;44:104–109. doi: 10.1002/jmv.1890440119. [DOI] [PubMed] [Google Scholar]
- 3.Hall W W, Ishak R, Zhu S W, Novoa P, Eiraku N, Takahashi H, Ferreira M C, Azevedo V, Ishak M O, Ferreira O C, Monken C, Kurata T. Human T lymphotropic virus type II (HTLV-II): epidemiology, molecular properties, and clinical features of infection. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:S204–S214. doi: 10.1097/00042560-199600001-00031. [DOI] [PubMed] [Google Scholar]
- 4.Kleinman S H, Kaplan J E, Khabbaz R F, Calabro M A, Thomson R, Busch M the Retrovirus Epidemiology Donor Study Group. Evaluation of a p21e-spiked Western blot (immunoblot) in confirming human T-cell lymphotropic virus type I or II infection in volunteer blood donors. J Clin Microbiol. 1994;32:603–607. doi: 10.1128/jcm.32.3.603-607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maloney E M, Biggar R J, Neel J V, Taylor M E, Hahn B H, Shaw G M, Blattner W A. Endemic human T cell lymphotropic virus type II infection among isolated Brazilian Amerindians. J Infect Dis. 1992;166:100–107. doi: 10.1093/infdis/166.1.100. [DOI] [PubMed] [Google Scholar]
- 6.Matutes E, Schulz T, Serpa M J, de Queiroz Campos Araujo A, de Oliveira M S. Report of the second international symposium on HTLV in Brazil. Leukemia. 1994;8:1092–1094. [PubMed] [Google Scholar]
- 7.McPherson M J, Hames B D, Taylor G R. PCR 2—a practical approach. Oxford, United Kingdom: IRL Press; 1995. [Google Scholar]
- 8.Varma M, Rudolph D L, Knuchel M, Switzer W M, Hadlock K G, Velligan M, Chan L, Foung S K, Lal R B. Enhanced specificity of truncated transmembrane protein for serologic confirmation of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2 infections by Western blot (immunoblot) assay containing recombinant envelope glycoproteins. J Clin Microbiol. 1995;33:3239–3244. doi: 10.1128/jcm.33.12.3239-3244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrielink H, Sisay Y, Reesink H W, Woerdeman M, Winkel C, de Leeuw S J, Lelie P N, van der Poel C L. Evaluation of a combined lysate/recombinant antigen anti-HTLV-I/II ELISA in high and low endemic areas of HTLV-I/II infection. Transfusion Med. 1995;5:135–137. doi: 10.1111/j.1365-3148.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 10.Vrielink H, Zaaijer H L, Cuypers H T, van der Poel C L, Woerdeman M, Lelie P N, Winkel C, Reesink H W. Evaluation of a new HTLV-I/II polymerase chain reaction. Vox Sang. 1997;72:144–147. doi: 10.1046/j.1423-0410.1997.7230144.x. [DOI] [PubMed] [Google Scholar]
- 11.Zaaijer H L, Cuypers H T, Dudok de Wit C, Lelie P N. Results of 1-year screening of donors in The Netherlands for human T-lymphotropic virus (HTLV) type I: significance of Western blot patterns for confirmation of HTLV infection. Transfusion. 1994;34:877–880. doi: 10.1046/j.1537-2995.1994.341095026973.x. [DOI] [PubMed] [Google Scholar]
- 12.Zehender G, Girotto M, De Maddalena C, Francisco G, Moroni M, Galli M. HTLV infection in ELISA-negative blood donors. AIDS Res Hum Retroviruses. 1996;12:737–740. doi: 10.1089/aid.1996.12.737. [DOI] [PubMed] [Google Scholar]
- 13.Zrein M, Louwagie J, Boeykens H, Govers L, Hendrickx G, Bosman F, Sablon E, Demarquilly C, Boniface M, Saman E. Assessment of a new immunoassay for serological confirmation and discrimination of human T-cell lymphotropic virus infections. Clin Diagn Lab Immunol. 1998;5:45–49. doi: 10.1128/cdli.5.1.45-49.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]