Summary
Indole is a molecule proposed to be involved in bacterial signaling. We find that indole secretion is induced by sublethal tobramycin concentrations and increases persistence to aminoglycosides in V. cholerae. Indole transcriptomics showed increased expression of raiA, a ribosome associated factor. Deletion of raiA abolishes the appearance of indole dependent persisters to aminoglycosides, although its overexpression leads to 100-fold increase of persisters, and a reduction in lag phase, evocative of increased active 70S ribosome content, confirmed by sucrose gradient analysis. We propose that, under stress conditions, RaiA-bound inactive 70S ribosomes are stored as “sleeping ribosomes”, and are rapidly reactivated upon stress relief. Our results point to an active process of persister formation through ribosome protection during translational stress (e.g., aminoglycoside treatment) and reactivation upon antibiotic removal. Translation is a universal process, and these results could help elucidate a mechanism of persistence formation in a controlled, thus inducible way.
Subject areas: Bacteriology, Microbial genomics, Viral microbiology
Graphical abstract
Highlights
-
•
Indole is produced under sub-MIC tobramycin stress in V. cholerae and upregulates raiA
-
•
RaiA is involved in indole-dependent formation of aminoglycoside specific persisters
-
•
RaiA overexpression allows faster growth restart and increases 70S ribosome content
-
•
RaiA-bound inactive 70S ribosomes form intact and reactivable sleeping ribosome pools
Bacteriology; Microbial genomics; Viral microbiology
Introduction
Antibiotic resistance is a major public health concern leading to increased health care costs and mortality (Opatowski et al., 2019; Touat et al., 2019). Although the majority of studies address the response of bacteria to lethal doses of antibiotics, the effect of low doses of antibiotics on bacteria has also recently started to draw attention. Antibiotic concentrations lower than the minimal inhibitory concentration (sub-MICs) have historically been proposed to serve as signaling molecules (Davies et al., 2006), provoking considerable changes in transcription and triggering a wide variety of cellular responses in different bacterial species (Andersson and Hughes, 2014) and mutagenesis (Gutierrez et al., 2013). In V. cholerae, sub-MIC aminoglycosides (AGs) are known to activate various stress response pathways, such as the SOS (Baharoglu et al., 2014; Baharoglu and Mazel, 2011) and RpoS stress responses (Baharoglu et al., 2013), allowing cells to cope with increased reactive oxygen species (ROS) levels and DNA breaks (Negro et al., 2019). AGs are bactericidal antibiotics that are known to enter the bacterial cell through the proton motive force (Fraimow et al., 1991; Herisse et al., 2017; Taber et al., 1987). AGs target the ribosome, leading to mistranslation and eventually cell death (Davis, 1987).
Interestingly, sub-MIC antibiotics, among which AGs, have been shown to stimulate the production of a small molecule, indole (Han et al., 2011). Indole is a byproduct of tryptophan degradation by tryptophanase TnaA (Evans et al., 1941) in both Gram+ and Gram− bacterial species (Lee and Lee, 2010), together with pyruvate and ammonia. While pyruvate and ammonia are respectively sources of carbon and nitrogen, the role of indole is not well understood. Indole is also found in plants and animals, and was linked with signaling and human diseases (for a review (Lee et al., 2015)). Common indole concentrations in the human gut are in the order of 250–1100 μM, and up to 200 μM in blood and other tissues. Regulation of indole production has been described, namely through carbon source utilization (Botsford and DeMoss, 1971) and catabolic repression (Yanofsky et al., 1991), amino acid availability (Newton and Snell, 1965), cold temperature (Lee et al., 2008), heat shock (Li et al., 2003) and growth phase (Kobayashi et al., 2006). Particularly, indole is produced during transition from exponential to stationary phase (Lelong et al., 2007).
In E. coli, indole is nontoxic at physiologic concentrations (below 1 mM) (Lee et al., 2007), and does not change the growth rate (Lee et al., 2008). At high concentrations however (above 1–3 mM), indole inhibits cell division (Chant and Summers, 2007; Chimerel et al., 2012). In V. cholerae, indole secretion reaches its maximum at 600 μM during transition from midlog to stationary phase (Howard et al., 2019; Mueller et al., 2009) and was not observed to have any effect on the polarity of the V. cholerae cell membrane at this concentration (Mueller et al., 2009).
Indole can pass across the cell membrane without the need for a transporter (Pinero-Fernandez et al., 2011), and was proposed to act as an interkingdom signaling molecule (Martino et al., 2003; Wang et al., 2001). An effect of indole in persistence to antibiotics has also been observed. At toxic concentrations (1–2 mM), where indole behaves as a membrane ionophore, it was observed to reduce persistence of stationary phase cultures to tested antibiotics (ciprofloxacin, ampicillin) (Hu et al., 2015). However, studies in E. coli show that lower concentrations of indole increase survival/persistence to lethal concentrations of ofloxacin, ampicillin, and kanamycin (Vega et al., 2012, 2013), suggesting that the protective effect of indole is not specific to one family of antibiotic. Studies also pointed to the involvement of indole secretion in the cooperation between antibiotic resistant and sensitive populations during antibiotic stress (Lee et al., 2010). Notably, a recent study identified indole production as a potential target for the increased activity of quinolones against persisters in E. coli (Zarkan et al., 2020). Because indole appears to be beneficial for bacteria in the presence of antibiotics, we addressed whether indole production is increased upon sub-MIC AG treatment in V. cholerae and whether this can lead to improved response to lethal antibiotic concentrations.
Importantly, we find that indole strongly increases persistence to AGs through the action of RaiA. We find that transcription from the raiA gene promoter is highly upregulated in the presence of indole in exponential phase V. cholerae cells. RaiA was shown to be a ribosome associated protein, in the same conditions as Rmf (ribosome modulation factor) and Hpf (hibernation promoting factor) (Maki et al., 2000). The two latter factors cause dimerization of vacant 70S into inactive 100S ribosome dimers, in a process called ribosome hibernation during stationary phase (Gohara and Yap, 2018), whereas RaiA mostly binds to free 70S monosomes (Maki et al., 2000; Sabharwal et al., 2015). E. coli mutants lacking these ribosome-associated factors do not show any growth defect during exponential growth, which is consistent with the fact that their expression is specific to stationary phase and stress (Prossliner et al., 2018). RaiA was observed to protect the 70S ribosome from degradation (Agafonov et al., 1999; Di Pietro et al., 2013), and was also observed to block the binding of tRNA to the ribosomal A site in a cell free translation system (Agafonov et al., 2001), and during cold shock (Vila-Sanjurjo et al., 2004). In the present study, characterization of the raiA deletion mutant shows that RaiA is instrumental in the appearance of persister cells to AGs. We propose here a new mechanism of induced persistence to AGs by which RaiA positively affects the intact ribosome content of the cell, and facilitates regrowth after removal of the antibiotic.
Results
Indole is produced in response to sub-MIC tobramycin and increases persistence to AGs
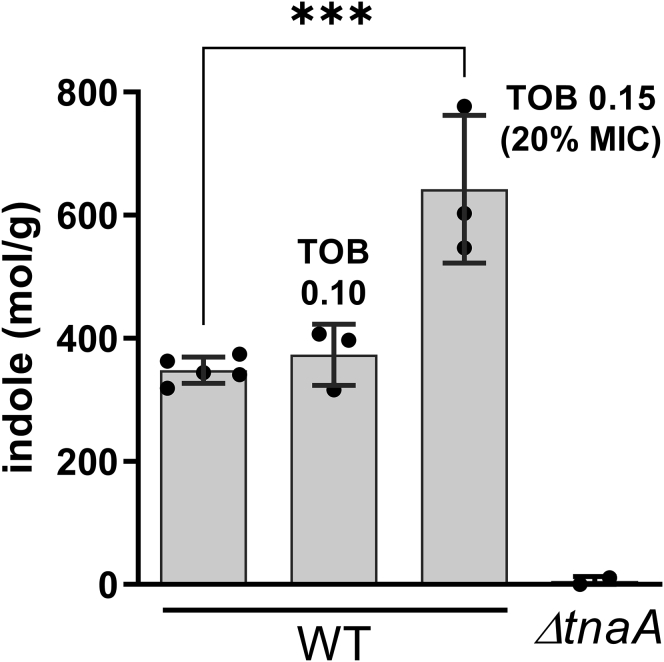
Because the antibiotics ampicillin and kanamycin increase indole levels in E. coli (Han et al., 2011), we measured indole secretion (Saint-Ruf et al., 2014) in V. cholerae to determine whether the aminoglycoside tobramycin also impacts indole levels in this case. We found increased extracellular indole concentrations in the presence of sub-MIC tobramycin (TOB 0.15 μg/mL, 20% of the MIC, Figure 1). We next addressed whether indole has an impact on the growth of V. cholerae, using an indole concentration of 350 μM. This concentration was previously shown to be physiologically relevant in V. cholerae and was observed to have no inhibitory effect on growth, and to complement the biofilm formation defect of a ΔtnaA mutant deficient for indole production (Mueller et al., 2009). We found that 350 μM indole does not affect growth in the absence of antibiotics, but improves growth in sub-MIC antibiotics tobramycin (Figure S1A).
Figure 1.
Indole is produced during growth in sub-MIC tobramycin and induces raiA
Measure of extracellular indole concentrations of bacterial cultures grown overnight in rich medium MOPS (Teknova EZ rich defined medium) with and without tobramycin at 0.10 μg/mL (TOB 0.10) or 0.15 μg/mL (TOB 0.15) using the Kovacs reagent (Saint-Ruf et al., 2014). ΔtnaA strain was used as a negative control without tobramycin. Experiments were performed in triplicates, and statistical analysis was performed (∗∗∗: p < 0.001). Error bars represent standard deviation.
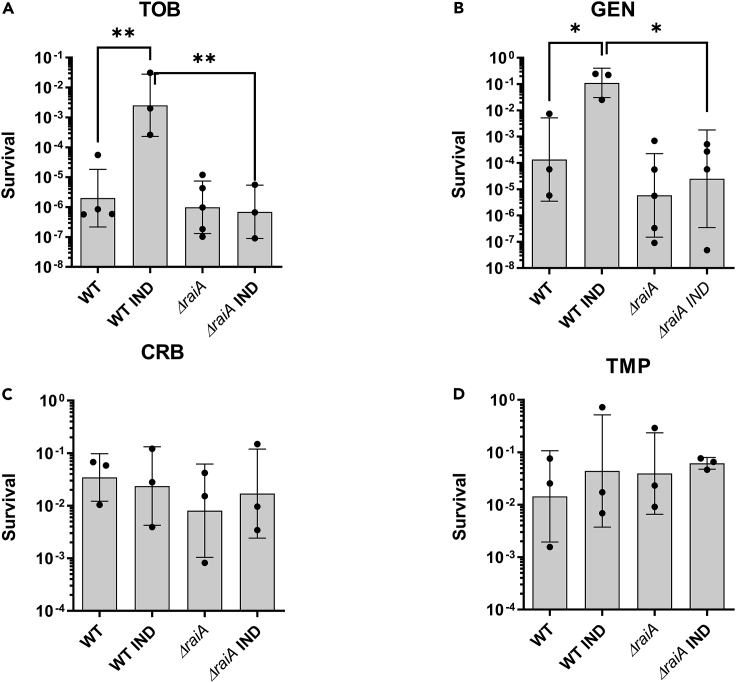
We next addressed the effect of indole in the response to lethal concentrations of antibiotics, by measuring persister cells formation in V. cholerae. To do so, we adapted to V. cholerae, a protocol developed for E. coli (I.M. and W.-L.S., personal communication). Early exponential phase cultures were treated with lethal doses of antibiotics (5–10 times the MIC) for 20 h. We first confirmed that cells surviving after 20 h of antibiotic treatment and which grow upon antibiotic removal were indeed persister cells, by performing survival curves (Figure S2A). The biphasic profiles of the killing curves we obtained were consistent with the formation of persister cells after 5 h (Brauner et al., 2016). Furthermore, these cells were not resistant to the antibiotic, as no growth was observed when streaked on antibiotic containing plates (not shown). We thus carried on with the quantification of persisters at 20 h of antibiotic treatment. We found that V. cholerae cultures grown in the presence of indole yielded higher numbers of persister cells to the AGs TOB and gentamicin (GEN) (Figures 2A and 2B), as previously observed for E. coli treated with kanamycin and ofloxacin (Vega et al., 2012), but we observed no effect for persistence to carbenicillin (CRB), or trimethoprim (TMP) (Figures 2C and 2D). Furthermore, a strain deleted for tnaA yielded less persisters to tobramycin than the WT strain (Figure S2B), and the effect was reversed by indole complementation, consistent with a link between indole and persistence.
