Abstract
We have recently shown that the orphan nuclear receptor Nur77 (NGFI-B) is most active in transcription when it is interacting with a cognate DNA sequence as a homodimer. Further, we have shown that the target for Nur77 dimers, the Nur response element (NurRE), is responsive to physiological stimuli in both endocrine and lymphoid cells, whereas other DNA targets of Nur77 action are not. The Nur77 subfamily also includes two related receptors, Nur-related factor 1 (Nurr1) and neuron-derived orphan receptor 1 (NOR-1). Often, more than one member of this subfamily is induced in response to extracellular signals. We now show that Nur77 and Nurr1 form heterodimers in vitro in the presence or absence of NurRE, and we have documented interactions between these proteins in vivo by using a two-hybrid system in mammalian cells. These heterodimers synergistically enhance transcription from NurRE reporters in comparison to that seen with homodimers. The naturally occurring NurRE from the pro-opiomelanocortin gene preferentially binds and activates transcription in the presence of Nur77 homo- or heterodimers, while a consensus NurRE sequence does not show this preference. Taken together, the data indicate that members of the Nur77 subfamily are most potent as heterodimers and that different dimers exhibit target sequence preference. Thus, we propose that a combinatorial code relying on specific NurRE sequences might be responsible for the activation of subsets of target genes by one of the members of the Nur77 subfamily of transcription factors.
Nur77 and the closely related proteins Nurr1 and NOR-1 form a subfamily (the Nur subfamily) of transcription factors belonging to the superfamily of nuclear receptors (NR) (7, 21). The Nur77 gene was the first to be cloned as a serum-inducible gene expressed during the G0/G1 transition phase in cultured mouse fibroblast cells (11, 16). It was also identified as a nerve growth factor (NGF)-inducible gene in differentiating rat PC12 cells; for this reason, Nur77 is also known as NGFI-B (for NGF-inducible factor B) (22). The Nur-related factor 1 (Nurr1) gene was cloned as a predominantly brain-specific gene whose expression is rapidly induced by membrane depolarization (17). Similarly, the neuron-derived orphan receptor 1 (NOR-1) gene was identified as a gene strongly expressed in apoptotic neuronal cells of the forebrain (26). Nur77 (NGFI-B), Nurr1, and NOR-1 are highly homologous in the zinc finger DNA binding domain (DBD), moderately homologous in the ligand binding domain, and somewhat divergent in the N termini (9).
Comparative tissue distribution studies revealed similarities and differences in the temporal and spatial patterns of expression of mRNAs for Nur subfamily members (26, 39, 45, 47). Nur77 and NOR-1 are fairly widely expressed, with Nur77 showing a later onset. They are both constitutively expressed in various peripheral tissues and in some regions of the brain, where their patterns of expression are quite similar (17, 26, 39). In contrast, basal expression of Nurr1 appears to be restricted to the central nervous system (17), where it has a pattern that is roughly complementary to that of Nur77 and NOR-1. Nurr1 plays an essential role in the development and maintenance of midbrain dopaminergic neurons, since inactivation of the Nurr1 gene results in agenesis of these neurons (2, 35, 46). Nurr1 is the only member of the Nur subfamily that is expressed in the affected dopaminergic neurons of the substantia nigra and in the ventral tegmental area. Nur subfamily members are also important in the development of the T-cell repertoire. Indeed, Nur77 and NOR-1 (but not Nurr1) are strongly induced following stimulation of the T-cell receptor (TCR), which leads to apoptosis of self-reactive immature thymocytes (3, 20, 43). Signals elicited by TCR activation and leading to negative selection by apoptosis appear to converge on the Nur signaling pathway, since either the overexpression of a dominant-negative Nur77 mutant or the use of an antisense Nur77 mRNA abrogates TCR-mediated apoptosis (20, 43). In this particular system, Nur77 and NOR-1 appear to play functionally redundant roles (3), which may explain the lack of a phenotype in Nur77-deficient mice (18).
Nur subfamily members are also implicated at multiple levels of the hypothalamic-pituitary-adrenal (HPA) axis. Indeed, Nur77 expression is strongly induced by a variety of stress stimuli in corticotropin-releasing hormone (CRH)-producing neurons of the hypothalamic paraventricular nucleus (12, 27). Nur77 and Nurr1 may be involved in CRH transcription, since they were both shown to transactivate a reporter plasmid driven by the CRH promoter (24). Stress-induced signals also strongly stimulate Nur factor expression in the pituitary and the adrenal cortex (6, 24, 31, 41). In adrenal-derived Y1 cells, adrenocorticotropin treatment induces Nur77 and Nurr1, leading to enhancement of transcription of the gene encoding steroid 21α-hydroxylase (6, 41), a rate-limiting enzyme in steroidogenesis. In the anterior pituitary, the stimulatory effect of CRH on pro-opiomelanocortin (POMC) gene transcription appears to be mediated through Nur77 and Nurr1 activation (24, 31). In addition, negative-feedback regulation of POMC transcription by glucocorticoids (Gc) and their receptors appears to be, at least in part, exerted on the Nur signaling pathway (24, 32).
The Nur subfamily members are orphan nuclear receptors because no ligand has yet been identified for them (9). The existence of such a ligand remains elusive, considering that these transcription factors are constitutively active in numerous cell lines, even in the absence of serum or any other exogenous agent (29). Nur77 was the first NR shown to bind DNA and to activate transcription as a monomer. The target binding site of Nur77, the NGFI-B response element (NBRE), was identified by genetic selection in Saccharomyces cerevisiae (40); it is an octanucleotide that contains the canonical nuclear receptor binding motif AGGTCA preceded by two adenines. Recognition of these adenines was shown to depend on non-zinc finger residues of NGFI-B, a domain called the A box (42). Nurr1 and NOR-1 were later shown to activate transcription upon binding the NBRE (3, 28, 47). In addition, Nur77 and Nurr1 (but not NOR-1) mediate retinoid signaling by heterodimerization with the retinoid X receptor (RXR) (10, 30, 47). These heterodimers bind and activate transcription through a DR-5 element, and unlike with other RXR heterodimerization events, in which RXR is a silent partner, transcriptional activation depends on the presence of 9-cis retinoic acid (9-cis RA). It has also been demonstrated that Nur77 interacts with COUP-TF, another orphan NR, and that this interaction modulates RA sensitivity in human lung cancer cells (44). We recently reported the identification of a novel Nur77 target sequence, the Nur response element (NurRE). This naturally occurring response element was identified in the POMC promoter, and it was shown to bind homodimers of Nur77 (31). The NurRE is much more responsive than the NBRE to Nur77 and to physiological stimuli, such as CRH treatment in POMC cells or TCR activation in T-cell hybridomas.
