Abstract
Hepatic gene transfer with adeno-associated viral (AAV) vectors shows much promise for the treatment of the X-linked bleeding disorder hemophilia B in multiple clinical trials. In an effort to further innovate this approach and to introduce alternative vector designs with potentially superior features into clinical development, we recently built a vector platform based on AAV serotype 3 because of its superior tropism for human hepatocytes. A vector genome with serotype-matched inverted terminal repeats expressing hyperactive human coagulation factor IX (FIX)-Padua was designed for clinical use that is optimized for translation using hepatocyte-specific codon-usage bias and is depleted of immune stimulatory CpG motifs. Here, this vector genome was packaged into AAV3 (T492V + S663V) capsid for hepatic gene transfer in non-human primates. FIX activity within or near the normal range was obtained at a low vector dose of 5 × 1011 vector genomes/kg. Pre-existing neutralizing antibodies, however, completely or partially blocked hepatic gene transfer at that dose. No CD8+ T cell response against capsid was observed. Antibodies against the human FIX transgene product formed at a 10-fold higher vector dose, albeit hepatic gene transfer was remarkably consistent, and sustained FIX activity in the normal range was nonetheless achieved in two of three animals for the 3-month duration of the study. These results support the use of this vector at low vector doses for gene therapy of hemophilia B in humans.
Keywords: adeno-associated virus, AAV, hemophilia, factor IX, liver, non-human primate, immune response, capsid, antibodies
Graphical abstract

Hepatic AAV gene transfer shows much promise for the treatment of hemophilia. Here, a vector with AAV3 serotype-matched inverted terminal repeats expressing coagulation factor IX-Padua was designed using hepatocyte-specific codon bias and depletion of CpG motifs. FIX activity near the normal range was obtained at low vector dose in non-human primates.
Introduction
Gene therapy for the X-linked bleeding disorder hemophilia is currently being evaluated in multiple phase I/II and III trials that are based on hepatic gene transfer with adeno-associated viral (AAV) vectors following intravenous delivery.1, 2, 3, 4 The various trials utilize a variety of different capsids with liver tropism and expression cassettes for coagulation factor VIII (FVIII, hemophilia A) or factor IX (FIX, hemophilia B) containing a hepatocyte-specific promoter for production and secretion of the coagulation factor transgene products from hepatocytes into the blood circulation. Curative levels of expression have been achieved, which, however, have not been sustained for hemophilia A.5,6 In contrast, sustained expression of FIX activity near the normal range for at least 3 years has been reported.4,7 These results highlight the potential of gene therapy to achieve sufficient sustained levels of clotting factor expression that prevents bleeding in persons with hemophilia without the traditional need for frequent injections of coagulation factor products or other drugs that restore hemostasis. The success in hemophilia B was facilitated by use of a naturally occurring Padua variant of FIX, in which a single amino acid change (R338L) results in an ∼1 log increased specific activity.4,8 Therefore, efficacy can be achieved at low levels of expression, permitting the use of reduced vector doses. Importantly, none of the clinical trials using the Padua variant of FIX has reported an immune response to FIX-Padua protein.
Besides gene therapy, diverse alternative approaches are dramatically changing treatment of hemophilia. Extended half-life coagulation factors have significantly reduced the frequency of injections required with standard half-life factor replacement therapy, and in the case of hemophilia A, the bispecific antibody emicizumab can be given subcutaneously weekly to monthly. Other alternatives to coagulation factors are being developed such as antibodies or small interfering RNA (siRNA) molecules that interfere with anti-coagulant pathways and may also allow for easier administration and reduced frequency of drug administration.1,9 However, unlike these other treatment options, only gene therapy can correct hemophilia over the long term without a need for repeated drug injections. Large animal studies and developing clinical trial data suggest that gene therapy may correct hemophilia for at least a decade following a single drug administration.2,10,11 In patients with hemophilia, coagulation activities >5% of normal correlate with mild disease, rarely requiring treatment with additional coagulation factor. Recent data from people with mild hemophilia have shown that levels above 30% factor activity have a bleeding profile close to those in the range of 50%–150% activity in plasma of hemostatically normal humans.12
To further innovate hepatic AAV gene therapy for hemophilia, we previously developed vectors that utilize the capsid of serotype 3 or variations thereof. Among the commonly used serotypes, AAV3 has the strongest tropism for human hepatocytes, which can be further enhanced by mutations such as AAV3-ST (AAV3 variant T492V + S663V).13, 14, 15 In order to produce a vector of highest potency, we utilized AAV3 instead of AAV2 inverted terminal repeats (ITRs) and deleted the terminal resolution sequence in one ITR to generate self-complementary AAV3 (scAAV3, which does not need to undergo second-strand synthesis to achieve transgene expression).16 Finally, Brown et al.17,18 optimized the expression cassette for human FIX (hFIX) by (1) combining a strong hepatocyte-specific enhancer from the human SERPINA1 untranslated region (termed HSh) with the transthyretin promoter, and (2) by synthesizing an F9 coding region that is CpG depleted and codon optimized for expression in human liver. This clinical candidate vector construct, scAAV-HSh/TTR-hFIX-Padua, is awaiting evaluation in phase I/II clinical trial in patients with severe hemophilia B after packaging into AAV3 capsid. In this study, we evaluate the efficacy and immunogenicity of scAAV3-ST-HSh/TTR-hFIX-Padua in non-human primates (NHPs). We find that levels of hFIX activity near normal or in the normal range can be achieved at low vector doses with no evidence for CD8+ T cell responses to capsid, albeit gene transfer is only successful in animals with low pre-existing neutralizing antibodies against the capsid. To the best of our knowledge, our study for the first time evaluates a vector with non-AAV2 serotype-matched ITRs in non-human primates. In addition, we determine the efficacy of a CpG-depleted, target organ-optimized expression cassette in non-human primates. We find high efficacy with highly consistent levels of hepatic gene transfer and expression unless pre-existing antibody titers against capsid are elevated. Levels and stability of circulating hFIX antigen and activity may be impacted by humoral immune responses and other unknown factors and thus result in inter- and intra-animal variabilities.