Figure 2.
Modulation of persistence of exponential phase WT and ΔraiA V. cholerae by indole
(A–D) Early exponential phase of wild-type (WT) and ΔraiA V. cholerae cultures were treated with lethal doses of the specified antibiotics for 20 h. The y axis represents survival, as the number of CFU growing after antibiotic treatment and removal divided by the total number of CFU at time zero (before antibiotic treatment). Bars represent geometric means and error bars represent geometric standard deviation. Tobramycin (TOB): 10 μg/mL, gentamicin (GEN): 5 μg/mL, carbenicillin (CRB): 100 μg/mL, indole (IND): 350 μM. Experiments were performed 3 to 6 times, and statistical analysis was performed (∗: p < 0.05; ∗∗: p < 0.01).
Indole, at concentrations allowing growth, does not affect AG entry
We next addressed whether the beneficial effect of indole treatment in the presence of AGs is because of modifications in membrane potential, which could lead to decreased AG entry into the cell. AG uptake by the bacterial cell is known to be linked to the proton motive force (PMF). At high concentrations (5 mM) indole is a proton ionophore which blocks cell division by dissipating the PMF (Kralj et al., 2011), and was observed to interact with the cell membrane and change its physical structure (Mitchell, 2009). To measure AG entry in the bacterial cell, we used the aminoglycoside neomycin coupled to the fluorophore Cy-5, which was previously synthesized for aminoglycoside uptake studies in bacteria and demonstrated to bear the properties of aminoglycosides for uptake, mode of action and activity against Gram negative bacteria (Sabeti Azad et al., 2020; S.A.P. et al., unpublished data). We found that indole at the physiological concentration of 350 μM does not affect AG entry into the bacterial cell (Figure S3A), ruling out the possibility of decreased AG entry because of modifications of PMF in the presence of indole. Moreover, the presence of 350 μM indole did not change the MIC of two aminoglycosides: tobramycin and gentamicin (Figure S3B). The beneficial effect of indole in the presence of AGs is thus not through reduced antibiotic entry.
Indole induces RaiA (VC0706) expression
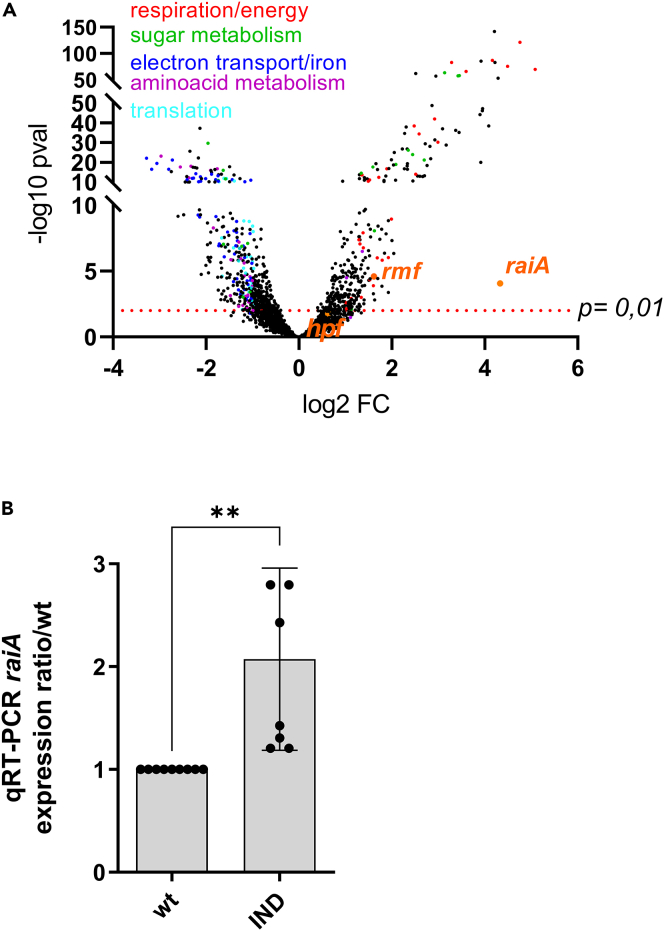
To shed light into mechanisms allowing for more efficient response to antibiotic stress upon indole treatment, we decided to study the transcriptomic changes of V. cholerae in response to 350 μM indole. mRNA sequencing of exponential phase cultures shows differential regulation of 260 genes shown in Table S1 and represented in Figure 3A (>2-fold change, adjusted p value < 0.01 as in Krin et al. (2018)). The most affected categories were respiration (31 genes upregulated), electron transfer and iron uptake (55 genes downregulated). Indole mediated protection against antibiotic killing was previously proposed to be through upregulation of efflux pumps (Blair et al., 2013; Hirakawa et al., 2005; Kobayashi et al., 2006; Lee and Lee, 2010; Nikaido et al., 2012), or through an increase in OxyR associated oxidative stress response (Vega et al., 2013). However, our RNA-seq data show no induction of oxidative stress response related genes in V. cholerae (oxyR, soxRS, katG) by indole, and rather suggest decreased expression of proteins linked to iron uptake, suggesting decreased iron and ROS levels upon indole treatment (Baharoglu et al., 2013; Mehi et al., 2014). The second most affected category belongs to translation related genes (31 genes, approximately 10% of total differentially regulated genes, with p < 3.00 × 10−4). Notably, one translation related gene was markedly upregulated: raiA (VC0706, 20-fold up), together with rmf (VC1484, 3-fold up), which are both described as factors associated with inactive ribosomes in stationary phase (Agafonov et al., 1999; Di Pietro et al., 2013). raiA expression is known to be triggered by transition to stationary phase (Maki et al., 2000), and RaiA is usually weakly expressed during exponential phase. RT-qPCR on raiA (Figure 3B) and fluorescence associated flow cytometry on cells carrying GFP fused to the raiA promoter (Figures 4A, S4A, and S4B) confirmed upregulation of raiA by indole during exponential phase, and increased expression of raiA in stationary phase. Because transcriptomic data pointed RaiA as one of the most differentially regulated genes by indole, we next decided to address the contribution of RaiA in the indole associated phenotypes in V. cholerae.
Figure 3.
Indole production during growth in sub-MIC tobramycin induces raiA
(A) Volcano plot showing differentially expressed genes upon indole treatment. X axis represents log 2-fold change, Y axis represents the negative log 10 of the p value. raiA, rmf, and hpf are indicated. The dashed line represents a p value of 0.01, all of the dots above thus show a p value < 0.01. Red dots indicate genes linked with respiration, green with sugar metabolism, dark blue with iron, purple with amino acid metabolism, and light blue with translation. RNA-seq was performed in triplicates for each condition. See also Table S1.
(B) raiA mRNA levels measured by RT-qPCR on exponential phase V. cholerae cultures in presence or absence of indole. Statistical analysis was performed (∗∗: p < 0.01). Error bars represent standard deviation.
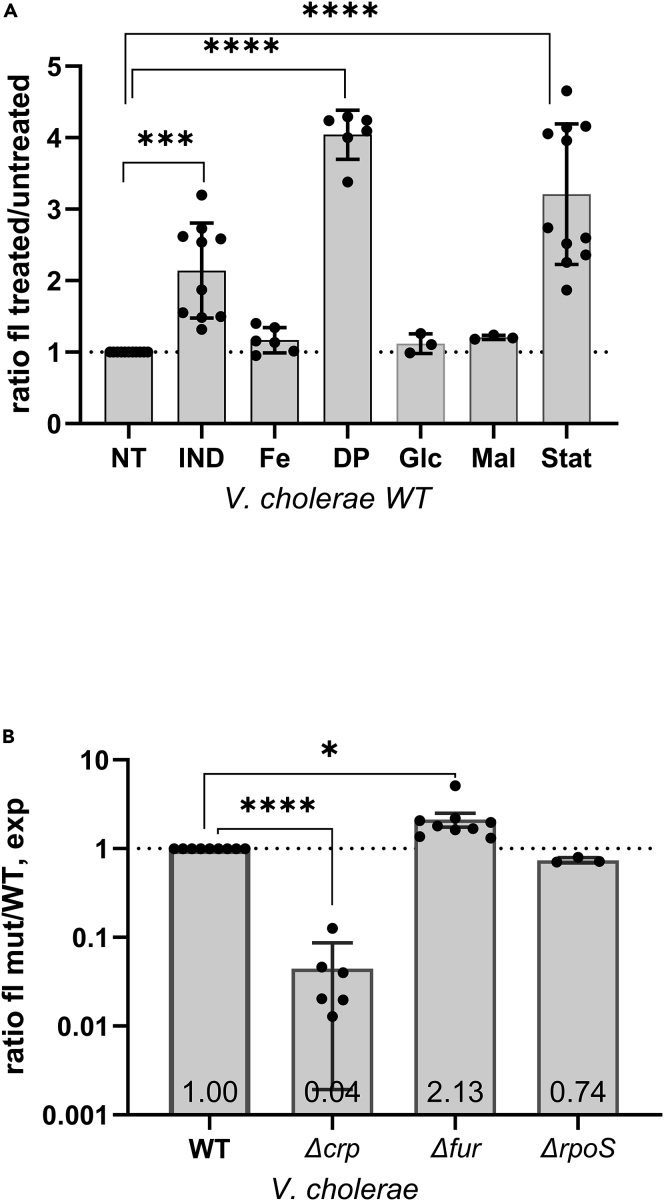
Figure 4.
Environmental stress induce raiA expression in exponential phase V. cholerae
Fluorescence quantification of GFP expression from the raiA promoter by flow cytometry in MH media in exponential phase (except for “Stat”).
(A) in WT V. cholerae. NT: Non-treated, IND: indole (350 μM), Fe: iron (18 μM), DP: 2,2′-Dipyridyl (500 μM), Glc: Glucose (1%), Mal: Maltose (1%), Stat: stationary phase. The y axis represents the fluorescence ratio of the treated over non-treated (NT) strain.
(B) in indicated V. cholerae deletion mutants. The y axis represents the fluorescence ratio of the mutant over wild type (WT) strain. Mean fold change values are indicated within histogram bars. Experiments were performed at least 3 times, and statistical analysis was performed (∗∗: p < 0.01; ∗∗∗∗: p < 0.0001). Error bars represent standard deviation. See also Figure S4.
