Abstract
Objective
Sweet taste receptors (STR) are expressed in the gut and other extra-oral tissues, suggesting that STR-mediated nutrient sensing may contribute to human physiology beyond taste. A common variant (Ile191Val) in the TAS1R2 gene of STR is associated with nutritional and metabolic outcomes independent of changes in taste perception. It is unclear whether this polymorphism directly alters STR function and how it may contribute to metabolic regulation.
Methods
We implemented a combination of in vitro biochemical approaches to decipher the effects of TAS1R2 polymorphism on STR function. Then, as proof-of-concept, we assessed its effects on glucose homeostasis in apparently healthy lean participants.
Results
The Ile191Val variant causes a partial loss of function of TAS1R2 through reduced receptor availability in the plasma membrane. Val minor allele carriers have reduced glucose excursions during an OGTT, mirroring effects previously seen in mice with genetic loss of function of TAS1R2. These effects were not due to differences in beta-cell function or insulin sensitivity.
Conclusions
Our pilot studies on a common TAS1R2 polymorphism suggest that STR sensory function in peripheral tissues, such as the intestine, may contribute to the regulation of metabolic control in humans.
Keywords: Sweet taste receptors, TAS1R2, rs35874116, Polymorphism, OGTT, Intestine, rs9701796
Abbreviations: sweet taste receptors, (STR); extracellular domain, (ECD); ligand binding domain, (LBD); fluorescence lifetime imaging microscopy, (FLIM); plasma membrane, (PM)
Highlights
-
•
Sweet taste receptors (STR) have functional roles beyond taste perception.
-
•
The TAS1R2 gene of STR is highly polymorphic, suggesting nutrient-dependent adaptive roles.
-
•
The Ile191Val variant of TAS1R2 gene causes a partial STR loss-of-function.
-
•
Val carriers have reduced glucose excursions mirroring TAS1R2-KO mice.
1. Introduction
Sweet taste receptors (STR) traditionally mediate sweet nutrient sensing on the tongue. These G-protein coupled receptors (GPCRs) are also expressed in a variety of other cells, suggesting a broader chemosensory function. For instance, STR regulate insulin [1,2] and incretin [3] secretion in mice and were shown to affect glucose absorption [4] and the development of diet-induced obesity [5]. Therefore, STR likely function as sugar sensors to coordinate adaptive responses to nutrient availability, but whether they contribute to human physiology and how, beyond taste perception, is still unknown.
STR belong to the T1R family of GPCRs and function as obligate heterodimers between TAS1R2 and TAS1R3 receptors. Notably, STR are highly polymorphic, with TAS1R2 being the most diverse, suggesting potential adaptive roles in response to nutrient availability [6]. Several variants in the TAS1R2 gene [7] were shown to alter sweet taste sensitivity or preference. Interestingly, the rs35874116 variant of the TAS1R2, which causes a nonsynonymous substitution (Ile191Val), is associated with sugar and carbohydrate intake [[8], [9], [10]], BMI [8], fasting insulin [8], and risk of hypertriglyceridemia [10]. Strikingly, these associations cannot be attributed to differences in taste perception [7], suggesting extra-oral contributions of STR signaling. The Ile191Val substitution resides in the N-terminal extracellular domain (ECD), which includes the ligand binding domain (LBD) and dimerization sites [11]. Therefore, we reasoned that the variant could potentially alter STR function. However, it is impossible to infer from the reported associations alone whether this is a gain- or loss-of-function variant of the TAS1R2 receptor. Understanding the functional properties of the variant is critical for evaluating the nature of known links with metabolic variables and formulating appropriate hypotheses that explore the clinical and physiological significance of extra-oral STR chemosensing.
2. Materials and methods
2.1. In vitro studies
Assay methods were performed exactly as previously described for a) functional expression, single-cell calcium imaging and analysis [12], b) surface or total expression of TAS1R2/TAS1R3 receptors and flow cytometry analysis [11], and c) fluorescence resonance energy transfer (FRET) microscopy and fluorescence lifetime imaging microscopy (FLIM) [13,14]. For details see Supp. Methods.
2.2. Clinical studies
The clinical studies were performed in accordance with the requirements of Good Clinical Practice and the Revised Declaration of Helsinki. Recruitment, enrollment, and all study-related visits, including specimen collection and point-of-care laboratory testing, took place at Advent-Health Translational Research Institute (TRI) Clinical Research Unit (CRU) as previously described (NCT02835859) [15]. The study was approved by the Institutional Review Board at Advent-Health and all participants signed an informed consent. Mathematical modeling was performed exactly as described for beta-cell function, insulin sensitivity, and insulin clearance [16]. For details see Supp. Methods.
