Abstract
Objective
The loss of forkhead box protein O1 (FoxO1) signaling in response to metabolic stress contributes to the etiology of type II diabetes, causing the dedifferentiation of pancreatic beta cells to a cell type reminiscent of endocrine progenitors. Lack of methods to easily model this process in vitro, however, have hindered progress into the identification of key downstream targets and potential inhibitors. We therefore aimed to establish such an in vitro cellular dedifferentiation model and apply it to identify novel agents involved in the maintenance of beta-cell identity.
Methods
The murine beta-cell line, Min6, was used for primary experiments and high-content screening. Screens encompassed a library of small-molecule drugs representing the chemical and target space of all FDA-approved small molecules with an automated immunofluorescence readout. Validation experiments were performed in a murine alpha-cell line as well as in primary murine and human diabetic islets. Developmental effects were studied in zebrafish and C. elegans models, while diabetic db/db mouse models were used to elucidate global glucose metabolism outcomes.
Results
We show that short-term pharmacological FoxO1 inhibition can model beta-cell dedifferentiation by downregulating beta-cell-specific transcription factors, resulting in the aberrant expression of progenitor genes and the alpha-cell marker glucagon. From a high-content screen, we identified loperamide as a small molecule that can prevent FoxO inhibitor-induced glucagon expression and further stimulate insulin protein processing and secretion by altering calcium levels, intracellular pH, and FoxO1 localization.
Conclusions
Our study provides novel models, molecular targets, and drug candidates for studying and preventing beta-cell dedifferentiation.
Keywords: Diabetes, Beta-cell dedifferentiation, FoxO1 inhibitor, Loperamide
Highlights
-
•
Pharmacological FoxO1 inhibition can model many aspects of beta cell dedifferentiation in vitro, including loss of beta cell identity and gain of progenitor and alpha cell markers.
-
•
Through a high content screen with this cellular dedifferentiation model, we identified the FDA-approved small molecule, loperamide, to counter FoxO inhibitor-mediated beta cell dedifferentiation.
-
•
Acute loperamide treatment increases FoxO1 nuclear translocation and insulin granule maturation through modulation of intracellular pH and free calcium levels.
-
•
Independently of pharmacological FoxO1 inhibition, loperamide treatment increases insulin secretion, protein and mRNA levels in human and murine diabetic pancreatic islets and strongly reduces the number of glucagon/insulin double positive cells in diabetic db/db mice.
1. Introduction
In type II diabetes, initial peripheral insulin resistance often progresses to insulin deficiency caused by beta-cell dedifferentiation and apoptosis. A key regulator of beta-cell dedifferentiation is the transcription factor forkhead box protein O1 (FoxO1). Prolonged and severe hyperglycemia results in the loss of FoxO1 expression and concomitant beta-cell failure [1,2]. The dedifferentiation of beta cells via inhibition of FoxO family proteins is not yet fully understood, but it has been linked with decreased metabolic flexibility [3]. Introduction of a metabolic stressor to beta cells lacking FoxO1 triggers a loss of function and a subsequent increase in the pancreatic endocrine progenitor marker neurogenin3 (Ngn3). Additionally, a subset of the dedifferentiated beta cells begins expressing the alpha-cell marker glucagon, supporting the multi-lineage potential of these progenitors [4]. Notably, dedifferentiated beta cells have been identified in human islets from diabetic donors [5].
Identifying mechanisms to inhibit or reverse beta-cell dedifferentiation represent key goals in current type II diabetes research. Robust models to study this process in vitro have yet to be established [6]. Existing efforts to identify compounds that improve beta-cell health have focused on differentiation models with knockouts of type II diabetes GWAS susceptibility genes DKAL1, KCNQ1, or KCNJ11 [7], or on FGF2-mediated beta-cell dedifferentiation [8]. Here, our aim was to establish an in vitro model for FoxO1-dependent beta-cell dedifferentiation that was highly reproducible, did not rely on precious materials, and was amenable for high-content screening.
We hypothesized that pharmacological FoxO1 inhibition in beta-cell lines could provide an in vitro model system for the beta-cell dedifferentiation process observed in mice following genetic knockout of the transcription factor FoxO1 [4]. Here, we show that pharmacological inhibition of FoxO1 in beta-cell lines mimics many aspects of in vivo dedifferentiation. Subsequently, using a high-content screening assay, we identify the small-molecule loperamide to reverse aspects of this FoxO1 inhibition in beta cells. We show that it modulates calcium signaling, intracellular pH, secretory granule maturation, and FoxO1 expression in pancreatic islet cells and causes corresponding physiological effects in vivo.
2. Results
2.1. A cellular model for beta-cell dedifferentiation
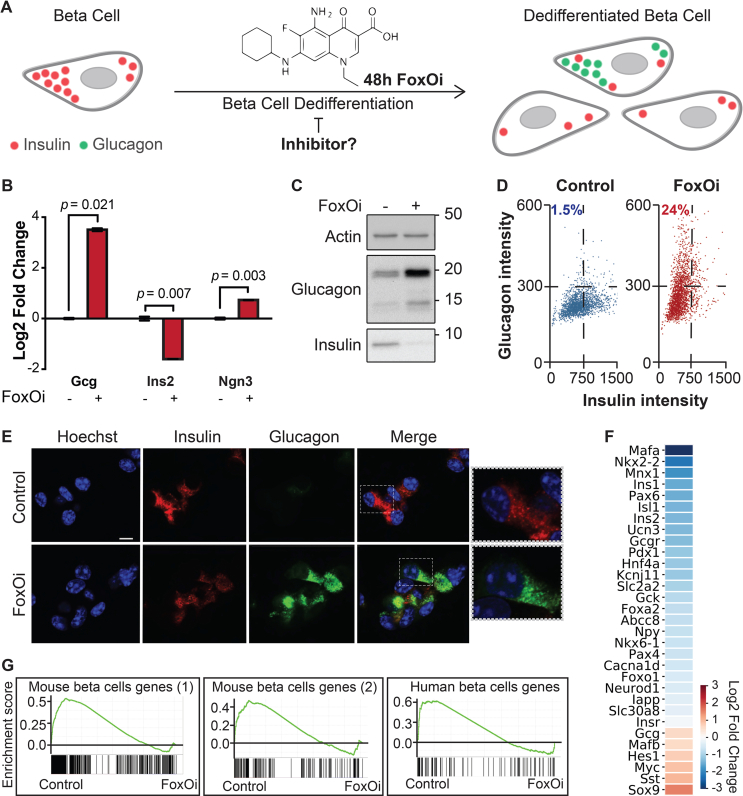
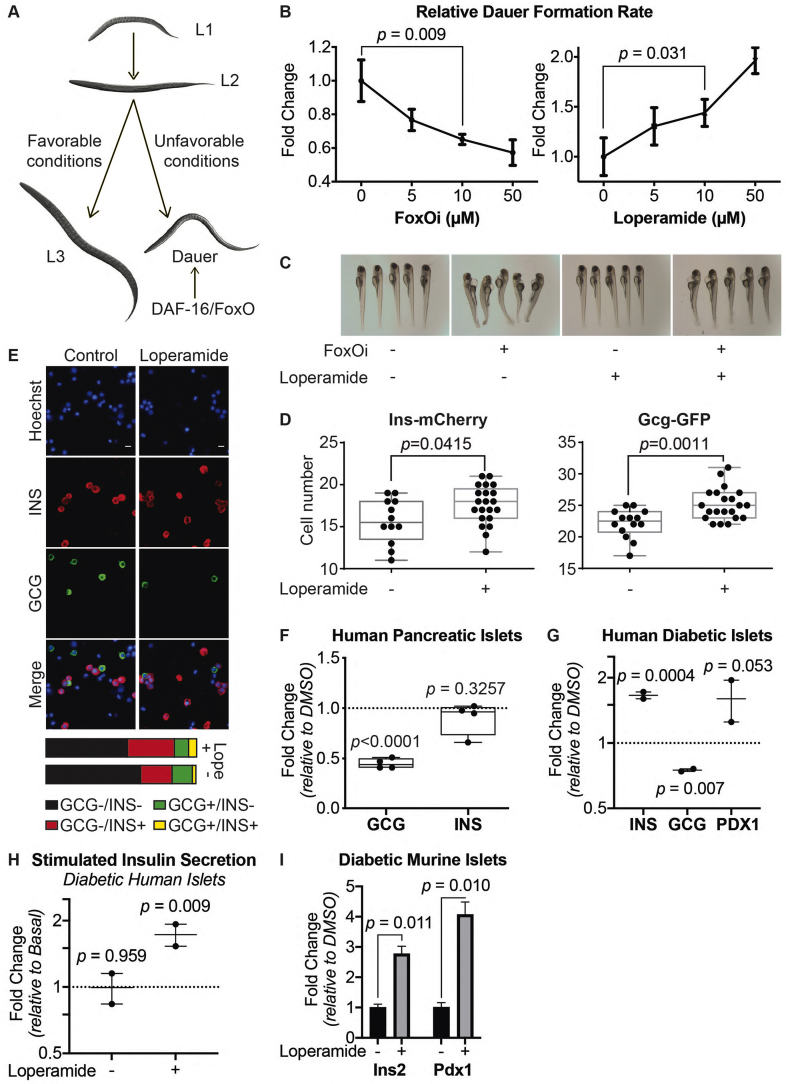
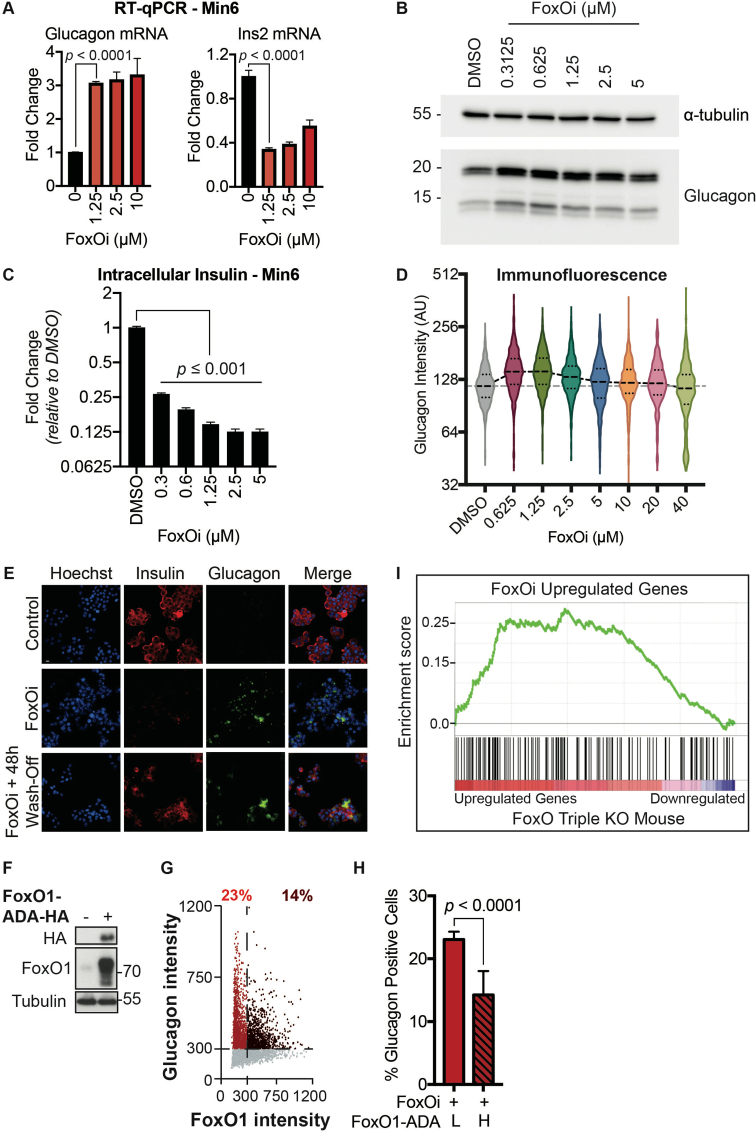
To establish a screenable model system for inhibitors of beta-cell dedifferentiation, we treated the murine beta-cell line Min6 with the selective small-molecule FoxO1 inhibitor AS1842856 (FoxOi) [9] for 48 h (Figure 1A). We observed an approximately threefold reduction in insulin mRNA, accompanied by an upregulation of the pancreatic progenitor marker Ngn3 and a strong induction of the alpha-cell marker glucagon (Figure 1B and SFig. 1A), with comparable changes at the protein level (Figure 1C, SFig. 1B and C). On a population level, most cells showed reduced insulin levels and in approximately a quarter of cells glucagon was strongly increased (Figure 1D, SFig. 1D). In these cells, glucagon staining was granular and partially overlapped with the remaining insulin staining (Figure 1E). Washout of the inhibitor for 48 h successfully reestablished insulin protein levels; however, the glucagon-positive subpopulation remained stable and converted to a double-positive phenotype (SFig. 1E). It is possible that later time points post-washout might reveal a complete reversibility of the phenotype. Overexpression of a constitutively active form of FoxO1 significantly decreased the fraction of FoxOi-induced glucagon-positive cells (SFig. 1F–H). Importantly, FoxOi caused gene expression changes comparable to the existing triple FoxO knockout mouse model [10], both in terms of FoxO1 target genes (Figure 1F) and genome-wide (SFig. 1I). Beta-cell-specific genes were downregulated, whereas progenitor (i.e., Sox9, Myc, Hes1) and alpha-cell markers (i.e., Gcg, Mafb) were enriched (Figure 1F,G). This correlates with depleted Pdx1, Nkx6-1, MafA, and Mafb mRNA levels observed in diabetic human islets [11] and inactivation of these transcription factors after simulation of a diabetic milieu [11]. These results support the hypothesis that chemical inhibition of FoxO1 in this beta-cell line is able to phenocopy the genetic depletion of FoxO in mice and generally mimic beta-cell dedifferentiation.