In view of the parallel induction of more than one Nur member in many systems, the aim of this study was to investigate the possibility of a concerted action by different Nur subfamily members. We have indeed found that Nur factors cooperate with each other to synergistically activate NurRE-dependent transcription and that this synergism likely results from heterodimerization between Nur subfamily members. Further, we have shown that the POMC NurRE sequence is a preferential target for Nur77 by comparison to a consensus NurRE which is equally sensitive to all three Nur subfamily members.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
The various reporter plasmids were constructed in pXP1-luc (25) containing the minimal (−34 to +63) POMC promoter. The −480 POMC promoter was previously described (36). Oligonucleotides corresponding to NBRE (5′-GATCCTCGTGCGAAAAGGTCAAGCGCTA-3′), NurREPOMC (5′-GATCGTGATATTTACCTCCAAATGCCA-3′), and NurRECON (5′-GATCCGTGACCTTTATTCTCAAAGGTCA-3′) were cloned in either one or three copies in the BamHI site of the minimal POMC–pXP1-luc plasmid. The DR5-luc (β-RE) and (UAS)4-TK-luc reporter plasmids as well as the CMX-GAL4, CMX-GAL4/Nur77, CMX-GAL4/Nurr1, and CMX-VP16 expression vectors were previously described (30). CMX-Nurr1, CMX-NOR-1, and CMX-Nur77 expression vectors contain complete cDNA sequences cloned into pCMX (38). CMX-Nur77ΔC-term encodes a C-terminal truncation (amino acids 1 to 380) mutant of Nur77. CMX-VP16/Nur77 and CMX-VP16/Nurr1 contain the entire Nur77 and Nurr1 coding sequences, respectively, cloned in phase into CMX-VP16 (30). MBP-Nur77 was obtained by insertion of the Nur77 coding sequences into the pMal-C (New England Biolabs) plasmid.
Cell culture and transfections.
CV1 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine fetal serum and maintained at 37°C in an atmosphere of 5% CO2. AtT20 D16v cells were grown under the same conditions, except that charcoal-stripped fetal bovine serum was used. CV1 cells were transfected by the calcium phosphate coprecipitation method, whereas AtT-20 cells were transfected by lipofection using LipofectAMINE (Gibco BRL), as previously described (32). Results are presented as the means of data from three to five experiments performed in duplicate. When reported as fold activation, the basal levels of activity were always at least 50-fold above background. Rous sarcoma virus-GH was used as an internal control for transfection efficiency.
Electrophoretic mobility shift assays.
The electrophoretic mobility shift assays were performed with proteins produced with the TNT coupled reticulocyte lysate system (Promega). Binding reactions were performed in a 20-μl volume containing 10 mM Tris-HCl (pH 8.0), 40 mM KCl, 1 mM dithiothreitol (DTT), 6% glycerol, 0.05% NP-40, 5 ng of poly(dI-dC), and about 10 ng of in vitro-synthesized Nur77, Nurr1, or NOR-1. We used, per reaction, 50,000 cpm (∼20 fmol) of double-stranded oligonucleotide probes, end labeled by filling in with the Klenow fragment of DNA polymerase in the presence of [α-32P]dATP and purified on a G-25 Sephadex column. The reaction mixtures were incubated for 10 min at 25°C prior to being loaded on gels. The samples were separated by electrophoresis on 5% polyacrylamide gels (29:1 acrylamide/bisacrylamide) in 0.5× Tris-borate-EDTA at 25°C for 2 to 2.5 h. For supershifting experiments, the antibodies were preincubated with the nuclear extracts for 15 min on ice prior to probe addition.
Recombinant protein production and pull-down assays.
The Nur77–maltose-binding protein (MBP) and MBP-LacZ fusion proteins were produced as described previously (37). 35S-labeled in vitro-synthesized Nurr1 and luciferase were obtained by using the TNT coupled reticulocyte lysate system (Promega). Protein-protein interaction assays were performed with 1 μg of fusion protein coupled to amylose beads (New England Biolabs) and about 80 ng of 35S-labeled protein as described in reference 37.
AtT-20 nuclear extracts and coimmunoprecipitations.
After 1 h of treatment with 10 μM forskolin, approximately 4 × 107 AtT-20 cells were washed once with cold phosphate-buffered saline and harvested in cold phosphate-buffered saline containing 1 mM EDTA. The cells were then centrifuged and resuspended in 500 μl of buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.1 mM EGTA, 20 mM NaF, 1 mM Na4P2O7 · 10H2O, 1 mM Na3VO4, 0.25 mM Na2MbO4, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM DTT, and 10 μg each of the protease inhibitors leupeptin, aprotinin, and pepstatin/ml). Cells were allowed to swell on ice for 15 min before addition of 50 μl of NP-40 followed by vigorous vortexing. After centrifugation, the nuclear pellet was resuspended in 400 μl of buffer B (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.1 mM EGTA, 0.4 M NaCl, 5% glycerol, 0.5 mM PMSF, 1 mM DTT, and 10 μg of each protease inhibitor/ml as above) and shaken vigorously at 4°C for 30 min. The extract was then centrifuged, and the supernatant was dialyzed against 100 volumes of buffer C (20 mM HEPES [pH 7.9], 75 mM NaCl, 0.1 mM EDTA, 20% glycerol, 1 mM DTT, and 0.5 mM PMSF) overnight at 4°C, with a change of buffer after 4 h. The dialyzed extract was then centrifuged, and the protein concentration of the supernatant was estimated by the Bradford assay. Coimmunoprecipitation experiments were performed essentially as described elsewhere (8), except that 300 μg of AtT-20 nuclear extract was used per sample and extracts were precleared of nonspecific interactions with 2 μg of purified rabbit immunoglobulin G (IgG; Sigma). One microgram of Nur77 antibody (N19; Santa Cruz Biotechnology) was used for the immunoprecipitation. Nurr1 was revealed by Western blotting with a Nurr1 antibody (a gift from T. Perlmann) and an anti-rabbit antibody–horseradish peroxidase conjugate (Sigma). Revelation was performed by chemiluminescence as described by the manufacturer (ECL+plus; Amersham Pharmacia).