Results
Expression of hFIX following hepatic gene transfer in non-human primates
As illustrated in Figure 1, six male non-human primates (cynomolgus macaques, 2–2.5 years old) were divided into two cohorts (n = 3) and injected intravenously with 5 × 1011 or 5 × 1012 vector genomes (vg)/kg of scAAV3ST-hFIX-Padua for liver-directed gene transfer and expression of hFIX (Padua variant) from a hepatocyte-specific enhancer/promoter combination.17 Subsequently, plasma samples were analyzed at different time points for hFIX expression levels. A dose-dependent expression of hFIX was observed in these animals, with each group reaching peak expression at 14–28 days after vector infusion (Figures 2A and 2B). In the low-dose group (Figure 2A), two of three animals had detectable levels of hFIX. In one low-dose animal (ID no. 1609755/teal circle), expression reached 170 ng/mL at 14 days. Thereafter, hFIX expression was maintained around 125 ng/mL throughout the course of study. In a second animal (ID no. 1609991/orange square), the peak of hFIX expression (75 ng/mL) was at 28 days after vector infusion, followed by a stable level of ∼55 ng/mL throughout the course of study. A third animal (ID no. 1610371/purple triangle) initially failed to show hFIX expression, albeit low levels (∼10 ng/mL) were detected late at day 84.
Figure 1.
Experimental scheme to evaluate hFIX expression in non-human primates after AAV gene transfer
(A) Vector genome design. HSh4/TTR, liver-specific enhancer/transthyretin promoter; MVM intron, intron derived from minute virus of mice; PA, polyadenylation signal; TR3, wild-type AAV3 inverted terminal repeat; mTR3, mutated AAV3 inverted terminal repeat (terminal resolution site deleted). (B) AAV capsid. (C) Animal groups, vector doses, and administration route. (D) Evaluation of hFIX expression, activity, immune responses, blood chemistry, and other factors at different time points.
Figure 2.
Dose-dependent expression of hFIX in plasma samples from two cohorts (n = 3) of non-human primates injected intravenously with AAV3-hFIX vector
(A and B) Expression levels of hFIX in animals injected with (A) 5 × 1011 vg/kg and (B) 5 × 1012 vg/kg AAV3-hFIX. Each line represents an individual animal.
All three animals in the cohort infused with 5 × 1012 vg/kg expressed hFIX. The animal with the highest expression maintained a level of ∼1,500 ng/mL for 70 days (ID no. 1610611/green circle) (Figure 2B). A sudden decline in expression (to ∼1,000 ng/mL) was observed at day 84; however, expression levels rebounded to ∼1,300 ng/mL by day 90. A second animal (ID no. 1609933/red square) in this cohort had peak expression of ∼750 ng/mL at 28 days, after which a steady decline in systemic expression was observed. By day 70, plasma hFIX became undetectable. A steady level of expression of ∼200 ng/mL was observed in the third animal (ID no. K999/magenta triangle) of this cohort throughout the course of the study.
Coagulation activity of hFIX in plasma
The coagulation activity of hFIX in plasma samples from non-human primates was determined by a one-stage activated partial thromboplastin time (aPTT) assay. Endogenous FIX activity was determined from plasma collected prior to vector infusion. hFIX-specific activity for experimental animals was calculated by deducting average endogenous FIX activity from total FIX activity determined after vector administration (total FIX activity data are provided in Table S1). In general, hFIX activity correlated with expression levels of hFIX (Figures 2A, 2B, 3A, and 3B). In the low-dose cohort (Figure 3A), the animal with the highest hFIX expression (ID no. 1609755/teal circle) achieved 100% of normal hFIX activity at 14 days after vector infusion followed by a modest decline to a level of 50%–80% of normal that was sustained throughout the remainder of study. The animal with the second highest hFIX expression (ID no. 1609991/orange square) had peak specific activity of ∼20% at 28 days after gene transfer, which declined to ∼1%–2% at the 56-day time point. However, by 84 days after vector infusion, hFIX activity in this animal had returned to ∼15%. Consistent with hFIX antigen levels, no hFIX coagulation activity was detected in the third animal (ID no. 1610371/purple triangle) in this cohort until the last two time points tested in the study, when an activity of 10%–15% was measured.
Figure 3.
hFIX activity in plasma samples from non-human primates collected at different time points after AAV3-hFIX administration
(A and B) Vector doses were (A) 5 × 1011 vg/kg and (B) 5 × 1012 vg/kg. Values are baseline corrected to eliminate endogenous FIX activity. Each line represents an individual animal.
In the high-dose cohort, all animals initially showed greater than 50% hFIX coagulation activity, and greater than 100% of normal in two animals (reaching nearly 200% in one animal). However, the two animals with >100% levels experienced a sudden decline in activity by day 42 (ID no. 1610611/green circle and no. 1609933/red square) (Figure 3B). One animal (ID no. 1610611/green circle) recovered from this loss in activity after 56 days after vector infusion and achieved nearly the initial peak activity by the end of the experiment. In contrast, no recovery in activity was observed in the second animal (ID no. 1609933/red square). The third animal (ID no. K999/magenta triangle) in this cohort maintained its activity level of 50%–70% throughout the course of study.
Development of antibodies against hFIX
In order to determine whether the experimental animals mounted a humoral response to hFIX, plasma from non-human primates was screened for anti-hFIX immunoglobulin G (IgG) by an enzyme-linked immunosorbent assay (ELISA). None of the animals in the low-dose cohort developed antibodies against hFIX (Figure 4A), while two (ID no. 1610611/green circle and no. 1609933/red square) out of three animals in the high-dose cohort had anti-hFIX IgG (Figure 4B). Of these, one animal had a stable antibody titer of 1 μg/mL, while the second animal’s titer increased steadily to ∼5 μg/mL by day 84. To determine whether these antibodies were inhibitory, an aPTT based one-stage Bethesda assay was performed on plasma from these two animals (Figure 4C). One of these animals (ID no. 1609933/red square) developed an inhibitor by 42 days after vector infusion, which correlated with the loss of hFIX expression and activity. The inhibitor titer in this animal increased by 10-fold (4–40 Bethesda units [BU]/mL) between day 70 and 91. In the second animal (ID no. 1610611/green circle), a low-titer inhibitor (2.7 BU/mL) at day 56 was observed, which coincided with the decline in hFIX activity. Although hFIX activity rebounded in this animal, the inhibitor titer stayed consistent (around 2–4 BU/mL) throughout the course of study. Since none of the animals experienced a decline in endogenous FIX activity, antibody formation was likely specific to hFIX. To test whether the presence or absence of circulating anti-hFIX IgG correlated with cellular immune responses, we performed a B cell enzyme-linked immunospot (ELISpot) assay on splenocytes collected at the end of experiment. Out of six animals, only those two animals (ID no. 1610611 and no. 1609933) that had inhibitors had detectable anti-hFIX-producing cells (Figures 4D and 4E), while the other four animals that had neither IgG nor inhibitor titers against hFIX also had no evidence of a B cell response to hFIX.