Absence of raiA reduces persistence to AGs and abolishes induction by indole
To address the involvement of RaiA in indole induced persistence, we constructed a V. cholerae ΔraiA mutant, and measured the frequency of persistence to tobramycin (TOB), gentamicin (GEN), carbenicillin (CRB), and trimethoprim (TMP) in exponential phase cultures, grown in the absence and presence of indole (Figures 2A–2D). We found that V. cholerae ΔraiA generally formed slightly less persister cells to TOB, although the difference was not statistically significant when we compare persistence frequency, probably because at exponential phase raiA expression is not strong enough in the untreated WT strain, and because of variability between experiments. However, the ratio of persisters calculated separately for each experiment shows a significant 10-fold decrease in ΔraiA compared to WT (Figure S5A). Strikingly, the deletion of raiA completely abolished induction of persistence by indole to both tested AGs (TOB, GEN, Figures 2A and 2B), pointing to a role of RaiA in persister cell formation. No effect of neither indole nor raiA was observed in the formation of persisters to CRB or TMP (Figures 2C and 2D), suggesting that the effect of RaiA in persister formation is specific to AGs. The involvement of RaiA in persistence to AGs also appears to be conserved in E. coli as the raiA deficient mutant yielded less persisters to TOB (Figure S5B). Finally, as performed above in the WT V. cholerae strain, we confirmed that the presence of indole does not affect the AGs MIC of the ΔraiA strain, and that deletion of raiA does not affect the MIC and AG uptake (Figures S3A and S3B), meaning that the phenotypes we observe are not because of reduced entry of AGs or increased resistance. On the other hand, RaiA is dispensable for growth improvement by indole (Figure S1B), because indole still improves growth in TOB when raiA is deleted, showing that the mechanism of AG persister induction by indole (antibiotic concentration > MIC) is different than the mechanism of growth improvement in sub-MIC AGs. It is worth mentioning here that the growth of ΔraiA strain appeared to be slightly slower than the WT strain, suggesting that RaiA may also have a role during exponential growth, despite its low level of expression.
RaiA overexpression increases persistence to AGs and promotes earlier exit from stationary phase
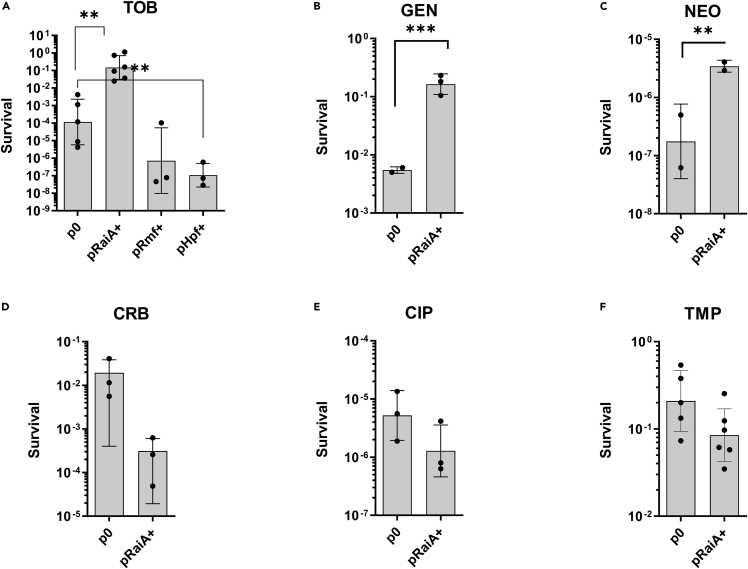
To mimic conditions of RaiA induction, we cloned it under a controlled Para promoter, which is repressed by glucose and induced by arabinose. Since the presence of different carbon sources may differentially affect growth and the response to aminoglycosides (S.A.P. et al., unpublished data), and also because arabinose was shown to have an impact on growth in V. cholerae, we compared the persistence levels of cells carrying the empty vector (p0 in Figures 5A–5F) or the pBAD-RaiA plasmid, in the presence of arabinose (RaiA overexpression conditions). Overexpression of RaiA strongly increased persisters formation in three aminoglycosides: TOB, GEN, and NEO (Figures 5A–5C), but not in three non-aminoglycoside antibiotics from different families: CRB, TMP and ciprofloxacin (CIP) (Figures 5D–5F), indicating that RaiA is directly and specifically involved in the persistence mechanism to aminoglycosides. In addition, the MIC showed no difference between the raiA overexpression strain compared to the strain with empty plasmid p0 and pRaiA+ (MIC = 0.75 μg/mL for both on MH plates containing arabinose), ruling out a potential effect on resistance.
Figure 5.
Modulation of persistence of exponential phase V. cholerae by RaiA
(A-F) Early exponential phases of wild-type (WT) V. cholerae carrying either the empty pBAD vector (p0) or with specified gene (pGene) cultures were treated with lethal doses of the specified antibiotics for 20 h. The y axis represents survival, as the number of CFU growing after antibiotic treatment and removal divided by the total number of CFU at time zero (before antibiotic treatment). (A) Tobramycin (TOB): 10 μg/mL. (B) Gentamicin (GEN): 5 μg/mL. (C) Neomycin (NEO): 30 μg/mL. (D) Carbenicillin (CRB): 100 μg/mL. (E) Ciprofloxacin (CIP): 0.025 μg/mL. (F) Trimethoprim (TMP): 50 μg/mL.. Experiments were performed 3 to 6 times, and statistical analysis was performed (∗: p < 0.05; ∗∗: p < 0.01). Error bars represent standard deviation.
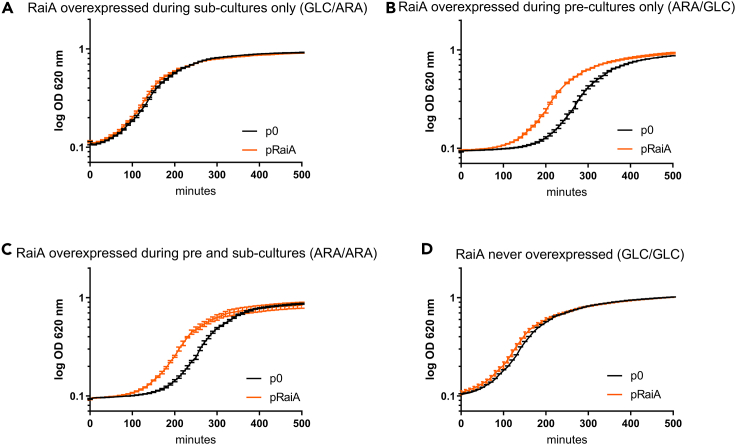
We next asked whether such increased persistence could be because of slower growth when RaiA is overexpressed. We monitored growth in conditions where (1) RaiA is not overexpressed previous to inoculation and only overexpressed during the growth curve and (2) RaiA is previously overexpressed in cells used for inoculation (Figure S6A). First, our results show no difference in growth rate (slope) in presence or absence of RaiA overexpression (Figures 6A, 6D, and S6B), indicating that RaiA overexpression does not cause a slow growth phenotype, which discards the hypothesis linking RaiA-mediated persistence to slow growth/dormancy. Interestingly, we observe in cells where RaiA overexpression was pre-induced, that these cells start growing faster because of a reduction in lag phase (Figures 6B, 6C, and S6B). Such a reduction in lag phase is reminiscent of increased active ribosome content which allows faster resumption of growth at the exit of the stationary phase (Condon et al., 1995). As a corollary, when raiA was deleted, the lag phase was increased (Figures 7A and S6C), suggesting that RaiA levels affect inactive but “ready to use” ribosome content, which we called sleeping ribosomes, in stationary phase.
Figure 6.
RaiA influences lag phase upon growth restart after the stationary phase
(A–D) Growth is measured on a TECAN plate reader. The curves of wild type V. cholerae with the empty plasmid are compared with plasmid carrying raiA under inducible promoter in MH media containing either glucose or arabinose. The promoter is repressed using glucose (GLC) and induced using arabinose (ARA). Growth was performed as specified, with RaiA expression repressed or induced in the overnight culture used for inoculum (overnight culture supplemented or not with ARA; indicated as “during pre-cultures”), or/and with RaiA expression induced or not during growth in the microplate reader (growth media supplemented or not with ARA in the microplate; indicated as “during sub-cultures”): (A) GLC/ARA, (B) ARA/GLC, (C) ARA/ARA, and (D) GLC/GLC. A scheme of the experimental setup is found on Figure S5A. Experiments were performed in triplicates and geometric means are represented. Error bars represent the geometric standard deviation. See also Figure S6B.
Figure 7.
The effect of RaiA on lag phase is independent of ribosome hibernation factors Rmf/Hpf
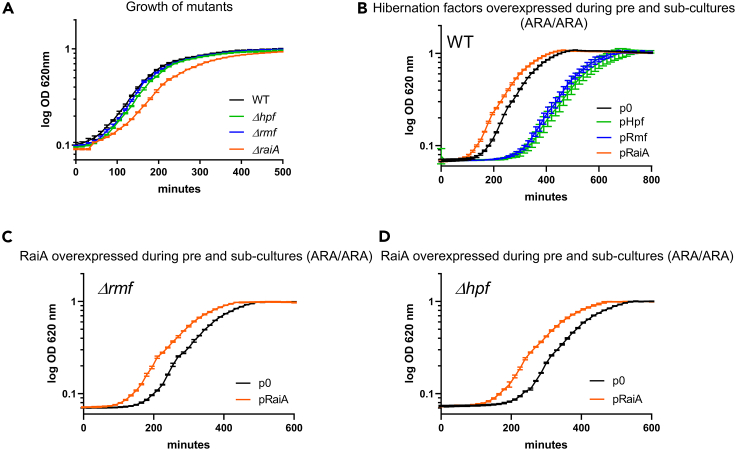
(A) Growth in MH media of WT and mutant strains.
(B) Growth of V. cholerae overexpressing hibernation factors or empty plasmid in MH media containing arabinose (ARA/ARA).
(C and D) Growth of hibernation factor mutants Δrmf and Δhpf overexpressing RaiA in MH media containing arabinose (ARA/ARA). Experiments were performed in triplicates and geometric means are represented. Error bars represent the geometric standard deviation. See also Figure S6C.
The effect of RaiA on lag phase and persistence is independent of ribosome hibernation factors Rmf/Hpf
RaiA was previously found to be associated with the inactive ribosomes at stationary phase, in the same conditions as Rmf and Hpf factors. Rmf and Hpf are known to dimerize ribosomes into so called hibernating 100S ribosomes, whereas RaiA associates with monomeric 70S ribosomes (Gohara and Yap, 2018; Maki et al., 2000; Prossliner et al., 2018). To address whether Hpf/Rmf dependent ribosome hibernation also favors rapid exit from stationary phase, we performed deletion and overexpression experiments similar to what we did for RaiA. Deletion of rmf or hpf has no effect on lag phase nor growth (Figures 7A and S6C). Our results showed that in contrast to overexpression of RaiA which decreases the lag phase, overexpression of Rmf or Hpf rather increases lag phase (Figures 7B and S6C). These findings are consistent with a model where increased ribosome dimerization by Hpf/Rmf would require action of dissociation factors to resume growth whereas spontaneous dissociation of RaiA from inactive 70S ribosomes (Agafonov et al., 2001; Maki et al., 2000), is sufficient for growth restart. Furthermore, when we overexpressed RaiA in Δhpf or Δrmf mutants, we observed a reduction of lag phase similar to what is observed upon overexpression of RaiA in the WT strain (Figures 7C and 7D), meaning that RaiA action is not dependent of Hpf/Rmf mediated ribosome hibernation.