Figure 1.
Cellular model for FoxO-mediated beta-cell dedifferentiation. A. Schematic overview of our cellular model for FoxO-mediated beta-cell dedifferentiation, with potential to screen for inhibitors. B. Transcription of Ins2, Gcg, and Ngn3 in Min6 cells after two-day FoxO inhibitor (FoxOi, 1 μM unless otherwise stated) treatment, as measured by qPCR. C. Western blot of glucagon and insulin protein expression in Min6 cells following two-day FoxO inhibitor treatment. D. Measurement of insulin and glucagon intensity in Min6 cells treated with FoxO inhibitor at the single cell level, as quantified by immunofluorescence. E. Representative images of Min6 cells stained for insulin and glucagon after two-day treatment. Scale bars = 10 μm. F. RNAseq transcriptional expression changes in pancreatic endocrine factors after two-day FoxOi treatment versus control DMSO treatment in Min6 cells. All RNAseq results are representative of three biological replicates (n = 3). G. Gene set enrichment analysis (GSEA) showing downregulation of mouse (left) [58,59] and human (right) [61] beta-cell-specific genes with FoxO inhibitor treatment.
2.2. A high-content screen identifies inhibitors of beta-cell dedifferentiation
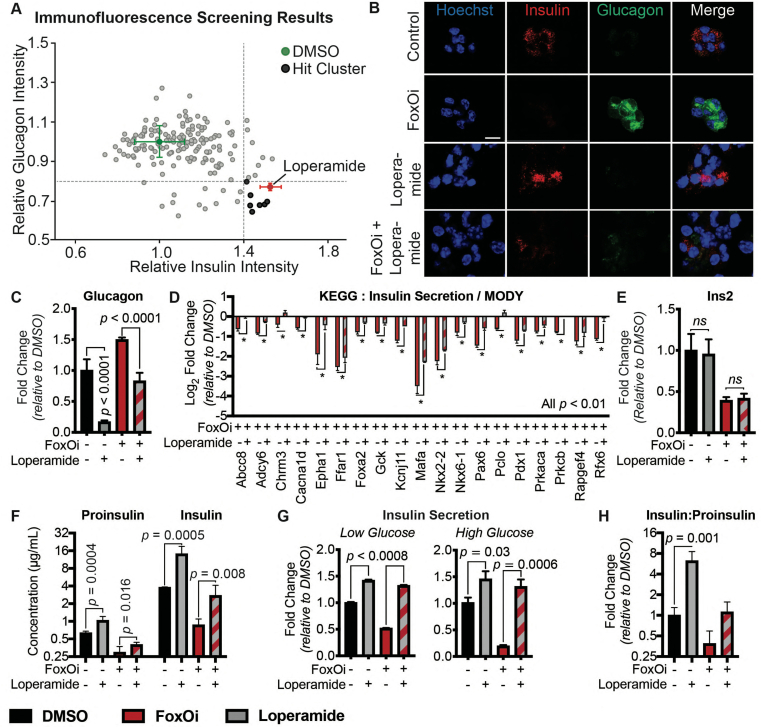
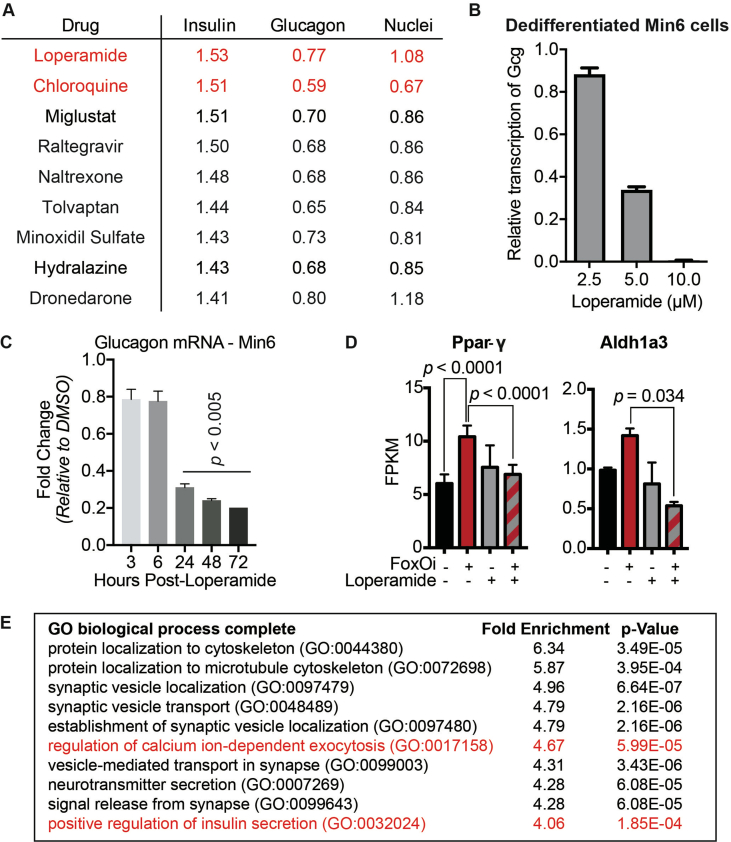
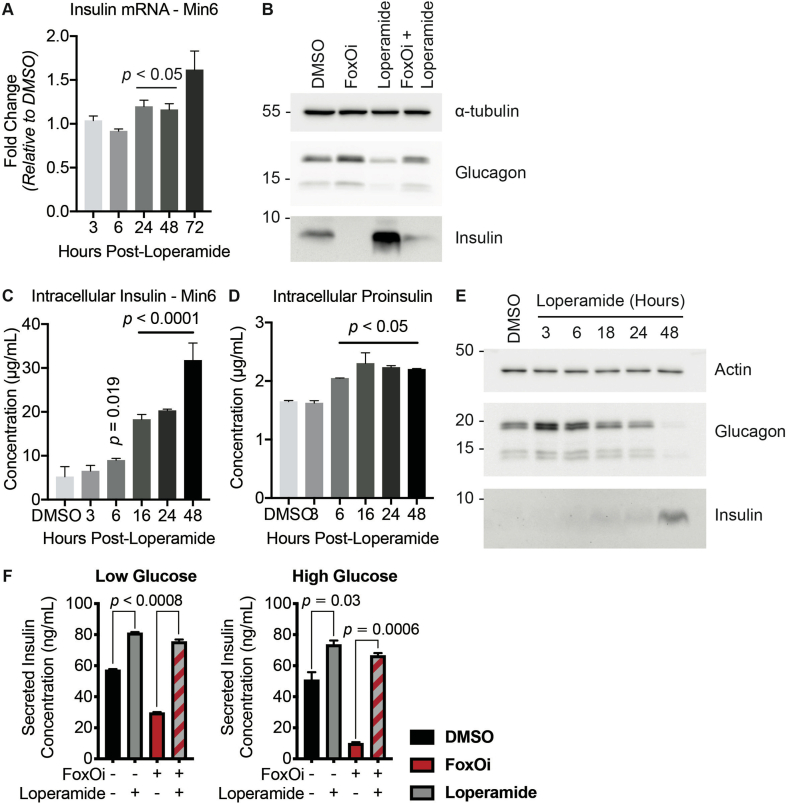
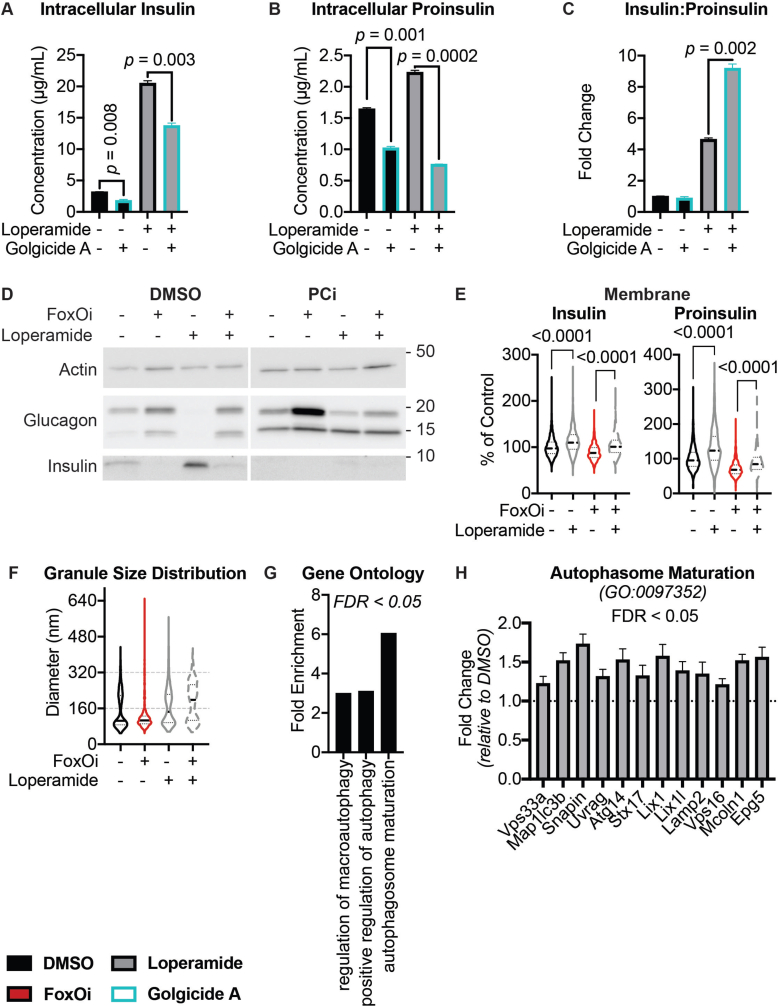
We next wanted to test the suitability of our cellular system for the identification of small-molecule dedifferentiation inhibitors. To do so, we used an immunofluorescence assay for glucagon and insulin on an automated microscope. The cells were treated with FoxOi for 48 h in combination with a library of 283 representative, clinically approved drugs covering the diversity of clinically used compounds with regards to chemical structure and molecular targets [12]. We found that a subset of these compounds caused an increase in insulin staining and a reduction in glucagon intensity compared to FoxOi treatment alone (Figure 2A and SFig. 2A). Among them, loperamide treatment was the most effective at antagonizing the actions of FoxOi without affecting cell numbers (Figure 2A,B, SFig. 2A and B). When we analyzed gene expression changes caused by loperamide, we observed that the compound decreased glucagon mRNA levels regardless of FoxOi treatment (Figure 2C and SFig. 2C). Globally, loperamide countered FoxOi-mediated transcriptional effects on transcription factors important for beta-cell development (Figure 2D), genes linked to beta-cell metabolic inflexibility and dysfunction [3,10] (SFig. 2D), and insulin secretion (Figure 2D and SFig. 2E). Interestingly, loperamide treatment had no effect on insulin mRNA levels (Figure 2E and SFig. 3A), yet resulted in a strong upregulation of intracellular insulin protein levels (Figure 2F and SFig. 3B-E). Enhanced insulin biosynthesis (i.e., translation, processing) or hindered secretion could account for such changes. Glucose-stimulated insulin secretion assays, however, revealed an increase in secreted insulin following loperamide treatment in both low- and high-glucose conditions (Figure 2G and SFig. 3F), which was able to fully counter the decrease induced by FoxOi. The most striking difference was noted when we compared the ratio of mature insulin to precursor proinsulin protein levels (SFig. 3C and D). Forty-eight-hour loperamide treatment induced a sixfold increase compared to controls, suggesting a strong stimulation of proinsulin processing (Figure 2H). Combined, these initial results propose a role for loperamide in the reinforcement of beta-cell character and function.
Figure 2.