Figure 4.
Anti-hFIX IgG and Bethesda titers in plasma samples from non-human primates after AAV3-hFIX administration
(A and B) Anti-hFIX IgG antibodies level in (A) 5 × 1011 vg/kg cohort and (B) 5 × 1012 vg/kg cohort. (C) Inhibitory antibodies against hFIX as detected by Bethesda assay. Each line represents an individual animal. (D) Frequency of B cells secreting antibodies to hFIX in splenocytes of each non-human primate as determined by B cell ELISpot assay. Data are presented as spots per million splenocytes. (E) Representative wells from B cell ELISpot assay performed on the splenocytes of each non-human primate collected at the end of experiment.
Antibody formation against AAV3-ST capsid
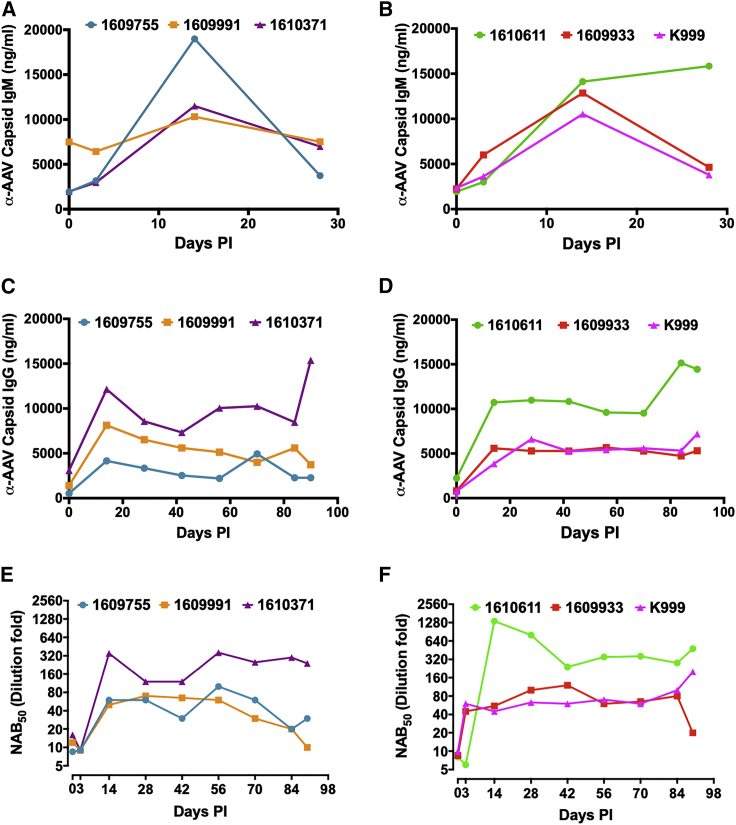
All animals were pre-screened by an in vitro neutralizing antibody (NAb) assay to exclude animals with an NAb titer >1:10. Prior to vector administration and biweekly thereafter, anti-AAV capsid-specific IgG and IgM levels were determined in plasma. IgG binding to capsid was observed in all animals prior to gene transfer (Figures 5C and 5D). A substantial elevation in anti-capsid-IgG titers was evident at 14 days after vector infusion, and these higher titers were maintained in all animals. Interestingly, three animals, two in the low-dose cohort (ID no. 1610371/purple triangle, and to lesser extent no. 1609991/orange square) and one in the high-dose cohort (ID no. 1610611/green circle), had higher pre-existing IgG antibodies to the AAV capsid than did the other animals. However, only the animals in the low-dose cohort showed elevated pre-existing NAbs, which correlated with a failure to achieve hFIX expression in the animal with the highest titer (ID no. 1610371), suggesting a block of gene transfer (Figure 5E). This elevation of NAb titers occurred between the initial screen and gene transfer. Furthermore, animal no. 1610371 developed a higher NAb titer after gene transfer than did the other two animals in this dose cohort (Figure 5E). In the high-dose cohort animal mentioned above (ID no. 1610611/green circle), elevated pre-existing IgG against capsid did not correlate with elevated pre-exiting NAb titer (Figures 5D and 5F) or prevent reaching high levels of hFIX expression (Figures 2B and 3B). However, NAb titers were substantially higher after gene transfer in this animal compared to the other two in the high-dose cohort (Figure 5F). Interestingly, both low- and high-dose cohorts developed similar IgG titers against capsid after gene transfer, and NAb titers were also comparable at least for those animals that had low pre-existing immunity. Similar to anti-AAV-capsid IgG, animals in both cohorts developed anti-AAV-capsid IgM that typically declined between 14 and 28 days (Figures 5A and 5B). Animal no. 1609991 in the low-dose cohort had higher titer pre-existing anti-AAV-capsid IgM than did the other animals and a slightly higher NAb and IgG titer compared to animal no. 1609755, which may explain the approximately 2- to 3-fold difference in hFIX expression between the two animals.
Figure 5.
Antibody response to AAV capsid
(A and B) Anti-AAV3 capsid-specific IgM antibodies levels in (A) 5 × 1011 vg/kg and (B) 5 × 1012 vg/kg cohort. (C and D) Anti-AAV3 capsid-specific IgG antibodies levels in (C) 5 × 1011 vg/kg and (D) 5 × 1012 vg/kg cohort. (E and F) AAV3 neutralizing antibody (NAb) profiles in the serum from animals before and after administration of AAV3-hFIX in (E) 5 × 1011 vg/kg and (F) 5 × 1012 vg/kg cohort. Each line represents an individual animal.
Cellular immune response to capsid
In order to assess whether a capsid-specific CD8+ T cell response was induced after AAV3-ST administration to non-human primates, peripheral blood mononuclear cells (PBMCs, collected at 14 days after vector infusion) and splenocytes (collected at the end of experiment) from non-human primates were stimulated with three pools (a total of 146 overlapping peptides) of an AAV3-ST capsid library. Following stimulation, interferon (IFN)-γ secretion was measured by an ELISpot assay. However, both PBMCs and splenocytes failed to secrete IFN-γ upon stimulation with either peptide pool (Figure S1). These results corroborate a recent study by Bertolini et al.,19 which showed that depletion of CpG motifs from vector genome significantly reduced capsid-specific as well as transgene-specific CD8+ T cell responses. Serum levels of alanine transaminase (ALT), a liver damage marker, also showed no elevations during the study period (Figure S2).