In addition, we addressed the effect of hibernation factors on persistence in the exponential phase. Unlike for RaiA overexpression, no increase in persistence to TOB was detected upon overexpression of Hpf or Rmf, excluding an effect of 100S ribosome dimer formation on persistence to AGs in exponential phase (Figure 5A). Unexpectedly, persistence levels even decreased upon overexpression of Hpf, suggesting that 70S-RaiA (sleeping ribosome) and 100S-Rmf/Hpf (hibernating ribosome) complexes may have opposite effects on persistence to AGs in exponentially growing bacteria. We also tested the persistence levels of rmf and hpf deletion mutants. Surprisingly again, and consistent with overexpression results, we found that deletion of rmf or hpf hibernation factors increases persistence to AGs (Figure S7). The increase of persistence because of deletion of rmf is dependent on the presence of raiA, as the double mutant raiA rmf shows reduced persistence compared to WT, and similar to the raiA mutant. This can be because of the observed increased expression of raiA in the absence of rmf (Sabharwal et al., 2015) or amplified ribosome-RaiA complex formation in the absence of rmf, owing to decreased 100S formation (Ueta et al., 2005). Finally, it is important to note that the hpf raiA double mutant could not be constructed despite the use of two different strategies, implying synthetic lethality. Overall these results suggest an equilibrium between Rmf/Hpf and RaiA actions, consistent with previous literature that showed a combined role for these proteins in ribosome hibernation and antagonizing regulation of rmf/hpf and RaiA in V. cholerae (Sabharwal et al., 2015).
RaiA overexpression increases 70S ribosome proportion over 50S and 30S subunits in stationary phase
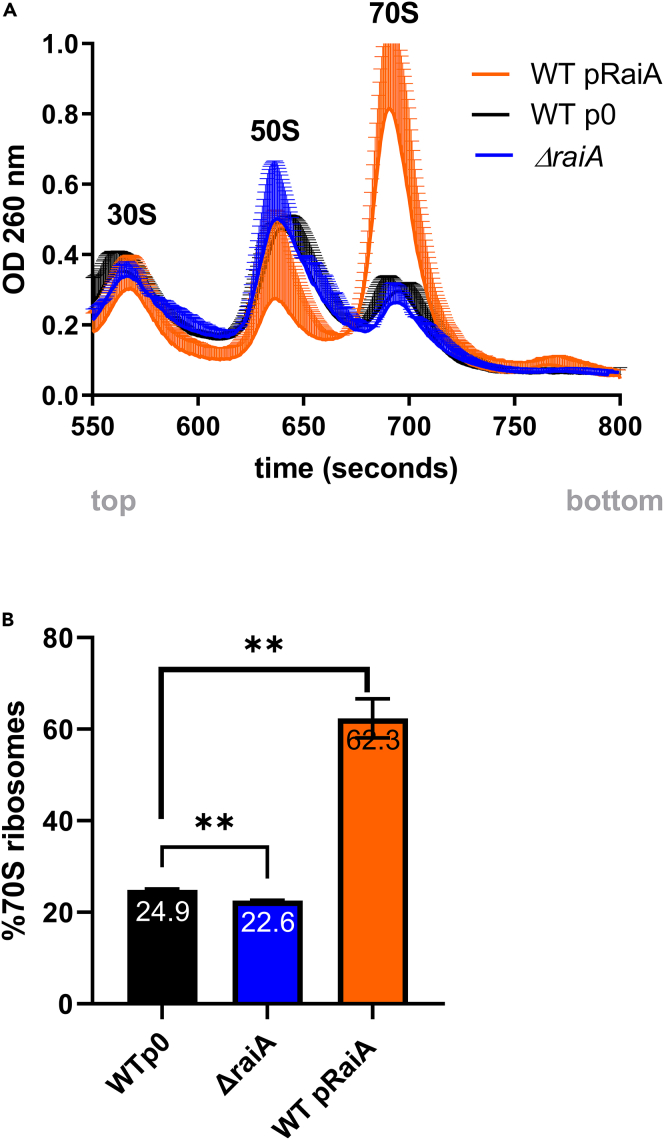
To address whether intact 70S ribosomes are protected/stored upon RaiA overexpression, we measured the ribosome contents of WT and RaiA overexpressing cells, as well as the strain deleted for raiA, by performing 10–50% sucrose gradients on cellular extracts from 24h stationary phase cultures. The profiles obtained were consistent with well described peaks for 30S and 50S subunits followed by a third peak corresponding to the 70S ribosome (Ueta et al., 2013). No clear 100S peak was observed in any of the experiments. Deletion of raiA leads to a slight increase in the proportion of dissociated subunits compared to the 70S ribosome. We found that the proportion of 70S ribosomes is increased compared to 30S + 50S dissociated subunits upon RaiA overexpression (Figures 8A and 8B). This agrees with the hypothesis that RaiA would stabilize the intact 70S ribosome and increase functional ribosome pools, reducing the lag phase of the population upon growth restart.
Figure 8.
RaiA levels influence stationary phase 70S ribosome content relative to 50S + 30S subunits in V. cholerae
(A) Cellular extracts of 24 h cultures (ΔraiA and WT V. cholerae carrying the p0/pBAD-RaiA vector, in MH media containing spectinomycin and arabinose) were separated on 10–50% sucrose density gradient. Ribosomal RNA content was measured at OD 260 nm using a spectrometer coupled to a pump and time on the X axis represents samples from less dense (upper fragments, smaller complexes) to denser (bottom of the tube, heavier complexes). Lysis was performed in the presence of 10 mM MgCl2. Cell debris eluting before 550 s are not shown. Graphs are normalized to total OD 260 nm = 1 for each sample. Mean values are indicated within histogram bars. Error bars represent standard deviation.
(B) Percentage of 70S ribosomes over total ribosome subunits (70S)/(70S + 50S + 30S). Error bars represent standard deviation (∗∗: p < 0.01).
RaiA expression is under environmental control and linked to the Fur iron sensing regulon in V. cholerae
RaiA is known to be expressed in stationary phase and upon temperature stress (Agafonov et al., 1999, 2001; Maki et al., 2000; Sabharwal et al., 2015; Slamti et al., 2007). Regulation of raiA was also described to occur through carbon catabolite control (CRP-cAMP) (Manneh-Roussel et al., 2018; Shimada et al., 2013). Our transcriptomic data suggests that indole affects genes from the Fur regulon (Table S1). Fur responds to iron levels and is generally known to be a repressor of iron/heme transport genes (such as hutA, hutXW, tonB, fbpA, and viuB among others) and also activates a small number of genes (namely the napABC operon and menB in V. cholerae) (Mey et al., 2005). The V. cholerae raiA gene promoter, appears to carry sequences similar to Fur boxes described in V. cholerae (Davies et al., 2011). To shed light on the means by which indole induces expression from the raiA promoter in exponential phase, we constructed deletion mutants for fur, crp and for the stationary phase sigma factor rpoS. We used our PraiA-gfp transcriptional fusion to measure expression in the presence and absence of indole in the mutants compared to wild type strain.
As expected, in WT V. cholerae, fluorescence was triggered by indole at 3 h of culture which corresponds to early exponential phase (OD 0.2 to 0.3), and accumulated in stationary phase (Figures 4A, S4A, and S4B). Deletion of crp strongly decreases raiA expression (25x in exponential phase, Figures 4B, S4A, and S4B), confirming that CRP is a prevailing activator of V. cholerae raiA. On the other hand, deletion of rpoS had no major effect on V. cholerae raiA expression (1.25 x decreases). In the Δfur strain, raiA expression was increased 2-fold (Figures 3B, S4A, and S4B), suggesting Fur dependent repression of the raiA promoter. No major effect on raiA promoter was observed upon treatment with iron, possibly because iron is already in excess levels for the cells during growth. Strikingly, treatment with dipyridyl (DP), an iron chelator which mimics conditions of iron starvation strongly induced fluorescence (Figure 4A). To confirm that these effects were specific to the raiA promoter, we introduced constitutively expressed gfp using the same plasmid vector in the strains WT, Δcrp and Δfur and observed no effect on fluorescence (Figure S8). This was also the case upon indole treatment of the WT strain. DP appears to decrease fluorescence for the constitutive promoter, while increasing it 4x from the raiA promoter (Figure 3B). Overall, these data confirm that decreased fluorescence in Δcrp and increased fluorescence in Δfur or upon indole and DP treatments are specific to an effect on transcription from the raiA promoter.
These results show a link between extracellular iron levels and RaiA expression. Together with the CRP-cAMP control, RaiA expression appears to be under environmental control, highlighting a link between bacterial persistence and environmental stress.
The effect of RaiA overexpression is conserved in the Gram-negative pathogen Pseudomonas aeruginosa
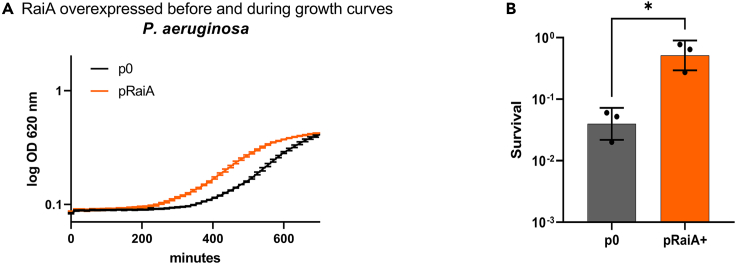
We next asked whether RaiA could have a similar function in other Gram-negative pathogens such as Pseudomonas aeruginosa, an organism associated with antibiotic resistance and persistence (Koeva et al., 2017; Spoering and Lewis, 2001) and of high concern regarding resistant infections. P. aeruginosa RaiA exhibits 37% protein identity with RaiA from V. cholerae. We overexpressed V. cholerae RaiA in P. aeruginosa and assessed lag phase and persistence, as we performed for V. cholerae. We found that upon RaiA overexpression, lag phase is also decreased in P. aeruginosa (Figures 9A and S6D), and strikingly, persistence to tobramycin is increased (Figure 9B). These results show that RaiA-protected sleeping ribosomes can be involved in persistent infections by various pathogenic bacteria.
Figure 9.
RaiA overexpression in Pseudomonas aeruginosa increases tolerance to tobramycin in exponential phase and promotes earlier exit from stationary phase
(A) Growth is measured on a TECAN plate reader. The curves of WT P. aeruginosa with the empty plasmid (p0) are compared with the plasmid carrying raiA of V. cholerae under inducible promoter in MH media containing arabinose. See also Figure S6D.
(B) Early exponential phases of WT P. aeruginosa cultures were treated with lethal doses of tobramycin (TOB: 10 μg/mL) for 20 h. The y axis represents survival, as the number of CFU growing after antibiotic treatment and removal divided by the total number of CFU at time zero (before antibiotic treatment) (∗: p < 0.05). Experiments were performed in triplicates. Error bars represent standard deviation.
Discussion
We show here that indole is produced upon sub-MIC aminoglycoside treatment in V. cholerae and at physiological concentrations and increases the appearance of persister cells in lethal concentrations of AGs. We find that such increase in persistence occurs through an inducible mechanism involving RaiA (previously called pY or yfiA in E. coli, and vrp in V. cholerae). Although indole improved growth in the presence of sub-MIC antibiotics seems to be nonspecific to an antibiotic class and independent of RaiA, increased persistence involving RaiA is specific to AGs in exponentially growing bacteria. Because RaiA is regulated by several environmental cues and signaling molecules, our findings highlight a new, inducible mechanism of persistence, based on increased protection of ribosomes during stress, rather than slowdown of the metabolism.