2.3. Statistical analysis
All data are represented as mean +/− standard error and plotted with Prism 9 (GraphPad Software). Calcium responses were analyzed by linear or 4-parameter logistic regression. All other variables were analyzed by the two-tailed t-test or paired t-test, as appropriate. All participants were retrospectively assigned to two groups based on TAS1R2 genotypes. For clinical data, allele equilibrium, frequency, and SNP linkage were analyzed by Chi-square using jamovi 1.6 (jamovi team). Plasma excursions of glucose and related hormones was analyzed by two-way repeated measures ANOVA using Prism 9 (Graphpad Software). Baseline characteristics and metabolic responses to the OGTT were analyzed through a general linear model using jamovi 1.6 (jamovi team). Sex was used as a covariate. Non-parametric variables were log-transformed prior to analysis.
2.4. Data and resource availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request. No applicable resources were generated or analyzed during the current study.
3. Results and discussion
3.1. The TAS1R2-(Ile191Val) substitution reduces plasma membrane (PM) availability of STR
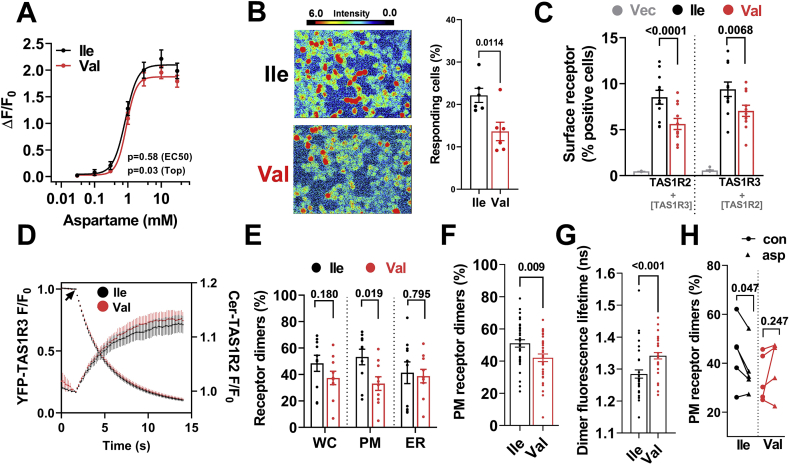
First, we expressed TAS1R2-(Ile) or TAS1R2-(Val) with TAS1R3 by transient transfection in HEK293 cells and monitored single-cell calcium mobilization in response to STR ligands [12]. In cells that responded to the stimuli, we found similar dose–response curves for aspartame (Figure 1A; Aspartame EC50 Ile: 0.84 mM vs. Val: 0.90 mM, p = 0.58) or sucralose (Supp.Figure.1A) between variants, excluding major differences in ligand binding affinity or signaling. However, we noted that the percentage of cells responding in the Val variant was significantly lower (Figure 1B) despite identical transfection efficiencies (368 ± 9 vs. 374 ± 10 copies/cell, p = 0.68). These findings, along with the small downward trend at max stimulation (Top, p = 0.03; Figure 1A), may suggest a mechanism for reduced function of the Val variant.
Figure 1.
The TAS1R2-(Val) variant reduces plasma membrane (PM) availability of STR dimer. A) Calcium mobilization in response to aspartame concentrations in transfected HEK293 cells with TAS1R2-(Ile) or TAS1R2-(Val) along with TAS1R3 and Gα16-gust44 (n = 6, ∼30 cells). B) Calcium response images (color scale indicates the fluorescence intensity) with quantitation of the percent of responding cells to a saturating concentration (30 mM) of aspartame. C) Analysis of surface expression of TAS1R2 (left bars) and TAS1R3 (right bars) shown as normalized percent positive cells in Expi293F cells co-expressing FLAG-TAS1R2 constructs (Ile or Val) with Myc-TAS1R3. Vec, vector-only control. D) Whole-cell progressive acceptor photobleaching showing TAS1R2/TAS1R3 hetero-oligomerization using the TAS1R2-(Ile) and TAS1R2-(Val) variants. E) Quantification of FRET in subcellular regions (WC, whole cell; PM, plasma membrane; ER, endoplasmic reticulum). F) Single-point lifetime measurements of PM-localized receptors, shown as percentage of receptors in dimeric complexes for each of the TAS1R2 variants. G) Single-point acquisition of fluorescence lifetime (ns) for the TAS1R2-(Ile)/TAS1R3 and TAS1R2-(Val)/TAS1R3 dimers indicative of the donor–acceptor distance of the complexes. H) Single-point lifetime measurements of PM-localized STR before and after the addition of aspartame, shown as percentage of receptors in dimeric complexes. Con, control; asp, after aspartame. Four-parameter logistic regression (A); t-test (B,C,E,F,G); Paired t-test (H).