Northern blotting.
Total AtT-20 RNA was extracted and used in Northern blotting experiments as previously described (15). Hormone treatment was performed with 10−7 M CRH, 10−7 M dexamethasone (DEX), or both.
RESULTS
CRH induces all three Nur factors.
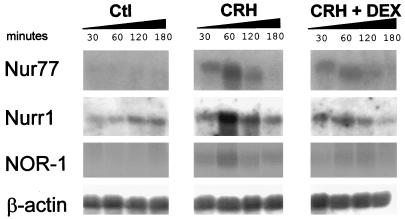
Since previous studies have implicated Nur77 in the regulation of the HPA axis (6, 12, 27, 41), particularly at the pituitary level (Nur77 mRNA is rapidly and transiently induced in response to CRH in both isolated pituitary cells [24] and POMC-expressing At-T20 cells [31]), we tested whether Nurr1 and NOR-1 are also inducible in this system. All three Nur family members were found to be rapidly and transiently induced upon CRH treatment (Fig. 1), reaching maximum expression around 1 h after stimulation. Treatment with the synthetic Gc DEX severely reduced the induction of both NOR-1 and Nurr1 mRNAs by CRH (Fig. 1) but only blunted the Nur77 increase (Fig. 1 and reference 32). Under basal conditions, only Nurr1 mRNA could be detected by Northern blot analysis, suggesting that Nurr1 may contribute to basal activity whereas all three factors may be involved in the CRH response.
FIG. 1.
Induction of Nur77, Nurr1, and NOR-1 mRNAs by CRH in pituitary-derived AtT-20 cells. The effect of CRH (10−7 M), alone or in combination with DEX (10−7 M), on Nur77, Nurr1, and NOR-1 mRNAs was measured by Northern blotting. RNA was extracted from cells treated or untreated (Ctl) for the indicated time periods. β-Actin mRNA was used as a loading control.
The NurREPOMC is highly responsive to all Nur factors.
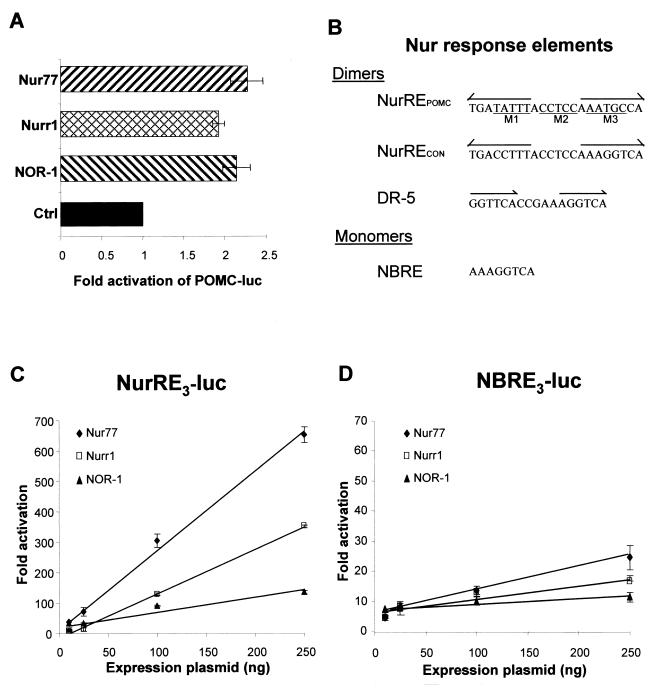
Since all three Nur family members are induced by CRH, we tested whether they can activate POMC transcription. Forced expression of all three Nur factors activated transcription of a luciferase reporter driven by the POMC promoter (Fig. 2A). Previous analyses of the POMC promoter had identified a target sequence responsive to both CRH and Nur77 (31). This sequence, the POMC NurRE (NurREPOMC) (Fig. 2B), is highly responsive to Nur77, up to 50 times more responsive under our conditions than the NBRE, the previously described target for Nur monomers (31). The NurREPOMC is an everted repeat of an octameric motif separated by 6 nucleotides. Each motif is related to the NBRE, with two mismatches in each half-site evident upon comparison to a consensus NBRE (Fig. 2B). The ability of Nurr1 and NOR-1 to activate the NurREPOMC and NBRE reporters (31) was compared to that of Nur77 (Fig. 2C and D). All three Nur factors activated both reporters, the NurREPOMC reporter (Fig. 2C) being at least 10 times more sensitive than the NBRE (Fig. 2D; note the 10-fold difference in scale). Nur77 was the most potent activator of both reporters, followed by Nurr1 and then NOR-1. Similar results were obtained in POMC-expressing AtT-20 cells (data not shown). These data suggested that Nurr1 and NOR-1 might also bind the NurRE as homodimers.
FIG. 2.
Transcriptional effects of Nur77 (NGFI-B), Nurr1, and NOR-1 on various promoter targets. (A) The effect of the three Nur subfamily members (7) on transcription driven from the POMC (bp −480) was assessed by lipofection into AtT-20 cells, using expression vectors for each transcription factor and the POMC-luc reporter as described previously (31). Results are shown, relative to basal expression, as the means ± standard errors of the means of data from three experiments, each performed in duplicate. (B) DNA sequences of various Nur response elements. The POMC NurRE (31) sequence is present in the rat POMC promoter at bp −382. The positions of three NurREPOMC mutants (labeled M1 to M3) used in the present work are shown under the sequence. The NurRECON is composed of two NBRE canonical sites organized as in the NurREPOMC. The NBRE sequence was described by Wilson et al. (40), and the Nur-responsive DR-5 was described by Perlmann and Jansson (30). (C) Dose-response curves of NurREPOMC-luc activation by the three Nur family members. The reporter was cotransfected with expression vectors for each Nur factor in CV-1 cells as described previously (31). (D) Dose-response curves similar to those in panel C, except that an NBRE reporter was used. Both sets of data were obtained in the same experiments.