High-content screen identifies loperamide to counter aspects of FoxOi-mediated beta-cell dedifferentiation.A. Overview of the small-molecule screening results. All the results were normalized to the DMSO control. Each dot represents the mean value of three replicates. Results whose standard deviations were more than 20% of the mean values were removed. Compounds that decreased mean cell numbers by more than 20% were considered cytotoxic and were also removed. Hit cluster = all compounds that yielded a greater than 1.5-fold increase in insulin intensity. B. Representative images of Min6 cells treated with/without FoxO inhibitor in combination with loperamide (5 μM unless otherwise stated). Scale bar = 10 μm. C. Loperamide suppresses FoxOi-induced glucagon transcription in Min6 cells, as measured by RNAseq with fragments per kilobase of transcript per million mapped reads (FPKM). D. Rescue of the expression of genes involved in insulin secretion with loperamide in Min6 cells treated with FoxOi for 48h, as measured by RNAseq. E. Loperamide does not affect insulin mRNA levels, as measured by RNAseq. F. Intracellular insulin and proinsulin protein concentrations in Min6 cells following 48h FoxOi/loperamide treatment, as measured by ELISA. G. Insulin secretion from Min6 cells in low (0.3 g/L) and high (3 g/L) glucose medium pretreated with FoxOi and loperamide for 48h. H. Insulin to proinsulin total protein ratio following 48h FoxOi/loperamide treatment, relative to DMSO control, measured by ELISA.
2.3. Loperamide increases FoxO1 expression and nuclear localization promoting changes in calcium signaling
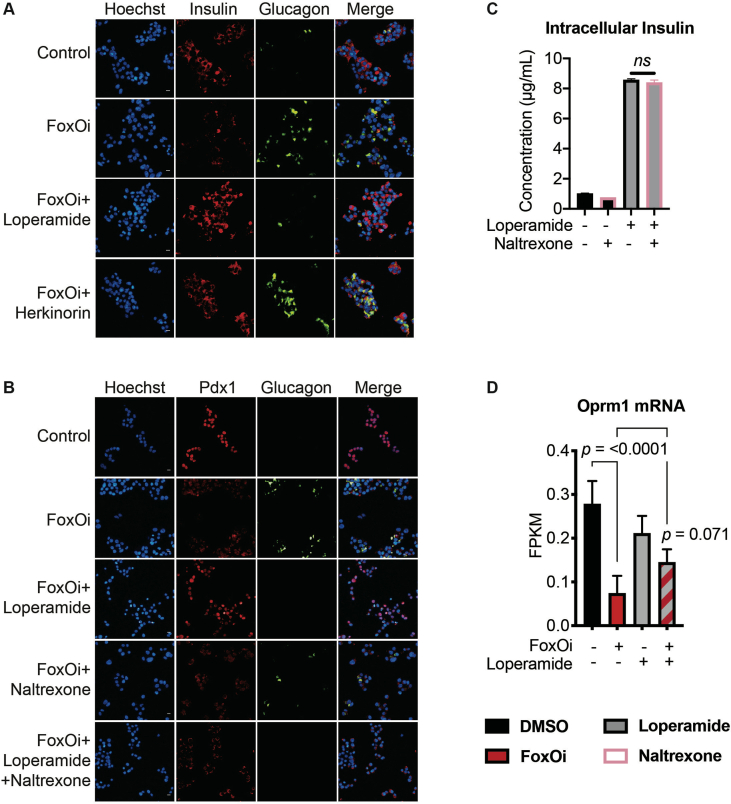
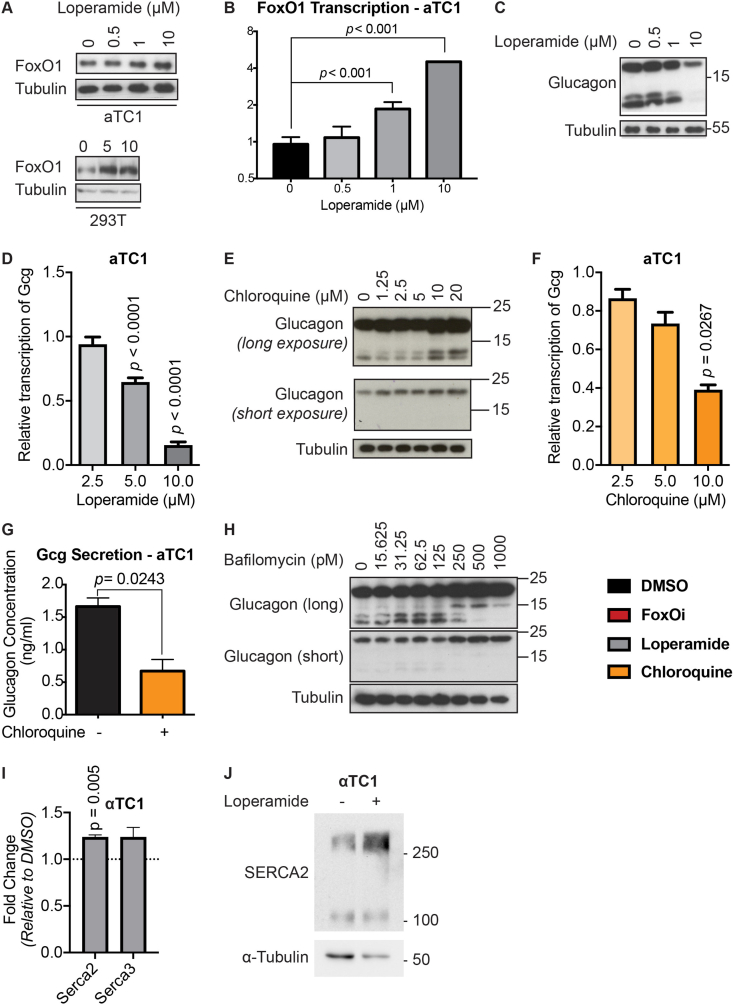
Loperamide is a mu-opioid receptor agonist [13] which has been used for the treatment of diarrhea since the 1970s (FDA application No. 017694). In contrast to other opioid receptor agonists, loperamide does not have effects on the central nervous system. To test if activation of mu-opioid receptors could inhibit beta-cell dedifferentiation and phenocopy loperamide's actions, we used a novel mu-opioid receptor agonist, herkinorin, in combination with FoxOi [14,15]. Like loperamide, herkinorin prevented the decrease of insulin levels following FoxO inhibition. However, herkinorin did not inhibit the upregulation of glucagon in Min6 cells (SFig. 4A). To further investigate on-vs. off-target effects of loperamide, the opioid receptor antagonist naltrexone was used in combination with loperamide. While naltrexone did not prevent the loperamide-mediated decrease in glucagon or increase in insulin levels, the beta-cell master regulatory transcription factor Pdx1 was repressed (SFig. 4B and C). In summary, loperamide's effects on dedifferentiation cannot fully be explained by the activation of mu-opioid receptors, supported by their noticeably low expression level in Min6 cells (SFig. 4D).
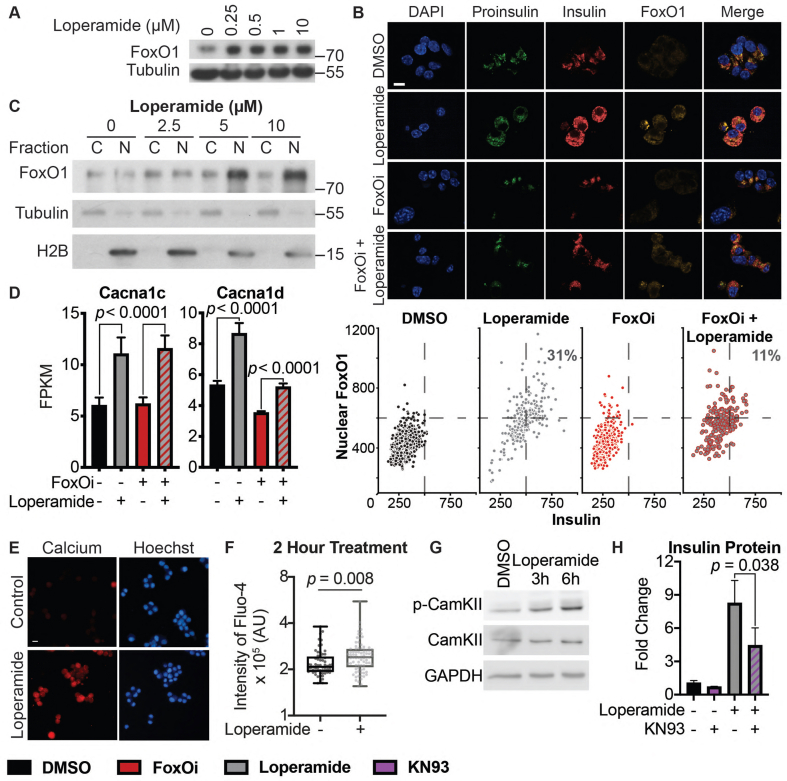
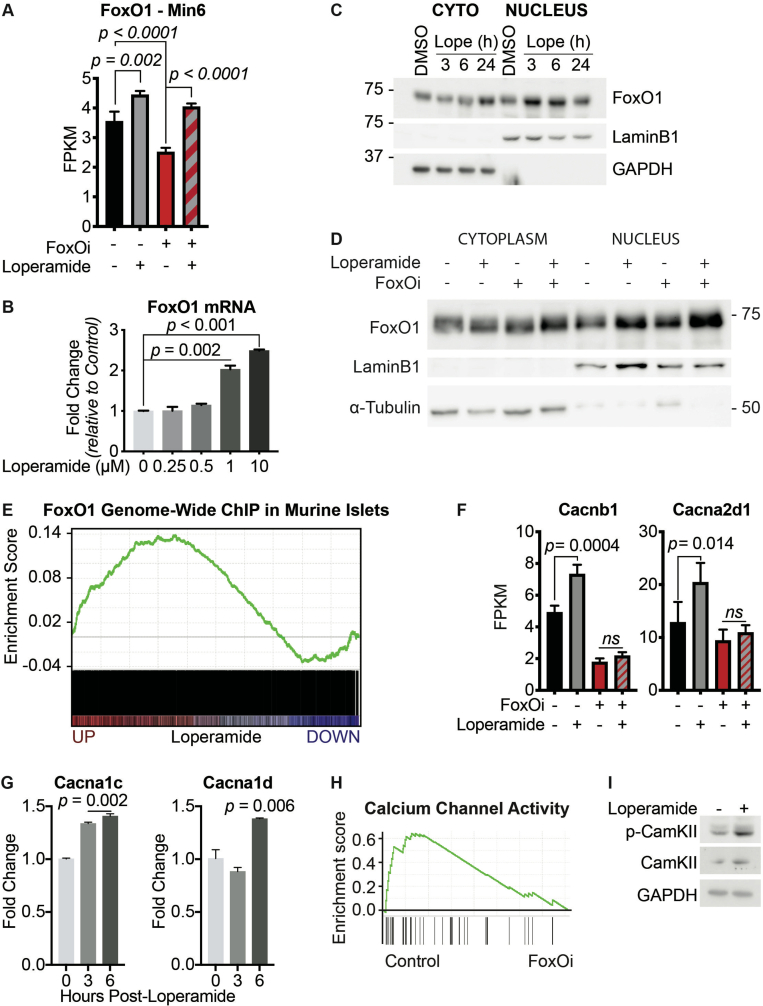
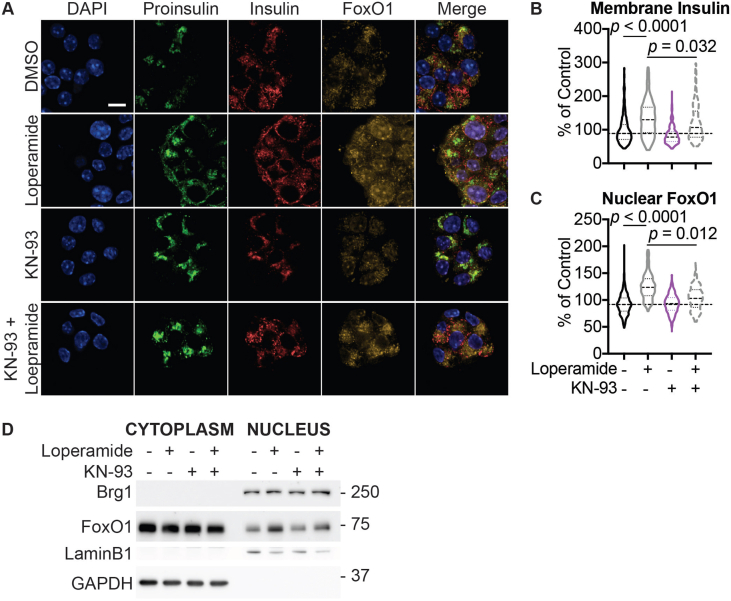
Based on FoxOi's published role as a selective FoxO1 inhibitor [9], we tested whether loperamide treatment had any effect on the expression levels or activity of FoxO1. Remarkably, we observed an increase in FoxO1 mRNA and protein levels (Figure 3A,B, SFig 5A and B). Moreover, loperamide was able to completely rescue FoxOi's suppression of FoxO1 transcription (SFig. 5A). Further experiments by subcellular fractionation of Min6 cells treated with loperamide uncovered that the increase in FoxO1 protein is mostly restricted to the nucleus (Figure 3B,C), indicating that loperamide induces an increase in active nuclear FoxO1 protein. This increase occurs within hours and is maintained when loperamide is co-treated with FoxOi (Figure 3B, SFig 5C and D). Immunofluorescence experiments highlight a positive cellular correlation between nuclear FoxO1 and insulin expression levels upon loperamide treatment, with higher levels of both evident in approximately one-third of cells compared to control (Figure 3B).
