Abstract
Methylmercury is an environmental pollutant that induces potent neurotoxicity. We previously identified transcription factor 3 (TCF3) as a transcription factor that is activated in the brains of mice treated with methylmercury, and reported that methylmercury sensitivity was increased in cells in which TCF3 expression was suppressed. However, the mechanisms involved in the activation of TCF3 by methylmercury and in the reduction of methylmercury toxicity by TCF3 remained unclear. We found that treatment of mouse neuronal C17.2 cells with methylmercury increased TCF3 protein levels and promoted the binding of TCF3 to DNA consensus sequences. In cells treated with actinomycin D, a transcription inhibitor, an increase in TCF3 protein levels was also observed under methylmercury exposure. However, in the presence of cycloheximide, a translation inhibitor, methylmercury delayed the degradation of TCF3 protein. In addition, treatment with MG132, a proteasome inhibitor, increased TCF3 protein levels, and there was not significant increase in TCF3 protein levels by methylmercury under these conditions. These results suggest that methylmercury may activate TCF3 by increasing its levels through inhibition of TCF3 degradation by the proteasome. It has been previously reported that the induction of apoptosis in neurons is involved in methylmercury-induced neuronal damage in the brain. Although apoptosis was induced in C17.2 cells treated with methylmercury, this induction was largely suppressed by overexpression of TCF3. These results indicate that TCF3, which is increased in the brain upon exposure to methylmercury, may be a novel defense factor against methylmercury-induced neurotoxicity.
Keywords: Methylmercury, Transcription factor 3 (TCF3), Neurotoxicity, Apoptosis
Introduction
Methylmercury is a heavy metal that is widely present in the environment and is eventually taken up by humans through bioaccumulation in fish and shellfish [1]. In the body, methylmercury takes on a methionine-like structure when bound to cysteine and can cross the blood–brain barrier via a neutral amino acid transporter [2]. High concentrations of methylmercury in the brain cause Minamata disease, which includes central nervous system disorders as a main symptom [3, 4]. However, the mechanisms involved in methylmercury-induced neurotoxicity and the defense response against it remain largely unknown.
When an organism is exposed to an exogenous stressor, a variety of transcription factors are activated. The activation of these transcription factors plays a central role in the induction of apoptosis and stress response by regulating the expression of downstream genes. We have shown that methylmercury activates transcription factors such as cAMP response element binding protein (CREB), NF-E2-related factor 2 (Nrf2), and heat shock factor 1 (HSF1), and that these transcription factors play a role in reducing methylmercury toxicity by inducing the expression of genes involved in apoptosis inhibition and methylmercury excretion [5–8]. We have also found that the methylmercury-activated transcription factor HOXB13 is involved in enhancing the toxicity of methylmercury by promoting the induction of the cytokine oncostatin M [9]. We also reported that the transcription factor NF-kB, which is activated by methylmercury in mouse neural C17.2 cells, is involved in enhancing the toxicity of methylmercury via induction of TNF-α expression [10]. Methylmercury has been reported to inhibit cell proliferation and induce apoptosis through the activation of the transcription factor p53 [11]. Thus, investigation of transcription factor activation may provide important clues to understanding methylmercury toxicity and the defense response against it.
Recently, we conducted a comprehensive search for transcription factors activated in the cerebellum of methylmercury-treated mice using a protein–DNA binding assay and found that transcription factor 3 (TCF3), paired box 4 (PAX4), paired box 6 (PAX6) and high mobility group AT-hook 1 (HMGA1) were significantly activated [12]. We found that knockdown of TCF3 expression increased the sensitivity of C17.2 cells to methylmercury, suggesting that TCF3 activation may be part of the defense response against methylmercury toxicity. However, the mechanisms involved in the methylmercury-induced TCF3 activation and the reduction of methylmercury toxicity remain unclear. TCF3, a basic helix loop helix (bHLH) transcription factor, is highly expressed during early neurogenesis and is involved in neuronal differentiation and other processes [13, 14]. It has been reported that doxorubicin-induced apoptosis is increased in cancer cells when TCF3 expression is repressed, while pre-renewal line carcinoma cells with high TCF3 expression show resistance to doxorubicin [15]. It has also been reported that TCF3 exerts an anti-apoptotic effect by negatively regulating the expression of p53 upregulated modulator of apoptosis (PUMA), which is an apoptosis-inducing agent [16]. Since methylmercury is known to induce neurological damage by inducing apoptosis, we suspected that TCF3 might be a factor that reduces methylmercury toxicity by inhibiting apoptosis in neurons. Therefore, we aimed to investigate these relationships in the present study using C17.2 cells.
