Abstract
In the last decade, several advancements have been made in omics technologies and they have been applied extensively in diverse research areas. Especially in toxicological research, omics technology can efficiently and accurately generate relevant data on the molecular dynamics associated with adverse outcomes. Toxicomics is defined as the combination of toxicology and omics technologies and encompasses toxicogenomics, toxicoproteomics, and toxicometabolomics. This paper reviews the trend of applying omics technologies to evaluate cadmium (Cd) toxicity in zebrafish (D. rerio). Cd is a toxic heavy metal posing several environmental concerns; however, it is being used widely in everyday life. Zebrafish embryos and larvae are employed as standard models for many toxicity tests because they share 71.4% genetic homology with humans. This study summarizes the toxicity of Cd on the nerves, liver, heart, skeleton, etc. of zebrafish and introduces detailed omics techniques to understand the results of the toxicomic studies. Finally, the trend of toxicity evaluation in the zebrafish model of Cd based on omics technology is presented.
Keywords: Cadmium, Zebrafish, Toxicogenomics, Toxicoproteomics, Toxicometabolomics
Introduction
Cadmium (Cd) is widely used in objects such as batteries and in processes such as plating and can also leak around factories manufacturing these products. In our daily lives, we may be exposed to Cd through food and air, but the actual amounts are negligible and have little effect on the human body. However, if harmful metals from factory by-products or household waste pollute the atmosphere, soil, and water and accumulate in animals and plants, they may be absorbed in large amounts by humans and cause deleterious health effects. For example, the Itai-Itai disease ranks among the four major pollution-related diseases in Japan; its symptoms are pain and osteomalacia, and it was caused by the consumption of rice contaminated with Cd from a factory leakage [1]. In Korea, high concentrations of Cd in groundwater and rivers have been reported; the source was a smelter located upstream of the Nakdong River [2]. Water pollution with Cd leads to soil accumulation, and finally affects agricultural and marine products consumed by humans. Fine dust, an air pollution source, can also be a source of exposure to Cd [3]. In addition to environmental problems, exposure to Cd is caused by problems in the processing of pharmaceuticals, cosmetics, toys, and household goods or because of occupational reasons [4].
Omics technologies produce large-scale datasets with information on genes, proteins, metabolites and/or protein modification by measuring the global, qualitative, and quantitative changes at the molecular, cell, tissue, and individual levels [5, 6]. In the last decade, omics technologies have advanced tremendously and have been applied extensively in research. From a toxicological perspective, omics can efficiently and accurately generate relevant data on molecular dynamics associated with adverse outcomes [7]. Compared with previous approaches to precisely measure toxicant-induced molecular alteration, omics technologies have the potential to improve chemical safety assessment and reduce animal testing in regulatory toxicology [8].
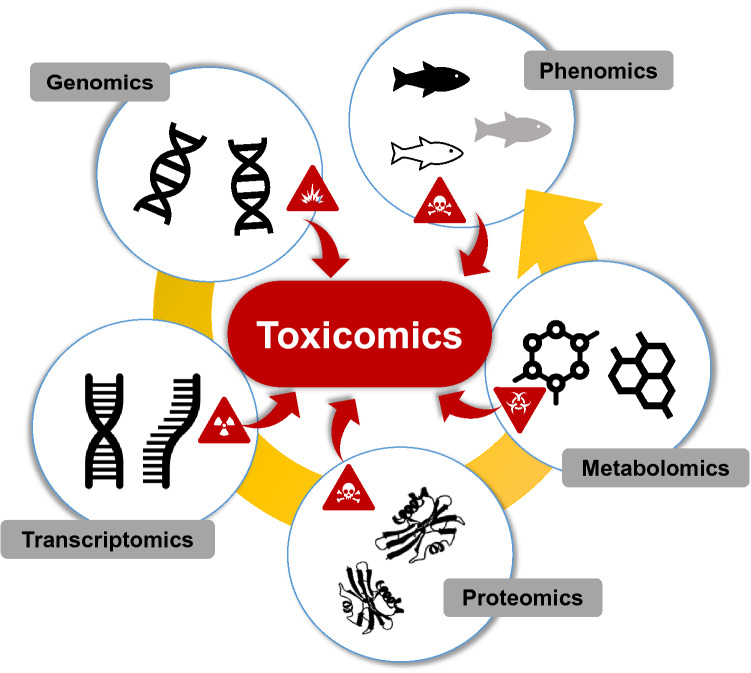
The term toxicomics is not defined in the existing literature, but this study proposes it to refer to the omics applied to toxicology. Specialized omics terms exist for toxicology research fields based on a single omics technology, such as toxicogenomics, toxicoproteomics, and toxicometabolomics [7–9]. These three terms can be collectively defined as toxicomics, just as omics includes genomics, epigenomics, transcriptomics, proteomics, and metabolomics (Fig. 1). Toxicogenomics generally refers to transcriptomics, the techniques used to study genomic-scale changes in RNA, which are mainly detected via a known set of differentially expressed target genes [5, 10]. Toxicoproteomics applies global protein expression technologies to toxicology testing and clinical research [11]. Toxicometabolomics is the systematic study of endogenous metabolites and biochemical processes in the cell, tissue, or organism to identify and characterize the end products of toxic reactions [12]. Genomics is based on the sequence information of genes and proteins, while transcriptomics, proteomics, and metabolomics provide information about the biological function of genetic information.
Fig. 1.
Overview of harmonized toxicomics. Molecules that change according to the exposure to toxic substances are collected based on each omics technology and the toxic mechanism is identified through integrative omics analysis
Zebrafish (Danio rerio) is a standard model animal for omics-based toxicity assessment. Its embryos and larvae are used as models in many toxicity tests. Embryos can be obtained in large quantities, and the fact that they are huge and transparent enables the easy visualization of toxicity-related changes. Zebrafish is a vertebrate that shares 71.4% genetic homology with humans. Besides, the major organs such as the heart, liver, and kidneys are comparable between both species [10]. This study describes and compares the omics- and zebrafish model-based Cd toxicity assessments. Based on the research findings from the zebrafish model, a new, promising research trend that uses the omics technology in the identification of the toxicity mechanism is emerging.