In vitro characterization of RaiA in E. coli, has previously shown association with ribosomes during cold shock and stationary phase, but not during growth at 37°C, suggesting that binding of RaiA is prompted by stress (Agafonov et al., 2001). Based on crystal structures, RaiA was suggested to arrest translation (Vila-Sanjurjo et al., 2004). However, our results in V. cholerae do not support such a role, because there is no impact of RaiA overexpression on growth, and rather point to a protective effect of RaiA under ribosomal stress caused by AG treatment.
Alternatively, since RaiA is able to stabilize the 70S ribosome monomers against dissociation in vitro (Agafonov et al., 1999; Di Pietro et al., 2013), it was proposed to constitute a pool of inactive 70S ribosomes preserved from degradation in bacteria (Giuliodori, 2016; Giuliodori et al., 2007; Gualerzi et al., 2011).
In vivo effects of RaiA on bacterial phenotypes are less well described. A protective effect of RaiA during stress, such as starvation, was previously observed in the Gram-positive species Mycobacterium tuberculosis (Li et al., 2018) and Lactococcus lactis (Puri et al., 2014). Yet, the mechanisms remained enigmatic. Our results show a role of RaiA on survival to antibiotic stress in Gram-negative pathogens.
One known mechanism of protection of non-translating ribosomes is ribosome hibernation. The ribosome hibernation factors, Rmf (ribosome modulation factor) and Hpf (hibernation promoting factor) (Maki et al., 2000), dimerize 70S ribosomes (monosomes) into 100S hibernating ribosome dimers. The importance of ribosome hibernation in stress survival is well established in various bacteria (McKay and Portnoy, 2015; Tkachenko et al., 2017), as 100S dimers are less susceptible to degradation by RNases (Feaga et al., 2020; Prossliner et al., 2018, 2021; Wada et al., 2000; Yamagishi et al., 1993). Ribosome hibernation factors were even proposed as potential new targets for antibiotics (Matzov et al., 2019). However, in some cases, 70S particles appear to be more robust during heating than 100S dimers which dissociate into 30S and 50S subunits more rapidly (Niven, 2004).
RaiA was previously identified as bound to the ribosome together with Rmf and Hpf, but its role in relation with ribosome hibernation is unclear. In V. cholerae, RaiA shows a synergistic effect with Hpf for survival to starvation (Sabharwal et al., 2015), and we observe a synthetic lethal phenotype for the deletion of raiA and hpf. Despite such apparent synergy with hibernation factors, RaiA was shown to inactivate 70S ribosomes without forming 100S dimers (Polikanov et al., 2012; Ueta et al., 2005). RaiA thus appears to act in a process different than ribosome hibernation, maybe by blocking breaking down of ribosomes into 30S and 50S subunits by ribosome recycling factors (Rrf/EF-G) (Agafonov et al., 1999; Janosi et al., 1996).
In E. coli, RaiA can even prevent 100S dimer formation by Hpf and Rmf (Maki et al., 2000; Ueta et al., 2005). According to cryo-EM data, RaiA can compete with Rmf for ribosome binding, hence shifting the ribosome content from a Rmf-mediated dimeric inactive form to a RaiA-bound monomeric inactive form (Franken et al., 2017). Recent work has identified RaiA as a player in persister formation together with hibernation factors, through a dormancy related mechanism linked to stringent response/ppGpp (Song and Wood, 2020; Wood and Song, 2020). In these studies, persister formation was assessed on near stationary cell cultures (OD 0.8), where these factors are highly expressed, and persistence is largely because of dimerization of inactive ribosomes, leading to a general dormancy state which also favors persister formation to other antibiotics like ampicillin and ciprofloxacin. In contrast, the present study shows in exponentially growing cells and without nutrient limitation, a specific mechanism involving the induction of RaiA, triggering persistence specifically to aminoglycosides (and not to other antibiotics), without affecting growth (Figure 10A). The mechanism we describe is also different from the ppGpp-independent persistence caused in slow growing bacteria (Pontes and Groisman, 2019), because increased RaiA does not affect growth rate in our study. In this regard, the RaiA-dependent persistence mechanism observed in the present study is not a dormancy-like or slow growth mechanism, unlike the increased persistence triggered by ppGpp to antibiotics from various families in the stationary phase. Moreover, no effect of hibernation factors Rmf/Hpf is observed, suggesting different persistence mechanisms than ribosome dormancy formation in growing cells. Recent studies also show the existence of mechanisms triggering persistence independently of ppGpp or hibernation factors in bacteria (Hossain et al., 2021; Wood et al., 2021). We show here that RaiA is one player enhancing persister formation in the Gram-negative bacterium V. cholerae.
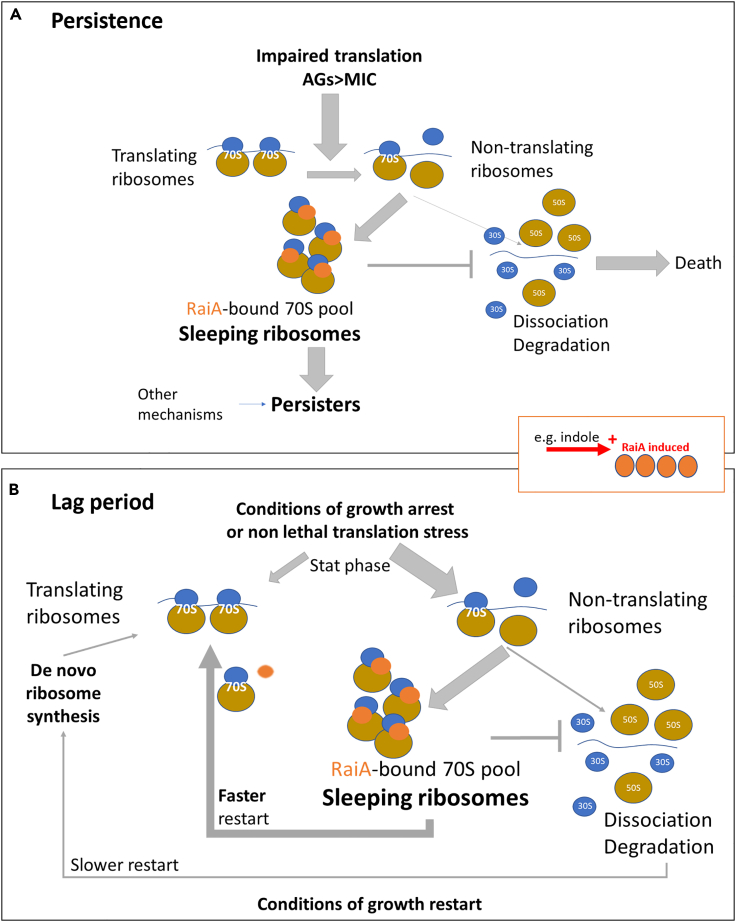
Figure 10.
Summary model: RaiA mediated ribosome protections increases persistence to aminoglycosides and decreases lag phase during growth restart
Diagrams depict RaiA induced conditions.
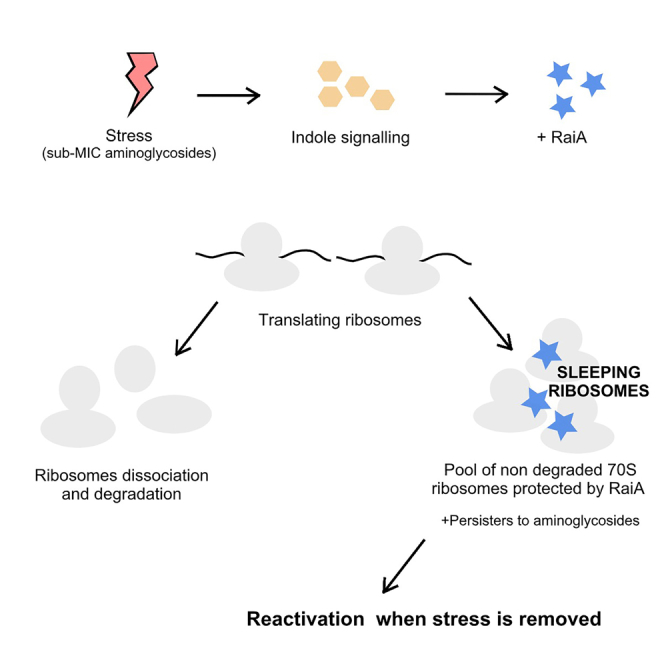
(A) Persistence. During exponential growth, treatment with lethal doses of aminoglycosides leads to disruption of translation and stalling of translating 70S ribosomes, and eventually to their dissociation into 50S + 30S subunits and degradation of these ribosomes, resulting in cell death. Upon induction of RaiA (e.g., by indole signaling), we propose that RaiA does not affect translating ribosomes, but binds non-translating 70S ribosomes to form sleeping ribosomes, and protects them from dissociation into subunits and degradation. This results in increased persistence.
(B) Length of lag phase. In a stationary phase culture (e.g. overnight culture), translation is highly decreased, leading to dissociation from the mRNA ribosome degradation. Upon growth restart, a lag phase thus occurs where ribosomes are resynthesized. We propose that when RaiA production is increased before entering the stationary phase, an increased proportion of non-translating ribosomes are bound and are preserved as 70S by RaiA, rather than being degraded. The presence of such an increased pool of intact 70S ready-to-use sleeping ribosomes gives an advantage on translation reactivation, whereas degraded ribosomes have to be recycled or de novo synthetized, decreasing the lag phase necessary for the synthesis of a sufficient number of ribosomes.
Although RaiA, Hpf and Rmf are rapidly released from ribosomes when normal growth conditions are restored (Agafonov et al., 2001; Maki et al., 2000), ribosome reactivation necessitates dissociation of hibernating 100S ribosome dimers into monomers by HflX and other factors (Basu and Yap, 2017), whereas no dissociation factor is needed for the reactivation of the RaiA inactivated 70S ribosome. There may thus be an interplay and equilibrium between hibernating 100S-Hpf/Rmf and “sleeping” 70S-RaiA forms.
Such synergy or antagonism between inactive 70S and 100S ribosome pools can however depend on the nature of the stress and the bacterial species. We show here that persistence to aminoglycosides is better achieved in the presence of RaiA (70S), rather than hibernation factors (100S) in V. cholerae.
Here, we propose that increased production of RaiA (e.g., upon stress) leads to preservation of non-translating ribosomes in a pool of inactive and intact 70S “sleeping” ribosomes. In that scenario, bacteria can keep growing in the absence of the antibiotic, because RaiA will not bind to actively translating ribosomes. Upon stress affecting translation (e.g., aminoglycosides), ribosomes may stall or stop, which would lead to the dissociation from the mRNA. If RaiA is induced, we propose that such non-translating 70S ribosomes will be bound and protected by RaiA, and avoid death upon antibiotic treatment, not by entering a dormant state, but by inducing protection of ribosomes. We propose that, in contrast to hibernating ribosomes which need recycling factors to dissociate into 70S monosomes, such sleeping ribosomes can be rapidly reactivated upon stress relief through spontaneous dissociation of RaiA from the intact 70S monosome (Agafonov et al., 2001; Maki et al., 2000), thus conferring an advantage for stress survival (Figures 10A and 10B).