Materials and methods
Animals
The present study was performed in accordance with the recommendations of Regulations for Animal Experiments and Related Activities at Tohoku University. All mice used in the study were 6-week-old male C57BL/6 mice that were purchased from Japan SLC Inc. (Shizuoka, Japan). Prior to the experiments, mice were housed in plastic cages at 22 ± 2 °C and at a relative humidity of 55 ± 20% with a 12 h light–dark cycle for 1 week. Food (F-2, Oriental yeast, Tokyo, Japan) and filtrated tap water were provided ad libitum. Methylmercuric chloride dissolved in saline (25 mg/kg, single administration) was injected subcutaneously. The mice were dissected after the indicated time course.
Cells and cell cultures
Mouse neural C17.2 cells were maintained in DMEM supplemented with 10% FBS, 5% horse serum (Thermo Fisher Scientific, Waltham, MA, USA), 2 mmol/l l-glutamine, and antibiotics (100 IU/ml penicillin and 100 mg/ml streptomycin). Before starting the experiments, cells (2.5 × 105 cells/well) were seeded on a 12-well plate and cultured for 24 h. These cells were maintained at 37 °C in a humidified incubator in an atmosphere of CO2 (5%) and ambient air (95%).
Immunoblotting
Cells or organs were harvested or homogenized in a sodium dodecyl sulfate (SDS) buffer (1 mM trisaminomethane-hydrochloride (pH 7.4), 2% SDS, 150 mM sodium chloride (NaCl), and 1 mM ethylenediaminetetraacetic acid) supplemented with a protease inhibitor cocktail (Roche, Basel, Switzerland). The lysates were then incubated at 95 °C for 10 min. Protein concentration of each lysate was examined using the DC protein assay kit (Bio-Rad, Hercules, CA, USA). Aliquots of samples (20 µg) were subjected to SDS–polyacrylamide gel electrophoresis. The gel was transferred to an Immobilon-P polyvinylidene fluoride membrane (Millipore, Burlington, MA, USA) and stained with antibodies against TCF3 (Thermo Fischer Scientific), lamin, cleaved-caspase 3 (Cell Signaling Technology, Danvers, MA), PA-tag, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Fujifilm-Wako, Osaka, Japan). Densitometry was measured by ImageJ software 10.2 (https://imagej.nih.gov/ij/).
Quantitative PCR (qPCR)
Total RNA was isolated using Isogen II (Nippon Gene, Tokyo, Japan), according to the manufacturer’s instructions. Reverse transcription was performed using PrimeScript® RT reagent kit (Takara, Shiga, Japan). Quantitative PCR was performed using KAPA SYBR (Nippon Genetics, Tokyo, Japan) by Thermal Cycler Dice® (Takara) with the following primers: TCF3, F: 5′-TTGACCCTAGCCGGACATAC-3′, R: 5′-TGCCAACACTGGTGTCTCTC-3′; and GAPDH, F: 5′-ATCACCATCTTCCAGGAGCGA-3′, R: 5′-AGGGGCCATCCACAGTCTT-3′. GAPDH was used as an internal control. The data are presented as values corrected by GAPDH.
Construction of TCF3-PA expressing plasmid
Complementary DNA prepared from C17.2 cells was amplified by KOD-plus neo (Toyobo, Osaka, Japan) with the following primers that targeted TCF3 with a c-terminal PA-tag, F: 5′-TTCCTCGAGACTAGTTCTAGAATGATGAACCAGTCTCAGA-3′, R: 5′- GGTAGAATTGGATCCGCGGCCGCTCAAACAACGTCGTCCT-3′. The resulting PCR product was ligated into pLVSIN-CMV pur (Takara) by using the Infusion HD cloning kit (Takara). The ligation products were transformed into E. coli (XL-1 blue, GM biolab, Dali, Taiwan) and the full sequence was evaluated by the Sanger method (Fasmac, Kanagawa, Japan). We cloned multiple transcript variants from C17.2 cells and chose the clone of transcript variant 3 (NP_001157621.1) for this study, because transcript variant 3 may be predominantly expressed in the cells, as the number of the clones encoding transcript variant 3 were dominant.