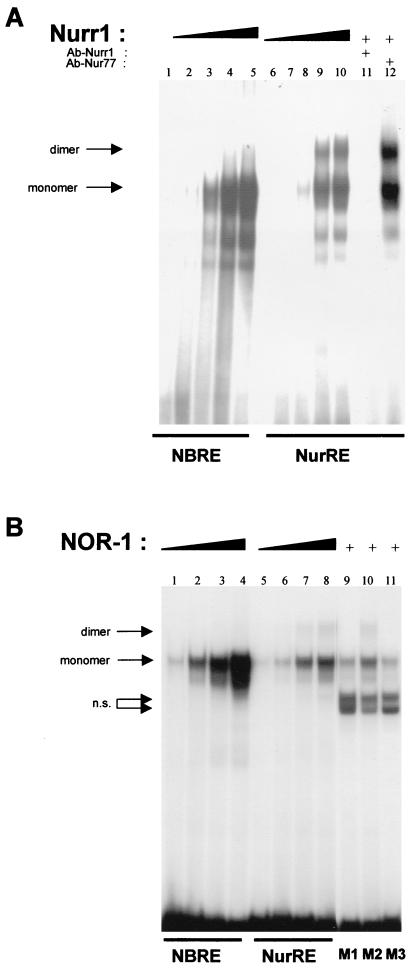
Nurr1 and NOR-1 bind the NurRE as homodimers.
We previously showed that homodimers of Nur77 bound the NurREPOMC and that this binding appeared cooperative, in contrast to the binding of monomers to the NBRE (31). Gel retardation experiments showed that Nurr1 forms two major complexes with a NurREPOMC probe (Fig. 3A, lanes 6 to 10), one that comigrated with Nurr1 monomers bound to a NBRE probe (Fig. 3A, lanes 1 to 5) and the other, more slowly migrating complex that likely consisted of Nurr1 homodimers. Both complexes were blocked by pretreatment with a Nurr1 antibody but not a Nur77 antibody (Fig. 3A, lanes 11 and 12). Interestingly, Nurr1 appeared to form fewer dimer than monomer complexes, in comparison to Nur77 (31). Similarly, NOR-1 formed monomer complexes with both NBRE (Fig. 3B, lanes 1 to 4) and NurREPOMC (Fig. 3B, lanes 5 to 8). However, it formed even fewer dimer complexes with the NurREPOMC than Nurr1. Nonetheless, each NurREPOMC half-site was bound by NOR-1, and both were required for dimer binding. This was shown by using mutated NurREPOMC oligonucleotides (31) in which a mutation in either half-site abolished dimer formation (mutants M1 and M3) (Fig. 3B, lanes 9 and 11), whereas mutation of the spacer nucleotides (mutant M2) did not affect NOR-1 dimer binding to the NurRE (Fig. 3B, lane 10). Similar results were obtained with Nur77 (31) and Nurr1 (data not shown).
FIG. 3.
Nurr1 and NOR-1 can also form homodimers bound to a NurRE DNA sequence. As for Nur77 (31), both in vitro-translated Nurr1 (A) and NOR-1 (B) form monomeric complexes with the NBRE (lanes 1 to 5 and 1 to 4, respectively). Both also form homodimers with the NurREPOMC probe (lanes 6 to 10 and 5 to 8, respectively). The Nurr1 antiserum (panel A, lane 11), but not the Nur77 antiserum (panel A, lane 12), specifically blocks formation of Nurr1-dependent complexes. As for Nur77 (31), formation of NOR-1 homodimers required each half-site, as indicated by the absence of dimeric complexes when probes containing a mutation of either half-site (mutants M1 and M3) (panel B, lanes 9 and 11), but not a mutation of the intervening nucleotides (mutant M2) (panel B, lane 10), were used; the mutant sequences are shown in Fig. 2B. n.s., nonspecific binding.
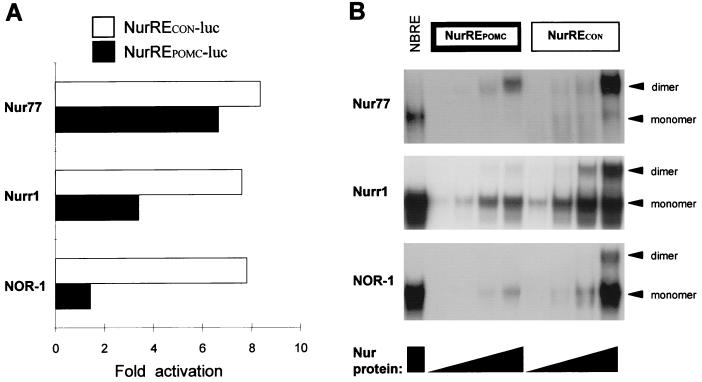
The NurREPOMC is preferentially bound and activated by Nur77 homodimers.
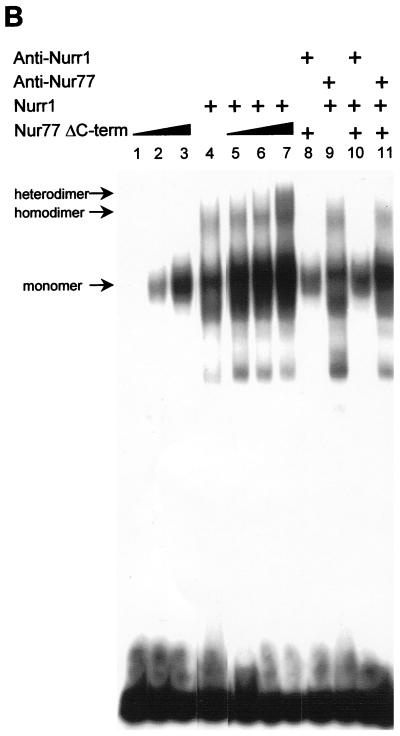
Unlike Nur77, Nurr1 and NOR-1 less readily formed homodimer complexes on the NurREPOMC (31) (Fig. 3). This correlates with their weaker potency in activation of transcription of the NurREPOMC reporter (Fig. 2C). We wondered whether this reflects an intrinsic difference between Nur members or is a peculiarity of the NurREPOMC target sequence. To address this question, we constructed reporter plasmids containing a single NurRE target sequence corresponding either to the NurREPOMC or to a synthetic NurRE (NurRECON) that has two perfect consensus half-sites identical to the NBRE (Fig. 2B). In contrast to the NurREPOMC reporter, which is less responsive to Nurr1 and NOR-1 (Fig. 2C and 4A), the NurRECON was activated by all three Nur factors as effectively as Nur77 activated the NurREPOMC reporter (Fig. 4A). The greater transcriptional responsiveness of NurRECON correlated with an increased ability to form homodimers in vitro. Indeed, gel retardation experiments showed that all three receptors effectively formed homodimers with the NurRECON whereas Nur77 was the only one to readily form homodimers with the NurREPOMC (Fig. 4B).