To shed light on this possibility, we tested whether the Ile191Val substitution structurally alters TAS1R2, as saturation mutagenesis studies have shown that missense mutations in the LBD of TAS1R2 can alter its surface localization and co-trafficking with TAS1R3 [11]. We assessed surface protein expression using flow cytometry of Expi293F cells co-transfected with c-myc-tagged TAS1R3 and either FLAG-tagged TAS1R2-(Ile) or TAS1R2-(Val) [11]. The TAS1R2-(Val) variant reduced the fraction of cells presenting TAS1R2 at the cell surface and also induced a proportional reduction of surface TAS1R3 (Figure 1C). The localization of TAS1R3 at the PM was strictly dependent on its co-expression with TAS1R2 (Supp.Figure 1B, right bars), confirming previous findings [11,17]. Interestingly, expression of TAS1R2-(Val) alone, without TAS1R3, also had reduced surface expression (Supp.Figure 1B, left bars), suggesting an intrinsic mechanism that affects TAS1R2 availability. Indeed, using the same assay with permeabilized cells, we noted that the total cell signal of the TAS1R2 receptor was also mildly reduced in the Val variant (Supp.Figure 1C). Thus, the substitution could possibly destabilize TAS1R2, altering receptor trafficking.
As a result, we evaluated the physical interaction of TAS1R2 with TAS1R3 and the relative distribution of the heterodimer in intracellular compartments using FRET microscopy [13]. We observed an exponential decrease in YFP-TAS1R3 fluorescence during photobleaching with a concomitant increase in Cer-TAS1R2-(Ile) fluorescence (Figure 1D). This is consistent with loss of donor fluorescence quenching, as FRET was abolished by destruction of the YFP acceptor. Similar FRET was also observed using the TAS1R2-(Val) variant (Figure 1D). Comparison of the donor and acceptor fluorescence intensities revealed linear Cer/YFP relationships for both variants, consistent with a single YFP acceptor in the TAS1R2/TAS1R3 complex (Supp.Figure.1D). This linear relationship is compatible with a dimeric TAS1R2/TAS1R3 hetero-oligomer complex [11]. Next, we performed more precise localization-specific quantification of FRET using FLIM [14]. Performing 2-exponent fitting of the fluorescence lifetime decay, we observed that the TAS1R2-(Val) variant reduced the number of STR dimer complexes at the PM, but not in the ER (Figure 1E), suggesting that the TAS1R2-(Val) variant is unlikely to be trapped in this compartment.
To improve the precision of the fluorescence lifetime analysis, we performed extended (1 min) single-point acquisition at the PM, yielding increased photon counts for decay analysis [14]. We found the percentage of PM-localized receptor dimers decreased for the Val variant (Figure 1F), consistent with our analysis of FLIM image data (Figure 1E). In addition, we observed a longer dimer fluorescence lifetime for the TAS1R2-(Val)/TAS1R3 dimers (Figure 1G), indicative of weaker interaction between the TAS1R2 and TAS1R3 receptors of the complex. Thus, the reduced availability of STR in the PM may be linked to structural changes that affect the stability of the dimer at the PM without directly altering the efficiency of the signaling cascade per se (Figure 1A) or dimer trafficking from the ER to the PM (Figure 1E). For instance, upon ligand binding, GPCRs can be rapidly desensitized through internalization [18]. So, we evaluated TAS1R2/TAS1R3 dimerization and quaternary conformation in response to a ligand. We performed paired experiments by acquiring control fluorescence decay data from individual cells using the Ile or Val variant and measured a second fluorescence decay from the same cell after addition of aspartame, a potent TAS1R2 agonist. For the TAS1R2-(Ile) variant, aspartame caused a reduction in the apparent percentage of dimeric TAS1R2/TAS1R3 at the PM, but these effects were absent using the TAS1R2-(Val) variant (Figure 1H). The TAS1R2/TAS1R3 dimer structure remained unchanged for both variants (Supp.Figure.1E). Thus, the Val variant may cause ligand-independent receptor internalization, contributing to the observable reduction of the STR dimer in the PM.