In line with this, a recent study showed that the greater the ribosome content of the cell, the faster persister cells resuscitate (Kim et al., 2018). In parallel, protein synthesis was shown to be necessary for persister cell formation in V. cholerae (Paranjape and Shashidhar, 2019), consistent with the fact that we see no effect of RaiA overexpression on growth rates, thus on protein synthesis during growth. However, our results do not exclude another role for RaiA on translating ribosomes.
Increased RaiA levels thus allow higher persistence, but what is controlling RaiA expression? RaiA is expressed in the stationary phase and during cold shock (Agafonov et al., 1999, 2001; Maki et al., 2000; Sabharwal et al., 2015). Other known factors are the heat shock in V. cholerae (Slamti et al., 2007), stringent response (Prossliner et al., 2018), envelope stress and the carbon catabolite response (Manneh-Roussel et al., 2018; Shimada et al., 2013). We additionally show that raiA expression is linked to iron levels and responds to Fur regulation. Interestingly, iron is associated with ribosomes (Bray et al., 2018) and a link between iron and modulation of ribosome function during stress has been described (Zinskie et al., 2018). Iron control of RaiA may thus allow protection of ribosomes from iron related damage. Altogether, RaiA appears to be part of the bacterial response to environmental stress.
Results of the present study constitute a link between bacterial signaling, ribosome protection, and persistence. Moreover, because RaiA action appears to be conserved in various pathogens (Gram-negatives and positives, such as S. aureus and M. tuberculosis), it may be one factor involved in the failure of treatment in persistent infections. It would be interesting to ask now whether RaiA can be used as an early indicator of persistence, which would allow isolation of persisters within a heterogeneous population and further studies using single cell approaches.
Limitation of the study
This study shows that indole is secreted under sub-MIC aminoglycosides treatment and increases raiA expression. It also shows that indole is implicated in raiA-dependent persister formation under lethal aminoglycosides treatment. However, we do not address whether indole is also produced under lethal treatment because it is challenging to quantify indole production or tnaA expression on dying cells. Development of this method in our lab could be the purpose of future work.
In addition, raiA-dependent persister formation is shown here to be specific to aminoglycosides (bactericidal), which target translation. Bacteriostatic antibiotics such as chloramphenicol or tetracycline also target the ribosome, but persistence assays are usually not performed using these antibiotics. Our study does not exclude a role for raiA protection on sleeping ribosomes in the response to such translation targeting antibiotics.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Vibrio cholerae El Tor N16961 hapR+ | Gift from Mélanie Blokesh. | N/A |
| ΔraiA | This paper | N/A |
| Δrmf | This paper | N/A |
| Δhpf | This paper | N/A |
| ΔraiA Δrmf | This paper | N/A |
| Δcrp | This paper | N/A |
| Δfur | This paper | N/A |
| ΔrpoS | Baharoglu et al. (2013) | N/A |
| ΔtnaA | This paper | N/A |
| Escherichia coli K12 subst. MG1655 | Laboratory collection | ATCC 47076 |
| ΔraiA | This paper | N/A |
| Pseudomonas aeruginosa PAO1 | Laboratory collection | RRID: SCR_006590 |
| Chemicals, peptides, and recombinant proteins | ||
| Carbenicillin | Sigma-Aldrich | Cat#C1389 |
| Chloroform | VWR Chemicals | Cat#22711.290 |
| Ciprofloxacin | Sigma-Aldrich | Cat#Y0000198 |
| Gentamicin | Sigma-Aldrich | Cat#G1397 |
| Indole | Acros Organics | Cat#120-72-9 |
| Isopropanol | VWR Chemicals | Cat# 20839.297 |
| Neomycin | Sigma-Aldrich | Cat#N1142 |
| Tobramycin | Sigma-Aldrich | Cat#T4014 |
| Trimethoprim | Sigma-Aldrich | Cat#T7883 |
| TriZOL | Thermofischer Scientific | Cat#15596026 |
| Critical commercial assays | ||
| pTOPO-TA Cloning kit | Thermofischer Scientific | Cat#451641 |
| SuperScript III First Strand | Thermofischer Scientific | Cat#18080051 |
| SYBR Green Master Mix | Thermofischer Scientific | Cat#4309155 |
| Ribolock RNase | Thermofischer Scientific | Cat#E00382 |
| Deposited data | ||
| RNA-seq | This paper | GEO: GSE182561 |
| Oligonucleotides | ||
| See Table S2 | This paper | N/A |
| Recombinant DNA | ||
| pBAD43 | Laboratory collection | N/A |
| pBAD43-Hpf+ | This paper | N/A |
| pBAD43-RaiA+ | This paper | N/A |
| pBAD43-Rmf+ | This paper | NA |
| pMP7-Δcrp::aad7 | Baharoglu et al. (2013) | N/A |
| pMP7-Δfur::aph | This paper | N/A |
| pMP7-Δhpf::aph | This paper | N/A |
| pMP7-Δrmf::aph | This paper | N/A |
| pSC101-Pc-gfp | This paper | N/A |
| pSC101-PraiA-gfp | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Magellan | Life Science Tecan | https://lifesciences.tecan.com/software-magellan |
| Other | ||
| Etest® gentamicin | Biomérieux | Cat #412368 |
| Etest® tobramycin | Biomérieux | Cat #533100 |
| Neocy5 | Sabeti Azad et al., 2020 | N/A |
Resource availability
Lead contact
Further informations and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Zeynep Baharoglu (baharogl@pasteur.fr).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Bacterial strains and plasmids
All V. cholerae strains used in this study are derivative of V. cholerae N16961 hapR+, and were constructed by allelic exchange. All E. coli strains used in this work are derivatives of E. coli MG1655, and were constructed by transduction using E. coli Keio knockouts strains. Strains and plasmids are listed in key resources table and Table S2 for more details.
Media and growth conditions
Vibrio cholerae: Colonies on plates grew at 37°C, in MH media. Plates are conserved at room temperature and should not be placed at 4°C. Liquid cultures grew at 37°C in appropriate media (see STAR Methods), in aerobic conditions, with 180 rotations per minute. E. coli and P. aeruginosa: Colonies on plates grew at 37°C, in MH media, and are conserved at 4°C. Liquid cultures grew 37°C in aerobic condition in appropriate media (see STAR Methods), with 180 rotations per minute.
Method details
Persistence tests
Persistence tests were performed on early exponential phase cultures. In order to clear the culture from previously non-growing cells that could potentially be present from the stationary phase inoculum, we performed a two-step dilution protocol, before antibiotic treatment. For overexpression experiments, glucose 1% was added to the overnight cultures to repress the pBAD promoter. Overnight V. cholerae cultures were first diluted 1000x in 4 ml fresh Mueller-Hinton (MH) medium, without indole (with the antibiotic allowing to maintain the plasmid and incubated at 37°C with shaking. For overexpression experiments, arabinose 0.2% was added at this first dilution step to the fresh MH media to induce the pBAD promoter. When the OD 620 nm reached ∼0.2, cultures were diluted 1000x a second time, in order to clear them from non-growing cells, in Erlenmeyers containing 25 ml fresh MH medium, without or with indole at 350 μM (with the antibiotic allowing to maintain the plasmid, when the strain carried a plasmid), and were allowed to grow at 37°C. For overexpression experiments, arabinose 0.2% was added again at this second dilution step to the fresh MH media to induce the pBAD promoter. When cultures reached an OD 620 nm between 0.25 and 0.3 (early exponential phase), appropriate dilutions were plated on MH plates to determine the total number of CFUs in time zero untreated cultures. Note that for V. cholerae, it was important to treat cultures at the precise OD 620 nm 0.25-0.3, as persistence levels seem to be particularly sensitive to growth phase in this species, where they decline in stationary phase, and because we wanted to avoid any stationary phase protein expression such as raiA or rpoS at later growth. 5 ml of cultures were collected into 50 ml Falcon tubes and treated with lethal doses of desired antibiotics (5-10 times the MIC: tobramycin 10 μg/ml, gentamicin 5 μg/ml, neomycin 30 μg/ml, carbenicillin 100 μg/ml, ciprofloxacin 0.025 μg/ml, trimethoprim 50 μg/ml) for 20 hours at 37°C with shaking in order to guarantee oxygenation. The same protocol was used for persistence assays on P. aeruginosa. Appropriate dilutions were then plated on MH agar without antibiotics and proportion of growing CFUs were calculated by doing a ratio with total CFUs at time zero. Experiments were performed 3 to 6 times.
Quantification of extracellular indole concentrations
Extracellular indole concentration was measured on bacterial cultures grown overnight with and without antibiotics using the Kovacs reagent (Saint-Ruf et al., 2014). First, we established an indole concentration standard curve using 1 ml culture medium (without bacteria) supplemented with indole 0 to 1000 μM (100 μM steps). After adding 500 μl KOVACS reagent, 100 μl of the top layer of the reaction was mixed with 800 μl isoamyl-HCl and OD was read at 570 nm. For indole measurement on bacterial samples, 1 ml of culture was subjected at the same protocol. Measured indole concentration was normalized to the bacterial dry mass based on the assumption that for an OD 600 nm = 1 bacterial dry mass is 0.3 mg/ml (Soini et al., 2008). We detected no indole production in MH medium in any condition. In order to quantify secreted indole in the presence and absence of sub-MIC antibiotics, we thus used the defined rich MOPS transparent medium (Teknova EZ rich defined medium), where MIC TOB is 0.75 μg/ml. All the following experiments (persistence, growth) were conducted in MH, as it allows to study the impact of defined indole concentrations added to the media.
Quantification of fluorescent neomycin uptake was performed as described (S.A.P. et al., unpublished data; Okuda, 2015; Sabeti Azad et al., 2020). Neo-cy5 is an aminoglycoside coupled to the fluorophore Cy5, and has been shown to be active against Gram- bacteria (Okuda, 2015; Sabeti Azad et al., 2020). Briefly, overnight cultures were diluted 100-fold in rich MOPS (Teknova EZ rich defined medium). When the bacterial strains reached an OD 620 nm of ∼0.25, they were incubated with 0.4 μM of Cy5 labeled Neomycin for 15 minutes at 37°C. 10 μl of the incubated culture were then used for flow cytometry, diluting them in 250 μl of PBS before reading fluorescence. WT V. cholerae, was incubated simultaneously without Neo-Cy5 as a negative control. Flow cytometry experiments were performed as described (Baharoglu et al., 2010) and repeated at least 3 times. For each experiment, 100,000 events were counted on the Miltenyi MACSquant device.
MIC determination using etests
Stationary phase cultures were diluted 20 times in PBS, and 300 μL were plated on MH plates and dried for 10 minutes. etests (Biomérieux) were placed on the plates and incubated overnight at 37°C.
RNA-seq
Overnight cultures of the O1 biovar El Tor N16961 hapR+ V. cholerae strain were diluted 100x and grown in triplicate in MH medium until an OD 620 nm ∼0.4 with or without 350 μM indole. Sample collection, total RNA extraction, library preparation, sequencing and analysis were performed as previously described (Krin et al., 2018).
raiA qRT-PCR
Total RNA was extracted and purified from exponential phase cultures in MH in presence or absence of indole, as previously described (Krin et al., 2018). Reverse transcription (RT) was performed on 100 ng total RNA using SuperScript® III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative PCR was performed on 2 μl RT sample diluted 10-fold using SYBR Green PCR Master Mix (APPLIED) and QuantStudio 6. Quantification was performed using standard range. Expression values were normalized against gyrA as previously described in V. cholerae (Liu et al., 2010; Lo Scrudato and Blokesch, 2012).