Figure 3.
Loperamide increases FoxO1 expression and nuclear localization promoting changes in calcium signaling.A. Western blot showing an increase of FoxO1 protein with loperamide treatment in Min6 cells. B. Top: Representative immunofluorescence images of Min6 cells treated with loperamide and FoxOi for 48h. Scale bar = 10 μm. Bottom: Quantification of insulin and nuclear FoxO1 intensities at the single cell level. C. Western blot of FoxO1 protein expression in the cytoplasm vs. nucleus of Min6 cells following different doses of loperamide treatment. D. mRNA levels of L-type voltage-gated calcium channels Cacna1c and Cacna1d in Min6 cells upon 48h loperamide and FoxOi treatment, as measured by RNAseq. E. Fura-2 staining in Min6 cells pretreated with loperamide for 18h. F. Quantification of Fluo-4 staining at the single cell level in Min6 cells treated with loperamide for 2h. G. Western blot highlighting an increase in p-CamKII relative to total CamKII protein levels in Min6 cells when treated with loperamide for the stated hours. H. Total mature insulin protein levels in Min6 cells treated with loperamide and 1 μM KN-93 for 24h.
To determine whether the increase in nuclear FoxO1 also corresponded with an increase in genomic binding and activity, we compared our loperamide RNAseq dataset to published global FoxO1 ChIPseq results from murine islets [16]. A core positive enrichment was observed between genes bound by FoxO1 and upregulated with loperamide (SFig. 5E). Within those genes, a 26-fold enrichment in high-voltage-gated calcium channels (VGCC) was identified (GO: 0061577) (Figure 3D, SFig 5F and G). Notably on the list were CaV1.2 (Cacna1c) and CaV1.3 (Cacna1d), the two major L-type calcium channel isoforms necessary for insulin secretion [17,18]. Interestingly, FoxOi treatment appears to globally downregulate genes related to calcium channel activity (SFig. 5H). Their loss is an early marker of beta-cell dysfunction, but loperamide appears capable of rescuing their levels independently of FoxOi (Figure 3D). In effect, previous studies on loperamide have suggested it increases free intracellular calcium levels [62,63]. Coupled with this knowledge, we utilized the Fura-2 and Fluo-4 calcium stains to confirm a strong increase in intracellular calcium levels with loperamide (Figure 3E,F).
Increased intracellular calcium is able to trigger FoxO1 nuclear translocation in a CamKII-dependent pathway in hepatocytes [19]. We observe an accumulation of active, phosphorylated CamKII in Min6 cells within hours of loperamide exposure, sustained during prolonged treatment (Figure 3G, SFig. 5I). Treatment with KN-93, a CamKII inhibitor, significantly halted loperamide's increase in insulin protein and FoxO1 nuclear localization (Figure 3H, SFig. 6A-D), supporting a role for the kinase downstream of loperamide and reinforcing a key role for calcium in beta-cell maintenance.
2.4. Loperamide counters FoxOi-mediated changes in pH
It is widely accepted that the regulation of intracellular pH and Ca2+ concentration play vital roles in pancreatic hormone processing and secretion [20,21]. Calcium and pH levels facilitate and dictate every stage of insulin's biosynthesis process. Starting from proper proinsulin folding in the neutral rER, to proinsulin processing in the acidic and Ca2+-rich secretory granules. More specifically, the primary insulin processing enzymes, PCSK1/3 and PCSK2, require a minimum of 1 mM Ca2+ and an acidic pH of 5.0–5.5 to properly exert their function [22]. Furthermore, the final secreted hormone has an important autocrine function in regulating future transcription and secretion [23,24]. Perturbations to this delicate feedback loop via aberrant intracellular pH and Ca2+ changes could explain FoxOi's role in effecting beta-cell dysfunction and loperamide's counteraction.
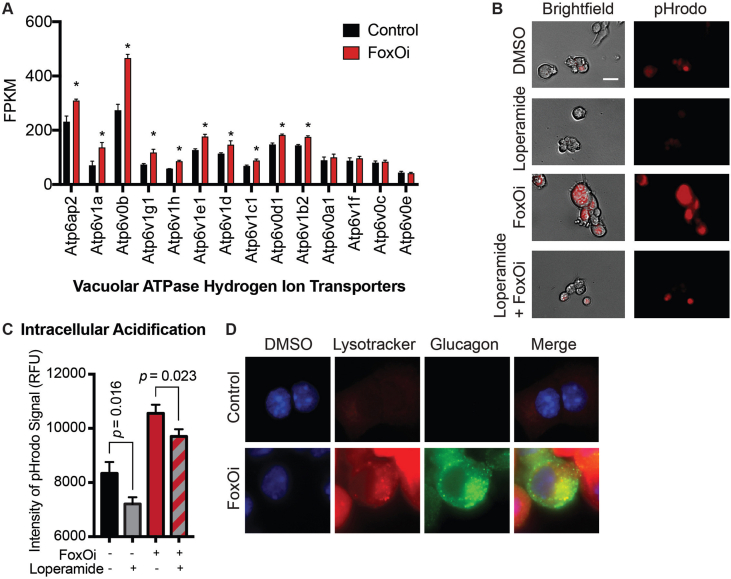
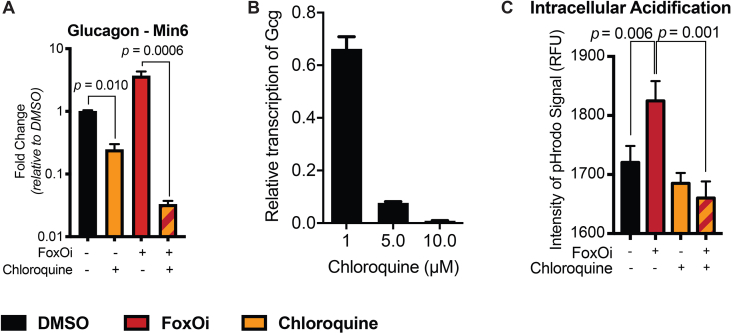
Consequently, we observed an increase in the majority of vacuolar ATPase subunits upon FoxO inhibition (SFig. 7A). This was accompanied by a global and glucagon granule-specific increase in intracellular acidification (SFig. 7B-D). Treatment with loperamide reduced this FoxOi-induced intracellular acidification (SFig. 7B and C). Interestingly, another hit compound from our screen (SFig. 2A), chloroquine, is known to increase lysosomal pH [25]. We show that chloroquine treatment phenocopies many aspects of loperamide's effects in the FoxOi-mediated beta-cell dedifferentiation model (SFig. 8), reaffirming the role pH plays in hormone processing.
2.5. Loperamide alters the ER proteome, rescuing FoxOi-induced arrest of insulin granule maturation and promotes autophagy
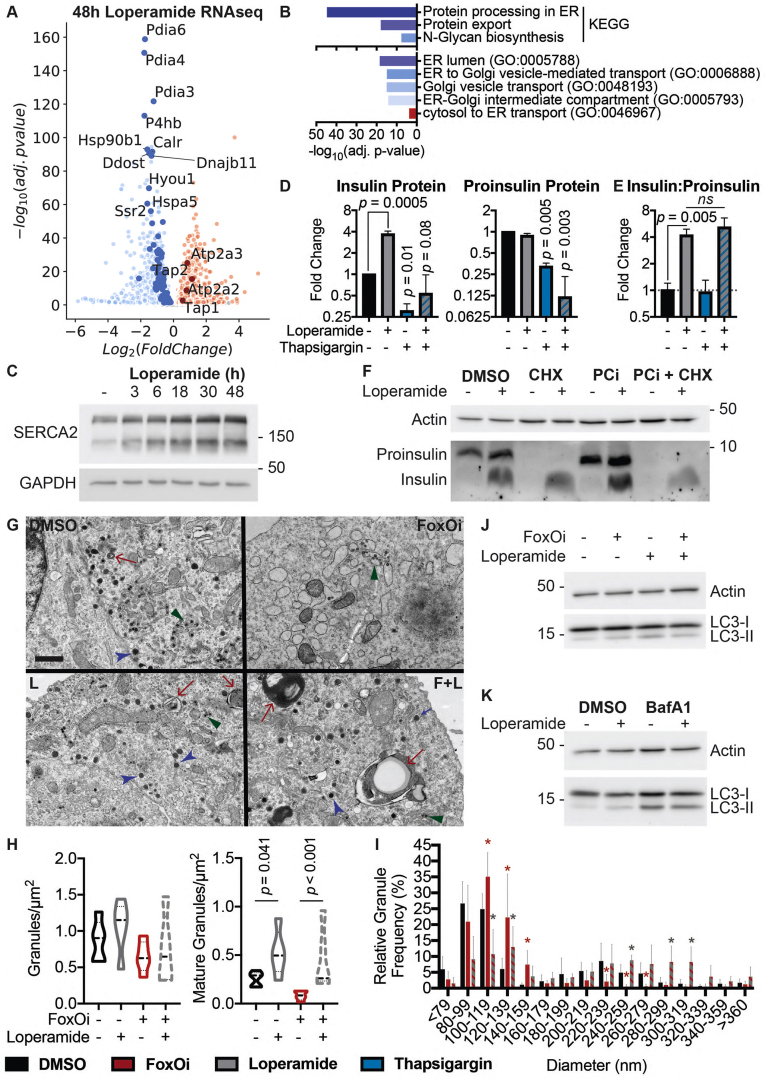
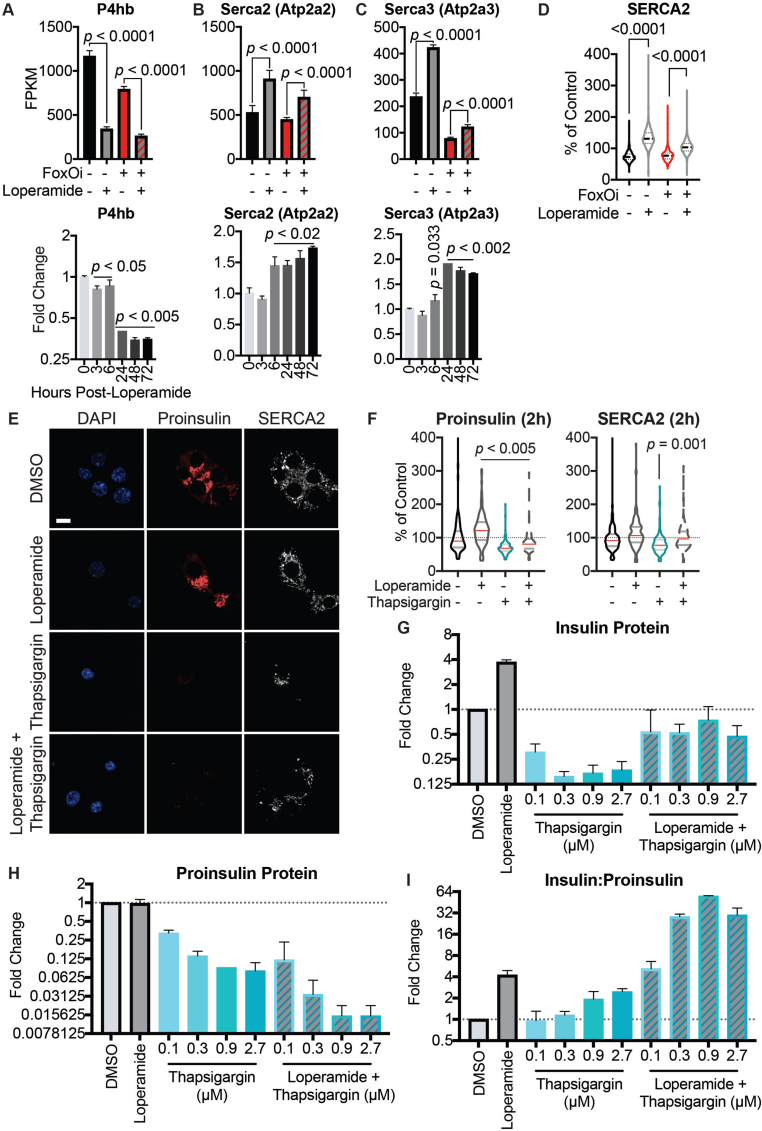
Analysis of our gene expression datasets for other deregulated pathways found a large subset of ER proteins involved in protein processing and transport to be downregulated following treatment of Min6 cells with loperamide (Figure 4A,B). Notably, the most significantly decreased genes all corresponded to protein disulfide isomerases (i.e., P4hb, Pdia3, Pdia4, and Pdia6) (Figure 4A, SFig. 9A). While necessary for the proper folding of proinsulin, knockdown of P4hb induces faster export, maturation, and secretion of newly synthesized proinsulin with an increased insulin to proinsulin ratio [26], highly reminiscent of loperamide's phenotype (Figure 2F–H), and a possible explanation for the increased proinsulin turnover. Interestingly, loperamide also upregulated both ER calcium ATPases: Serca2 and Serca3 (Figure 4A, SFig. 9B-D), with increases in Serca2 protein and mRNA observed as early as 3 h post-loperamide treatment (Figure 4C and SFig. 9B). Serca2's locus is bound by FoxO1 in multiple tissues [16,27,28], suggesting its upregulation is downstream of FoxO1's translocation. To understand SERCA's role in loperamide's phenotype, we treated our beta-cell line with the SERCA inhibitor, thapsigargin. Thapsigargin induced a significant decrease in proinsulin and SERCA protein levels already within 2 h (SFig. 9E and F). Twenty-four-hour thapsigargin treatment further decreased both proinsulin and mature insulin protein levels (Figure 4D, SFig. 9G and H). Interestingly, while co-treatment of loperamide with thapsigargin incited a decrease in the total levels of proinsulin and insulin, the ratio of insulin to proinsulin remained significantly increased (Figure 4E, SFig. 9I). This suggests that while proinsulin generation and export require ER calcium stores, loperamide's role in granule maturation does not. Disruption of the trans-Golgi network (TGN), the site of secretory granule packaging and proinsulin processing initiation, perfectly mirrored this result (SFig. 10A-C). Contrastingly, the prolonged loss of prohormone convertase activity depleted all insulin stores and countered loperamide's glucagon decrease (SFig. 10D).