FIG. 4.
The NurREPOMC is preferentially activated and bound by Nur77, whereas a consensus NurRE does not exhibit this preference. (A) The abilities of the three Nur subfamily members to activate transcription from luciferase reporters containing one copy of either the POMC gene NurRE (NurREPOMC) or a consensus NurRE (NurRECON) composed of two half-sites of canonical NBREs were tested by transfection in CV1 cells. (B) DNA binding of increasing amounts of in vitro-translated Nur subfamily proteins to either NBRE, NurREPOMC, or NurRECON probe. The positions of homodimer and monomer complexes are indicated.
Nur dimers exhibit transcriptional synergy on the NurREPOMC target.
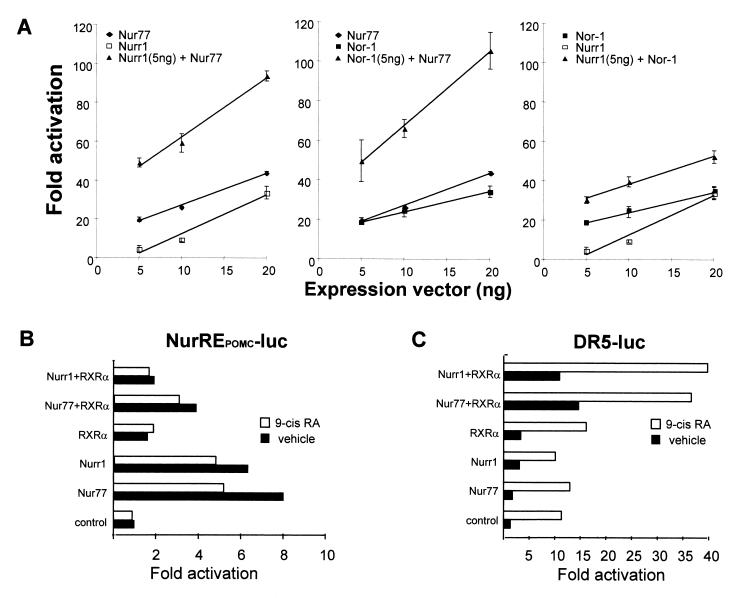
Since all three Nur subfamily members are induced in response to CRH, and given that all three can bind the naturally occurring POMC promoter NurRE, we tested whether Nur subfamily members exhibit transcriptional synergy. When combining pairs of receptors in dose-response experiments, we observed a synergistic response when Nur77 was present. Indeed, the dose-response curve to Nur77 was enhanced and steeper when either Nurr1 or NOR-1 was present at a minimally active concentration (Fig. 5A, left and center panels). The synergy between Nurr1 and NOR-1 was somewhat less striking (Fig. 5A, right panel). No synergy was observed when the same experiments were performed with the NBRE-luciferase reporter (data not shown). Since it has been reported that both Nur77 and Nurr1 can heterodimerize with RXR to activate transcription driven by a DR5 response element (30) or by an NBRE (10), we tested whether these heterodimers could activate NurRE-dependent transcription. Rather than enhance Nur77- or Nurr1-dependent activity, the addition of RXR decreased reporter activity (Fig. 5B), presumably because of Nur factor squelching by RXR. Thus, Nur-RXR heterodimers did not activate the NurREPOMC target as they do on a reporter containing a DR5 (Fig. 5C).
FIG. 5.
Synergistic activation of NurRE reporter by pairs of Nur subfamily factors. (A) The activities of the three possible pairs of Nur77 subfamily members were assessed in comparison to that of each factor alone, using the same NurRE reporter as was used for the experiment shown in Fig. 2, after cotransfection in CV1 cells. (B) The effect of RXR, with or without its ligand 9-cis RA, on the activity of the NurRE reporter was assessed in the presence of Nur77 or Nurr1. (C) Effect of RXR with and without 9-cis RA, together with Nur77 or Nurr1, on a DR5 reporter.
Heterodimerization between Nur77 and Nurr1.
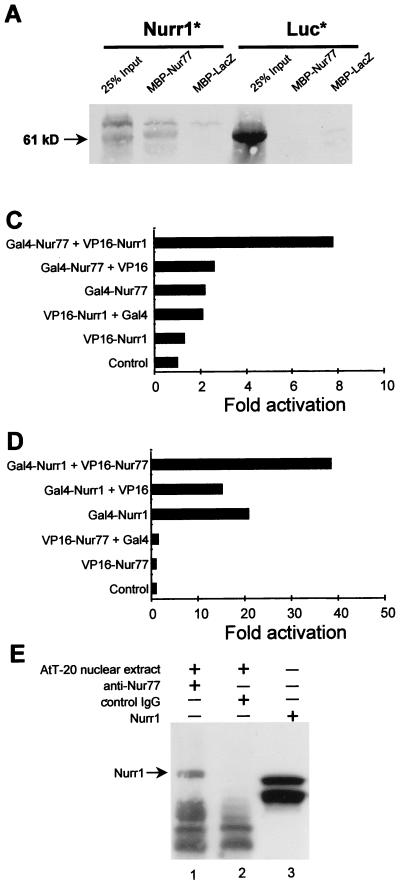
The formation of heterodimers binding the NurRE is the simplest way to explain the transcriptional synergy exerted by Nur subfamily members on NurRE reporters (Fig. 5A). We therefore investigated the possibility of direct protein-protein interaction by using an in vitro pull-down assay (Fig. 6A) in which a resin-bound MBP-Nur77 fusion protein was tested for interaction with in vitro-translated Nurr1 (left panel) or luciferase as a negative control (right panel). Nurr1 specifically bound the MBP-Nur77 column but not the control MBP-LacZ column. Next, the abilities of Nur77 and Nurr1 to form heterodimers on the NurRE were analyzed by gel retardation. Initial attempts at heterodimer formation with the intact proteins were difficult to interpret because of similar electrophoretic migration patterns (data not shown). We could separate heterodimer complexes from homodimer complexes by using a mutant Nur77 deleted of its ligand binding domain (Fig. 6B). This mutant Nur77 formed less dimer (lanes 1 to 3) than Nurr1 (lane 4). When mixed together, they formed a new complex that contained heterodimers of the two factors (lane 7). The presence of both Nur77 and Nurr1 in the new complex was verified by using specific antisera against Nur77 (lane 11) and Nurr1 (lane 10). Since the pull-down assay suggested interaction even in the absence of NurRE, we set up a mammalian two-hybrid system to ascertain protein interaction in vivo (Fig. 6C and D). Using a UAS-containing reporter, we observed that the activity of a Gal4 DBD-Nur77 fusion protein was greatly enhanced in the presence of a VP16-Nurr1 chimera which contains the VP16 activation domain fused to Nurr1 (Fig. 6C). Conversely, the activity of a Gal4 DBD-Nurr1 fusion was specifically enhanced in the presence of a VP16-Nur77 chimera (Fig. 6D). To determine whether Nur proteins exist as heterodimers in vivo, we performed coimmunoprecipitation experiments using AtT-20 cells nuclear extracts and antibodies against Nur77 (Fig. 6E). The immunoprecipitates were analyzed by Western blotting with an antiserum to Nurr1. This experiment clearly showed that Nurr1 was precipitated with Nur77 from AtT-20 nuclei (Fig. 6E, lane 1) but not with control IgG (lane 2). Taken together, these results indicate that Nur77 and Nurr1 can form heterodimers by direct protein-protein interactions and that these heterodimers are present in cell nuclei.