Biodistribution studies
At the end of the experiment, the vector genome copies and hFIX mRNA were determined by quantitative PCR in multiple tissues. For each animal, samples from all four liver lobes were analyzed. As shown in Figures 6A and 6B, gene copy numbers and corresponding mRNA levels were vector dose-dependent and 4- to 6-fold higher in high-dose than in low-dose animals. Gene transfer was undetectable in low-dose animal no. 1610371 that largely lacked hFIX expression in plasma and had an elevated pre-existing NAb titer. This animal also lacked hFIX protein in the liver (Figure 6C). The 2- to 3-fold difference in circulating hFIX levels (and corresponding activity) in animal no. 1609755 compared to no. 1609991 corresponded to a 2-fold difference in gene copy numbers and a similar difference in hFIX antigen in the liver (Figures 2B, 3B, and 6). Within the high-dose cohort, all three animals had similar gene copy numbers and mRNA as well as hFIX protein levels. Consequently, the substantially lower hFIX levels in animal no. K999 compared to the other two animals treated with the same vector dose can unlikely be explained by a difference in gene transfer or hepatic transgene expression. In further biodistribution studies, we failed to detect vector genomes in PBMCs or various organs, except for low vector copy numbers in the spleen (Figure S3). No mRNA was detected in non-hepatic tissues (Figure S3).
Figure 6.
Quantification of hepatic gene transfer and transgene expression
(A–C) Quantification of (A) AAV genome, (B) hFIX mRNA transcript normalized to actin mRNA, and (C) hFIX protein levels (ng/mg liver tissue) in different liver lobes of individual animal from both cohorts. Each dot represents an individual lobe, and each bar represents an individual animal. Error bar indicate standard deviation.
Discussion
Owing to the combination of a viral capsid optimized for transduction of human hepatocytes, a therapeutic cassette optimized for hepatic transgene expression, and use of the hyperactive hFIX-Padua variant, we were able to achieve hFIX activity in the mild to normal range at low vector doses of 5 × 1011 vg/kg in non-human primates.
Development of clinical candidate vector
Our vector design takes advantages of multiple features that we previously identified and optimized using in vitro and murine models.16 These include a serotype-matched ITR, which improves vector production yields and also modestly enhances transduction. Others documented that a switch in ITRs does not alter the vector’s tissue tropism, which is determined by the capsid.20 Furthermore, to improve translational efficiency of the transgene produced in hepatocytes, we applied our previously described target cell-specific codon optimization algorithm to the hFIX cDNA sequence.18 This algorithm attempts to match the codon usage bias (CUB) of F9 cDNA to the CUB of genes highly and specifically expressed in human hepatocytes. We have previously shown this tissue-directed codon optimization approach to confer greater benefit than the standard genome-wide CUB strategies commonly used in gene therapy vector designs. The algorithm also avoids any instances of C-G dinucleotides within the coding sequence, a motif that has been implicated in loss of transgene expression in previous AAV gene therapy trials. This optimized cDNA is driven by our compact but powerful hepatocyte-directed promoter HSh-TTR. This promoter, which is a fusion of a regulatory element taken from the human SERPINA1 promoter region and the transthyretin promoter, has been previously described and shown to confer high-level expression from hepatocytes in vitro and in vivo.17
Our measurements in the non-human primate plasma samples showed that hFIX activity levels were ∼20-fold higher than hFIX antigen levels would predict for wild-type (WT) hFIX activity. Given that the animals did not have hemophilia B, the measurements on the primate coagulation background was complicated, so it is possible that our activity levels were somewhat overestimated, or antigen levels were underestimated (albeit our ELISA is hFIX-specific and fails to detect simian FIX). Published studies using hFIX-Padua in gene transfer reported an increase in coagulation activity of 5- to 10-fold compared to WT FIX.21 However, results depend on the assay, with aPTT-based measurements, as we performed here, typically showing higher activity than chromogenic assays.22 Regardless, the efficacy that we achieved in the cynomolgus macaques is comparable to that reported by Spark Therapeutics in hemophilia B patients receiving a similar dose of AAV-FIX-Padua vector with an engineered capsid, which has resulted in stable expression and normalization of annual bleeding rates for at least 3 years.4,7 Levels in non-human primates do not exactly predict levels in humans, which can only be determined by a clinical trial. Nonetheless, our results are encouraging, offering a potential alternative vector to that developed by Spark Therapeutics. The vector construct, optimized for transcription and translation in hepatocytes, packaged into AAV3 capsid, has received regulatory approval for testing in an academic clinical trial for severe hemophilia B patients in India following successful pre-clinical toxicology testing in non-human primates. Our prior non-human primate study in rhesus macaques suggested that the AAV3-ST capsid variant offers an additional 3- to 5-fold increase in transgene expression compared to AAV3 (although this was tested with a different secreted protein, a higher vector dose, and using AAV2 ITRs).15
Effects of pre-existing humoral immunity on gene transfer
Consistent with multiple other studies, we find that pre-existing NAbs can prevent gene transfer.23 This is likely a greater problem for low-vector doses, which are desirable for improved patient safety and decreased production costs but may be more easily neutralized even by low-titer antibodies. All animals had at least low-titer pre-existing NAbs and also showed pre-existing binding antibodies against the capsid by ELISA. It is possible that these represent cross-reactivity resulting from similar serotypes infecting the primate colony. However, we had to screen a large number of cynomolgus macaques (nearly 400 animals) to identify a sufficient number of animals that could be enrolled in the study. Even then, two animals developed increased titers within a 6-week period just prior to gene transfer. These observations suggest that AAV3 or a related serotype is prevalent at least in this particular colony. In the low-dose cohort, gene transfer failed in the animal with the highest pre-existing NAb level (titer of ∼1:20). The animal with the lowest titer achieved the highest level of expression, while the third animal with intermediate titer of >1:10 (and elevated pre-existing IgM) showed intermediate levels of gene transfer hFIX expression. Traditionally, a NAb of 1:5 has been viewed as sufficient to prevent hepatic gene transfer. However, this cutoff depends on the sensitivity of the assay and vector dose. In our assay, a titer of >1:5 but <1:10 did not prevent gene transfer. We previously found that approximately 40% of human serum samples had NAb titers >1:5 against AAV3, which could, however, be further reduced by capsid modifications.24
It is unclear why evidence for low levels of hFIX expression emerged by both ELISA and an activity assay at a late time point (∼3 months after gene transfer) in the animal that initially failed to express and had no evidence for hepatic gene transfer or transgene expression. This raises the possibility of either delayed transduction or a low level of vector integration and expansion at a site in the liver that was not sampled, or, alternatively, extrahepatic gene transfer, although the liver-specific promoter should have prevented such expression, and no transgene mRNA was detected in other tissues. Nguyen et al.25 recently found an increase in FVIII gene expression starting ∼4 years after hepatic AAV gene transfer in hemophilia A dogs. The authors further provided evidence for clonal expansion, but recovered integrated vector genomes contained deletions or rearrangements, so that the origin of FVIII expression remained uncertain.