Figure 4.
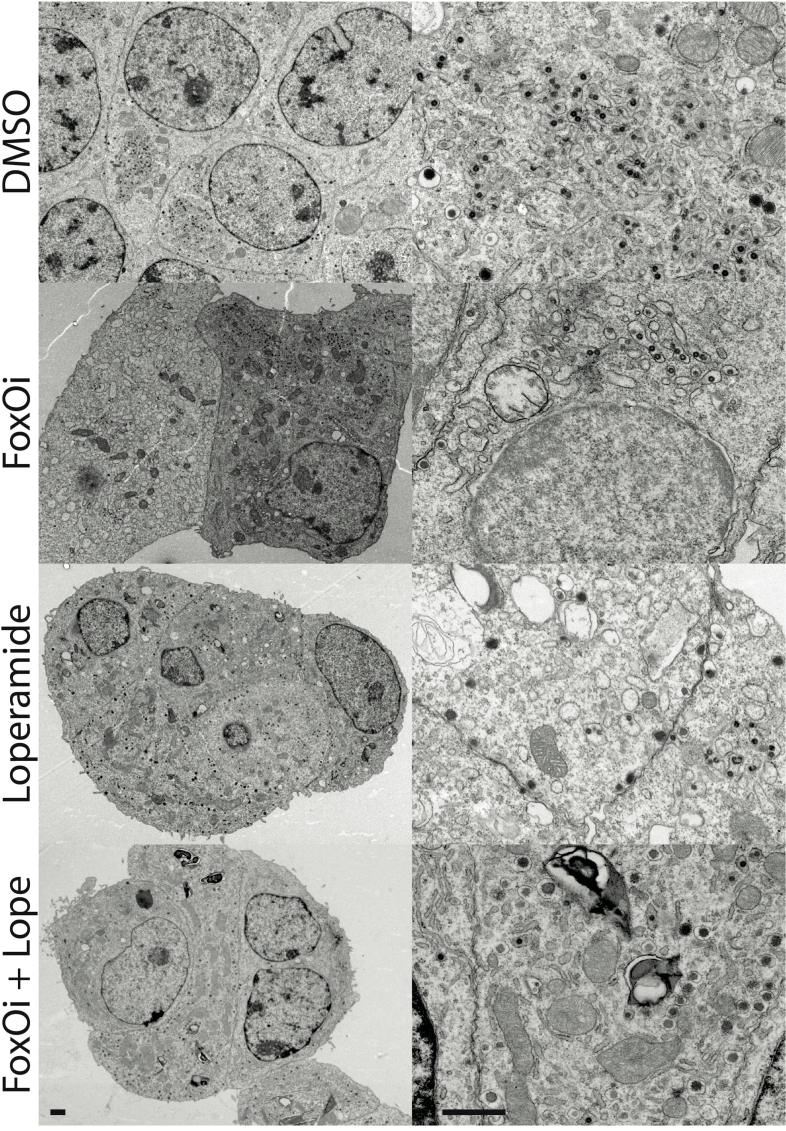
Loperamide alters ER proteome, rescuing FoxOi-induced arrest in insulin granule maturation and promotes autophagy.A. Volcano plot of gene expression changes upon 48h loperamide treatment, as measured by RNAseq. B. Overview of top enrichment terms for genes both significantly altered by loperamide relative to control and significantly rescued when co-treated with FoxOi relative to FoxOi. Volcano plot highlights the top KEGG term (protein processing in ER) in blue and GO:0046967 in red. C. Western blot showing increase in SERCA2 protein upon loperamide treatment in Min6 cells. D. Total intracellular insulin and proinsulin protein levels in Min6 cells treated with loperamide and 100 nM thapsigargin for 24h, as quantified by ELISA. E. Ratio of mature insulin to proinsulin protein. F. Western blot of proinsulin and insulin protein levels following 48h treatment of Min6 cells with loperamide +/− 200 μg/mL cycloheximide (CHX) or 10 μM prohormone convertase inhibitor (PCi) for the final 24h. G. Representative electron microscopy images of Min6 cells treated with FoxOi or loperamide. L = Loperamide; F + L = FoxOi + Loperamide. Green arrows pointing to immature secretory granules. Red arrows highlighting (autophago-)lysosomes. Blue arrows indicate mature insulin secretory granules. Scale bar = 1 μm. H. Quantification of insulin granule density in electron microscopy images. Total number of granules per μm2 (right) and number of mature (>160 μm in diameter) granules per μm2. N = 5 cells per treatment condition. I. Histogram of insulin granule size distribution upon FoxOi/loperamide treatment. DMSO = black; FoxOi = red; FoxOi + Loperamide = red/gray stripes. N = 5 cells per treatment condition. J. Levels of LC3B in Min6 cells treated with FoxOi and loperamide for 48h. K. Levels of LC3B in Min6 cells treated with loperamide for 48h +/− 200 nM bafilomycin A1 for the final 2h.
The importance of a processable proinsulin pool for loperamide's action was further strengthened upon co-inhibiting translation and prohormone convertase activity. While translation inhibition predictably diminished proinsulin levels, downregulation of mature insulin protein was only observed following prohormone convertase co-inhibition (Figure 4F). This reinforces the notion that while active translation is necessary to maintain the proinsulin pool, loperamide sustains mature insulin levels via a different mechanism, i.e., enhanced processing within secretory granules, which relies heavily on prohormone convertases.
In line with increased export and proinsulin processing, the analysis of FoxOi and loperamide-treated Min6 cells revealed a discrepancy in the localization of insulin and proinsulin. Upon FoxOi treatment, insulin and proinsulin stains are highly colocalized and restricted to regions in the nuclear periphery, whereas loperamide induces an increase in granules near the membrane (Figure 3B, SFig. 10E).
Ultrastructural studies reinforce previous findings, revealing an overall decrease in insulin secretory granules with FoxOi, a phenotype prevented upon loperamide addition (Figure 4G and SFig. 11). Interestingly, an abundance of smaller (80–120 nm) granules with an electron-lucent core is evident, specifically in DMSO and FoxOi-treated Min6 cells (Figure 4G–I and SFig. 11; green arrows). These most likely represent very early stage or stalled secretory granules [29]. Strikingly, loperamide treatment, regardless of FoxOi, minimized the number of these small nascent granules in favor of an observable increase in larger (200–300 nm) mature insulin granules with a concomitant increase in (autophago-)lysosomes (Figure 4G–I and SFig. 10F and 11; blue and red arrows). This is corroborated by loperamide's transcriptional enrichment of genes involved in autophagosome maturation (SFig. 10G and H). To further validate these observations, we tested whether loperamide promotes autophagy by blotting for the autophagosome marker LC3B. On its own, loperamide induces a mild increase in LC3B-II and decrease in LC3B–I levels (Figure 4J). Co-treatment with bafilomycin A1 (BafA1), a potent inhibitor of autophagolysosomal fusion, revealed no synergistic increase in LC3B-II levels, indicative of loperamide stimulating autophagic flux (Figure 4K). Active autophagy is essential for the maintenance of functional healthy beta cells and optimal insulin content [[30], [31], [32]]. Hence, it appears loperamide stimulates both the maturation of insulin granules, while equally regulating their concentration by promoting healthy lysosomal recycling.
Many of these pathways are conserved in the different endocrine cell types, and we therefore tested drug effects in the alpha-cell line aTC1 (SFig. 12). We observed that also in alpha-cells loperamide represses glucagon and increases FoxO1 and Serca2 levels.
2.6. Loperamide counters FoxO inhibition in multiple model systems
FoxO proteins are known to promote dauer formation in C. elegans [33] (Figure 5A). Therefore, we used it as a model organism to test the effects of FoxOi and loperamide. Interestingly, the chemical inhibition of FoxO decreased the dauer formation rate (Figure 5B, left), as expected from loss of FoxO, while loperamide increased it (Figure 5B, right). These results support the conclusion that loperamide's effects on FoxO proteins are conserved in different organisms.
Figure 5.
Loperamide counters FoxO inhibition in different model systems.A. Overview of C. elegans development in favorable or unfavorable environmental conditions. B. Effects of FoxOi and loperamide treatment on the FOXO-dependent developmental process of Dauer formation in C. elegans. Eggs of CB1370 animals were hedged and grown at 23 °C (left panel) or 22 °C (right panel) in the presence of FoxOi or loperamide at the indicated concentrations. In the absence of compounds (0 μM), the fraction of animals forming Dauer was 20.2% (left panel) and 3.2% (right panel). Compound-induced fold changes of this fraction are shown. N = 3; 100 animals per condition and replicate. C. Representative images of developmental defects in zebrafish larvae following 48h FoxOi treatment and rescue with loperamide. D. Quantification of insulin or glucagon-positive cells following loperamide treatment in zebrafish larvae. Ncontrol = 12; Nloperamide = 21. E. Representative images of pancreatic human islets stained for insulin and glucagon. Scale bars = 10 μm. On the right: Summary of the population distribution change in the human islets. F. Measurement of INS and GCG transcription in human islets treated with loperamide by RT-qPCR. All the data points are normalized to DMSO control, n = 4. G. Transcription of GCG, INS, and PDX1 in pancreatic islets from human diabetic donors treated for 48h with loperamide, as measured by RT-qPCR, n = 2. H. Glucose-stimulated insulin secretion assay on diabetic human islets pretreated with loperamide for 48 h, n = 2. Basal = 0.3 g/L glucose; stimulated = 3 g/L glucose. I. Relative insulin and Pdx1 mRNA levels in islets from db/db mice pretreated with loperamide for 48h.
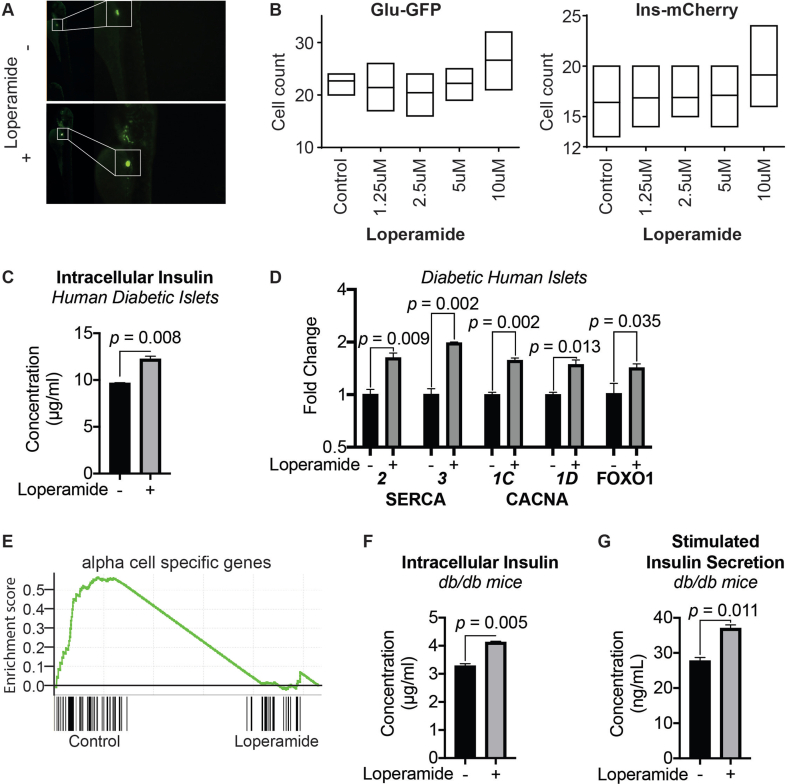
We next tested the compounds in a zebrafish model. Interestingly, loperamide was able to rescue a FoxOi-induced developmental defect in the zebrafish (Figure 5C), underscoring its ability to counteract FoxOi's global effects. To observe the effects of loperamide during islet development, we utilized zebrafish larvae carrying double reporters: Gcg-GFP and Ins-mCherry. High concentrations of loperamide resulted in increased alpha and beta-cell numbers (Figure 5D, SFig. 13A and B).