TUNEL assay
MEBSTAIN Apoptosis TUNEL Kit II (Medical and Biological Laboratories, Nagoya, Japan) was used for TUNEL staining, according to the manufacturer’s instructions. Cells were fixed with 4% paraformaldehyde in PBS for 30 min at 4 °C. After removal of the supernatant, terminal deoxynucleotidyl transferase (TdT) with biotinylated dNTP was added to the cells and incubated at 37 °C for 30 min for the addition of biotin to fragmented DNA. Cells were incubated with avidin-FITC II solution for 30 min at 25 °C. Fluorescence was measured by FV1000 confocal microscope (Olympus, Tokyo, Japan) at wavelengths of 488 nm for excitation and 530 nm for emission. Data analysis was performed by Fluoview (Olympus).
Statistical analysis
Statistical significance was analyzed using a one-way ANOVA and Tukey’s post hoc test. Analyses were performed with GraphPad Prism 8 (GraphPad Software, CA, USA).
Results
Effects of methylmercury on TCF3 protein levels and TCF3 activity
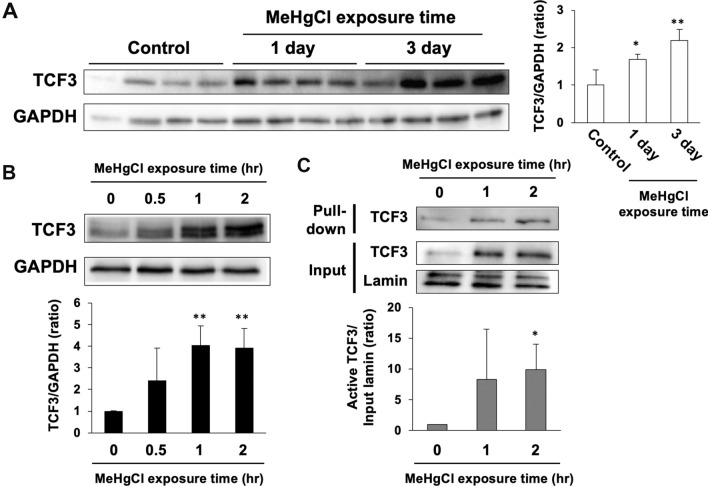
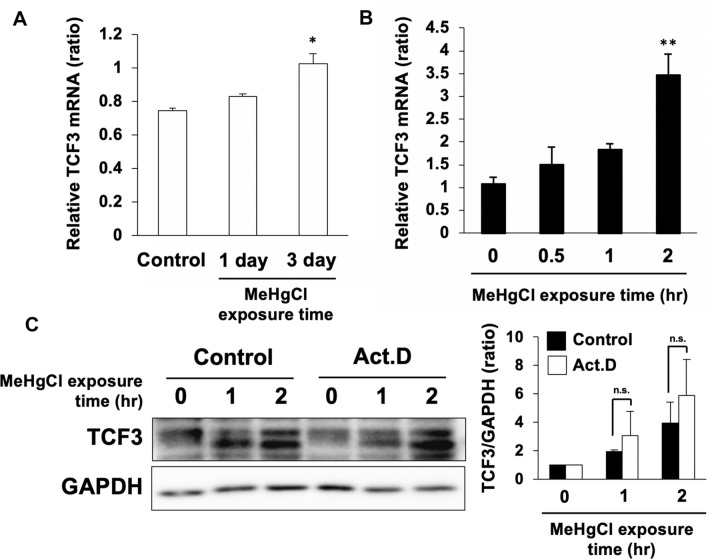
We previously found that TCF3 was activated in the cerebellum of methylmercury-treated mice. Therefore, we subcutaneously administered methylmercury (25 mg/kg) to mice and excised the cerebellum at 1 or 3 days post-administration to examine the levels of TCF3 protein. We found that TCF3 levels in the cerebellum of methylmercury-treated mice were significantly increased in a time-dependent manner compared with the control group (Fig. 1a). In C17.2 cells, TCF3 was found in the nuclear fraction only and was not detected in the cytoplasm (data not shown), suggesting that TCF3 is constantly present in the nucleus and functions as a transcription factor. In addition, since multiple transcriptional variants in the TCF3 gene are known to exist, it is likely that endogenous TCF3 was detected as multiple or smear bands in C17.2 cells. After treatment of C17.2 cells with methylmercury, an increase in TCF3 protein levels was observed in a time-dependent manner (Fig. 1b). Under these conditions, we examined the activation of TCF3 by DNA pull-down assay and found that binding of TCF3 to the consensus sequence increased in response to increased TCF3 expression (Fig. 1c). This suggests that methylmercury may be involved in the activation of TCF3 by increasing TCF3 protein levels. TCF3 mRNA levels were also significantly increased in the brains of methylmercury-treated mice and in C17.2 cells (Fig. 2a, b). However, pretreatment with actinomycin D (Act. D), a transcription inhibitor, had little effect on the increase in TCF3 protein levels by methylmercury (Fig. 2c). These results suggest that methylmercury may increase TCF3 protein levels through post-translational modifications, such as the repression of degradation, rather than through transcriptional promotion.