3.2. TAS1R2-val carriers have reduced glucose excretions during an OGTT
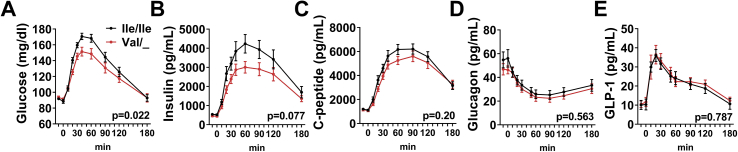
In taste buds, STR are abundant, so a lower number of functional STR due to reduced PM availability may not cause a strong phenotype. In contrast, STR expression in intestinal L-cells or pancreatic beta-cells is less pronounced; hence, a reduced number of functional STR could have consequences leading to observable physiological effects. Indeed, we have previously shown that genetic loss of function of TAS1R2 in mice (TAS1R2-KO) causes reduced glucose excursions in response to an oral glucose tolerance test (OGTT) [4], but whether these findings are applicable to human physiology is unknown. Thus, as a proof-of-concept, we tested whether partial loss of function of STR through Ile191Val polymorphism similarly alters glucose and hormonal responses to an OGTT in a cohort of healthy lean participants (Table.1). Participants were genotyped for the Ile191Val (rs35874116) variant of TAS1R2 and assigned to either the homozygous major allele (Ile/Ile) or the carrier of minor allele (Val/_) group. The genotype distribution did not deviate from the Hardy–Weinberg equilibrium and the minor allele frequency was not different from the recorded aggregate (Supp.Table.1). There were no differences in the fasting metabolic characteristics between genotypes (Table.1). Strikingly, we observed a notable reduction in glucose excursions in Val carriers during the OGTT (Figure 2A and Table.1). Insulin responses in Val carriers also trended to be lower, but the effect was not statistically significant (Figure.1B). No genotype differences were noted in C-peptide, glucagon, or active GLP-1 concentrations (Figure 1C-E and Table.1). The lower plasma glucose excursions in Val carriers mirrors the phenotype of TAS1R2-KO mice [4] and further corroborates our biochemical data, suggesting that the Ile191Val substitution leads to partial loss of function of STR in humans.
Table.1.
Baseline and metabolic responses to an OGTT in healthy lean adults grouped by two common TAS1R2 polymorphisms.
| Ile/Ile | Val/_ | p | Ser/_ | Cys/Cys | p | |
|---|---|---|---|---|---|---|
| Baseline variables | ||||||
| Total n (Male/Female) | 26 (9/17) | 20 (5/15) | 16 (7/9) | 30 (7/23) | ||
| Age (years) | 29.19 ± 1.52 | 31.00 ± 2.18 | 0.281 | 32.88 ± 2.12 | 27.88 ± 1.54 | 0.062 |
| Height (cm) | 1.68 ± 0.02 | 1.67 ± 0.02 | 0.591 | 1.67 ± 0.02 | 1.68 ± 0.02 | 0.766 |
| Weight (kg) | 62.68 ± 1.79 | 62.16 ± 1.85 | 0.808 | 63.72 ± 2.22 | 61.78 ± 1.57 | 0.899 |
| BMI (kg/m2) | 22.07 ± 0.41 | 22.31 ± 0.44 | 0.650 | 22.76 ± 0.53 | 21.86 ± 0.35 | 0.190 |
| Glucose (mg/dL) | 88.90 ± 1.41 | 91.75 ± 1.76 | 0.208 | 89.91 ± 1.99 | 90.27 ± 1.36 | 0.861 |
| Insulin (μU/L) | 7.17 ± 0.91 | 7.67 ± 1.08 | 0.838 | 5.37 ± 0.56 | 8.46 ± 0.96 | 0.056 |
| HbA1c (%) | 4.90 ± 0.06 | 4.92 ± 0.06 | 0.773 | 4.93 ± 0.06 | 4.90 ± 0.06 | 0.984 |
| Triglycerides (mg/dL) | 73.81 ± 7.31 | 76.45 ± 8.12 | 0.849 | 66.00 ± 7.29 | 79.73 ± 7.20 | 0.263 |
| Total Cholesterol (mg/dL) | 167.80 ± 6.67 | 165.8 ± 5.33 | 0.681 | 158.60 ± 7.00 | 171.30 ± 5.50 | 0.294 |
| HDL (mg/dL) | 62.27 ± 2.77 | 63.65 ± 2.86 | 0.994 | 59.56 ± 3.31 | 64.63 ± 2.45 | 0.547 |