Antibody formation against the transgene product
At the high vector dose, all animals had remarkably similar levels of gene transfer. In contrast to the low-dose cohort, none of the three animals had elevated pre-existing NAbs. Therefore, it is unknown to what extent higher titers might have interfered with gene transfer at the 10-fold higher vector dose. At this dose, two of three animals developed inhibitors against hFIX. One was low titer and transiently decreased coagulation activity while failing to clear the hFIX antigen. Another animal developed a high-titer inhibitor that cleared hFIX antigen (among the low-dose animals, one showed a somewhat similar phenomenon of transient reduction in hFIX activity despite persistent hFIX antigen expression, although no antibody was detected in this case). The third animal had no detectable anti-hFIX but expressed substantially lower levels than did the other two despite similar vector gene copy numbers and equal levels of transgene mRNA and hFIX protein in the liver. We initially speculated that the animal developed a low-titer antibody that partially cleared hFIX and that we failed to detect hFIX-antibody complexes. However, B cell ELISpots were negative for antibody-producing cells in the spleen of this animal, consistent with our inability to detect an antibody against hFIX in circulation. Results in the other two animals suggest that the high dose had a greater tendency to induce formation of inhibitors against hFIX. This is in contrast to studies in mice that have shown greater capacity for immune tolerance induction to hFIX at higher vector doses/levels of expression.26,27 Inhibitors against hFIX have been described for hepatic AAV gene transfer studies in non-human primates.28 However, these were rare, and most animals did not form inhibitors over a wide range of circulating hFIX antigen expression levels.28,29 One difference is that published studies had been carried out in rhesus macaques, raising the possibility of a species-specific difference. Furthermore, we did not detect anti-hFIX formation at the lower vector dose, which produced hFIX activity within the target of clinical gene therapy (near or at the lower range of normal activity).
Lack of cellular immune response to capsid
Sensing of the AAV genome by TLR9 is a requirement for innate immune responses that happen in the liver early after vector administration.30 Moreover, TLR9 signaling in plasmacytoid dendritic cells serves as an activation signal for cross-priming of AAV capsid-specific CD8+ T cells through type I IFN (IFN I) and CD4+ T help-dependent mechanism.30, 31, 32, 33 CD8+ T cell responses against capsid have been implemented in hepatotoxicity and loss of AAV-FIX gene therapy in patients, prompting the inclusion of IFN-γ ELISpot assays and immune suppression in clinical trials.34, 35, 36 In particular, increased propensity for loss of expression in clinical trials could be correlated with the extent of presence of immune stimulatory CpG motifs in the vector construct.2,37, 38, 39 Viral DNA containing hypomethylated CpG motifs is a strong agonist for TLR9.40 In non-human primate and mouse studies, CD8+ T cell responses against capsid were observed by day 7 after gene transfer, especially for capsid with heparin binding sites, as is the case for AAV3.31,41 However, our vector with an expression cassette that lacks CpG motifs within the hFIX cDNA did not elicit a detectable IFN-γ+ T cell response to capsid. Although we previously found increased TLR9-dependent responses for scAAV vectors, elimination of CpG motifs within the transgene sequence likely countered this effect.30 While we did not formally evaluate a CD8+ T cell response to the hFIX transgene product, the persistence of hFIX antigen expression (except in the animal with the high-titer inhibitor) and maintenance of vector copy and mRNA (except for an animal with higher pre-existing NAbs) argue for a lack of such a response.
In conclusion, the data presented herein further corroborate our contention that the use of an AAV serotype vector with a natural tropism for human hepatocytes, coupled with a further optimized (and de-immunized) transgene expression cassette, is likely to lead to safe, efficacious, and long-term phenotypic correction of hemophilia B in humans with a reduced need for post-vector administration of immune suppression.
Materials and methods
Non-human primate experiments
Male cynomolgus macaques (2.0–2.5 kg) from the Alpha Genesis Primate Research Center were screened for AAV NAbs by an in vitro neutralizing antibody assay as previously described.42 Animals with NAb titers of ≤1/10 were selected for the study. Animal studies were performed at the Animal Medicine Department, University of Massachusetts Medical School under Institutional Animal Care and Use Committee (IACUC) approval. Vector was injected intravascularly via saphenous vein infusion. Monkey sera were collected at different time points for detection of parameters including, ALT and NAbs.
Production of recombinant AAV vector
Recombinant AAV3B mutant (T492V-S663V) vector expressing codon-optimized hFIX was produced by the Viral Vector Core, University of Massachusetts Medical School, using transient transfection of human embryonic kidney (HEK)293 cells followed by CsCl density gradient purification as previously described.43 The self-complementary vector genome, containing a CpG-depleted expression cassette (liver-specific enhancer/promoter combination, intron, hepatic codon-optimized human F9 coding sequence, and a short synthetic poly(A) signal) and AAV3 ITRs, was as published.17 Purity of vector preparation was assessed by 4%–12% SDS-polyacrylamide gel electrophoresis and silver staining (Invitrogen). Vector preparation was quantified by Droplet Digital polymerase chain reaction (ddPCR). Endotoxin levels were undetected (<1.0 endotoxin units [EU]/mL) by the chromogenic limulus amebocyte lysate (LAL) method (Lonza).