Fig. 1.
Effects of methylmercury on TCF3 protein levels and activity. a C57BL6 mice were administered with 25 mg/kg of methylmercuric chloride (MeHgCl) by subcutaneously (s.c.) for 1 or 3 days and the protein levels of TCF3 and GAPDH in the cerebellum were examined by immunoblotting (n = 4). Right panel indicates quantification of TCF3 band intensities and those corrected by GAPDH are shown in the left panel. b C17.2 cells were treated to 20 µM of MeHgCl for the indicated time course and protein levels of TCF3 and GAPDH were determined by immunoblotting. A representative blot is shown in the upper panel. Lower panel indicates quantification of TCF3 band intensities that has been corrected by GAPDH (n = 3). c C17.2 cells were treated with 20 µM of MeHgCl for the indicated time course and nuclear fractions were collected. Activated TCF3 was measured by DNA pull-down assay using the TCF3 binding sequence. A representative blot is shown in the upper panel. Lower panel indicates quantification of activated TCF3 band intensities that have been corrected by lamin (n = 3). The data are represented as mean ± SD. *p < 0.05 vs. control, **p < 0.01 vs. control
Fig. 2.
Contribution of TCF3 transcription to increase in TCF3 protein levels by methylmercury. a C57BL6 mice were administrated with 25 mg/kg of methylmercuric chloride (MeHgCl) by s.c. for 1 or 3 days and mRNA levels of TCF3 in the cerebellum were examined by qPCR. Values corrected by GAPDH are shown (n = 4). b C17.2 cells were treated with 20 µM of MeHgCl for the indicated time course and mRNA levels of TCF3 were determined by qPCR. Values corrected by GAPDH are shown (n = 3). c C17.2 cells were pretreated with 2 µM of actinomycin D (Act. D) for 1 h and treated with 20 µM of MeHgCl for the indicated time course. Protein levels of TCF3 and GAPDH were determined by immunoblotting. A representative blot is shown in the left panel. Right panel indicates quantification of TCF3 band intensities that have been corrected by GAPDH (n = 3). The data are represented as mean ± SD. *p < 0.05 vs. control, **p < 0.01 vs. control. n.s. indicates not significant
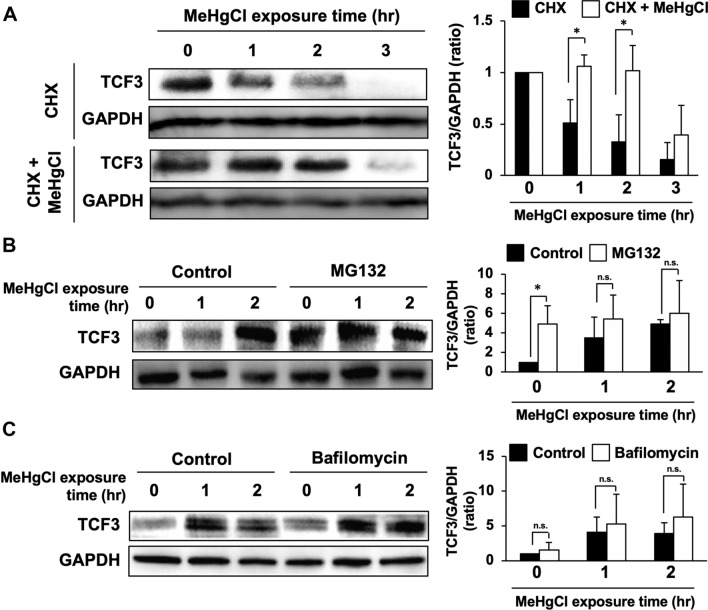
Effects of proteasome inhibitor or lysosome inhibitor on the increase in TCF3 protein levels by methylmercury
When C17.2 cells were treated with cycloheximide, a translation inhibitor, TCF3 protein expression was mostly eliminated over a 3 h period, with a half-life of 58.5 min (Fig. 3a), whereas methylmercury treatment prolonged the half-life of TCF3 to 144.8 min (Fig. 3a). This suggests that inhibition of protein degradation may contribute to the increase in TCF3 protein levels by methylmercury. TCF3 is known to undergo degradation by the proteasome following ubiquitination by Ubc9, a ubiquitin-conjugating enzyme (E2) [17]. Therefore, we treated C17.2 cells with MG132, a proteasome inhibitor, and examined the effects of methylmercury on TCF3 protein levels. We found that treatment