FIG. 6.
Nur77 and Nurr1 form heterodimers. (A) The abilities of Nur77 and Nurr1 to interact in vitro were assessed by using a pull-down assay. Fusion proteins consisting of MBP and either Nur77 or LacZ (as a control) were bound to a maltose column, and either in vitro-translated Nurr1 or luciferase (Luc; as a control) was tested for binding. The position of full-length Nurr1 (61 kDa) is indicated by an arrow. (B) Formation of Nurr1-Nur77 heterodimer complexes upon binding of a NurRECON probe in a gel retardation assay. Since native proteins comigrated in gel retardation experiments (data not shown), we used a mutant Nur77 with a truncated C-terminal domain for the experiment; this mutant preferentially binds as a monomer on its own (lanes 1 to 3), whereas Nurr1 binds both as a monomer and as a homodimer (lane 4). In the presence of increasing amounts of mutant Nur77 (lanes 5 to 7) and Nurr1, a new complex (labeled heterodimer) appears (lane 7). Formation of these heterodimers is blocked by an antiserum specific for Nurr1 (lane 10) or Nur77 (lane 11). The anti-Nurr1 does not recognize Nur77 (lane 8), and the anti-Nur77 does not recognize Nurr1 (lane 9). (C and D) Two-hybrid assays were performed in CV1 cells to show that the chimeric proteins Gal4-Nur77 and VP16-Nurr1 (C) or Gal4-Nurr1 and VP16-Nur77 (D) interact in vivo. Cells were cotransfected with a UAS-containing luciferase reporter plasmid. (E) Presence of Nur77-Nurr1 complexes, as revealed by coimmunoprecipitation of AtT-20 cell nuclear extracts. Extracts were immunoprecipitated with antibodies against Nur77 (lane 1) or control IgG (lane 2) and analyzed by Western blotting with Nurr1 antiserum. In vitro-translated Nurr1 protein was used as a reference (lane 3).
DISCUSSION
The present work demonstrates for the first time that Nur subfamily members form heterodimers (Fig. 6) and that these heterodimers can be more potent transcriptional activators than homodimers of the same factors (Fig. 5). We have also shown that Nur dimers (homo- or heterodimers) are far more active than monomers on their respective cognate target sequences, the NurRE or NBRE (31) (Fig. 2C and D). Significantly, we showed that Nur dimers exhibit target sequence preference (Fig. 4). Thus, a combinatorial code for downstream-gene-specific effects may result from both the tissue-specific expression of Nur subfamily members and the presence of specific NurRE sequences on subsets of target genes.
Consistent with this model, Nur subfamily factors exhibit expression patterns that are partly overlapping and partly complementary. For example, exclusive expression of Nurr1 in the midbrain dopaminergic system is consistent with the absence of these neurons in Nurr1-deficient mice (2, 35, 46). Also, Nur77 is specifically induced by light in the suprachiasmatic nucleus, where the mammalian circadian clock is located (19, 23, 33). Recently, a wide screen of genes induced by serum in fibroblasts revealed that human NOR-1 (MINOR) is a major serum-responsive gene (14). In other tissues, Nur subfamily member expression patterns are overlapping, as in the case of Nur77 and NOR-1 in immature thymocytes, in which these factors seemingly play functionally redundant roles (3). We now report that all three Nur subfamily members are rapidly but transiently induced by CRH in POMC-expressing pituitary cells and that this induction is antagonized by Gc. These results may explain the lack of a pituitary and HPA axis phenotype in both Nur77- and Nurr1-null mice (4, 5).
We have recently shown the importance of dimer formation for Nur77-dependent transcriptional activation (31); that work suggested that Nur77 dimer action on the NurRE might constitute the physiological mechanism of action for this NR. Here we extended this model to Nurr1 and NOR-1, and we further showed that Nur factor heterodimers are even more potent transcriptional activators than homodimers. This was particularly true for heterodimers containing Nur77 and less so for Nurr1–NOR-1 heterodimers (Fig. 5A). The Nur77 preference appears to be unique to the NurREPOMC since it was not observed for the NurRECON, which contains consensus half-sites (Fig. 4). Indeed, the in vitro affinity of the three homodimers correlated well with their in vivo ability to activate the NurREPOMC. This observation is very interesting because it suggests that some NurREs could have evolved to respond preferentially to one specific member of the subfamily (Fig. 4) or to heterodimers containing this Nur factor (Fig. 5). Recent data suggest that this model may also apply for T-cell target genes. Indeed, forced expression of Nur77 or NOR-1 in the T cells of transgenic mice resulted in upregulation of CD25 in CD4+ CD8+ T cells and in thymocyte apoptosis (3). In contrast, this was not observed in transgenic mice overexpressing Nurr1. Thus, it appears that CD25 may respond specifically to Nur77 or NOR-1 but not to Nurr1 and that this specificity may be due to a NurRE preferentially activated by homo- or heterodimers of Nur77 and/or NOR-1.