Next, we treated intact human islets from healthy donors with loperamide for 48h to test the compound's effects on mature, ex vivo, human alpha and beta cells. Compared to the control population, loperamide treatment decreased the number of alpha cells and increased beta-cell and double-positive cell numbers (Figure 5E). Loperamide treatment also dramatically decreased glucagon transcription, however, without a significant effect on insulin transcription (Figure 5F), consistent with previous findings. Contrastingly, the analysis on human islets from diabetic donors treated with loperamide revealed an increase in insulin mRNA and protein levels (Figure 5G, SFig. 13C). This was accompanied by a decrease in glucagon and increase in PDX1, SERCA2, SERCA3, CACNA1C, CACNA1D, and FOXO1 expressions (SFig. 13D). At the transcriptome-wide level, loperamide treatment led to a general suppressive effect on alpha-cell-specific genes (SFig. 13E). Loperamide also appeared to rescue the glucose responsiveness of these diabetic islets, sparking a significant increase in insulin secretion upon high-glucose treatment (Figure 5H). We further validated these results in a diabetic murine model, with similar increases in insulin mRNA, protein, and secretion observed upon loperamide treatment (Figure 5I, SFig. 13F and G). Overall, these results strengthen the notion that loperamide has a positive influence on beta-cell identity and function also in relevant models of diabetic islets.
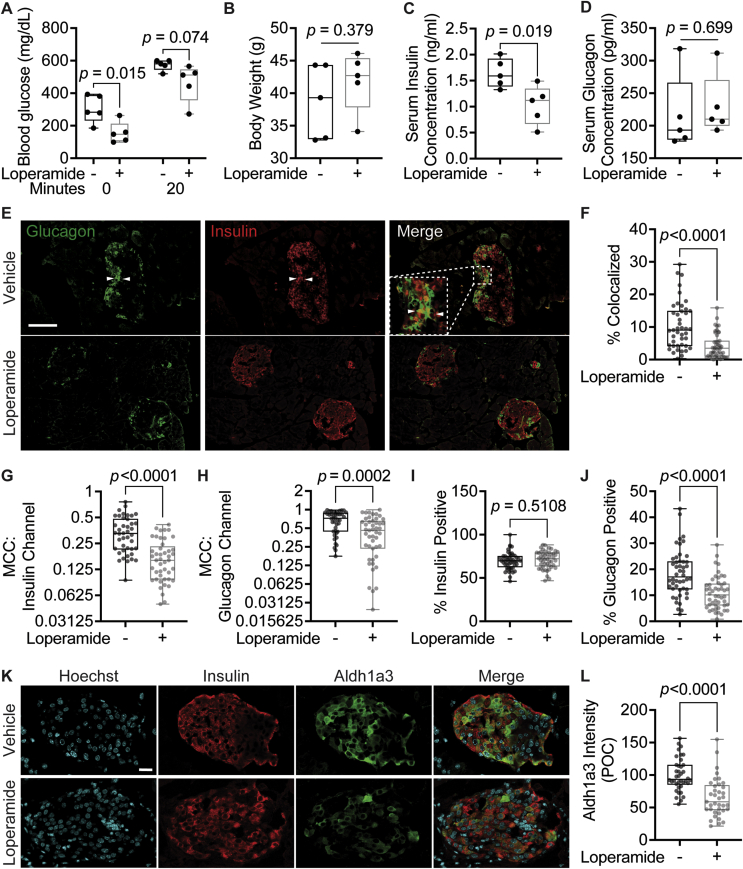
2.7. Loperamide decreases fasting blood glucose and double-positive islet cells in diabetic mice
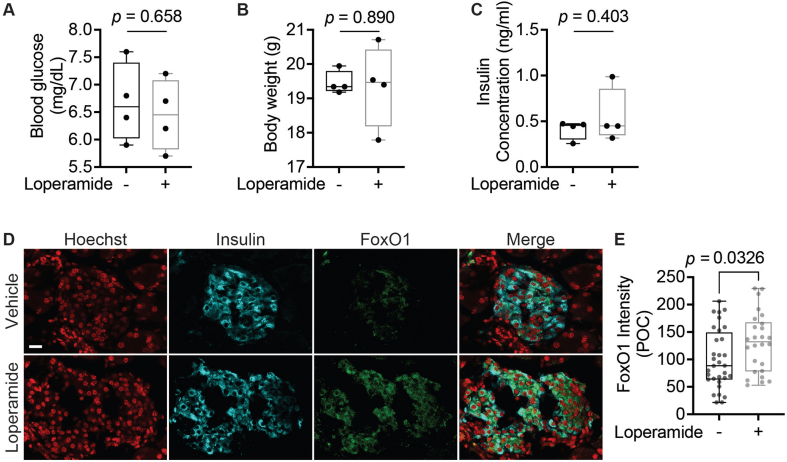
At present, no studies have focused on the long-term effects of loperamide treatment on glucose metabolism in a diabetic setting. Thus, we studied its systemic effects by treating 8-week-old db/db mice on a chow diet with loperamide or vehicle (a mix of DMSO, PEG300, Tween-80, and PBS at a 1:4:0.5:4.5 ratio) for 4 weeks. Loperamide treatment triggered a striking decrease in fasting blood glucose levels with no change in overall body weight (Figure 6A,B). The vehicle-treated mice had a high average fasting blood glucose concentration of 304.5 mg/dL characteristic of a diabetic mouse model. The loperamide-treated group, on the other hand, had an average fasting concentration of 156.4 mg/dL, almost half that of the control-treated mice, and well below the 250 mg/dL threshold generally used to stratify diabetic mice [34]. Furthermore, following a 20-minute IP glucose tolerance test (IPGTT), blood glucose concentrations remained lower in the loperamide-treated mice (Figure 6A). Interestingly, serum insulin concentrations post-IPGTT were also significantly decreased with loperamide (Figure 6C). Based on the observed decrease in blood glucose levels, one would expect a concomitant increase in circulating insulin. These results, therefore, suggest that loperamide treatment likely accelerates glucose clearance, indicative of improved insulin sensitivity. Conversely, glucagon serum levels remain unchanged relative to control (Figure 6D). These changes were specific to diabetic mice, as no significant effects were observed in wild-type-treated animals (SFig. 14A-C).
Figure 6.
Loperamide has positive systemic and islet-specific in vivo effects in diabetic mice.A. Serum glucose concentrations after overnight fast (0 min) and 20 min after IP injection with 1 g/kg glucose (20 min). N = 5 mice/treatment. B. Body weight measured after 4-week treatment. C. Serum insulin concentration measured by ELISA. D. Serum glucagon concentration measured by ELISA. E. Representative immunofluorescence panel of pancreas sections stained with insulin and glucagon antibodies. White arrows point to insulin/glucagon double-positive cells. Scale bar = 100 μm. F-J. Quantifications of immunofluorescence images. N = 45 islets/treatment from five different mice/treatment. F. Percent of total islet pixel number representing colocalized red/green channel pixels. G-H. Thresholded Manders’ correlation coefficients for the insulin (red) channel or glucagon (green) channel per islet. I-J. Percent of total islet volume occupied by the insulin (red) channel or glucagon (green) channel pixels. K. Representative immunofluorescence panel of db/db mouse pancreas sections 4-week post-loperamide treatment, stained for insulin and Aldh1a3. Scale bar = 20 μm. L. Quantification of immunofluorescence images. Cytoplasmic Aldh1a3 intensity was only calculated within insulin-positive cells. POC = percent of control. N = 38 islets/treatment from five different mice/treatment.
Next, we wanted to identify whether long-term loperamide treatment had any islet-specific effects. Islet immunofluorescence analysis in the control mice revealed a large number of insulin/glucagon double-positive cells (Figure 6E,F), accounting for 10% of total islet volume, characteristic of diabetic islets [35,36]. Strikingly, this number was significantly reduced in the loperamide-treated animals (Figure 6F). Thresholded Manders' correlation coefficient (MCC) colocalization quantifications reiterated these results, with decreased colocalization observed in both the insulin and glucagon channels (Figure 6G,H). This effect was especially pronounced in the insulin channel, underscoring that loperamide treatment dramatically increased the percent of mono-hormonal insulin-positive cells (Figure 6G). Comparisons of total islet volume occupied by insulin or glucagon signal reinforced these findings, highlighting a decrease in glucagon area from 18% to just over 10% of the total with loperamide, with no change to insulin (Figure 6I,J). Hence, it would appear loperamide specifically represses the emergence of glucagon expression in insulin-positive cells. The high Aldh1a3 expression observed in the control mice is a further characteristic of dedifferentiated beta cells [10], also strikingly reduced by loperamide treatment (Figure 6K,L). Mechanistically, we also detected a noticeable increase in FoxO1 protein levels upon loperamide treatment (SFig. 14D and E), in line with observations in Min6 cells (Figure 3A,B) and in diabetic human islets (SFig. 13D). FoxO1 deficiency is another known driver and marker of beta-cell dedifferentiation [4]. Thus, loperamide's ability to enhance FoxO1 expression and minimize Aldh1a3 levels and double-positive cell numbers suggest loperamide's reinforcement of beta-cell character is conserved in vivo, and for the first time show that long-term loperamide treatment has globally positive effects on glucose metabolism.
3. Discussion
Beta-cell failure as a result of sustained insulin resistance is the key driver in the pathogenesis of type II diabetes. Beta-cell death, dysfunction, and dedifferentiation have been proposed as the main sources of this failure [6]. Interfering with this process might prevent, or at least delay, the progression of diabetes and therefore underscores an important therapeutic target.
Physical evidence supporting the presence of beta-cell dedifferentiation includes increased numbers of hormone-negative cells [5,37], double-positive cells [35,36], and degranulated beta cells [38] in islets from type II diabetic patients. The molecular basis for this loss of beta-cell identity remains to be fully elucidated. Several proposed mechanisms exist in addition to the loss of FoxO1 signaling, such as decreased PRC2-mediated progenitor gene silencing [39], oxidative-stress-mediated upregulation of microRNAs targeting beta-cell transcription factors [[40], [41], [42], [43]], and hypoxia-induced adaptive UPR inactivation [21].
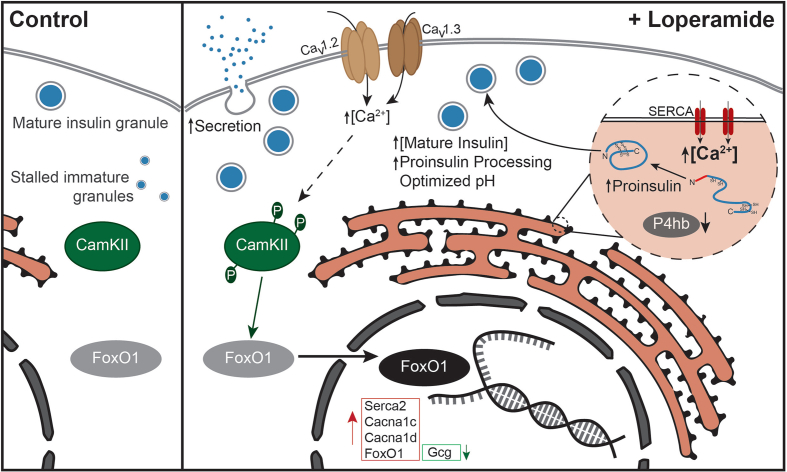
We developed an in vitro cell system that allowed us to specifically study and understand the molecular mechanisms underlying FoxO1-mediated beta-cell dysfunction. In our cellular system, beta-cell dedifferentiation can be robustly induced by a two-day treatment with a small-molecule FoxO inhibitor. This phenotype is not restricted to the cell line model, as we have recently shown that the compound also induces beta-cell dedifferentiation in primary human and murine pancreatic islets [44]. With this cellular beta-cell dedifferentiation model, we uncovered that increased intracellular acidification is a critical step in beta-cell dedifferentiation. By performing a high-content chemical screen, we identified the approved drug loperamide to counteract the observed dedifferentiation by neutralizing intracellular pH, mobilizing intracellular calcium ions, enhancing insulin processing, and elevating FoxO1 expression and activity (Figure 7).
Figure 7.
Overview of loperamide's effects in beta cells.
More specifically, loperamide triggers a calcium and CamKII-dependent nuclear shift of FoxO1 with a concomitant increase in the expression of over 1000 of its target genes. An important fraction of these genes is involved in calcium ion mobilization into the cytoplasm and the ER. The most notable of these: SERCA2, CACNA1C, and CACNA1D are reduced in the islets of diabetic mice and humans [17], highlighting their importance in beta-cell maintenance. Loperamide's increase in SERCA2, coupled with its decrease in protein disulfide isomerases, most likely provides the heightened pool of exported proinsulin near the membrane. In effect, glucose uptake triggers an increase in insulin processing, mature granule numbers, and secretion by invoking an initial increase in ER calcium levels [45], highlighting a parallel between glucose and loperamide. Secretion relies on heightened local concentrations of calcium, ten times higher than the cytoplasmic average. These are observed near clusters of CaV1.2, CaV1.3, and actively secreted insulin granules [18,20]. This clustering is disrupted in type II diabetic islets, indicative of their stunted secretion. Loperamide successfully counters this by increasing secretion and the expressions of CaV1.2 and CaV1.3 in diabetic islets.