with MG132 for 4 h prior to the addition of methylmercury increased TCF3 protein levels to approximately the same level as observed in the methylmercury treatment, but no additive increase was observed after methylmercury treatment (Fig. 3b). This result suggests that methylmercury increases the levels of TCF3 protein by inhibiting its degradation through the ubiquitin–proteasome pathway. Treatment of C17.2 cells with bafilomycin, a lysosome inhibitor, did not change TCF3 protein levels and had no effect on the increase in TCF3 protein levels by methylmercury, suggesting that the lysosomal pathway may not be involved in the increase in TCF3 protein levels in response to methylmercury (Fig. 3c).
Fig. 3.
Effects of inhibition of proteolytic pathways on increase in TCF3 protein levels by methylmercury. a C17.2 cells were pretreated with 40 µM of cycloheximide (CHX) for 1 h and 20 µM of methylmercuric chloride (MeHgCl) for the indicated time course. Protein levels of TCF3 and GAPDH were determined by immunoblotting. A representative blot is shown in the left panel. Right panel indicates quantification of TCF3 band intensities that have been corrected by GAPDH (n = 3). b C17.2 cells were pretreated with 30 µM of MG132 for 4 h and 20 µM of MeHgCl for the indicated time course. Protein levels of TCF3 and GAPDH were determined by immunoblotting. A representative blot is shown in the left panel. Right panel indicates quantification of TCF3 band intensities that have been corrected by GAPDH (n = 3). c C17.2 cells were pretreated with 0.5 µM of bafilomycin for 3 h and 20 µM of MeHgCl for the indicated time course. Protein levels of TCF3 and GAPDH were determined by immunoblotting. A representative blot is shown in the left panel. Right panel indicates quantification of TCF3 band intensities that have been corrected by GAPDH (n = 3). The data are represented as mean ± SD. *p < 0.05 vs. control. n.s. indicates not significant
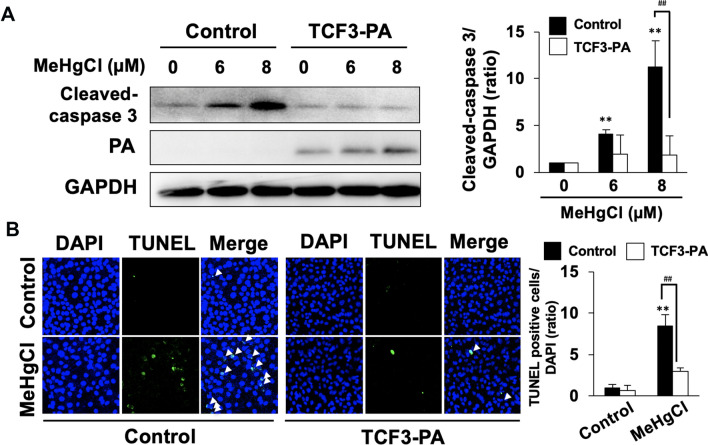
Effects of overexpression of TCF3 on the induction of apoptosis by methylmercury
Our previous study demonstrated that knockdown of TCF3 expression enhances methylmercury toxicity, suggesting that TCF3 may be a defense factor against methylmercury toxicity [12]. Therefore, we transfected a PA-tagged TCF3 expression plasmid (TCF3-PA) into C17.2 cells to generate TCF3-PA overexpressing cells and examined the effects of overexpression of TCF3 on the induction of apoptosis by methylmercury. We examined the cleaved-caspase 3 levels and DNA fragmentation by TUNEL assay as indicators of the induction of apoptosis. Treatment with methylmercury for 24 h increased the levels of cleaved-caspase 3 and the number of TUNEL-positive cells, but these increases were both inhibited by overexpression of TCF3-PA (Fig. 4a, b). These results indicate that the increase in TCF3 expression in response to methylmercury is a defense response that suppresses the induction of apoptosis.