Quantification of raiA expression by fluorescent flow cytometry using a gfp fusion
gfp was amplified by PCR using primers carrying the raiA promoter region and cloned into pTOPO-TA cloning vector. The PraiA-gfp fragment was then extracted using EcoRI and cloned into the low copy plasmid pSC101 (1 to 5 copies per cell). The plasmid was introduced into desired strains, and fluorescence was measured on indicated conditions, by counting 100,000 cells on the Miltenyi MACSquant device. Likewise, the control plasmid Pc-gfp (constitutive) was constructed using primers ZIP513/ZIP200 and similarly cloned in pSC101.
Growth curves
Overnight cultures were diluted 100x in fresh medium, on 96 well plates. Each well contained 200 μl. For overexpression lag experiments, glucose 1% was added to the media to repress the pBAD promoter while arabinose 0.2% was added to the media to induce the pBAD promoter, and same growth conditions (glucose or arabinose) were compared in order to avoid noise due to the effects of arabinose on growth and cells shape in V. cholerae (Espinosa et al., 2020). Spectinomycin was also added during each experiment to maintain the plasmid. Plates were incubated with shaking on TECAN plate reader device at 37°C, OD 620 nm was measured every 15 minutes.
Preparation of cell lysate for the analysis of ribosome content
The protocol was adapted from Qin and Fredrick (2013). Since we used stationary phase cultures instead of exponential phase, presence of polysomes is not expected. 10 ml of 20 hours cultures were centrifuged in ice cold 50 ml Falcon tubes for 15 minutes at 5000 rpm at 4°C. Pellets were resuspended in 500 μl lysis buffer (10 mM Tris-HCl, pH 8, 10 mM MgCl2, Lysozyme 1 mg/ml, protease inhibitor), transferred in ice cold 1.5 ml tubes and incubated with 12 μl Ribolock RNase inhibitor (Thermo scientific) and DNaseI (5 U/ml) at 4°C for 15 minutes. Cell lysis was performed through 3 cycles of flash-freezing in dry ice and thawing in a water bath at 4°C. 15 μl of 10% sodium deoxycholate were added and cell lysate was obtained after centrifugation at 10,000 rpm for 10 minutes at 4°C. The pellet containing cell debris was discarded. Lysate was kept at -80°C until sucrose gradient ultracentrifugation.
Sucrose gradient
10-50% sucrose gradient tubes (Beckman ULTRA CLEAR) were prepared. 2U of OD 260 nm of each cell extracts were deposited on sucrose gradient tubes. Ultracentrifugation was performed at 39,000 rpm at 4°C for 2 hours 45 minutes. Fractions were collected using a pump coupled to a spectrometer at OD 260 nm, and plotted as a function of time (seconds).
Quantification and statistical analysis
First an F-test was performed in order to determine whether variances are equal or different between comparisons. For comparisons with equal variance, Student’s t-test was used. For comparisons with significantly different variances, we used Welch’s t-test. For multiple comparisons, we used one-way ANOVA. We used GraphPad Prism to determine the statistical differences between groups. ∗∗∗∗ means p<0.0001, ∗∗∗ means p<0.001, ∗∗ means p<0.01, ∗ means p<0.05. Number of replicates for each experiment was 3<n<6. Means and geometric means for logarithmic values were also calculated using GraphPad Prism. For persistence tests, data were first log transformed in order to achieve normal distribution, and statistical tests were performed on these log-transformed data.
Acknowledgments
We are thankful to Micheline Fromont-Racine for her valuable help with the experiments for ribosome content analysis by sucrose density gradients. We thank Ivan Matic, Wei-Lin Su and Sébastien Fleurier for helpful discussions and Ivan Matic for critical reading of the manuscript. We also thank Dominique Fourmy for the gift of Neo-Cy5 and Sebastian Aguilar Pierlé for advice for Neo-Cy5 uptake experiments. We thank Louna Fruchard for help with persistence assays. This work was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS-UMR 3525), the Fondation pour la Recherche Médicale (FRM Grant No. DBF20160635736), ANR Unibac (ANR-17-CE13-0010-01) and Institut Pasteur grant PTR 245-19.
Author contributions
Experiments were designed by Z.B. Experiments were conducted by M.L., E. K., C.K. and Z. B., and results were interpreted by Z.B. and M.L. RNA-seq library preparation, sequencing and statistical analysis was performed by O. S., H.V. and J-Y. C. The manuscript was written and reviewed by Z.B., M.L. and D.M. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103128.
Contributor Information
Didier Mazel, Email: mazel@pasteur.fr.
Zeynep Baharoglu, Email: zeynep.baharoglu@pasteur.fr.
Supplemental information
Data and code availability
RNA-seq data have been deposited at GEO: GSE182561 and are publicly available as of the date of publication. Accession number is listed in the key resources table.
References
- Agafonov D.E., Kolb V.A., Nazimov I.V., Spirin A.S. A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12345–12349. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agafonov D.E., Kolb V.A., Spirin A.S. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2001;2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D.I., Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014;12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- Baharoglu Z., Babosan A., Mazel D. Identification of genes involved in low aminoglycoside-induced SOS response in Vibrio cholerae: a role for transcription stalling and Mfd helicase. Nucleic Acids Res. 2014;42:2366–2379. doi: 10.1093/nar/gkt1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z., Bikard D., Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. Plos Genet. 2010;6:e1001165. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z., Krin E., Mazel D. RpoS plays a central role in the SOS induction by sub-lethal aminoglycoside concentrations in Vibrio cholerae. Plos Genet. 2013;9:e1003421. doi: 10.1371/journal.pgen.1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z., Mazel D. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob. Agents Chemother. 2011;55:2438–2441. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Yap M.N. Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8165–E8173. doi: 10.1073/pnas.1709588114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J.M.A., Cloeckaert A., Nishino K., Piddock L.J.V. Alternative explanation for indole-induced antibiotic tolerance in Salmonella. P Natl. Acad. Sci. USA. 2013;110:E4569. doi: 10.1073/pnas.1318318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botsford J.L., DeMoss R.D. Catabolite repression of tryptophanase in Escherichia coli. J. Bacteriol. 1971;105:303–312. doi: 10.1128/jb.105.1.303-312.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner A., Fridman O., Gefen O., Balaban N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- Bray M.S., Lenz T.K., Haynes J.W., Bowman J.C., Petrov A.S., Reddi A.R., Hud N.V., Williams L.D., Glass J.B. Multiple prebiotic metals mediate translation. Proc. Natl. Acad. Sci. U S A. 2018;115:12164–12169. doi: 10.1073/pnas.1803636115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant E.L., Summers D.K. Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol. Microbiol. 2007;63:35–43. doi: 10.1111/j.1365-2958.2006.05481.x. [DOI] [PubMed] [Google Scholar]
- Chimerel C., Field C.M., Pinero-Fernandez S., Keyser U.F., Summers D.K. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim. Biophys. Acta. 2012;1818:1590–1594. doi: 10.1016/j.bbamem.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C., Liveris D., Squires C., Schwartz I., Squires C.L. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 1995;177:4152–4156. doi: 10.1128/jb.177.14.4152-4156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B.W., Bogard R.W., Mekalanos J.J. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Spiegelman G.B., Yim G. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Davis B.D. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 1987;51:341–350. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro F., Brandi A., Dzeladini N., Fabbretti A., Carzaniga T., Piersimoni L., Pon C.L., Giuliodori A.M. Role of the ribosome-associated protein PY in the cold-shock response of Escherichia coli. Microbiologyopen. 2013;2:293–307. doi: 10.1002/mbo3.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa E., Daniel S., Hernandez S.B., Goudin A., Cava F., Barre F.X., Galli E. L-arabinose induces the formation of viable non-proliferating spheroplasts in Vibrio cholerae. Appl. Environ. Microbiol. 2020;87:e02305-20. doi: 10.1128/AEM.02305-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W.C., Richard W., Handley C., Happold F.C. The tryptophanase-indole reaction: some observations on the production of tryptophanase by Esch. coli; in particular the effect of the presence of glucose and amino acids on the formation of tryptophanase. Biochem. J. 1941;35:207–212. doi: 10.1042/bj0350207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaga H.A., Kopylov M., Kim J.K., Jovanovic M., Dworkin J. Ribosome dimerization protects the small subunit. J. Bacteriol. 2020;202:e00009-20. doi: 10.1128/JB.00009-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraimow H.S., Greenman J.B., Leviton I.M., Dougherty T.J., Miller M.H. Tobramycin uptake in Escherichia coli is driven by either electrical potential or ATP. J. Bacteriol. 1991;173:2800–2808. doi: 10.1128/jb.173.9.2800-2808.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken L.E., Oostergetel G.T., Pijning T., Puri P., Arkhipova V., Boekema E.J., Poolman B., Guskov A. A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat. Commun. 2017;8:722. doi: 10.1038/s41467-017-00718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori A.M. In: Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. de Bruijn F.J., editor. Wiley Blackwell; 2016. Cold-shock response in Escherichia coli: a model system to study post-transcriptional regulation; pp. 859–872. [Google Scholar]
- Giuliodori A.M., Brandi A., Giangrossi M., Gualerzi C.O., Pon C.L. Cold-stress-induced de novo expression of infC and role of IF3 in cold-shock translational bias. RNA. 2007;13:1355–1365. doi: 10.1261/rna.455607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohara D.W., Yap M.F. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr. Genet. 2018;64:753–760. doi: 10.1007/s00294-017-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C.O., Giuliodori A.M., Brandi A., Di Pietro F., Piersimoni L., Fabbretti A., Pon C.L. In: Ribosomes: Structure, Function, and Dynamics. Rodnina M.V., Wintermeyer W., Green R., editors. Springer-Verlag; 2011. Translation initiation at the root of the cold-shock translational bias; pp. 143–154. [Google Scholar]
- Gutierrez A., Laureti L., Crussard S., Abida H., Rodriguez-Rojas A., Blazquez J., Baharoglu Z., Mazel D., Darfeuille F., Vogel J. beta-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013;4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.H., Lee J.H., Cho M.H., Wood T.K., Lee J. Environmental factors affecting indole production in Escherichia coli. Res. Microbiol. 2011;162:108–116. doi: 10.1016/j.resmic.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herisse M., Duverger Y., Martin-Verstraete I., Barras F., Ezraty B. Silver potentiates aminoglycoside toxicity by enhancing their uptake. Mol. Microbiol. 2017;105:115–126. doi: 10.1111/mmi.13687. [DOI] [PubMed] [Google Scholar]
- Hirakawa H., Inazumi Y., Masaki T., Hirata T., Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 2005;55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- Hossain T., Deter H.S., Peters E.J., Butzin N.C. Antibiotic tolerance, persistence, and resistance of the evolved minimal cell, Mycoplasma mycoides JCVI-Syn3B. iScience. 2021;24:102391. doi: 10.1016/j.isci.2021.102391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.F., Bina X.R., Bina J.E. Indole inhibits ToxR regulon expression in Vibrio cholerae. Infect. Immun. 2019;87:e00776-18. doi: 10.1128/IAI.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Kwan B.W., Osbourne D.O., Benedik M.J., Wood T.K. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ. Microbiol. 2015;17:1275–1285. doi: 10.1111/1462-2920.12567. [DOI] [PubMed] [Google Scholar]