Loperamide equally neutralizes intracellular pH which is known to further affect calcium signaling and secretion [46]. The acidification of secretory vesicles is an important event required for the posttranslational processing of proinsulin to the mature hormone [47]. In effect, loss of the a3 isoform of V-ATPase, which is highly expressed in alpha and beta cells, results in reduced insulin and glucagon secretion [48]. Typically, the effects of increasing granule pH on impaired prohormone convertase activity are studied. Since prohormone convertases have a relatively tight pH optimum in the range of pH 5–5.5, the lowered pH we observe following FoxO inhibition might also be responsible for impaired hormone processing and secretion. Finally, since glucose controls V-ATPase activity, which in turn is required for PKA signaling [49], it is clear that intracellular pH, calcium, and glucose signaling are all tightly interwoven and that all three are affected by FoxOi and loperamide.
Limited literature exists regarding loperamide's in vivo effects on glucose metabolism. Encouragingly, a few studies observed that IV-injected loperamide dramatically decreased serum glucose concentrations in streptozotocin (STZ)-induced diabetic rats within three days [[50], [51], [52]]. Interestingly, this decrease was only observed in STZ-induced and obese Zucker rats [53] but not in non-diabetic Wistar rats, suggesting it may functionally ameliorate organs affected in diabetes such as islets, liver, or muscle. Consistently, patent US20050234100 specifically attributes loperamide's effects to the improvement of insulin sensitivity in fructose-induced insulin-resistant rats. Unfortunately, all of these represented short-term treatments, from minutes to three days. Consequently, ours is the first study to focus on the long-term effects of loperamide on glucose metabolism. Excitingly, we could show that the decrease in blood glucose concentrations is maintained after 4-week treatment, as well as the potential increase in insulin sensitivity. In effect, our observations that (1) long-term in vivo loperamide treatment decreases double-positive islet cell numbers and Aldh1a3 levels in diabetic mice and (2) ex vivo loperamide treatment increases insulin secretion and expression in diabetic islets are the first to support the hypothesis that loperamide exerts its effects on glucose metabolism directly via the islets. However, the decrease in serum insulin levels post-loperamide coupled to the unchanged serum glucagon concentrations suggest the effect is more complex and likely relies on multiple tissues.
In summary, we established a novel screenable cellular assay to model beta-cell dedifferentiation, amenable to transcriptional studies, and high-throughput screening. With this model, we identified the small-molecule loperamide, which successfully increases mature insulin protein levels and secretion in various models of beta-cell failure. These results should help accelerate our understanding of the mechanisms underlying beta-cell dedifferentiation and aid in the development of future therapies for beta-cell failure in diabetes.
4. Materials and methods
4.1. Reagents
Antibodies used in this project are directed against insulin (Agilent Dako IR00261-2 and Abcam ab7842 (Figure 4F), glucagon (Abcam ab92517), histone H2B (Cell Signaling Technology 2934), beta-actin (Abcam ab8227), alpha-tubulin (Abcam ab7291), FoxO1 (Cell Signaling Technology 2880P), Aldh1a3 (Novus Biologicals NBP2–15339), p-CamKII (Cell Signaling Technology 12716T), CamKII (Santa Cruz Biotechnology sc-13141), and LC3B (Novus Biologicals NB100-2220). The FoxO inhibitor AS1842856 and proprotein convertase inhibitor (US1537076) were obtained from Calbiochem. Herkinorin (ab120147) and naltrexone (ab120075) were ordered from Abcam. Golgicide A (HY-100540) was acquired from MedChemExpress. Thapsigargin (BML-PE180-0001) came from Enzo Life Sciences. Chloroquine (C6628), loperamide (L4762), cycloheximide (01810), KN-93 (K1385), and primers were obtained from Sigma. All the fluorescently labeled secondary antibodies were purchased from Thermo Fischer Scientific.
4.2. Cell culture
Min6 and alpha-TC1 cell lines were obtained from ATCC. Min6 cells were grown in high-glucose DMEM supplemented with 15% Tet System Approved FBS (Biowest S181T), 71 μM 2-mercaptoethanol, 50 U/mL penicillin, and 50 μg/mL streptomycin. The mouse pancreatic cell line alpha-TC1 was grown in low-glucose DMEM medium supplemented with 10% FBS, 50 U/ml penicillin, and 50 μg/mL streptomycin.
4.3. Human islets
4.3.1. Donor information
Human islets were obtained through the Integrated Islet Distribution Program (IIDP; NIH Grant # 2UC4DK098085) and the European Consortium on Islet Transplantation (ECIT): Islets for Basic Research Program from approved brain-dead organ donors (JDRF awards 31-2012-783 and 1-RSC-2014-100-I-X). All studies were approved by the Ethics Committee of the Medical University of Vienna (EK-Nr. 1228/2015). No material or information for this study was procured from living individuals.
| Gender | Age (years) | BMI | HbA1c | Source | Figure | |
|---|---|---|---|---|---|---|
| 1 | Female | 42 | 27.7 | – | ECIT; University of Geneva |
5F |
| 2 | Female | 57 | 28.0 | 5.5% | IIDP; Southern California Islet Cell Resource Center |
5F |
| 3 | Female | 56 | 26.6 | – | IIDP; The Scharp-Lacy Research Institute |
5F |
| 4 | Female | 62 | 29.8 | 5.5% | IIDP; Southern California Islet Cell Resource Center |
5E-F |
| 5 | Male | 47 | 28.0 | 8.2% | IIDP; University of Wisconsin |
5G-H and SFig. 13E |
| 6 | Female | 62 | 28.5 | 7.6% | IIDP; Southern California Islet Cell Resource Center |
5G-H and SFig. 13C-D |
4.3.2. Human islet culture
Human islets were cultured in CMRL medium (Life Technology) supplemented with 10% FBS, 2 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Intact islets were treated with different drugs for two days. Half of the intact islets were collected for RT-qPCR or RNAseq. The other half of the islets were incubated in Accutase (Life Technology) at 37 °C for 20 min, neutralized by CMRL medium, seeded to a 384-well plate, and utilized for immunofluorescence assays.
4.4. High-content screening
Compounds (10 mM in 25 nL) were transferred to 384-well plates (Corning 3712) from DMSO stock plates using acoustic transfer (Labcyte Inc.). The screened library, CLOUD (CeMM's collection of clinically approved drugs with unique structures) [12] consisted of 283 compounds. Min6 cells (3000 cells per well) incubated with 10 μM FoxO inhibitor were plated in 50 μl media on top of the compounds. Two days after treatment, cells were fixed in 3.7% formaldehyde for 10 min at room temperature. Following PBS washing, cells were fixed with cold pure methanol in −20 °C for 10 min, permeabilized by 1% Triton X-100 in PBS for 30 min and blocked by 3% BSA in PBS for 30 min. Twenty microliters of primary anti-insulin antibody and anti-glucagon antibody, both diluted in 1:2000 in 1.5% BSA, were added per well and incubated in 4 °C overnight. After washing with PBS twice, 20 μl fluorescence-labeled secondary antibody diluted 1:1000 and 10 μg/mL Hoechst 3342 in PBS was added per well and incubated for 1 h. After two washes with PBS, plates were stored in 4 °C in the dark until analysis.
Images were taken by an automated microscope (Perkin Elmer) using a 20X objective. The screened library, CLOUD (CeMM's collection of clinically approved drugs with unique structures) [12] consisted of 283 compounds. Images were exposed for 10 ms in Hoechst channel and 400 ms in Alexa Fluor 488 and 546 channels. Images were analyzed by Harmony software. Nuclei were identified by Harmony Method C, and cytoplasm was defined based on the nuclei (Harmony Method C). Hits were selected based on the intensity of insulin in the Alexa Fluor 548 channel, intensity of glucagon in the Alexa Fluor 488 channel, and amount of DNA and cell numbers in the Hoechst channel. Cells whose Hoechst intensity was lower than 1000 were treated as dead cells and removed from the screening. All the other immunofluorescence assays were done and analyzed in the same way unless otherwise specified.
4.5. RNA sequencing
Cells were lysed and RNA isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. The libraries for RNAseq for 24h induction in Min6 cells were prepared with Ribo-zero Kit and Scriptseq v2 Kit obtained from Epicenter by following the manual from the provider. For the rest of the RNAseq libraries, the amount of total RNA was quantified using Qubit 2.0 Fluorometric Quantitation System (Life Technologies), and the RNA integrity number (RIN) was determined using Experion Automated Electrophoresis System (Bio-Rad). RNAseq libraries were prepared with TruSeq Stranded mRNA LT Sample Preparation Kit (Illumina) using Sciclone and Zephyr liquid handling robotics (PerkinElmer). Library amount was quantified using Qubit 2.0 Fluorometric Quantitation System (Life Technologies), and the size distribution was assessed using Experion Automated Electrophoresis System (Bio-Rad). For sequencing libraries were pooled and sequenced on Illumina HiSeq 2000 using 50 bp single-read. Reads were aligned with TopHat (v2.0.4) with the --no-novel-juncs --no-novel-indels options [54]. Gene expression was calculated as fragments per kilobase per million of reads (FPKMs) using FPKM_count.py from RSeQC package [55], and the NCBI RNA reference sequences collection (RefSeq) was downloaded from UCSC [56]. The RNAseq data have been deposited in NCBI's GEO repository (accession number: GSE184210). The enrichment calculation was done by gene set enrichment analysis [57]. The basal expressions of genes in alpha and beta cells were taken from microarray data in the literature [58,59].
4.6. RT-qPCR
After the RNA was isolated with the RNeasy Mini Kit (Qiagen), it was reverse transcribed with random primers using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) on LightCycler 480 qPCR machine (Roche). All the results were analyzed using the delta-delta-Ct method and normalized to beta-actin and representative of three biological replicates and two technical replicates. Primer sequences used are the same as previously published ones [60,61] except for mouse Ngn3 (F: 5′TCCGAAGCAGAAGTGGGTGACT; R: 5′CGGCTTCTTCGCTGTTTGCTGA), mouse FoxO1 (F: 5′GGGTGATTTTCCGCTCTTGC; R: 5′GGGTGATTTTCCGCTCTTGC), and mouse MafA (F: 5′TTCAGCAAGGAGGAGGTCAT; R: 5′CCGCCAACTTCTCGTATTTC).
4.7. Western blotting
Whole-cell extracts were generated by lysing cells in Triton lysis buffer containing 150 mM sodium chloride, 1 mM EDTA, 1.0% Triton X-100, 50 mM Tris, and pH 7.4 supplemented with Protease Inhibitor Cocktail (Roche). Twenty μg whole-cell lysates were loaded onto an SDS–polyacrylamide gel and then transferred by electrophoresis to a nitrocellulose membrane (GE Healthcare Life Science). All the blots were incubated with corresponding primary antibodies diluted 1:1000 in 5% milk at 4 °C overnight and in HRP-labeled secondary antibodies diluted 1:20,000 for 1h at RT. The signals were detected using ECL Prime Western Blotting Detection Reagent (Amersham).
4.8. Intracellular insulin and proinsulin content measurements
Min6 cells were pretreated with FoxOi and loperamide for 48 h. Cell pellets were lysed using Triton lysis buffer, and intracellular insulin and proinsulin levels were measured using an Insulin ELISA (Alpco 80-INSMS-E01) or Proinsulin ELISA (Mercodia 10-1232-01). All results are representative of three biological replicates and two technical replicates.
4.9. Insulin secretion assay in mouse beta-cell line and murine islets
Min6 cells were pretreated with FoxOi and loperamide for 48 h. Cells were incubated in low-glucose (0.3 g/L glucose) KRB buffer for 1 h followed by high-glucose (3 g/L glucose) KRB buffer for another hour. Supernatants after both the low- and high-glucose challenges were collected to measure insulin content using a Mouse Insulin ELISA (Alpco 80-INSMS-E01). All results are representative of three biological replicates and two technical replicates.
Islets from db/db mice (GemPharmatech, Nanjing) were purified and cultured in high-glucose DMEM supplemented with 10% FBS (Gibco), 50 U/mL penicillin, and 50 μg/mL streptomycin. The islets were pretreated with loperamide for 48 h. Islets were incubated in low-glucose (0.3 g/L glucose) KRB buffer for 1 h followed by high-glucose (3 g/L glucose) KRB buffer for another hour. Supernatants after both the low and high-glucose challenges were collected. The islets were also collected after high-glucose challenges and lysed with Triton lysis buffer for intracellular insulin content measurement. To measure insulin content, a Mouse Ultrasensitive Insulin ELISA Kit was used (Alpco 80-INSMSU-E01).