Fig. 4.
Effects of overexpression of TCF3 on apoptosis induced by methylmercury. a C17.2 cells were transfected with control vector or TCF3-PA-expressing plasmid for 24 h. Cells were then treated with the indicated concentration of methylmercuric chloride (MeHgCl) for 24 h. Protein levels of cleaved-caspase 3, TCF3-PA, and GAPDH were determined by immunoblotting. A representative blot is shown in the left panel. Right panel indicates quantification of cleaved-caspase 3 band intensities that have been corrected by GAPDH (n = 3). b After transfection and MeHgCl exposure, cells were subjected to TUNEL assay. Representative pictures are shown and TUNEL positive cells are indicated by white arrows. Right panel indicates quantification of TUNEL positive cells that are corrected by DAPI (n = 3). The data are represented as mean ± SD. **p < 0.01 vs. MeHgCl (0 µM), ##p < 0.01 vs. control
Discussion
The present study revealed that the mechanisms related to the activation of TCF3 by methylmercury involve an increase in TCF3 protein levels through inhibition of protein degradation by the ubiquitin–proteasome pathway. Our results suggest that the increase in TCF3 expression may be a defense response that inhibits the induction of apoptosis by methylmercury.
Some transcription factors that are activated in response to various stresses, such as hypoxia inducible factor 1 (HIF-1) and Nrf2, have relatively short half-lives of 5–8 min and 15–30 min, respectively [18, 19]. However, upon sensing the stressor, the degradation of these proteins via the ubiquitin–proteasome pathway is inhibited, protein levels increase, and rapid translocation into the nucleus and induction of the expression of downstream genes occurs. TCF3 is endemic to the nucleus and is likely to be similarly regulated because it responds rapidly, although the half-life of TCF3 is relatively long compared with these transcription factors. As mentioned above, Ubc9 is known to be involved in TCF3 ubiquitination [17], and it is assumed that methylmercury may inhibit TCF3 ubiquitination mediated by Ubc9. It will be necessary to clarify the effect of methylmercury on TCF3 ubiquitination and its mechanisms in future studies.
It has been reported that regulation of TCF3 activation involves the binding of transcriptional repressors, and that Id3 represses TCF3 activity by forming heterodimers with TCF3 [20]. However, when arsenic binds to a cysteine residue at the N-terminus of Id3, Id3 is translocated from the nucleus to the cytoplasm [21]. It is possible that methylmercury also binds to the cysteine residues of Id3 and decreases its nuclear levels, resulting in increased transcriptional activity of TCF3. When TCF3 and Id3 were co-expressed in C17.2 cells and the binding between them was examined by immunoprecipitation, methylmercury had little effect on TCF3–Id3 binding (data not shown). This result suggests that the binding of methylmercury to Id3 is not involved in the activation of TCF3 by methylmercury. TCF3 is known to exhibit anti-apoptotic effects by inhibiting the induction of PUMA expression, which is involved in the induction of apoptosis [22]. However, an increase in PUMA mRNA levels by methylmercury treatment was still observed even when TCF3 was overexpressed (data not shown), suggesting that TCF3 inhibits the induction of apoptosis in a PUMA-independent manner. We found that methylmercury induces expression of TNF-α, an inflammatory cytokine, and that TNF-α released extracellularly binds to TNF receptor 1 and induces apoptosis by death receptor signaling pathway [10]. It has also been reported that methylmercury induces apoptosis by inducing mitochondrial stress through inhibition of the electron transfer system and endoplasmic reticulum stress through inhibition of protein-disulfide isomerase (PDI) [23, 24]. Thus, methylmercury induces apoptosis through various pathways, and identification of the pathways inhibited by TCF3 is expected to lead to the elucidation of the detailed molecular mechanisms involved in methylmercury-induced apoptosis.