| LDL (mg/dL) | 90.69 ± 5.80 | 86.75 ± 5.15 | 0.617 | 85.88 ± 6.72 | 90.63 ± 4.92 | 0.587 |
| LDL/HDL ratio | 1.55 ± 0.14 | 1.46 ± 0.15 | 0.792 | 1.54 ± 0.20 | 1.49 ± 0.12 | 0.871 |
| OGTT variables | ||||||
| Baseline glucose (mg/dL) | 91.06 ± 1.50 | 90.84 ± 1.86 | 0.928 | 92.21 ± 1.75 | 90.10 ± 1.49 | 0.342 |
| 2h glucose (mg/dL) | 118.89 ± 5.32 | 125.43 ± 6.60 | 0.437 | 116.79 ± 6.22 | 124.71 ± 5.30 | 0.318 |
| Baseline insulin (pg/mL∗10−3) | 0.52 ± 0.04 | 0.46 ± 0.05 | 0.343 | 0.46 ± 0.05 | 0.52 ± 0.04 | 0.237 |
| 2h insulin (pg/mL∗10−3) | 3.22 ± 0.44 | 2.77 ± 0.54 | 0.520 | 2.62 ± 0.51 | 3.34 ± 0.43 | 0.262 |
| Glucose peak (mg/dL) | 176.98 ± 4.11 | 162.14 ± 4.82 | 0.019 | 167.88 ± 5.40 | 172.75 ± 4.36 | 0.479 |
| Glucose time of peak (min) | 141.21 ± 13.37 | 120.47 ± 14.29 | 0.237 | 132.70 ± 16.08 | 132.61 ± 13.04 | 0.996 |
| Insulin peak (pg/mL∗10−3) | 4.76 ± 0.47 | 3.57 ± 0.47 | 0.075 | 4.18 ± 0.58 | 4.32 ± 0.48 | 0.844 |
| Insulin time of peak (min) | 149.85 ± 9.51 | 134.99 ± 10.57 | 0.288 | 141.11 ± 11.56 | 145.38 ± 9.46 | 0.774 |
| C-peptide peak (pg/mL∗min∗10−3) | 6.75 ± 0.43 | 6.03 ± 0.50 | 0.264 | 6.63 ± 0.54 | 6.34 ± 0.44 | 0.665 |
| C-peptide time of peak (min) | 138.62 ± 8.22 | 133.01 ± 9.48 | 0.649 | 135.84 ± 10.14 | 136.60 ± 8.17 | 0.953 |
| AUC glucose (mg/dL∗min∗10−3) | 23.92 ± 0.62 | 22.19 ± 0.58 | 0.048 | 23.31 ± 0.72 | 23.10 ± 0.57 | 0.752 |
| AUC insulin (pg/mL∗min∗10−3) | 543.59 ± 62.26 | 417.56 ± 41.62 | 0.119 | 445.82 ± 43.35 | 511.71 ± 57.34 | 0.462 |
| AUC C-peptide (pg/mL∗min∗10−3) | 872.05 ± 56.34 | 785.21 ± 55.86 | 0.244 | 827.25 ± 48.45 | 838.05 ± 56.43 | 0.940 |
| AUC glucagon (pg/mL∗min∗10−3) | 4.99 ± 0.57 | 4.74 ± 0.48 | 0.830 | 5.43 ± 0.80 | 4.59 ± 0.40 | 0.389 |
| AUC active GLP1 (pg/mL∗min∗10−3) | 4.34 ± 0.56 | 4.02 ± 0.40 | 0.585 | 4.32 ± 0.43 | 4.14 ± 0.50 | 0.649 |
| OGTT modeling analysis | ||||||
| Basal glucose (mmol/L) | 5.00 ± 0.08 | 5.12 ± 0.10 | 0.341 | 5.04 ± 0.10 | 5.05 ± 0.08 | 0.927 |
| Mean glucose (mmol/L) | 7.35 ± 0.18 | 6.80 ± 0.22 | 0.048 | 7.18 ± 0.24 | 7.08 ± 0.19 | 0.752 |
| Basal insulin (pmol/L) | 90.10 ± 6.64 | 78.30 ± 7.80 | 0.237 | 79.10 ± 8.28 | 89.20 ± 6.68 | 0.343 |
| Mean insulin (pmol/L) | 515.32 ± 52.52 | 391.84 ± 61.71 | 0.119 | 426.29 ± 67.59 | 488.72 ± 53.74 | 0.462 |
| Glucose sensitivity (pmol/min/m2/mM) | 65.60 ± 6.06 | 67.10 ± 7.12 | 0.871 | 70.40 ± 7.48 | 63.50 ± 6.03 | 0.473 |
| Insulin secretion rate (pmol/min/m2) | 69.40 ± 7.52 | 76.20 ± 8.84 | 0.545 | 63.30 ± 9.20 | 77.90 ± 7.42 | 0.214 |
| Potentiation factor ratio | 1.38 ± 0.10 | 1.48 ± 0.12 | 0.530 | 1.32 ± 0.13 | 1.49 ± 0.10 | 0.312 |
| Rate sensitivity (pmol/m2/mM) | 656.44 ± 101.37 | 710.65 ± 119.11 | 0.719 | 788.97 ± 123.99 | 608.22 ± 100.06 | 0.255 |
| Insulin sensitivity (μmol/min/kg) | 7.67 ± 0.46 | 8.45 ± 0.55 | 0.262 | 8.08 ± 0.58 | 7.93 ± 0.47 | 0.830 |
| Insulin clearance (L/min/m2) | 0.51 ± 0.04 | 0.60 ± 0.05 | 0.171 | 0.62 ± 0.05 | 0.50 ± 0.04 | 0.105 |
All values are mean ± SEM. P-value for genotype effect was obtained after sex adjustment using a general linear model. BMI, body mass index; HDL, high density lipoproteins; LDL, low density lipoproteins; VLDL, very-low density lipoproteins; HbA1c, glycated hemoglobin A1c; OGTT, oral glucose tolerance test; AUC, area under curve; GLP1, glucagon-like peptide 1.