ELISA for circulating hFIX, anti-hFIX, and anti-capsid antibodies
Citrated plasma collected at different time points after vector injection was screened for circulating hFIX levels using an ELISA kit (LSBio, Seattle, WA, USA) as per the manufacturer’s instructions. Liver lysates were also screened for hFIX levels. 100 μg of total protein was used to detect hFIX levels in liver lysates. Plasma was also analyzed for the development of anti-hFIX IgG and anti-capsid IgM and IgG as previously described.27,44 Intact AAV3 virus particles were used (2.5 × 109 vg/well) to coat ELISA plates for anti-capsid antibodies. Concentrations of unknown samples were extrapolated from respective standard curves using a four-parameter logistic curve fit.
hFIX activity assay
Activity of hFIX in non-human primate plasma was determined by an aPTT assay using the Diagnostica Stago STart hemostasis analyzer (Diagnostica Stago, Parsippany, NJ, USA). A standard curve was generated using 2-fold serial dilutions of imidazole-buffered TriniCHECK (Diagnostica Stago, Mount Olive, NJ, USA). A standard curve was used to calculate percent activity of hFIX in non-human primate plasma. To calculate hFIX-specific activity, endogenous FIX activity (determined from plasma collected prior to vector infusion) was subtracted from the total FIX activity of non-human primate plasma collected after vector administration.
Inhibitory antibody titers against hFIX were measured in heat-inactivated citrated non-human primate plasma by a one-stage aPTT-based Bethesda assay. One BU represents the reciprocal of the dilution of plasma required to inhibit the FIX activity of pooled normal human plasma by 50%. Percent residual activity of plasma samples was calculated from a standard curve generated by aPTT on 2-fold serial dilutions of imidazole-buffered TriniCHECK (Diagnostica Stago, Mount Olive, NJ).
ELISpot assays
B cell ELISpot
To determine the frequencies of B cells that secreted antibodies against AAV-encoded hFIX, we performed a B cell ELISpot assay.45,46 Briefly, 0.1 U of BeneFIX (Pfizer) in PBS was overnight coated onto surfactant-free multiscreen filter plates (Millipore). As a positive control, monkey IgG (whole molecule) (Rockland, Limerick, PA, USA) was coated at 500 ng/mL in PBS. Only medium was used as a negative control. Cryopreserved splenocytes from each non-human primate were revived in RMPI 1640 (Life Technologies) containing 10% fetal calf serum (Atlanta Biologicals, Norcross, GA, USA), 10,000 U/mL penicillin, and 100 mg/mL streptomycin (Life Technologies). One million splenocytes were added to each coated well and incubated for 16 h. IgG bound to hFIX was probed with horseradish peroxidase-conjugated goat anti-monkey-IgG (Rockland, Limerick, PA, USA). Spots were developed using BD ELISpot AEC substrate set (BD Biosciences, San Diego, CA, USA) and were counted using the ImmunoSpot analyzer (Cellular Technology, Shaker Heights, OH, USA).
T cell ELISpot
To determine the frequencies of AAV3 capsid-specific CD8+ T cells, an ELISpot assay was performed using a monkey IFN-γ ELISpot kit (MabTech, Cincinnati, OH, USA) as per the manufacturer’s instructions. A peptide library of AAV3 capsid constituting 146 peptides was synthesized (Mimotopes, Mulgrave, VIC, Australia). Each peptide was 15 aa long and had an overlap of 10 aa. Peptides were divided into three pools. PBMCs or splenocytes were plated, respectively, at 100,000 or 250,000 cells/well and stimulated with media alone, phorbol myristate acetate (PMA) (Invitrogen), or AAV3 capsid peptide pools. Each peptide was used at a concentration of 10 μg/mL. Analyses were performed in duplicate for individual animals. After around 18 h of stimulation, plates were developed and IFN-γ spot-forming units (SFU) were detected and counted using the ImmunoSpot analyzer (Cellular Technology, Shaker Heights, OH, USA).
Analysis of vector genome and human F9 mRNA in tissue samples by ddPCR
Detection and quantification of vector genomes and human F9 transcripts were performed by ddPCR. DNA was isolated from tissue samples collected at the end of experiment using QIAGEN DNeasy kit (QIAGEN, Valencia, CA, USA). Ten nanograms of DNA was subjected to ddPCR using primer/probe targeting F9 (hFIX forward, 5′-AAAGGACACCAGCACTCATAG-3′; hFIX probe, 5′-6-FAM/CTGTCTGAATGGAGGGTCTTGCAAAGAT-3′IBFQ; hFIX reverse, 5′-TGGAAGCAGTATGTGGATGG-3′) and endogenous RNase P (Thermo Fisher Scientific, catalog no. 4403326). For quantification of F9 transcripts, total RNA was isolated from tissue samples using TRIzol (Thermo Fisher Scientific) and treated with DNase I. Total RNA (0.2–0.5 μg) was primed with random hexamers and reverse transcribed using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific). cDNA was also subjected to ddPCR using primers and probes targeting hFIX and endogenous β-actin (Thermo Fisher Scientific, assay ID no. Hs01060665-g1).
Acknowledgments
This study was supported by National Institutes of Health (NIH) grants R01 NS076991, P01 HL131471, UG3 HL147367, and U19 AI149646 and by a grant from the University of Massachusetts Medical School to G.G.; by NIH grant R01 HL097088 to A.S., G.G., and R.W.H.; and by NIH grants R01 AI51390 and R01 HL131093, the Indiana Collaborative Initiative for Talent Enrichment (INCITE) funds provided by the Lilly Endowment, and by support from the Riley Children's Foundation to R.W.H.
Author contributions
S.R.P.K., J.X., S.H., J.K., Q.H., and H.C.B. performed experiments. S.R.P.K., J.X., H.C.B., C.B.D., H.T.S., Arun Srivastava, G.G., and R.H. designed, analyzed, and interpreted experiments. S.R.P.K., J.X., H.C.B., Alok Srivastava, D.M.M., C.B.D., H.T.S., Arun Srivastava, G.G., and R.W.H. wrote the manuscript. Arun Srivastava, G.G., and R.W.H. supervised the study.
Declaration of interests
G.G. is a scientific co-founder of Voyager Therapeutic, Adrenas Therapeutics, and Aspa Therapeutics and holds equity in these companies. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics, Aspa Therapeutics, and other biopharmaceutical companies. Arun Srivastava is a co-founder of, and holds equity in, Lacerta Therapeutics. Arun Srivastava, G.G., and R.W.H. are inventors on several issued patents on recombinant AAV vectors that have been licensed to various gene therapy companies. C.B.D. and H.T.S. are co-founders of Expression Therapeutics and own equity in the company. Expression Therapeutics owns the intellectual property associated with the LCO-FIX transgene. H.C.B. is an inventor of the technology and an employee of Expression Therapeutics. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict-of-interest policies. The remaining authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.08.001.