Cadmium toxicity in zebrafish
The toxicity results of Cd in zebrafish, which were identified without using omics technology, are summarized in Table 1. Cd mainly causes developmental abnormalities in zebrafish embryos and damages the nervous system. In particular, Cd inhibits the expression of glial fibrillary acidic protein; induces the expression of mpz, a specific myelin gene, which changes the glial cells; inhibits the expression of neurexin protein; and inhibits neuronal development [12, 13]. In the differentiation process, proneural gene transcription is reduced, thereby inhibiting neuron differentiation, which affects the activity of specific enzymes such as ATPase in the brain, and inhibits the estrogen signaling pathway [14–16].
Table 1.
Summary of toxic effects of cadmium in zebrafish
| Toxicity | Ref. | |
|---|---|---|
| Embryos | ||
| Liver | Hepatic lipid accumulation | [20] |
| Nerve | Neuroglia alterations | [12] |
| Increased ATPase activity in brain | [15] | |
| Reduction of neuronal differentiation and axonogenesis | [14] | |
| Interference of neural development | [13] | |
| Anti-estrogen in brain | [16] | |
| Abnormal somite patterning | [19] | |
| Myoskeletal retina | Eye hypoplasia and hypopigmentation | [18] |
| Cardiovascular organ | Heart edema and increased pericardial area | [17] |
| Activation of cell death pathway in olfactory epithelium | [40] | |
| Olfactory organ | Delay in hatching time | [17] |
| Others | Tail and axis malformation | [17] |
| Larvae | ||
| Nerve | Circadian rhythms disruption | [22] |
| Others | Cell death and structural alterations in olfactory epithelium | [21] |
| Adults | ||
| Liver | Carcinogenesis | [45] |
| Hepatic lipid accumulation | [23] | |
| Oxidative damage | [24, 25] | |
| Nerve | Oxidative damage | [25, 26] |
| Myoskeletal | Structural disorganization, disassembly of muscular myofibrils | [28] |
| Reproductive organ | Pair spawning reduction and teratogenicity | [27] |
| Ovary: oxidative damage | [25] | |
| Retina | Nerve fiber thickening and vacuolating | [29] |
Besides provoking neurological disorders, Cd delays hatching and damages the other organs [17]. The heavy metal inhibits the development of neural functions in pigment cells, causing abnormalities in the eyes. Furthermore, it leads to an abnormal expression of the genes necessary for the formation of the musculoskeletal system, causing larvae morphology [17–19]. It also induces apoptosis in the olfactory organs and damages the olfactory epithelium. In the liver, it alters the structure and function of HDL3, which is required for lipid metabolism [20].
Cd also inhibits metabolism and may damage the cardiovascular system [17]. In larvae, Cd affects the olfactory organs (in the same way it affects those of the embryo) and also causes an immune response which alters the circadian rhythm [21, 22]. In adults, the site of toxic reactions is similar to that of embryos, and the liver shows similar abnormalities in lipid metabolism [23]. The typical toxicity seen in adults is oxidative damage, which occurs in the liver, nerves, and ovaries [24–26], and exposure to Cd lowers the spawning success rate of female zebrafish and decreases the fertility of the born larvae [27]. It also causes structural abnormalities of the musculoskeletal system and retina [28, 29]. Reviewing the literature, we found that Cd exerts various toxic effects, including developmental disorders in several organs of zebrafish, and is involved in areas from transcription to enzyme activity. We then attempted to relate the omics result to the traditional Cd toxicity mechanisms.
Research technologies in omics
Omics research is subdivided into various fields such as genomics, transcriptomics, proteomics, and metabolomics (Fig. 1). Omics research emerged as a new tool in environmental toxicity assessment to identify the adverse outcome pathways, point of departure (PoD), etc. [6, 30]. Omics analysis is an effective technical tool for the qualitative and quantitative analysis of biological molecules which require sensitive analytical techniques [31]. When compared with conventional toxicity assessment, the advantage of omics technology is the ability to understand changes at the molecular level based on the abundance of useful information.
Genomics has provided fundamental information by identifying the nucleotide sequence, structure, and function of the genome [32]. Especially, the research applications of genomics in zebrafish are broad, and functional genomics plays a central role. At the DNA level, studies on mutagenesis typically modify zebrafish genes to produce specific diseases, such as Parkinson’s disease and cancer, or to utilize them in toxicity assessment models [33–35]. Epigenomics, (emerging toxicological indicator), investigates the inheritance phenomenon by altering the expression pattern of genes without changing the nucleotide sequence after birth [36]. It confirmed that DNA alterations (methylation, histone modifications, and miRNA expression) could be induced by external stimuli such as toxic chemical exposure [37].
In the translation process, mRNA is involved as a transcript, and the abundance of the transcriptome can be confirmed by analyzing the nucleotide sequence of the expressed mRNA through transcriptomics [38]. Microarray or new generation sequencing (NGS), such as RNA-sequencing technology, can provide information about the transcriptome [39].
NGS determined the mRNA expression of over 20,000 genes related to embryonic development in zebrafish much faster than any other methods [40]. Ontological validation methods, such as quantitative polymerase chain reaction, are commonly used to reinforce the reliability of volumonous data [41–44]. The function of genes corresponding to the expressed mRNAs or the relationship between genes can be associated with variations in metabolic pathways using databases such as the Kyoto Encyclopedia of Genes and Genomes pathways and Gene Ontology (GO) [45]. Meanwhile, progressive research is going on to derive PoD from the reduced zebrafish transcriptome approach and apply it to regulatory toxicology [46].