Coupled with the existence of factor-specific target sequences, the multiple forms (monomers, homodimers, or heterodimers) by which Nur factors affect transcription provide for great versatility in fine-tuning target cell and/or gene responses to particular stimuli. For example, Nurr1 is constitutively expressed in the hypothalamic paraventricular nucleus, which suggests that it participates in basal expression of some genes (34). In response to stress, Nur77 is rapidly induced (12), and it could therefore either specifically activate transcription of other genes or, together with Nurr1, synergistically activate transcription of common target genes. Our results for the POMC-expressing AtT-20 cells (Fig. 1) suggest that similar mechanisms could take place in the anterior pituitary, since only Nurr1 mRNA is detected under basal conditions whereas all three Nur subfamily members are induced upon CRH treatment. Transcriptional regulation by differential expression of Nur members could also take place in PC12 cells, in which Nur77 and Nurr1 are induced upon membrane depolarization but only Nur77 is induced in response to NGF (17). The finding that RXR represses Nur-dependent activation of the NurRE was somewhat surprising, since transcriptional cooperativity between the two subfamilies had been demonstrated (10, 30). However, this could provide an additional mechanism for cross talk between retinoid signaling and the Nur signaling pathway. For example, retinoids are known to inhibit TCR-induced apoptosis (1, 13, 33), which is absolutely dependent on Nur activation (20, 43). Thus, it is plausible that RXR could heterodimerize with Nur77 to block NurRE-dependent activation of proapoptotic genes downstream of the Nur pathway, while the same heterodimers could activate cell survival-promoting genes whose promoters contain DR5 elements. Despite the critical importance of the Nur signaling pathway in T-cell apoptosis, very little is known about the target genes lying downstream of it. The search for NurREs, whether degenerate or not, in the regulatory regions of putative target genes may prove to be instructive.
The demonstration in this work that dimers of the Nur subfamily exhibit strong, target sequence-specific transcriptional activity further highlights the importance of NurRE targets for gene regulation. In addition, the ability of Nur factors to heterodimerize with other NR, like RXR and COUP-TF, provides the basis for cross talk between the Nur signaling pathway and retinoid action. Additionally, we have already shown antagonism between Gc and the Nur signaling pathway. Taken together, these multiple interactions offer hypotheses to account for the diverse effects of the Nur signaling pathway on the hormone response, proliferation, differentiation, and programmed cell death.
ACKNOWLEDGMENTS
We are very thankful to Thomas Perlmann, Stockholm, Sweden, for providing antisera against Nur77 and Nurr1 as well as the UAS reporter and the GAL-4/Nur77 and GAL-4/Nurr1 constructs. We are grateful to O. Conneeley and N. Ohkura for the Nurr1 and NOR-1 cDNAs, respectively. J. Milbrandt generously provided the C-terminally deleted NGFI-B, and Vincent Giguère provided the NGFI-B expression vector and the DR-5 reporter plasmid. The help of Michel Chamberland in Northern blotting was appreciated. We also thank Lise Laroche for expert secretarial assistance.
This work was funded by the Medical Research Council of Canada, and M. Maira is the recipient of a doctoral research award from the Medical Research Council of Canada.
REFERENCES
- 1.Bissonnette R P, Brunner T, Lazarchik S B, Yoo N J, Boehm M F, Green D R, Heyman R A. 9-cis retinoic acid inhibition of activation-induced apoptosis is mediated via regulation of Fas ligand and requires retinoic acid receptor and retinoid X receptor activation. Mol Cell Biol. 1995;15:5576–5585. doi: 10.1128/mcb.15.10.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castillo S O, Baffi J S, Palkovits M, Goldstein D S, Kopin I J, Witta J, Magnuson M A, Nikodem V M. Dopamine biosynthesis is selectively abolished in substantia nigra/ventral tegmental area but not in hypothalamic neurons in mice with targeted disruption of the Nurr1 gene. Mol Cell Neurosci. 1998;11:36–46. doi: 10.1006/mcne.1998.0673. [DOI] [PubMed] [Google Scholar]
- 3.Cheng L E C, Chan F K M, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997;16:1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conneely O M, Satyamoorthy K, Saucedo-Cardenas O, De Mayo F. Abstracts of the 79th annual meeting of the Endocrine Society. Bethesda, Md: The Endocrine Society Press; 1997. Neurodevelopmental role of the orphan nuclear receptor, Nurr1, abstr. S40-3; p. 53. [Google Scholar]
- 5.Crawford P A, Sadovsky Y, Woodson K, Lee S L, Milbrandt J. Adrenocortical function and regulation of the steroid 21-hydroxylase gene in NGFI-B-deficient mice. Mol Cell Biol. 1995;15:4331–4336. doi: 10.1128/mcb.15.8.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis I J, Lau L F. Endocrine and neurogenic regulation of the orphan nuclear receptors Nur77 and Nurr-1 in the adrenal glands. Mol Cell Biol. 1994;14:3469–3483. doi: 10.1128/mcb.14.5.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drouin J, Maira M, Philips A. Novel mechanism of action for Nur77 and antagonism by glucocorticoids: a convergent mechanism for CRH activation and glucocorticoid repression of POMC gene transcription. J Steroid Biochem Mol Biol. 1998;65:59–63. doi: 10.1016/s0960-0760(97)00180-5. [DOI] [PubMed] [Google Scholar]
- 8.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enmark E, Gustafsson J A. Orphan nuclear receptors—the first eight years. Mol Endocrinol. 1996;10:1293–1307. doi: 10.1210/mend.10.11.8923456. [DOI] [PubMed] [Google Scholar]
- 10.Forman B M, Umesono K, Chen J, Evans R M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 11.Hazel T G, Nathans D, Lau L F. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci USA. 1988;85:8444–8448. doi: 10.1073/pnas.85.22.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honkaniemi J, Kononen J, Kainu T, Pyykonen I, Pelto-Huikko M. Induction of multiple immediate early genes in rat hypothalamic paraventricular nucleus after stress. Brain Res. 1994;25:234–241. doi: 10.1016/0169-328x(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 13.Iwata M, Mukai M, Nakai Y, Iseki R. Retinoic acids inhibit activation-induced apoptosis in T cell hybridomas and thymocytes. J Immunol. 1992;149:3302–3308. [PubMed] [Google Scholar]