Contributor Information
Arun Srivastava, Email: aruns@peds.ufl.edu.
Guangping Gao, Email: guangping.gao@umassmed.edu422.
Roland W. Herzog, Email: rwherzog@iu.edu.
Supplemental information
References
- 1.Butterfield J.S.S., Hege K.M., Herzog R.W., Kaczmarek R. A molecular revolution in the treatment of hemophilia. Mol. Ther. 2020;28:997–1015. doi: 10.1016/j.ymthe.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathwani A.C. Gene therapy for hemophilia. Hematology (Am. Soc. Hematol. Educ. Program) 2019;2019:1–8. doi: 10.1182/hematology.2019000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrin G.Q., Herzog R.W., Markusic D.M. Update on clinical gene therapy for hemophilia. Blood. 2019;133:407–414. doi: 10.1182/blood-2018-07-820720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George L.A., Sullivan S.K., Giermasz A., Rasko J.E.J., Samelson-Jones B.J., Ducore J., Cuker A., Sullivan L.M., Majumdar S., Teitel J. Hemophilia B gene therapy with a high-specific-activity factor IX variant. N. Engl. J. Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierce G.F. Gene therapy for hemophilia: Are expectations matching reality? Mol. Ther. 2020;28:2097–2098. doi: 10.1016/j.ymthe.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce G.F., Kaczmarek R., Noone D., O’Mahony B., Page D., Skinner M.W. Gene therapy to cure haemophilia: Is robust scientific inquiry the missing factor? Haemophilia. 2020;26:931–933. doi: 10.1111/hae.14131. [DOI] [PubMed] [Google Scholar]
- 7.Mendell J.R., Al-Zaidy S.A., Rodino-Klapac L.R., Goodspeed K., Gray S.J., Kay C.N., Boye S.L., Boye S.E., George L.A., Salabarria S. Current clinical applications of in vivo gene therapy with AAVs. Mol. Ther. 2021;29:464–488. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Von Drygalski A., Giermasz A., Castaman G., Key N.S., Lattimore S., Leebeek F.W.G., Miesbach W., Recht M., Long A., Gut R. Etranacogene dezaparvovec (AMT-061 phase 2b): Normal/near normal FIX activity and bleed cessation in hemophilia B. Blood Adv. 2019;3:3241–3247. doi: 10.1182/bloodadvances.2019000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancuso M.E., Mahlangu J.N., Pipe S.W. The changing treatment landscape in haemophilia: From standard half-life clotting factor concentrates to gene editing. Lancet. 2021;397:630–640. doi: 10.1016/S0140-6736(20)32722-7. [DOI] [PubMed] [Google Scholar]
- 10.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niemeyer G.P., Herzog R.W., Mount J., Arruda V.R., Tillson D.M., Hathcock J., van Ginkel F.W., High K.A., Lothrop C.D., Jr. Long-term correction of inhibitor-prone hemophilia B dogs treated with liver-directed AAV2-mediated factor IX gene therapy. Blood. 2009;113:797–806. doi: 10.1182/blood-2008-10-181479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soucie J.M., Monahan P.E., Kulkarni R., Konkle B.A., Mazepa M.A., US Hemophilia Treatment Center Network The frequency of joint hemorrhages and procedures in nonsevere hemophilia A vs B. Blood Adv. 2018;2:2136–2144. doi: 10.1182/bloodadvances.2018020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling C., Lu Y., Kalsi J.K., Jayandharan G.R., Li B., Ma W., Cheng B., Gee S.W., McGoogan K.E., Govindasamy L. Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum. Gene Ther. 2010;21:1741–1747. doi: 10.1089/hum.2010.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vercauteren K., Hoffman B.E., Zolotukhin I., Keeler G.D., Xiao J.W., Basner-Tschakarjan E., High K.A., Ertl H.C., Rice C.M., Srivastava A. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol. Ther. 2016;24:1042–1049. doi: 10.1038/mt.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S., Ling C., Zhong L., Li M., Su Q., He R., Tang Q., Greiner D.L., Shultz L.D., Brehm M.A. Efficient and targeted transduction of nonhuman primate liver with systemically delivered optimized AAV3B vectors. Mol. Ther. 2015;23:1867–1876. doi: 10.1038/mt.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling C., Yin Z., Li J., Zhang D., Aslanidi G., Srivastava A. Strategies to generate high-titer, high-potency recombinant AAV3 serotype vectors. Mol. Ther. Methods Clin. Dev. 2016;3:16029. doi: 10.1038/mtm.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown H.C., Doering C.B., Herzog R.W., Ling C., Markusic D.M., Spencer H.T., Srivastava A., Srivastava A. Development of a clinical candidate AAV3 vector for gene therapy of hemophilia B. Hum. Gene Ther. 2020;31:1114–1123. doi: 10.1089/hum.2020.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown H.C., Zakas P.M., George S.N., Parker E.T., Spencer H.T., Doering C.B. Target-cell-directed bioengineering approaches for gene therapy of hemophilia A. Mol. Ther. Methods Clin. Dev. 2018;9:57–69. doi: 10.1016/j.omtm.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertolini T.B., Shirley J.L., Zolotukhin I., Li X., Kaisho T., Xiao W., Kumar S.R.P., Herzog R.W. Effect of CpG depletion of vector genome on CD8+ T cell responses in AAV gene therapy. Front. Immunol. 2021;12:672449. doi: 10.3389/fimmu.2021.672449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm D., Pandey K., Nakai H., Storm T.A., Kay M.A. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samelson-Jones B.J., Finn J.D., Raffini L.J., Merricks E.P., Camire R.M., Nichols T.C., Arruda V.R. Evolutionary insights into coagulation factor IX Padua and other high-specific-activity variants. Blood Adv. 2021;5:1324–1332. doi: 10.1182/bloodadvances.2019000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson M.M., George L.A., Carr M.E., Samelson-Jones B.J., Arruda V.R., Murphy J.E., Rybin D., Rupon J., High K.A., Tiefenbacher S. Factor IX assay discrepancies in the setting of liver gene therapy using a hyperfunctional variant factor IX-Padua. J. Thromb. Haemost. 2021;19:1212–1218. doi: 10.1111/jth.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verdera H.C., Kuranda K., Mingozzi F. AAV vector immunogenicity in humans: A long journey to successful gene transfer. Mol. Ther. 2020;28:723–746. doi: 10.1016/j.ymthe.