An example is an experiment that evaluated the toxicity in a zebrafish model through transcriptomics. Dibenzazepine, one of the representative polyhalogenated carbazoles that are structurally similar to dioxin, disrupted the aryl hydrocarbon receptor activation genes, such as AhR1 and CYP1A [47]. As a result, the metabolic pathway related to protein processing in the endoplasmic reticulum (ER) and taste transduction was disrupted. Bisphenol A, a well-known environmental hormone, was identified for the adverse effects of retinol and glutathione metabolism and lipid transport by the upregulation of steroid hormone biosynthesis genes (such as cyp19a1b) and lipid transport protein genes (apoa1a, apobb1, and apoa4a) [48].
The proteome encompasses all the products of gene translation, which have a plethora of structures and functions [49]. The goal of proteomics is to identify the structure and function of proteins and their modifications [50, 51]. Toxicoproteomics applies global protein expression analysis technologies to toxicological and clinical research [11]. Especially, mass spectrometry (MS)-based toxicoproteomics has measured the quantitative changes in proteins, which are the adverse effects of toxicants, by identifying the protein through MS [52]. To apply MS, the samples must be digested to denature the protein and label the specific peptide site. Three labeling methods are available: (1) stable isotope labeling with amino acids in cell culture (SILAC), (2) isobaric tags for relative and absolute quantitation (iTRAQ), and (3) tandem mass tag (TMT) [53–55]. MS-based methods can identify thousands of proteins within hours using a single analysis by overcoming the limitations of the classical methods such as Western blot assay [56, 57]. These techniques can help identify differentially expressed proteins, and the quantitative results can be analyzed using bioinformatics tools to obtain information on the motif and protein–protein interaction networks [58].
There are previous reports in which the mechanism of toxicity has been elucidated through proteomic studies in a zebrafish model. In 3,4-dichloroaniline toxicity, non-detachment of the tail, lack of somite formation, absence of heartbeat, pericardial edema, abnormal curvature of the spine, and yolk sac edema were observed. These effects were presumed to be due to the disruption of hormone-related pathways and lipid metabolism [59]. Following proteomic analysis, β-methyl-amino-l-alanine showed increased protein biosynthesis and RNA processing proteins, such as eIF3a/c and CPSF5, and decreased 40S ribosomal protein S21 and phenylalanine-tRNA ligase α, which are linked to the disruption of pathways for glutamate receptor activity/recycling, ER stress, protein biosynthesis, and neural cell death [60].
Metabolites (endogenous small-molecule substances) are products of physiological processes. Metabolomics aims to obtain meaningful information on metabolic processes by analyzing changes in organisms at the metabolome level [61]. The method for metabolite analysis is either called target analysis or non-target analysis depending on whether the specific metabolite is targeted [62, 63]. Based on the research objectives, the metabolites are extracted by proper pretreatment methods, such as liquid–liquid extraction or solid-phase extraction, and then identified through MS-based analysis [64]. Since even very low concentrations (< pg ml−1) of metabolites can be detected, changes in the molecular level can be understood using a single analysis without the need for several detection kits [65]. Changes in the identified metabolite levels are visualized by statistical methods such as hierarchical clustering analysis, principal component analysis, and partial least squares discriminant analysis [66]. Similar to the other omics described thus far, toxicometabolomics analyzes the metabolic pathways and summarizes the results to obtain clear information on changes due to toxicity [67].
In previous studies, the mechanism of toxicity was identified through metabolomic analysis. Haloperidol, a common butyrophenone-derived antipsychotic drug, was identified to have adverse effects on vitamin B12 metabolism, neurotransmission, insulin signaling, and mammalian target of rapamycin pathway, which are linked to pericardial edema, curvature of the spinal cord, and heart sac edema [68]. Perfluorooctanoic acid, an alternative of perfluorooctane sulfonate, showed similar adverse effects on peroxisome proliferator-activated receptor-γ-regulated signaling pathways and mitochondrial pathways, and hepatoxicity and neurotoxicity were observed in exposed subjects [69].
Since changes in biological substances are closely related to biological processes, results derived from each omics need to be correlated with those from other omics [70, 71]. Hence, the multi-omics approach is gaining attention; it can satisfy this need and provide more reliable biomarkers, thanks to a wide field of view encompassing several layers of omics [72]. The overlapping goal of this multi-omics approach is to identify the levels of RNA, proteins, and metabolites through non-target analysis and link them to physiological changes [73]. Besides, a study employing multi-omics obtained significant information even at low concentrations, a level at which embryos do not show lethality [74]. Even at low concentrations, substances such as environmental hormones, which have similar toxic mechanisms, showed disruptive effects on transcriptomic, proteomic, and metabolomic profile in zebrafish in similar patterns. Therefore, it is reasonable to expect that detecting in vivo changes at the level of omics will allow the evaluation and prediction of toxicity in the near future [75, 76].
Omics-based cadmium toxicity
We confirmed the results from previous studies on zebrafish exposed to Cd with omics technologies, namely a detoxification mechanism in addition to the toxic mechanism of Cd (Table 2). The transcription of proteins mainly involved in metal homeostasis was increased. Exposition to Cd increases the expression of the mt2 gene, which codes for the metallothionein-2. This protein plays a role in homeostasis and detoxification of heavy metals and is a molecular marker for metal contamination [77]. Besides, hsp70.1 and hsp70l, whose expression is increased, induce the synthesis of heat shock protein 70, a chaperone [78]. Furthermore, exposure to Cd increases the expression or activity of antioxidant glutathione-S-transferase (GST), catalase (CAT), and glutathione reductase (GR) [79, 80]. Specifically, concerning GST, the expression of gstm3 at 48 hpf, and gstp1and gstp2 at 96 hpf increased depending on the CdCl2 concentration (at 0.9, 1.8, and 3.3 mg/L) and decreased when exposed to 4 µM for 5 days. CAT also showed a complex pattern. When exposed to 4 µM CdCl2 for 5 days, the expression decreased and the activity was reported to increase, but when exposed to 1.78 µM Cd for 7 days, the activity decreased [80, 81]. These omics approach results confirmed that additional studies on GST and CAT are needed.