Figure 2.
The TAS1R2-(Val) variant is associated with reduced glucose excursions in humans. Plasma excursions of (A) glucose, (B) insulin, (C) C-peptide, (D) glucagon, and (E) active GLP-1 in response to an oral glucose challenge in healthy lean Ile/Ile (black) and Val carriers (red) with normal glucose control (n = 20–26/group). Two-way ANOVA repeated measures, p-value of time x genotype effect.
In TAS1R2-KO mice, the reduced glucose excursions to an OGTT could not be explained by altered beta-cell function or insulin sensitivity [1,2], but were mainly attributed to a reduced rate of glucose absorption [4]. To shed light on the relative contributions of these regulatory mechanisms in Val carriers, we performed mathematical modeling analysis of the OGTT [19]. No genotype effects were noted in insulin sensitivity (Stumvoll index) and clearance or in beta-cell function through the evaluation of various insulin secretion parameters and indices (Table.1 and Supp. Methods). These data exclude major contributions of insulin secretion and/or action and suggest that the genotype effect on glucose excursions during the OGTT is consistent with altered glucose absorption. Indeed, pharmacological inhibition of STR reduced glucose flux in fresh human intestinal explants, recapitulating the effects of genetic ablation of STR in mouse intestines [4]. Sex-dependent differences in glucose absorption rates can also affect OGTT responses [20], but in our study the magnitude of the genotype effect on glucose responses was similar between male and female participants (p = 0.633), so no significant interaction between genotype and sex (p = 0.752) was noted. Finally, we assessed fecal microbiota composition because of known interactions with glucose regulation [21], but found similar microbial alpha and beta diversity between genotypes (Supp.Table.2), confirming findings in mice [15].
Our data collectively support a functional role of STR in postprandial glucose regulation in humans, as shown before in mice [3,4,22]. However, it is still unclear whether partial loss of function of TAS1R2 affects other glycemic variables or alters the risk for the development of metabolic diseases. Although our study was not designed or intended to expose such associations, we explored the Ile191Val polymorphism in the TAS1R2 gene (rs35874116) using phenome-wide association studies (PheWAS) at the “T2D Knowledge Portal” (https://t2d.hugeamp.org), which specifically enables the search and analysis of traits linked to type 2 diabetes. Because postprandial glucose excursion is not a measured trait in large-scale PheWAS for type 2 diabetes, a direct validation of our findings in heterogeneous populations could not be appropriately assessed. Nevertheless, the rs35874116 was significantly (p < 0.05) associated with phenotypes relevant to renal function, lipids, and other metabolic and glycemic indices (all traits with p < 0.05 as shown in Supp. Table.3). These findings suggest that partial loss of function of STR in peripheral tissues may affect phenotypic outcomes associated with metabolic control and disease.