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas M., Marsic D., Li N., Zou C., Gonzalez-Aseguinolaza G., Zolotukhin I., Kumar S.R.P., Rana J., Butterfield J.S.S., Kondratov O. Engineering and in vitro selection of a novel AAV3B variant with high hepatocyte tropism and reduced seroreactivity. Mol. Ther. Methods Clin. Dev. 2020;19:347–361. doi: 10.1016/j.omtm.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen G.N., Everett J.K., Kafle S., Roche A.M., Raymond H.E., Leiby J., Wood C., Assenmacher C.A., Merricks E.P., Long C.T. A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nat. Biotechnol. 2021;39:47–55. doi: 10.1038/s41587-020-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markusic D.M., Hoffman B.E., Perrin G.Q., Nayak S., Wang X., LoDuca P.A., High K.A., Herzog R.W. Effective gene therapy for haemophilic mice with pathogenic factor IX antibodies. EMBO Mol. Med. 2013;5:1698–1709. doi: 10.1002/emmm.201302859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mingozzi F., Liu Y.L., Dobrzynski E., Kaufhold A., Liu J.H., Wang Y., Arruda V.R., High K.A., Herzog R.W. Induction of immune tolerance to coagulation factor IX antigen by in vivo hepatic gene transfer. J. Clin. Invest. 2003;111:1347–1356. doi: 10.1172/JCI16887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathwani A.C., Rosales C., McIntosh J., Rastegarlari G., Nathwani D., Raj D., Nawathe S., Waddington S.N., Bronson R., Jackson S. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 2011;19:876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mingozzi F., Hasbrouck N.C., Basner-Tschakarjan E., Edmonson S.A., Hui D.J., Sabatino D.E., Zhou S., Wright J.F., Jiang H., Pierce G.F. Modulation of tolerance to the transgene product in a nonhuman primate model of AAV-mediated gene transfer to liver. Blood. 2007;110:2334–2341. doi: 10.1182/blood-2007-03-080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martino A.T., Suzuki M., Markusic D.M., Zolotukhin I., Ryals R.C., Moghimi B., Ertl H.C., Muruve D.A., Lee B., Herzog R.W. The genome of self-complementary adeno-associated viral vectors increases Toll-like receptor 9-dependent innate immune responses in the liver. Blood. 2011;117:6459–6468. doi: 10.1182/blood-2010-10-314518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers G.L., Shirley J.L., Zolotukhin I., Kumar S.R.P., Sherman A., Perrin G.Q., Hoffman B.E., Srivastava A., Basner-Tschakarjan E., Wallet M.A. Plasmacytoid and conventional dendritic cells cooperate in crosspriming AAV capsid-specific CD8+ T cells. Blood. 2017;129:3184–3195. doi: 10.1182/blood-2016-11-751040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirley J.L., de Jong Y.P., Terhorst C., Herzog R.W. Immune responses to viral gene therapy vectors. Mol. Ther. 2020;28:709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shirley J.L., Keeler G.D., Sherman A., Zolotukhin I., Markusic D.M., Hoffman B.E., Morel L.M., Wallet M.A., Terhorst C., Herzog R.W. Type I IFN sensing by cDCs and CD4+ T cell help are both requisite for cross-priming of AAV capsid-specific CD8+ T cells. Mol. Ther. 2020;28:758–770. doi: 10.1016/j.ymthe.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 35.Mingozzi F., Maus M.V., Hui D.J., Sabatino D.E., Murphy S.L., Rasko J.E., Ragni M.V., Manno C.S., Sommer J., Jiang H. CD8+ T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 36.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright J.F. Codon modification and PAMPs in clinical AAV vectors: The tortoise or the hare? Mol. Ther. 2020;28:701–703. doi: 10.1016/j.ymthe.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright J.F. Quantification of CpG motifs in rAAV genomes: Avoiding the Toll. Mol. Ther. 2020;28:1756–1758. doi: 10.1016/j.ymthe.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konkle B.A., Walsh C.E., Escobar M.A., Josephson N.C., Young G., von Drygalski A., McPhee S.W.J., Samulski R.J., Bilic I., de la Rosa M. BAX 335 hemophilia B gene therapy clinical trial results: Potential impact of CpG sequences on gene expression. Blood. 2021;137:763–774. doi: 10.1182/blood.2019004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faust S.M., Bell P., Cutler B.J., Ashley S.N., Zhu Y., Rabinowitz J.E., Wilson J.M. CpG-depleted adeno-associated virus vectors evade immune detection. J. Clin. Invest. 2013;123:2994–3001. doi: 10.1172/JCI68205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandenberghe L.H., Wang L., Somanathan S., Zhi Y., Figueredo J., Calcedo R., Sanmiguel J., Desai R.A., Chen C.S., Johnston J. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat. Med. 2006;12:967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- 42.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sena-Esteves M., Gao G. Introducing genes into mammalian cells: Viral vectors. Cold Spring Harb. Protoc. 2020;2020:095513. doi: 10.1101/pdb.top095513. [DOI] [PubMed] [Google Scholar]
- 44.Rogers G.L., Martino A.T., Zolotukhin I., Ertl H.C., Herzog R.W. Role of the vector genome and underlying factor IX mutation in immune responses to AAV gene therapy for hemophilia B. J. Transl. Med. 2014;12:25. doi: 10.1186/1479-5876-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biswas M., Palaschak B., Kumar S.R.P., Rana J., Markusic D.M. B cell depletion eliminates FVIII memory B cells and enhances AAV8-coF8 immune tolerance induction when combined with rapamycin. Front. Immunol. 2020;11:1293. doi: 10.3389/fimmu.2020.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hausl C., Ahmad R.U., Sasgary M., Doering C.B., Lollar P., Richter G., Schwarz H.P., Turecek P.L., Reipert B.M. High-dose factor VIII inhibits factor VIII-specific memory B cells in hemophilia A with factor VIII inhibitors. Blood. 2005;106:3415–3422. doi: 10.1182/blood-2005-03-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.