Table 2.
Toxicity of Cd in zebrafish based on omics analysis
| Toxicity | Omics | Ref. | |
|---|---|---|---|
| Detoxification | Metal ion binding gene (mt2, klf11a, klf11b) upregulation | Transcriptomics | [79, 88] |
| Matrix metalloproteinases gene (mmp9, mmp13a) upregulation | |||
| Detoxication gene (gstm3, gstp1, gstp2) upregulation | |||
| Oxidant stress gene (hsp70.1, hsp70l, mt2) upregulation | |||
| Metallothionein, glutathione reductase upregulation | Proteomics | [80] | |
| Nerve | uqcrfs1 and rpsa upregulation | Proteomics | [84] |
| Related to neuromast gene (cldnb, stat3) upregulation | Transcriptomics | [88] | |
| Liver: oxidative stress | Catalase downregulation | Transcriptomics | [81] |
| ATP7A and metallothionein downregulation | |||
| Downregulation of gene expression because of GpG methylation of HSP70 upregulation | |||
| GSH, SOD, catalase downregulation | Proteomics | [80] | |
| GSH, SOD downregulation | |||
| Immune | IL-1β, iNOS, TNF-α downregulation | Transcriptomics | [81] |
| Metal homeostasis | Calcium homeostasis gene (stc1l) downregulation | Transcriptomics | [79] |
| Skeletal muscle |
Pro-apoptotic gene (bax, mt1) upregulation Cytoglobin gene (cyt) upregulation Pyruvate carboxylase gene (pyc) upregulation ABC transporter gene (tap) upregulation |
Transcriptomics | [87] |
The omics approach results on the toxic effects of exposure to Cd are as follows: in zebrafish, Cd inhibits the activity and synthesis of antioxidant enzymes and proteins. Consequently, reactive oxygen species (ROS) in various organs cause several problems. For instance, the liver accumulates excessive fat through lipid peroxidation [82]. Cardiac toxicity of Cd is associated with the decreased expression of stanniocalcin 1, which is involved in the regulation of calcium and phosphate homeostasis [79]. Since stanniocalcin 1 protects the cells from ventricular dysfunction and ROS hyperplasia, its downregulation by Cd leads to heart edema or increased pericardial area [83]. Uqcrfs1, which is highly expressed in the Cd-induced zebrafish brain and encodes ubiquinol-cytochrome C reductase, Rieske iron-sulfur polypeptide I, constitutes an electron transport system and is involved in ATP synthesis. Abnormal expression of uqcrfs1 affects the electron transport system, and it is also used as a biomarker because it is related to cancerous conditions [52, 84, 85]. In the musculoskeletal system, the pro-apoptotic gene bax is upregulated by Cd [66]. Indeed, exposure to Cd causes abnormal apoptosis in zebrafish embryos [86]. Considering these two results together, the abnormal apoptosis caused by Cd may affect the development of nerves and muscles in embryos.
More toxicity can be inferred based on what was learned through omics research. We believe that additional studies can be conducted on immune adverse reactions based on the decreased expression of IL-1β, iNOS, and TNF-α and on blood coagulation disorders based on the overexpression of the cytoglobin gene (cyt) [81, 87].
Perspectives
In this paper, we investigated the toxicity that occurs when zebrafish are exposed to Cd and linked it to gene and protein expression. Cd mainly causes abnormalities in the development of embryos and larvae, and toxicity was also observed in the liver and nervous system. Omics research provided additional information about the toxicity of Cd. Regarding Cd-induced oxidative stress, toxicity was traditionally determined by evaluating the expression of only designated markers related to ROS after exposure to Cd [24, 25], whereas proteomics-based studies suggested proteins that respond to the ‘Response to Stress category’ even with designated markers [60]. A detailed mechanism of Cd-derived oxidative stress was suggested, providing insights for further research.
Although this study compared the Cd toxicity in the zebrafish model, the toxicity assessment in adults, larvae, and embryos was subdivided, and due to the difference in detailed methods in the omics-based and non-omics-based experiments, accurate comparisons could not be made. Since the two approaches have their own strengths and weaknesses and research goals, it may not be appropriate to consider comparative advantage. Overall, the technical basis of omics research is a great advantage when exploring detailed mechanisms for toxic phenomena. It goes beyond exploring a few mechanisms at a time and allows the quantification of several unspecified markers simultaneously. Since the accuracy and reproducibility of the omics research technology has rapidly increased in recent years, its utility in toxicity studies will surely receive attention. This study presents the results of Cd toxicity evaluation in the zebrafish model and the trend of toxicity evaluation research based on Omics technology. Although the number of omics-based studies is still insufficient, we expect the development of omics technologies to soon allow further clarification of the Cd toxicity mechanisms.
Acknowledgements
This study was partially supported by the National Institute of Environmental Research, Republic of Korea.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to disclose.
Footnotes
Eun Ki Min and Ahn Na Lee contributed equally to this work.