3.3. The effects of Ile191Val substitution are not linked to the Ser9Cys high-frequency variant
Finally, the allele frequencies of most missense variants in the translated region of TAS1R2 are low (<10%), but the rs9701796 allele (Ser9Cys) is comparable to Ile191Val [23]. This substitution is located in the putative signal peptide of TAS1R2 and has also been associated with dietary and anthropometric variables in children [24]. Therefore, to exclude the possibility that clinical interactions associated with the Ile191Val are linked to Ser9Cys polymorphism, we analyzed all clinical variables of the human cohorts based on this TAS1R2 variant. We found no associations between Ser9Cys and the assessed variables (Supp.Figure 2, Table.1, and Supp.Tables 1 and 2). Although we cannot exclude the possibility of linkage disequilibrium with another causal polymorphism, the effects of Ile191Val are independent from this high-allele-frequency TAS1R2 variant.
3.4. Conclusions
Using a combination of in vitro biochemical approaches, we identified that the Ile191Val substitution in the TAS1R2 gene reduces the availability of STR dimer in the PM, likely causing a partial loss of function of STR. This notion was further confirmed by proof-of-concept clinical findings. We demonstrated that in a cohort of healthy lean participants, carriers of the Val allele had similar phenotypic responses to those seen in mice with a genetic loss of function of TAS1R2 (TAS1R2-KO). Thus, our clinical observations corroborate our previous pre-clinical findings, which demonstrated contributions of STR in postprandial glucose excursions [4], and highlight that, beyond taste perception, STR can also act as peripheral sugar sensors in humans.
Funding
This work was supported by the National Institute of Food and Agriculture (NIFA-2018-67001-28246 to GAK), the National Institutes of Health (DK127444 to GAK; HL092321 and HL143816 to SLR), the Japan Society for the Promotion of Science KAKENHI (JP21K09818 to KS), and institutional support from the Ohio State University (to GAK), AdventHealth (to GAK and REP), and the Loyola Stritch School of Medicine Cardiovascular Research Institute (to SLR).
Acknowledgments
Author contributions: JoS, MP, JS, JP, KS, NS, and VS performed experiments; JoS, MP, JS, JP, KS, NS, FY, AM, EP, SLR, and GAK analyzed data; JoS, EP, REP, SLR, and GAK designed experiments and interpreted data; JoS, KS, AM, EP, REP, and SLR edited the manuscript; GAK wrote the manuscript, conceived the studies, and is the guarantor of work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101339.
Conflict of interest
None.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kyriazis G.A., Soundarapandian M.M., Tyrberg B. Sweet taste receptor signaling in beta cells mediates fructose-induced potentiation of glucose-stimulated insulin secretion. ProcNatlAcadSciUSA. 2012;109:E524–E532. doi: 10.1073/pnas.1115183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyriazis G.A., Smith K.R., Tyrberg B., Hussain T., Pratley R.E. Sweet taste receptors regulate basal insulin secretion and contribute to compensatory insulin hypersecretion during the development of diabetes in male mice. Endocrinology. 2014;155:2112–2121. doi: 10.1210/en.2013-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang H.J., Kokrashvili Z., Theodorakis M.J., Carlson O.D., Kim B.J., Zhou J. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. ProcNatlAcadSciUSA. 2007;104:15069–15074. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith K., Karimian Azari E., LaMoia T.E., Hussain T., Vargova V., Karolyi K. T1R2 receptor-mediated glucose sensing in the upper intestine potentiates glucose absorption through activation of local regulatory pathways. Mol Metab. 2018;17:98–111. doi: 10.1016/j.molmet.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith K.R., Hussain T., Karimian Azari E., Steiner J.L., Ayala J.E., Pratley R.E. Disruption of the sugar-sensing receptor T1R2 attenuates metabolic derangements associated with diet-induced obesity. American Journal of Physiology - Endocrinology And Metabolism. 2016;310:E688–E698. doi: 10.1152/ajpendo.00484.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valente C., Alvarez L., Marques P.I., Gusmao L., Amorim A., Seixas S. Genes from the TAS1R and TAS2R families of taste receptors: looking for signatures of their adaptive role in human evolution. Genome Biol Evol. 2018;10:1139–1152. doi: 10.1093/gbe/evy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias A.G., Eny K.M., Cockburn M., Chiu W., Nielsen D.E., Duizer L. Variation in the TAS1R2 gene, sweet taste perception and intake of sugars. Journal of Nutrigenetics and Nutrigenomics. 2015;8:81–90. doi: 10.1159/000430886. [DOI] [PubMed] [Google Scholar]