- Janosi L., Hara H., Zhang S., Kaji A. Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys. 1996;32:121–201. doi: 10.1016/0065-227x(96)84743-5. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Yamasaki R., Song S., Zhang W., Wood T.K. Single cell observations show persister cells wake based on ribosome content. Environ. Microbiol. 2018;20:2085–2098. doi: 10.1111/1462-2920.14093. [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Hirakawa H., Hirata T., Nishino K., Yamaguchi A. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J. Bacteriol. 2006;188:5693–5703. doi: 10.1128/JB.00217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeva M., Gutu A.D., Hebert W., Wager J.D., Yonker L.M., O'Toole G.A., Ausubel F.M., Moskowitz S.M., Joseph-McCarthy D. An antipersister strategy for treatment of chronic Pseudomonas aeruginosa infections. Antimicrob. Agents Chemother. 2017;61:e00987-17. doi: 10.1128/AAC.00987-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J.M., Hochbaum D.R., Douglass A.D., Cohen A.E. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science. 2011;333:345–348. doi: 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- Krin E., Pierle S.A., Sismeiro O., Jagla B., Dillies M.A., Varet H., Irazoki O., Campoy S., Rouy Z., Cruveiller S. Expansion of the SOS regulon of Vibrio cholerae through extensive transcriptome analysis and experimental validation. BMC Genomics. 2018;19:373. doi: 10.1186/s12864-018-4716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.H., Molla M.N., Cantor C.R., Collins J.J. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Jayaraman A., Wood T.K. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhang X.S., Hegde M., Bentley W.E., Jayaraman A., Wood T.K. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Wood T.K., Lee J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015;23:707–718. doi: 10.1016/j.tim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Lelong C., Aguiluz K., Luche S., Kuhn L., Garin J., Rabilloud T., Geiselmann J. The Crl-RpoS regulon of Escherichia coli. Mol. Cell Proteom. 2007;6:648–659. doi: 10.1074/mcp.M600191-MCP200. [DOI] [PubMed] [Google Scholar]
- Li Y., Cole K., Altman S. The effect of a single, temperature-sensitive mutation on global gene expression in Escherichia coli. RNA. 2003;9:518–532. doi: 10.1261/rna.2198203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sharma M.R., Koripella R.K., Yang Y., Kaushal P.S., Lin Q., Wade J.T., Gray T.A., Derbyshire K.M., Agrawal R.K. Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 2018;115:8191–8196. doi: 10.1073/pnas.1804555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Beyhan S., Lim B., Linington R.G., Yildiz F.H. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J. Bacteriol. 2010;192:4541–4552. doi: 10.1128/JB.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Scrudato M., Blokesch M. The regulatory network of natural competence and transformation of Vibrio cholerae. Plos Genet. 2012;8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki Y., Yoshida H., Wada A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells. 2000;5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- Manneh-Roussel J., Haycocks J.R.J., Magan A., Perez-Soto N., Voelz K., Camilli A., Krachler A.M., Grainger D.C. cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization. MBio. 2018;9:e00966-18. doi: 10.1128/mBio.00966-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino P.D., Fursy R., Bret L., Sundararaju B., Phillips R.S. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 2003;49:443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- Matzov D., Bashan A., Yap M.F., Yonath A. Stress response as implemented by hibernating ribosomes: a structural overview. FEBS J. 2019;286:3558–3565. doi: 10.1111/febs.14968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay S.L., Portnoy D.A. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 2015;59:6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehi O., Bogos B., Csorgo B., Pal F., Nyerges A., Papp B., Pal C. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol. 2014;31:2793–2804. doi: 10.1093/molbev/msu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey A.R., Wyckoff E.E., Kanukurthy V., Fisher C.R., Payne S.M. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect Immun. 2005;73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.A. Indole adsorption to a lipid monolayer studied by optical second harmonic generation. J. Phys. Chem. B. 2009;113:10693–10707. doi: 10.1021/jp809528n. [DOI] [PubMed] [Google Scholar]
- Mueller R.S., Beyhan S., Saini S.G., Yildiz F.H., Bartlett D.H. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 2009;191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negro V., Krin E., Aguilar Pierle S., Chaze T., Giai Gianetto Q., Kennedy S.P., Matondo M., Mazel D., Baharoglu Z. RadD contributes to R-loop avoidance in sub-MIC tobramycin. MBio. 2019;10:e01173-19. doi: 10.1128/mBio.01173-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton W.A., Snell E.E. Formation and interrelationships of tryptophanase and tryptophan synthetases in Escherichia coli. J. Bacteriol. 1965;89:355–364. doi: 10.1128/jb.89.2.355-364.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido E., Giraud E., Baucheron S., Yamasaki S., Wiedemann A., Okamoto K., Takagi T., Yamaguchi A., Cloeckaert A., Nishino K. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog. 2012;4:5. doi: 10.1186/1757-4749-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven G.W. Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch. Microbiol. 2004;182:60–66. doi: 10.1007/s00203-004-0698-9. [DOI] [PubMed] [Google Scholar]
- Okuda M. Université Paris; 2015. Mechanism of Action of a Class of Antibiotics from Their Entry to Their Target in Bacteria: A Real Time Visualization. [Google Scholar]
- Opatowski M., Tuppin P., Cosker K., Touat M., De Lagasnerie G., Guillemot D., Salomon J., Brun-Buisson C., Watier L. Hospitalisations with infections related to antimicrobial-resistant bacteria from the French nationwide hospital discharge database. Epidemiol. Infect. 2019;147:e144. doi: 10.1017/S0950268819000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape S.S., Shashidhar R. Inhibition of protein synthesis eradicates persister cells of V. cholerae. 3 Biotech. 2019;9:380. doi: 10.1007/s13205-019-1916-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero-Fernandez S., Chimerel C., Keyser U.F., Summers D.K. Indole transport across Escherichia coli membranes. J. Bacteriol. 2011;193:1793–1798. doi: 10.1128/JB.01477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov Y.S., Blaha G.M., Steitz T.A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes M.H., Groisman E.A. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci. Signal. 2019;12:eaax3938. doi: 10.1126/scisignal.aax3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossliner T., Skovbo Winther K., Sorensen M.A., Gerdes K. Ribosome hibernation. Annu. Rev. Genet. 2018;52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- Prossliner T., Sorensen M.A., Winther K.S. Hibernation factors directly block ribonucleases from entering the ribosome in response to starvation. Nucleic Acids Res. 2021;49:2226–2239. doi: 10.1093/nar/gkab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P., Eckhardt T.H., Franken L.E., Fusetti F., Stuart M.C., Boekema E.J., Kuipers O.P., Kok J., Poolman B. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol. Microbiol. 2014;91:394–407. doi: 10.1111/mmi.12468. [DOI] [PubMed] [Google Scholar]
- Qin D., Fredrick K. Analysis of polysomes from bacteria. Methods Enzymol. 2013;530:159–172. doi: 10.1016/B978-0-12-420037-1.00008-7. [DOI] [PubMed] [Google Scholar]
- Sabeti Azad M., Okuda M., Cyrenne M., Bourge M., Heck M.P., Yoshizawa S., Fourmy D. Fluorescent aminoglycoside antibiotics and methods for accurately monitoring uptake by bacteria. ACS Infect. Dis. 2020;6:1008–1017. doi: 10.1021/acsinfecdis.9b00421. [DOI] [PubMed] [Google Scholar]
- Sabharwal D., Song T., Papenfort K., Wai S.N. The VrrA sRNA controls a stationary phase survival factor Vrp of Vibrio cholerae. RNA Biol. 2015;12:186–196. doi: 10.1080/15476286.2015.1017211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Ruf C., Garfa-Traore M., Collin V., Cordier C., Franceschi C., Matic I. Massive diversification in aging colonies of Escherichia coli. J. Bacteriol. 2014;196:3059–3073. doi: 10.1128/JB.01421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Yoshida H., Ishihama A. Involvement of cyclic AMP receptor protein in regulation of the rmf gene encoding the ribosome modulation factor in Escherichia coli. J. Bacteriol. 2013;195:2212–2219. doi: 10.1128/JB.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamti L., Livny J., Waldor M.K. Global gene expression and phenotypic analysis of a Vibrio cholerae rpoH deletion mutant. J. Bacteriol. 2007;189:351–362. doi: 10.1128/JB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soini J., Ukkonen K., Neubauer P. High cell density media for Escherichia coli are generally designed for aerobic cultivations - consequences for large-scale bioprocesses and shake flask cultures. Microb. Cell Fact. 2008;7:26. doi: 10.1186/1475-2859-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Wood T.K. ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. Biochem. Biophys. Res. Commun. 2020;523:281–286. doi: 10.1016/j.bbrc.2020.01.102. [DOI] [PubMed] [Google Scholar]
- Spoering A.L., Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H.W., Mueller J.P., Miller P.F., Arrow A.S. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 1987;51:439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko A.G., Kashevarova N.M., Tyuleneva E.A., Shumkov M.S. Stationary-phase genes upregulated by polyamines are responsible for the formation of Escherichia coli persister cells tolerant to netilmicin. FEMS Microbiol. Lett. 2017;364:fnx084. doi: 10.1093/femsle/fnx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touat M., Opatowski M., Brun-Buisson C., Cosker K., Guillemot D., Salomon J., Tuppin P., de Lagasnerie G., Watier L. A payer perspective of the hospital inpatient additional care costs of antimicrobial resistance in France: a matched case-control study. Appl. Health Econ. Health Policy. 2019;17:381–389. doi: 10.1007/s40258-018-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta M., Wada C., Daifuku T., Sako Y., Bessho Y., Kitamura A., Ohniwa R.L., Morikawa K., Yoshida H., Kato T. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells. 2013;18:554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- Ueta M., Yoshida H., Wada C., Baba T., Mori H., Wada A. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells. 2005;10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- Vega N.M., Allison K.R., Khalil A.S., Collins J.J. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega N.M., Allison K.R., Samuels A.N., Klempner M.S., Collins J.J. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Sanjurjo A., Schuwirth B.S., Hau C.W., Cate J.H. Structural basis for the control of translation initiation during stress. Nat. Struct. Mol. Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- Wada A., Mikkola R., Kurland C.G., Ishihama A. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J. Bacteriol. 2000;182:2893–2899. doi: 10.1128/jb.182.10.2893-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Ding X., Rather P.N. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 2001;183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.K., Song S. Forming and waking dormant cells: the ppGpp ribosome dimerization persister model. Biofilm. 2020;2:100018. doi: 10.1016/j.bioflm.2019.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W.N., Mohler K., Rinehart J., Ibba M. Deacylated tRNA accumulation is a trigger for bacterial antibiotic persistence independent of the stringent response. mBio. 2021;12:e0113221. doi: 10.1128/mBio.01132-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M., Matsushima H., Wada A., Sakagami M., Fujita N., Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Horn V., Gollnick P. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 1991;173:6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkan A., Matuszewska M., Trigg S.B., Zhang M., Belgami D., Croft C., Liu J., El-Ouisi S., Greenhalgh J., Duboff J.S. Inhibition of indole production increases the activity of quinolone antibiotics against E. coli persisters. Scientific Rep. 2020;10:11742. doi: 10.1038/s41598-020-68693-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinskie J.A., Ghosh A., Trainor B.M., Shedlovskiy D., Pestov D.G., Shcherbik N. Iron-dependent cleavage of ribosomal RNA during oxidative stress in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2018;293:14237–14248. doi: 10.1074/jbc.RA118.004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO: GSE182561 and are publicly available as of the date of publication. Accession number is listed in the key resources table.