References
- 1.Nordberg GF. Historical perspectives on cadmium toxicology. Toxicol Appl Pharmacol. 2009;238:192–200. doi: 10.1016/j.taap.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Choi JY, Choi UK, Baek SH, Choi HB, Lee JH. A study on chemical compositions of sediment and surface water in nakdong river for tracing contaminants from mining activities. J Korean Earth Sci Soc. 2016;37:211–217. doi: 10.5467/JKESS.2016.37.4.211. [DOI] [Google Scholar]
- 3.Kang D, Kim JE. Fine ultrafine and yellow dust: emerging health problems in Korea. J Korean Med Sci. 2014;29:621–622. doi: 10.3346/jkms.2014.29.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho MJ, Choi H, Kim HJ, Youn HJ. Monitoring and risk assessment of heavy metals in perennial root vegetables. Korean J Environ Agric. 2016;35:55–61. doi: 10.5338/kjea.2016.35.1.07. [DOI] [Google Scholar]
- 5.Zhang X, Xia P, Wang P, Yang J, Baird DJ. Omics advances in ecotoxicology. Environ Sci Technol. 2018;52:3842–3851. doi: 10.1021/acs.est.7b06494. [DOI] [PubMed] [Google Scholar]
- 6.Brockmeier EK, Hodges G, Hutchinson TH, Butler E, Hecker M, et al. The role of omics in the application of adverse outcome pathways for chemical risk assessment. Toxicol Sci. 2017;158:252–262. doi: 10.1093/toxsci/kfx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canzler S, Schor J, Busch W, Schubert K, Rolle-Kampczyk UE, et al. Prospects and challenges of multi-omics data integration in toxicology. Arch Toxicol. 2020;94:371–388. doi: 10.1007/s00204-020-02656-y. [DOI] [PubMed] [Google Scholar]
- 8.Buesen R, Chorley BN, da Silva LB, Daston G, Deferme L, et al. Applying omics technologies in chemicals risk assessment: report of an ECETOC workshop. Regul Toxicol Pharmacol. 2017;91:S3–S13. doi: 10.1016/j.yrtph.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrison N, Cochrane G, Faruque N, Tatusova T, Tateno Y, et al. Concept of Sample in OMICS Technology. J Integr Biol. 2006;10:127–137. doi: 10.1089/omi.2006.10.127. [DOI] [PubMed] [Google Scholar]
- 10.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubinska-Magiera M, Daczewska M, Lewicka A, Migocka-Patrzalek M, Niedbalska-Tarnowska J, et al. Zebrafish: a model for the study of toxicants affecting muscle development and function. Int J Mol Sci. 2016;17:1941. doi: 10.3390/ijms17111941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monaco A, Grimaldi MC, Ferrandino I. Neuroglial alterations in the zebrafish brain exposed to cadmium chloride. J Appl Toxicol. 2016;36:1629–1638. doi: 10.1002/jat.3328. [DOI] [PubMed] [Google Scholar]
- 13.Tu H, Fan C, Chen X, Liu J, Wang B, et al. Effects of cadmium, manganese, and lead on locomotor activity and neurexin 2a expression in zebrafish. Environ Toxicol Chem. 2017;36:2147–2154. doi: 10.1002/etc.3748. [DOI] [PubMed] [Google Scholar]
- 14.Chow ES, Hui MN, Lin CC, Cheng SH. Cadmium inhibits neurogenesis in zebrafish embryonic brain development. Aquat Toxicol. 2008;87:157–169. doi: 10.1016/j.aquatox.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 15.Senger MR, Rosemberg DB, Rico EP, de Bem AM, Dias RD, et al. In vitro effect of zinc and cadmium on acetylcholinesterase and ectonucleotidase activities in zebrafish (Danio rerio) brain. Toxicol In Vitro. 2006;20:954–958. doi: 10.1016/j.tiv.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Chouchene L, Pellegrini E, Gueguen MM, Hinfray N, Brion F, et al. Inhibitory effect of cadmium on estrogen signaling in zebrafish brain and protection by zinc. J Appl Toxicol. 2016;36:863–871. doi: 10.1002/jat.3285. [DOI] [PubMed] [Google Scholar]
- 17.Fraysse B, Mons R, Garric J. Development of a zebrafish 4-day embryo-larval bioassay to assess toxicity of chemicals. Ecotoxicol Environ Saf. 2006;63:253–267. doi: 10.1016/j.ecoenv.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Zhou XY, Ma XF, Liu JX. Mechanisms of cadmium-caused eye hypoplasia and hypopigmentation in zebrafish embryos. Aquat Toxicol. 2015;167:68–76. doi: 10.1016/j.aquatox.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Hen Chow ES, Cheng SH. Cadmium affects muscle type development and axon growth in zebrafish embryonic somitogenesis. Toxicol Sci. 2003;73:149–159. doi: 10.1093/toxsci/kfg046. [DOI] [PubMed] [Google Scholar]
- 20.Kim JY, Kim SJ, Bae MA, Kim JR, Cho KH. Cadmium exposure exacerbates severe hyperlipidemia and fatty liver changes in zebrafish via impairment of high-density lipoproteins functionality. Toxicol In Vitro. 2018;47:249–258. doi: 10.1016/j.tiv.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Matz CJ, Krone PH. Krone Cell death stress-responsive transgene activation and deficits in the olfactory system of larval zebrafish following cadmium exposure. Environ Sci Technol. 2007;41:5143–5148. doi: 10.1021/es070452c. [DOI] [PubMed] [Google Scholar]
- 22.Xiao B, Chen TM, Zhong Y. Possible molecular mechanism underlying cadmium-induced circadian rhythms disruption in zebrafish. Biochem Biophys Res Commun. 2016;481:201–205. doi: 10.1016/j.bbrc.2016.081. [DOI] [PubMed] [Google Scholar]
- 23.Pan YX, Luo Z, Zhuo MQ, Wei CC, Chen GH, et al. Oxidative stress and mitochondrial dysfunction mediated Cd-induced hepatic lipid accumulation in zebrafish Danio rerio. Aquat Toxicol. 2018;199:12–20. doi: 10.1016/j.aquatox.2018.03.017b. [DOI] [PubMed] [Google Scholar]