- 8.Eny K.M., Wolever T.M., Corey P.N., El-Sohemy A. Genetic variation in TAS1R2 (Ile191Val) is associated with consumption of sugars in overweight and obese individuals in 2 distinct populations. American Journal of Clinical Nutrition. 2010;92:1501–1510. doi: 10.3945/ajcn.2010.29836. [DOI] [PubMed] [Google Scholar]
- 9.Melo S.V., Agnes G., Vitolo M.R., Mattevi V.S., Campagnolo P.D.B., Almeida S. Evaluation of the association between the TAS1R2 and TAS1R3 variants and food intake and nutritional status in children. Genetics and Molecular Biology. 2017;40:415–420. doi: 10.1590/1678-4685-GMB-2016-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos-Lopez O., Panduro A., Martinez-Lopez E., Roman S. Sweet taste receptor TAS1R2 polymorphism (Val191Val) is associated with a higher carbohydrate intake and hypertriglyceridemia among the population of west Mexico. Nutrients. 2016;8:101. doi: 10.3390/nu8020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J., Selvam B., Sanematsu K., Shigemura N., Shukla D., Procko E. Structural architecture of a dimeric class C GPCR based on co-trafficking of sweet taste receptor subunits. Journal of Biological Chemistry. 2019;294:4759–4774. doi: 10.1074/jbc.RA118.006173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanematsu K., Kusakabe Y., Shigemura N., Hirokawa T., Nakamura S., Imoto T. Molecular mechanisms for sweet-suppressing effect of gymnemic acids. Journal of Biological Chemistry. 2014;289:25711–25720. doi: 10.1074/jbc.M114.560409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh D.R., Dalton M.P., Cho E.E., Pribadi M.P., Zak T.J., Seflova J. Newly discovered micropeptide regulators of SERCA form oligomers but bind to the pump as monomers. Journal of Molecular Biology. 2019;431:4429–4443. doi: 10.1016/j.jmb.2019.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bovo E., Nikolaienko R., Cleary S.R., Seflova J., Kahn D., Robia S.L. Dimerization of SERCA2a enhances transport rate and improves energetic efficiency in living cells. Biophysical Journal. 2020;119:1456–1465. doi: 10.1016/j.bpj.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serrano J., Smith K.R., Crouch A.L., Sharma V., Yi F., Vargova V. High-dose saccharin supplementation does not induce gut microbiota changes or glucose intolerance in healthy humans and mice. Microbiome. 2021;9:11. doi: 10.1186/s40168-020-00976-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimian Azari E., Smith K.R., Yi F., Osborne T.F., Bizzotto R., Mari A. Inhibition of sweet chemosensory receptors alters insulin responses during glucose ingestion in healthy adults: a randomized crossover interventional study. American Journal of Clinical Nutrition. 2017;105:1001–1009. doi: 10.3945/ajcn.116.146001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu M., Goto M., Kawai T., Yamashita A., Kusakabe Y. Distinct human and mouse membrane trafficking systems for sweet taste receptors T1r2 and T1r3. PloS One. 2014;9 doi: 10.1371/journal.pone.0100425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whalen E.J., Rajagopal S., Lefkowitz R.J. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends in Molecular Medicine. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mari A., Pacini G., Murphy E., Ludvik B., Nolan J.J. A model-based method for assessing insulin sensitivity from the oral glucose tolerance test. Diabetes Care. 2001;24:539–548. doi: 10.2337/diacare.24.3.539. [DOI] [PubMed] [Google Scholar]
- 20.Anderwald C., Gastaldelli A., Tura A., Krebs M., Promintzer-Schifferl M., Kautzky-Willer A. Mechanism and effects of glucose absorption during an oral glucose tolerance test among females and males. Journal of Clinical Endocrinology & Metabolism. 2011;96:515–524. doi: 10.1210/jc.2010-1398. [DOI] [PubMed] [Google Scholar]
- 21.Gerard C., Vidal H. Impact of gut microbiota on host glycemic control. Frontiers in Endocrinology. 2019;10:29. doi: 10.3389/fendo.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolskee R.F., Dyer J., Kokrashvili Z., Salmon K.S., Ilegems E., Daly K. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. ProcNatlAcadSciUSA. 2007;104:15075–15080. doi: 10.1073/pnas.0706678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith N.J., Grant J.N., Moon J.I., So S.S., Finch A.M. Critically evaluating sweet taste receptor expression and signaling through a molecular pharmacology lens. FEBS Journal. 2021 doi: 10.1111/febs.15768. [DOI] [PubMed] [Google Scholar]
- 24.Pioltine M.B., de Melo M.E., Santos A.S., Machado A.D., Fernandes A.E., Fujiwara C.T. Genetic variations in sweet taste receptor gene are related to chocolate powder and dietary fiber intake in obese children and adolescents. Journal of Personalized Medicine. 2018;8 doi: 10.3390/jpm8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.