In summary, although the present study employed relatively higher concentrations of methylmercury and artificial conditions including TCF3 overexpression in cultured cell experiments, our results clearly suggest that the increase in TCF3 protein levels observed in the brains of mice treated with methylmercury is a novel defense response against methylmercury-induced neurotoxicity.
Acknowledgements
This work was partially supported by JSPS KAKENHI Grant Number 15H05714 and 19H04276. We thank Emma Longworth-Mills, PhD, from Edanz Group (https://en-author-services.edanz.com/ac) for editing a draft of this manuscript.
Author contributions
TT, GWH wrote the manuscript. GWH designed the experiments. MSK, TT, and YW prepared Fig. 1 and 2. YW prepared Fig. 3. TT. and YW prepared Fig. 4. TT. and AN assisted in the experimental design and provided conceptual advice. TT. and GWH analyzed the data. All authors discussed the results and provided feedback on the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no actual or potential competing financial interests.
References
- 1.Antunes DSA, Appel HM, Culbreth M, Lopez-Granero C, Farina M, Rocha JB, Aschner M. Methylmercury and brain development: a review of recent literature. J Trace Elem Med Biol. 2016;38:99–107. doi: 10.1016/j.jtemb.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmons-Willis TA, Koh AS, Clarkson TW, Ballatori N. Transport of a neurotoxicant by molecular mimicry: the methylmercury-l-cysteine complex is a substrate for human l-type large neutral amino acid transporter (LAT) 1 and LAT2. Biochem J. 2002;367:239–246. doi: 10.1042/BJ20020841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada M. Minamata disease: methylmercury poisoning in Japan caused by environmental pollution. Crit Rev Toxicol. 1995;25:1–24. doi: 10.3109/10408449509089885. [DOI] [PubMed] [Google Scholar]
- 4.Harada M. Congenital minamata disease: intrauterine methylmercury poisoning. Teratology. 1978;18:285–288. doi: 10.1002/tera.1420180216. [DOI] [PubMed] [Google Scholar]
- 5.Unoki T, Abiko Y, Toyama T, Uehara T, Tsuboi K, Nishida M, Kaji T, Kumagai Y. Methylmercury, an environmental electrophile capable of activation and disruption of the Akt/CREB/Bcl-2 signal transduction pathway in SH-SY5Y cells. Sci Rep. 2016;6:28944. doi: 10.1038/srep28944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toyama T, Sumi D, Shinkai Y, Yasutake A, Taguchi K, Tong KI, Yamamoto M, Kumagai Y. Cytoprotective role of Nrf2/Keap1 system in methylmercury toxicity. Biochem Biophys Res Commun. 2007;363:645–650. doi: 10.1016/j.bbrc.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Toyama T, Shinkai Y, Yasutake A, Uchida K, Yamamoto M, Kumagai Y. Isothiocyanates reduce mercury accumulation via an Nrf2-dependent mechanism during exposure of mice to methylmercury. Environ Health Perspect. 2011;119:1117–1122. doi: 10.1289/ehp.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang GW, Ryoke K, Lee JY, Takahashi T, Naganuma A. siRNA-mediated silencing of the gene for heat shock transcription factor 1 causes hypersensitivity to methylmercury in HEK293 cells. J Toxicol Sci. 2011;36:851–853. doi: 10.2131/jts.36.851. [DOI] [PubMed] [Google Scholar]