- 24.Zheng JL, Yuan SS, Wu CW, Li WY. Chronic waterborne zinc and cadmium exposures induced different responses towards oxidative stress in the liver of zebrafish. Aquat Toxicol. 2016;177:261–268. doi: 10.1016/j.aquatox.2016.06.001c. [DOI] [PubMed] [Google Scholar]
- 25.Zheng JL, Yuan SS, Wu CW, Lv ZM. Acute exposure to waterborne cadmium induced oxidative stress and immunotoxicity in the brain ovary and liver of zebrafish (Danio rerio) Aquat Toxicol. 2016;180:36–44. doi: 10.1016/j.aquatox.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Zheng JL, Yuan SS, Wu CW, Lv ZM, Zhu AY. Circadian time-dependent antioxidant and inflammatory responses to acute cadmium exposure in the brain of zebrafish. Aquat Toxicol. 2017;182:113–119. doi: 10.1016/j.aquatox.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Wu SM, Tsai PJ, Chou MY, Wang WD. Effects of maternal cadmium exposure on female reproductive functions gamete quality and offspring development in zebrafish (Danio rerio) Arch Environ Contam Toxicol. 2013;65:521–536. doi: 10.1007/s00244-013-9909-1. [DOI] [PubMed] [Google Scholar]
- 28.Avallone B, Agnisola C, Cerciello R, Panzuto R, Simoniello P, et al. Structural and functional changes in the zebrafish (Danio rerio) skeletal muscle after cadmium exposure. Cell Biol Toxicol. 2015;31:273–283. doi: 10.1007/s10565-015-9310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avallone B, Crispino R, Cerciello R, Simoniello P, Panzuto R, et al. Cadmium effects on the retina of adult Danio rerio. C R Biol. 2020;338:40–47. doi: 10.1016/j.crvi.2014.005. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Wang Z, Xia P, Zhang X. Concentration-dependent transcriptome of zebrafish embryo for environmental chemical assessment. Chemosphere. 2020;245:125632. doi: 10.1016/j.chemosphere.2019.125632. [DOI] [PubMed] [Google Scholar]
- 31.Yan SC, Chen ZF, Zhang H, Chen Y, Qi Z, et al. Evaluation and optimization of sample pretreatment for GC/MS-based metabolomics in embryonic zebrafish. Talanta. 2020;207:120260. doi: 10.1016/j.talanta.2019.120260. [DOI] [PubMed] [Google Scholar]
- 32.Wixon J. Featured organism: Danio rerio, the zebrafish. Yeast. 2000;17:225–231. doi: 10.1002/1097-0061(20000930)17:3<225::AID-YEA34>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alestrom P, Holter JL, Nourizadeh-Lillabadi R. Zebrafish in functional genomics and aquatic biomedicine. Trends Biotechnol. 2005;24:15–21. doi: 10.1016/j.tibtech.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Bretaud S, Allen C, Ingha PW, Bandmann O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J Neurochem. 2007;100:1626–1635. doi: 10.1111/j.1471-4159.2006.04291.x. [DOI] [PubMed] [Google Scholar]
- 35.Shepard JL, Amatruda JF, Stern HM, Subramanian A, Finkelstein D, et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. PNAS. 2005;102:13194–13199. doi: 10.1073/pnas.0506583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams TD, Mirbahai L, Chipman JK. The toxicological application of transcriptomics and epigenomics in zebrafish and other teleosts. Brief Funct Genomics. 2014;13:157–171. doi: 10.1093/bfgp/elt053. [DOI] [PubMed] [Google Scholar]
- 37.Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Gomez-Skarmeta JL. The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods. 2013;62:207–215. doi: 10.1016/j.ymeth.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Zheng M, Lu J, Zhao D. Toxicity and transcriptome sequencing (RNA-seq) analyses of adult zebrafish in response to exposure carboxymethyl cellulose stabilized iron sulfide nanoparticles. Sci Rep. 2018;8:8083. doi: 10.1038/s41598-018-26499-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magalhães JP, Finch CE, Janssens G. Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions. Ageing Res Rev. 2010;9:315–323. doi: 10.1016/j.arr.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White RJ, Collins JE, Sealy IM, Wali N, Dooley CM, et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. eLIFE. 2017;6:e30860. doi: 10.7554/eLife.30860.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan Y, Zhang Q, Zhang ZJ, Song BF, Wang XM, et al. Transcriptome analysis reveals a ribosome constituents disorder involved in the RPL5 downregulated zebrafish model of Diamond-Blackfan anemia. BMC Med Genomics. 2016;9:13. doi: 10.1186/s12920-016-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ordas A, Hegedus Z, Henkel CV, Stockhammer OW, Butler D, et al. Deep sequencing of the innate immune transcriptomic response of zebrafish embryos to Salmonella infection. Fish Shellfish Immunol. 2011;31:716–724. doi: 10.1016/j.fsi.2008.022. [DOI] [PubMed] [Google Scholar]
- 43.Hartig EI, Zhu S, King BL, Coffman JA. Cortisol-treated zebrafish embryos develop into pro-inflammatory adults with aberrant immune gene regulation. Biol Open. 2016;5:1134–1141. doi: 10.1242/bio.020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sreenivasan R, Cai M, Bartfai R, Wang X, Christoffels A, et al. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS ONE. 2008;3:e1791. doi: 10.1371/journal.pone.0001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veneman WJ, Spaink HP, Brun NR, Bosker T, Vijver MG. Pathway analysis of systemic transcriptome responses to injected polystyrene particles in zebrafish larvae. Aquat Toxicol. 2017;190:112–120. doi: 10.1016/j.aquatox.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Wang PP, Xia P, Yang JH, Wang ZH, Peng Y, et al. A reduced transcriptome approach to assess environmental toxicants using zebrafish embryo. Test Environ Sci Technol. 2018;52:821–830. doi: 10.1021/acs.est.7b04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji C, Yan L, Chen Y, Yue S, Dong Q, et al. Evaluation of the developmental toxicity of 2,7-dibromocarbazole to zebrafish based on transcriptomics assay. J Hazard Mater. 2019;368:514–522. doi: 10.1016/j.jhazmat.2019.01.079. [DOI] [PubMed] [Google Scholar]