- 14.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 15.Lamonerie T, Tremblay J J, Lanctôt C, Therrien M, Gauthier Y, Drouin J. PTX1, a bicoid-related homeobox transcription factor involved in transcription of pro-opiomelanocortin (POMC) gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 16.Lau L F, Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J. 1985;4:3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law S W, Conneely O M, DeMayo F J, O’Malley B W. Identification of a new brain-specific transcription factor, NURR1. Mol Endocrinol. 1992;6:2129–2135. doi: 10.1210/mend.6.12.1491694. [DOI] [PubMed] [Google Scholar]
- 18.Lee S L, Wesselschmidt R L, Linette G P, Kanagawa O, Russell J H, Milbrandt J. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (NUR77) Science. 1995;269:532–535. doi: 10.1126/science.7624775. [DOI] [PubMed] [Google Scholar]
- 19.Lin J T, Kornhauser J M, Singh N P, Mayo K E, Takahashi J S. Visual sensitivities of nur77 (NGFI-B) and zif268 (NGFI-A) induction in the suprachiasmatic nucleus are dissociated from c-fos induction and behavioral phase-shifting responses. Brain Res Mol Brain Res. 1997;46:303–310. doi: 10.1016/s0169-328x(97)00005-3. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z G, Smith S W, McLaughlin K A, Schwartz L M, Osborne B A. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature. 1994;367:281–284. doi: 10.1038/367281a0. [DOI] [PubMed] [Google Scholar]
- 21.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1:183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 23.Morris M E, Viswanathan N, Kuhlman S, Davis F C, Weitz C J. A screen for genes induced in the suprachiasmatic nucleus by light. Science. 1998;279:1544–1547. doi: 10.1126/science.279.5356.1544. [DOI] [PubMed] [Google Scholar]
- 24.Murphy E P, Conneely O M. Neuroendocrine regulation of the hypothalamic pituitary adrenal axis by the nurr1/nur77 subfamily of nuclear receptors. Mol Endocrinol. 1997;11:39–47. doi: 10.1210/mend.11.1.9874. [DOI] [PubMed] [Google Scholar]
- 25.Nordeen S K. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques. 1988;6:454–456. [PubMed] [Google Scholar]
- 26.Ohkura N, Hijikuro M, Yamamoto A, Miki K. Molecular cloning of a novel thyroid/steroid receptor superfamily gene from cultured rat neuronal cells. Biochem Biophys Res Commun. 1994;205:1959–1965. doi: 10.1006/bbrc.1994.2900. [DOI] [PubMed] [Google Scholar]
- 27.Parkes D, Rivest S, Lee S, Rivier C, Vale W. Corticotropin-releasing factor activates c-fos, NGFI-B, and corticotropin-releasing factor gene expression within the paraventricular nucleus of the rat hypothalamus. Mol Endocrinol. 1993;7:1357–1367. doi: 10.1210/mend.7.10.8264665. [DOI] [PubMed] [Google Scholar]
- 28.Paulsen R E, Granas K, Johnsen H, Rolseth V, Sterri S. Three related brain nuclear receptors, NGFI-B, Nurr1, and NOR-1, as transcriptional activators. J Mol Neurosci. 1995;6:249–255. doi: 10.1007/BF02736784. [DOI] [PubMed] [Google Scholar]
- 29.Paulsen R E, Weaver C A, Fahrner T J, Milbrandt J. Domains regulating transcriptional activity of the inducible orphan receptor NGFI-B. J Biol Chem. 1992;267:16491–16496. [PubMed] [Google Scholar]
- 30.Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 31.Philips A, Lesage S, Gingras R, Maira M-H, Gauthier Y, Hugo P, Drouin J. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philips A, Maira M, Mullick A, Chamberland M, Lesage S, Hugo P, Drouin J. Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol Cell Biol. 1997;17:5952–5959. doi: 10.1128/mcb.17.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rusak B, McNaughton L, Robertson H A, Hunt S P. Circadian variation in photic regulation of immediate-early gene mRNAs in rat suprachiasmatic nucleus cells. Brain Res Mol Brain Res. 1992;14:124–130. doi: 10.1016/0169-328x(92)90019-8. [DOI] [PubMed] [Google Scholar]
- 34.Saucedo-Cardenas O, Conneely O M. Comparative distribution of NURR1 and NUR77 nuclear receptors in the mouse central nervous system. J Mol Neurosci. 1996;7:51–63. doi: 10.1007/BF02736848. [DOI] [PubMed] [Google Scholar]
- 35.Saucedo-Cardenas O, Quintana-Hau J D, Le W D, Smidt M P, Cox J J, DeMayo F, Burbach J P H, Conneely O M. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Therrien M, Drouin J. Pituitary pro-opiomelanocortin gene expression requires synergistic interactions of several regulatory elements. Mol Cell Biol. 1991;11:3492–3503. doi: 10.1128/mcb.11.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tremblay J J, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson M A, Milbrandt J. Expression of the nerve growth factor-regulated NGFI-A and NGFI-B genes in the developing rat. Development. 1990;110:173–183. doi: 10.1242/dev.110.1.173. [DOI] [PubMed] [Google Scholar]
- 40.Wilson T E, Fahrner T J, Johnston M, Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 41.Wilson T E, Mouw A R, Weaver C A, Milbrandt J, Parker K L. The orphan nuclear receptor NGFI-B regulates expression of the gene encoding steroid 21-hydroxylase. Mol Cell Biol. 1993;13:861–868. doi: 10.1128/mcb.13.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson T E, Paulsen R E, Padgett K A, Milbrandt J. Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science. 1992;256:107–110. doi: 10.1126/science.1314418. [DOI] [PubMed] [Google Scholar]
- 43.Woronicz J D, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367:277–281. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- 44.Wu Q, Li Y, Liu R, Agadir A, Lee M O, Liu Y, Zhang X. Modulation of retinoic acid sensitivity in lung cancer cells through dynamic balance of orphan receptors nur77 and COUP-TF and their heterodimerization. EMBO J. 1997;16:1656–1669. doi: 10.1093/emboj/16.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Q, Castillo S O, Nikodem V M. Distribution of messenger RNAs for the orphan nuclear receptors Nurr1 and Nur77 (NGFI-B) in adult rat brain using in situ hybridization. Neuroscience. 1996;75:221–230. doi: 10.1016/0306-4522(96)00159-5. [DOI] [PubMed] [Google Scholar]
- 46.Zetterstrom R H, Solomin L, Jansson L, Hoffer B J, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 47.Zetterstrom R H, Solomin L, Mitsiadis T, Olson L, Perlmann T. Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol. 1996;10:1656–1666. doi: 10.1210/mend.10.12.8961274. [DOI] [PubMed] [Google Scholar]