- 9.ToyamaT XS, Nakano R, Hasegawa T, Endo N, Takahashi T, Lee JY, Naganuma A, Hwang GW. The nuclear protein HOXB13 enhances methylmercury toxicity by inducing oncostatin M and promoting its binding to TNFR3 in cultured cells. Cells. 2020;9:45. doi: 10.3390/cells9010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwai-Shimada M, Takahashi T, Kim MS, Fujimura M, Ito H, Toyama T, Naganuma A, Hwang GW. Methylmercury induces the expression of TNF-alpha selectively in the brain of mice. Sci Rep. 2016;6:38294. doi: 10.1038/srep38294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gribble EJ, Hong SW, Faustman EM. The magnitude of methylmercury-induced cytotoxicity and cell cycle arrest is p53-dependent. Birth Defects Res A. 2005;73:29–38. doi: 10.1002/bdra.20104. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi T, Yanjiao W, Toyama T, Kim MS, Kuge S, Hwang GW, Naganuma A. Small interfering RNA-mediated knockdown of the transcription factor TCF3 enhances sensitivity to methylmercury in mouse neural stem cells. Fundam Toxicol Sci. 2017;4:41–43. doi: 10.2131/fts.4.41. [DOI] [Google Scholar]
- 13.Patel D, Chinaranagari S, Chaudhary J. Basic helix loop helix (bHLH) transcription factor 3 (TCF3, E2A) is regulated by androgens in prostate cancer cells. Am J Cancer Res. 2015;5:3407–3421. [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto Y, Tsutsumi M, Myojin R, Maruta K, Onoda F, Tashiro F, Ohtsu M, Murakami Y. Interaction of Hand2 and E2a is important for transcription of Phox2b in sympathetic nervous system neuron differentiation. Biochem Biophys Res Commun. 2011;408:38–44. doi: 10.1016/j.bbrc.2011.03.113. [DOI] [PubMed] [Google Scholar]
- 15.Patel D, Chaudhary J. Increased expression of bHLH transcription factor E2A (TCF3) in prostate cancer promotes proliferation and confers resistance to doxorubicin induced apoptosis. Biochem Biophys Res Commun. 2012;422:146–151. doi: 10.1016/j.bbrc.2012.04.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrysik Z, Kim J, Tan AC, Espinosa JM. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep. 2013;3:1346–1354. doi: 10.1016/j.celrep.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loveys DA, Streiff MB, Schaefer TS, Kato GJ. The mUBC9 murine ubiquitin conjugating enzyme interacts with the E2A transcription factors. Gene. 1997;201:169–177. doi: 10.1016/S0378-1119(97)00444-7. [DOI] [PubMed] [Google Scholar]
- 18.Jewell UR, Kvietikova I, Scheid A, Bauer C, Wenger RH, Gassmann M. Induction of HIF-1 alpha in response to hypoxia is instantaneous. FASEB J. 2001;15:1312–1314. doi: 10.1096/fj.00-0732fje. [DOI] [PubMed] [Google Scholar]
- 19.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 20.Loveys DA, Streiff MB, Kato GJ. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucl Acid Res. 1996;24:2813–2820. doi: 10.1093/nar/24.14.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurooka H, Sugai M, Mori K, Yokota Y. The metalloid arsenite induces nuclear export of Id3 possibly via binding to the N-terminal cysteine residues. Biochem Biophys Res Commun. 2013;433:579–585. doi: 10.1016/j.bbrc.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Qiu W, Wang XW, Leibowitz B, Yang WC, Zhang L, Yu J. PUMA-mediated apoptosis drives chemical hepatocarcinogenesis in mice. Hepatology. 2011;54:1249–1258. doi: 10.1002/hep.24516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usuki F, Fujita E, Sasagawa N. Methylmercury activates ASK1/JNK signaling pathways, leading to apoptosis due to both mitochondria- and endoplasmic reticulum (ER)-generated processes in myogenic cell lines. Neurotoxicology. 2008;29:22–30. doi: 10.1016/j.neuro.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Makino K, Okuda K, Sugino E, Nishiya T, Toyama T, Iwawaki T, Fujimura M, Kumagai Y, Uehara T. Correlation between attenuation of protein disulfide isomerase activity through S-mercuration and neurotoxicity induced by methylmercury. Neurotox Res. 2015;27:99–105. doi: 10.1007/s12640-014-9494-8. [DOI] [PubMed] [Google Scholar]