- 48.Martíneza RE, Codina AH, Nogareda LO, Villanueva E, Barata C, et al. Dose-dependent transcriptomic responses of zebrafish eleutheroembryos to Bisphenol A. Environ Pollut. 2018;243:988–997. doi: 10.1016/j.envpol.2018.09.043. [DOI] [PubMed] [Google Scholar]
- 49.Khoury G, Baliban R, Floudas C. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. 2011 doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lippok B, Song S, Driever W. Pou5f1 protein expression and posttranslational modification during early zebrafish development. Dev Dyn. 2014;243:468–477. doi: 10.1002/dvdy.24079. [DOI] [PubMed] [Google Scholar]
- 51.Wang M, Chan LL, Si M, Hong H, Wang D. Proteomic analysis of hepatic tissue of zebrafish (Danio rerio) experimentally exposed to chronic microcystin-LR. Toxicol Sci. 2010;113:60–69. doi: 10.1093/toxsci/kfp248. [DOI] [PubMed] [Google Scholar]
- 52.Ellinger J, Gromes A, Poss M, Bruggemann M, Schmidt D, et al. Systematic expression analysis of the mitochondrial complex III subunits identifies UQCRC1 as biomarker in clear cell renal cell carcinoma. Oncotarget. 2016;7:86490–86499. doi: 10.18632/oncotarget.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westman-Brinkmalm A, Abramsson A, Pannee J, et al. SILAC zebrafish for quantitative analysis of protein turnover and tissue regeneration. J Proteomics. 2011;75:425–434. doi: 10.1016/j.jprot.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Lü A, Hu X, Wang Y, Shen X, Li X, et al. (TRAQ analysis of gill proteins from the zebrafish (Danio rerio) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2014;36:229–239. doi: 10.1016/j.fsi.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Wu Y, Lou QY, Ge F, Xiong Q, et al. Quantitative proteomics analysis reveals novel targets of miR-21 in zebrafish embryos. Sci Rep. 2017;7:4022. doi: 10.1038/s41598-017-04166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mann M. Can proteomics retire the western blot? J Proteome Res. 2008;7:3065. doi: 10.1021/pr800463v7(5):981-94. [DOI] [PubMed] [Google Scholar]
- 57.Lucitt MB, Price TS, Pizarro A, Wu W, Yocum AK, et al. Analysis of the zebrafish proteome during embryonic development. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M700382-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao Y, Lee H, Kwon OK, Kim KT, Lee S, et al. Global proteomic analysis of lysine succinylation in Zebrafish (Danio rerio) J Proteome Res. 2019;18:3762–3769. doi: 10.1021/acs.jproteome.9b00462. [DOI] [PubMed] [Google Scholar]
- 59.Vieira LR, Hissa DC, de Souza TM, Sá CA, Evaristo JAM, et al. Proteomics analysis of zebrafish larvae exposed to 3, 4-dichloroaniline using the fish embryo acute toxicity test. Environ Toxicol. 2020;35:849–860. doi: 10.1002/tox.22921. [DOI] [PubMed] [Google Scholar]
- 60.Frøyset AK, Khan EA, Fladmark KE. Quantitative proteomics analysis of zebrafish exposed to sub-lethal dosages of β-methyl-amino-l-alanine (BMAA) Sci Rep. 2016;6:29631. doi: 10.1038/srep29631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang SS, Benskin JP, Chandramouli B, Butler H, Helbing CC, et al. Xenobiotics produce distinct metabolomic responses in zebrafish larvae (Danio rerio) Environ Sci Technol. 2016;50:6526–6535. doi: 10.1021/acs.est.6b01128. [DOI] [PubMed] [Google Scholar]
- 62.Xu N, Mu P, Yin Z, Jia Q, Yang S, et al. Analysis of the enantioselective effects of PCB95 in zebrafish (Danio rerio) embryos through targeted metabolomics by UPLC-MS/MS. PLoS ONE. 2016;11:e0160584. doi: 10.1371/journal.pone.0160584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sotto RB, Medriano CD, Cho Y, Kim H, Chung IY, et al. Sub-lethal pharmaceutical hazard tracking in adult zebrafish using untargeted LC-MS environmental metabolomics. J Hazard Mater. 2017;339:63–72. doi: 10.1016/j.jhazmat.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol Biosyst. 2012;8:470–481. doi: 10.1039/c1mb05350g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouyang Y, Tong H, Luo P, Kong H, Xu Z, et al. A high throughput metabolomics method and its application in female serum samples in a normal menstrual cycle based on liquid chromatography–mass spectrometry. Talanta. 2018;185:483–490. doi: 10.1016/j.talanta.2018.03.087. [DOI] [PubMed] [Google Scholar]
- 66.Mishra P, Gong Z, Kelly BC. Assessing biological effects of fluoxetine in developing zebrafish embryos using gas chromatography-mass spectrometry based metabolomics. Chemosphere. 2017;188:157–167. doi: 10.1016/j.chemosphere.2017.08.149. [DOI] [PubMed] [Google Scholar]
- 67.Ortiz-Villanueva E, Navarro-Martin L, Jaumot J, Benavente F, Sanz-Nebot V. Metabolic disruption of zebrafish (Danio rerio) embryos by bisphenol A. An integrated metabolomic and transcriptomic approach. Environ Pollut. 2017;231:22–36. doi: 10.1016/j.envpol.2017.07.095. [DOI] [PubMed] [Google Scholar]
- 68.Lin YC, Huang C, Huang HC, Liao MT, Lai YH. Metabolomics profiling of haloperidol and validation of thromboxane-related signaling in the early development of zebrafish. Biochem Biophys Res Commun. 2019;513:608–615. doi: 10.1016/j.bbrc.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 69.Gebreab KY, Eeza MNH, Bai T, Zuberi Z, Matysik J, et al. Comparative toxicometabolomics of perfluorooctanoic acid (PFOA) and next-generation perfluoroalkyl substances. Environ Pollut. 2020;265:114928. doi: 10.1016/j.envpol.2020.114928. [DOI] [PubMed] [Google Scholar]
- 70.Elie MR, Choi J, Nkrumah-Elie YM, Gonnerman GD, Stevens JF, et al. Metabolomic analysis to define and compare the effects of PAHs and oxygenated PAHs in developing zebrafish. Environ Res. 2015;140:502–510. doi: 10.1038/nprot.2017.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ortiz-Villanueva E, Jaumot J, Martinez R, Navarro-Martin L, Pina B, et al. Assessment of endocrine disruptors effects on zebrafish (Danio rerio) embryos by untargeted LC-HRMS metabolomic analysis. Sci Total Environ. 2018;635:156–166. doi: 10.1016/j.scitotenv.2018.03.369. [DOI] [PubMed] [Google Scholar]
- 72.Martinez R, Navarro-Martin L, Luccarelli C, Codina AE, Raldua D, et al. Unravelling the mechanisms of PFOS toxicity by combining morphological and transcriptomic analyses in zebrafish embryos. Sci Total Environ. 2019;674:462–471. doi: 10.1016/j.scitotenv.2019.04.200. [DOI] [PubMed] [Google Scholar]
- 73.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weckwerth W. Metabolomics in systems biology. Annu Rev Plant Biol. 2003;54:669–689. doi: 10.1146/annurev.arplant.54.031902.135014. [DOI] [PubMed] [Google Scholar]
- 75.Audouze K, Sarigiannis D, Alonso-Magdalena P, Brochot C, Casas M, et al. Integrative strategy of testing systems for identification of endocrine disruptors inducing metabolic disorders—an introduction to the OBERON project. Int J Mol Sci. 2020;21:2988. doi: 10.3390/ijms21082988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kroeger M. How omics technologies can contribute to the ‘3R’ principles by introducing new strategies in animal testing. Trends Biotechnol. 2006;24:343–346. doi: 10.1016/j.tibtech.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 77.Chen WY, John JA, Lin CH, Lin HF, Wu SC, et al. Expression of metallothionein gene during embryonic and early larval development in zebrafish. Aquat Toxicol. 2004;69:215–227. doi: 10.1016/j.aquatox.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 78.Feder ME, Hofmann GE. Heat-shock proteins molecular chaperones and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 79.Sonnack L, Klawonn T, Kriehuber R, Hollert H, Schafers C, et al. Concentration dependent transcriptome responses of zebrafish embryos after exposure to cadmium cobalt and copper. Comp Biochem Physiol Part D Genomics Proteomics. 2017;24:29–40. doi: 10.1016/j.cbd.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 80.Zhu JY, Chan KM. Mechanism of cadmium-induced cytotoxicity on the ZFL zebrafish liver cell line. Metallomics. 2012;4:1064–1076. doi: 10.1039/c2mt20134h. [DOI] [PubMed] [Google Scholar]
- 81.Zheng JL, Guo SN, Yuan SS, Xia H, Zhu QL, et al. Preheating mitigates cadmium toxicity in zebrafish livers: evidence from promoter demethylation gene transcription to biochemical levels. Aquat Toxicol. 2017;190:104–111. doi: 10.1016/j.aquatox.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 82.Huang X, Li Y, Wang T, Liu H, Shi J, et al. Evaluation of the oxidative stress status in zebrafish (Danio rerio) liver induced by three typical organic UV filters (BP-4 PABA and PBSA) Int J Environ Res Public Health. 2020;17:651. doi: 10.3390/ijerph17020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawabata M, Umemoto N, Shimada Y, Nishimura Y, Zhang B, et al. Downregulation of stanniocalcin 1 is responsible for sorafenib-induced cardiotoxicity. Toxicol Sci. 2015;143:374–384. doi: 10.1093/toxsci/kfu235. [DOI] [PubMed] [Google Scholar]
- 84.Ling XP, Lu YH, Huang HQ. Differential protein profile in zebrafish (Danio rerio) brain under the joint exposure of methyl parathion and cadmium. Environ Sci Pollut Res Int. 2012;19:3925–3941. doi: 10.1007/s11356-012-1037-3. [DOI] [PubMed] [Google Scholar]
- 85.Kaneko SJ, Gerasimova T, Smith ST, Lloyd KO, Suzumori K, et al. CA125 and UQCRFS1 FISH studies of ovarian carcinoma. Gynecol Oncol. 2003;90:29–36. doi: 10.1016/s0090-8258(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 86.Chan PK, Cheng SH. Cadmium-induced ectopic apoptosis in zebrafish embryos. Arch Toxicol. 2003;77:69–79. doi: 10.1007/s00204-002-0411-1. [DOI] [PubMed] [Google Scholar]
- 87.Gonzalez P, Baudrimont M, Boudou A, Bourdineaud JP. Comparative effects of direct cadmium contamination on gene expression in gills liver skeletal muscles and brain of the zebrafish (Danio rerio) Biometals. 2006;19:225–235. doi: 10.1007/s10534-005-5670-x. [DOI] [PubMed] [Google Scholar]
- 88.Sonnack L, Klawonn T, Kriehuber R, Hollert H, Schafers C, et al. Comparative analysis of the transcriptome responses of zebrafish embryos after exposure to low concentrations of cadmium cobalt and copper. Comp Biochem Physiol Part D Genomics Proteomics. 2018;25:99–108. doi: 10.1016/j.cbd.2017.12.001. [DOI] [PubMed] [Google Scholar]