4.10. Intracellular pH measurement
To measure the intracellular pH change in Min6 cells, they were pretreated with FoxOi with or without chloroquine/loperamide for two days. The measurement was performed using pHrodo® Red AM Intracellular pH Indicator (Life Technologies) following the manufacturer's guidelines. The images were taken by an automated microscope (Operetta, Perkin Elmer) using a 20X objective, and the relative intensity was quantified using the Harmony software. All results are representative of three biological replicates.
4.11. Calcium staining
The calcium staining was done in live Min6 cells with a Fura-2 Kit (F-1201, Life Technologies). Min6 cells were seeded and pretreated with loperamide for 18h before the assay. The staining was performed according to the manufacturer's guidelines. Min6 cells were also stained with 2 μM Fluo-4 AM (ab241082, Abcam) in KRB buffer supplemented with 0.1% Pluronic® F127 and 2 mM probenecid for 30 min at 37 °C, followed by 30min at RT prior to 2h loperamide treatment at 37 °C. All results are representative of three biological replicates.
4.12. Electron Microscopy
Min6 cells were grown on Aclar® fluoropolymer discs (Aclar® 33C, 199 μm thickness; EMS, Hatfield, USA) after sterilizing and pre-soaking the discs in media. After a 48h treatment with either DMSO, FoxOi, Loperamide, or the combination of FoxOi and Loperamide, the cells reached a confluency of about 80–90%. The discs were immersed in EM-grade glutaraldehyde (Agar Scientific, Essex, UK) diluted to 2.5% in 0.1M cacodylate buffer pH 7.4 and fixed for 1h at RT. After rinsing in 0.1M cacodylate buffer, post-fixation was done in 0.5% osmium tetroxide (prepared from crystals; EMS, Hatfield, USA) diluted in 0.1M cacodylate buffer for 1 h on ice, followed by further washing steps. Samples were dehydrated in a graded series of acetones: 40%, 60%, 80%, 95%, and 2 × 100% for 5min each, done on ice. Infiltration with epoxy resin (Agar 100; Agar Scientific, Essex, UK) was done in mixtures of acetone and resin in the following ratios: 2:1, 1:1, and 1:2 for 30min each at RT, followed by 1h in pure, freshly thawed resin. Polymerization took place at 60 °C for 48h. Ultra-thin sections with a nominal thickness of 70 nm were cut on a Leica UCT ultramicrotome (Leica Microsystems, Vienna, Austria) and picked up on 100mesh Cu/Pd grids (Agar Scientific, Essex, UK), previously coated with a formvar support film. For enhanced contrast, the sections were post-stained with 2% aqueous uranyl acetate pH 4 (Merck, Darmstadt, Germany) and Reynold's lead citrate. Inspection of the grids was done in a FEI Morgagni 268D TEM (Thermo Fisher, Eindhoven, The Netherlands) and operated at 80 kV. Examination regions on the sections were selected randomly. Digital images were acquired using an 11-megapixel Morada CCD camera from Olympus-SIS (Muenster, Germany). Granule measurement analysis was performed using Fiji/ImageJ.
4.13. Dauer formation
C. elegans strain CB1370 carrying the daf-2(e1370) allele (hypomorphic allele of the insulin/IGF-like receptor that makes animals prone to form Dauer larvae) was seeded by egg-lay onto plates containing FoxOi or loperamide at concentrations of 0, 5, 10, or 50 μM. For testing the effect of FoxO1 inhibitor on the suppression of dauer formation, worms were then grown at 23 °C. For testing the effect of loperamide on the promotion of Dauer formation, worms were grown at 22 °C. Animals were evaluated after 7 days by scoring their survival after a 30 min treatment with 1% (w/v) SDS.
4.14. Zebrafish larvae assay
26hpf (hours post fertilization) aged larvae of AB∗ wildtype zebrafish were treated with FoxO1 inhibitor and/or loperamide. For documentation of developmental defects after 48 h, anaesthetized fish (Tricaine, Sigma-Aldrich) were imaged under the stereo microscope (Leica, M125). Larvae of intercrosses of Tg(gcga:GFP)ia1 and Tg(ins:NTR-mcherry)ml10 Zebrafish were treated with loperamide from 26hpf until 5dpf. Double-positive larvae were embedded in low-gelling agarose (Sigma-Aldrich) and imaged on the confocal microscope (Leica, SP8, 25x water objective). 3D models of islets were established and numbers of GFP and mCherry positive cells counted in the Leica Software quantification tool.
4.15. Mouse models
All the animal experiments were performed in the SPF level animal facility of Fudan University School of Life Science, according to procedures approved by the experimental animal ethics committee of Fudan University School of Life Sciences. The 8-week-old db/db mice or wild-type C57 B6/J mice (GemPharmatech, Nanjing) were maintained under a 12 h light/12 h dark cycle at constant temperature (23 °C) with free access to chow diet and water. Loperamide (MedChemExpress, China) was freshly prepared daily as DMSO: PEG300: Tween 80: Saline in a ratio of 1:4:0.5:4.5. The mice were treated with vehicle or loperamide at 1 mg/kg via intraperitoneal injection daily. The mice were fasted overnight, and IPGTT was performed by injecting 1 g/kg glucose in PBS. After 4-week treatment, the mice were sacrificed in a CO2 chamber, and the pancreata were collected into a 4% PFA solution for fixation. Immunofluorescence was performed as previously described [61]. Colocalization quantifications were performed using the ImageJ plugin “Colocalization Threshold”. Quantifications were done on individual islets isolated within each image using a specified region of interest. Signal thresholds were determined using the Costes method. The resultant thresholded Manders’ correlation coefficients were then used. Total volume was determined based on total pixel number per islet, consequent percent volume colocalized references the number of pixels positive for both the red and green channels above the set threshold as a fraction of the total islet pixels, whereas the percent insulin and glucagon values correspond to pixels positive for the red or green channel, respectively. Mean FoxO1 and Aldh1a3 intensities per cell were calculated using CellProfiler, with Hoechst and insulin counterstains used to specify nuclei, cytoplasm, and beta cells.
4.16. Statistical methods
All the p-values were calculated by Student's t-test, unless otherwise specified.
Author contributions
Conception and design: SK and JL. Cell line provision: TF and JH. Next-generation sequencing: TP and CB. RNAseq analysis: FP, TC, and JL. Electron microscopy sample preparation and imaging: MB and TH. Human islet preparations: CB and EB. Zebrafish assays: CT and MD. Dauer formation assay: CR and IS. Mouse experiments: JL, XL, YZ, and SH. Performing experiments: TC, JL, CHL, XL, YZ, and SH. Manuscript writing: TC, JL, and SK with input from all co-authors.
Funding
This work was funded by JDRF grants 3-SRA-2015-20-Q-R and 17-2011-258 (Generation of beta cells from alternative pancreatic subtypes). Human islets were provided through the JDRF awards 31-2012-783 and 1-RSC-2014-100-I-X (ECIT: Islet for Research program). Li lab is funded by MOST 2018YFA0801300, 2020YFA0803600, SKLGE-2118, and NSFC 32071138. C.G.R. and his lab were supported by the Swedish Research Council (VR) grants 2015-03740, 2017-06088, and 2019-04868, the Swedish Cancer Society (Cancerfonden) grant 20 1034 Pj, the COST grant BM1408 (GENiE), and an ICMC project grant.
Acknowledgements
The drug library was designed by Freya Klepsch and Patrick Markt from the PLACEBO team. Next-generation sequencing was performed by the Biomedical Sequencing Facility at CeMM. Electron microscopy was performed by the Electron Microscopy Facility at the Vienna BioCenter Core Facilities (VBCF), member of the Vienna BioCenter (VBC), Austria. Robert Pazdzior from the Kubicek lab assisted in the development of the CellProfiler image analysis pipelines. This work was supported by JDRF grants 3-SRA-2015-20-Q-R and 17-2011-258. Research in the Kubicek lab is supported by the Austrian Federal Ministry of Science, Research and Economy, the National Foundation for Research, Technology, and Development, and the Marie Curie Career Integration Grant EPICAL.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101329.
Contributor Information
Jin Li, Email: li_jin_lifescience@fudan.edu.cn.
Stefan Kubicek, Email: skubicek@cemm.oeaw.ac.at.
Conflict of interest
The authors declare that they have no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure 1.
A. Relative mRNA levels of glucagon and insulin following 48 h incubation with increasing doses of FoxOi, as measured by RT-qPCR. B. Western blot highlighting the increase in glucagon protein levels after 48 h incubation with increasing concentrations of FoxOi. C. Intracellular insulin protein concentrations in Min6 cells following 48 h FoxOi treatment, at different concentrations, as measured by ELISA. D. Measurement of glucagon immunofluorescence intensity in single Min6 cells treated with increasing concentrations of FoxOi for 48 h. E. Representative images of Min6 cells stained for insulin and glucagon after 48h FoxOi treatment followed by 48 h washing off of the compound. Scale bars = 10 μm. F. Western blot showing the overexpression of FoxO1. G. Glucagon-positive cell fractions in Min6 cells treated with FoxO inhibitor, with and without overexpression of FoxO1 constitutively active construct, as measured by immunofluorescence. H. Quantification of SFig. 1G. L = low levels of FoxO1-ADA; H = high levels of FoxO1-ADA I. GSEA showing general co-upregulation of similar genes in both FoxOi treated Min6 cells and the FoxO triple knockout mouse[10].
Supplementary Figure 2.
A. List of top candidate compounds from immunofluorescence screen with their respective fold changes for insulin and glucagon expression, and for the number of DAPI-positive nuclei, as measured via immunofluorescence intensity. B. Loperamide dose ressponse suppression of glucagon transcription in Min6 cells treated with FoxO inhibitor. C. Glucagon mRNA levels in Min6 cells following loperamide time course, as measured by RT-qPCR. D. PPAR-gamma and Aldh1a3 transcription in Min6 cells treated with FoxOi/loperamide for 48h, measured by RNAseq. E. Gene ontology enrichment terms for RNAseq dataset of genes significantly downregulated by FoxOi and significantly rescued by loperamide after 48h treatment in Min6 cells.
Supplementary Figure 3.
A. Insulin mRNA levels in Min6 cells following loperamide time course, as measured by RT-qPCR. B. Western blot highlighting the increase in insulin and decrease in glucagon protein levels after 48h incubation with loperamide and the rescue of glucagon and insulin protein upon combination treatment with FoxOi and loperamide. C-D. Intracellular insulin and proinsulin protein levels in Min6 cells following loperamide time course, as measured by ELISA. E. Simultaneous decrease in glucagon and increase in insulin protein levels with increasing loperamide treatement time in Min6 cells. F. Insulin secretion from Min6 cells in low (0.3 g/L) and high (3 g/L) glucose medium pretreated with FoxOi and loperamide for 48h.
Supplementary Figure 4.
A. Representative images of Min6 cells stained for insulin and glucagon. Herkinorin was used at 15 μM for two days. Scale bars 10 μm. B. Representative images of Min6 cells stained for Pdx1 and glucagon. Naltrexone was used at 20 μM for two days. Scale bars 10 μm. C. Intracellular insulin protein levels in Min6 cells following 48h loperamide and naltrexone treatment, as measured by ELISA. D. Oprm1 gene expression in Min6 cells, as measured by RNAseq.
Supplementary Figure 5.
A. FoxO1 transcription in Min6 cells following 48h FoxOi/loperamide treatment, measured via RNAseq. B. FoxO1 transcription at different concentrations of loperamide in Min6 cells, measured via RT-qPCR. C. Western blot of FoxO1 protein expression in the cytoplasm vs. nucleus of Min6 cells following treatment with loperamide for the indicated hours. D. Western blot of FoxO1 protein expression in the cytoplasm vs. nucleus of Min6 cells following 48h loperamide and FoxOi treatments. E. GSEA showing general enrichment of FoxO1 genomic binding sites [16] in loperamide-upregulated genes. FWER p-value = 0.001. F. mRNA levels of Cacnb1 and Cacna2d1 in Min6 cells upon 48h loperamide and FoxOi treatment, as measured by RNAseq. G. RT-qPCR time course of Cacna1c and Cacna1d expressions upon loperamide treatment. H. GSEA results for genes involved in calcium channel activity in DMSO vs. FoxOi-treated Min6 cells. I. Western blot of p-CamKII increase after 48h loperamide treatment in Min6 cells.
Supplementary Figure 6.
A. Immunofluorescence panel of Min6 cells treated with loperamide and 1 μM KN-93 for 48h. Scale bar = 10 μm. B–C. Quantification of immunofluorescence signal intensities of: (B) insulin at plasma membrane and (C) FoxO1 in the nucleus. Plasma membrane and nucleus defined using Harmony 4 (PerkinElmer) software. D. Western blot of fractionated Min6 cells pretreated with loperamide and 1 μM KN-93 for 24h, highlighting KN-93's prevention of loperamide-induced FoxO1 nuclear translocation.
Supplementary Figure 7.
A. FoxO inhibitor treatment upregulates the transcription of lysosomal hydrogen ion transporters in Min6 cells, as measured by RNA-seq. B–C Intracellular pH indicator shows increased intracellular acidification after two days of FoxO inhibitor treatment in Min6 cells. B. Representative images of pHrodo live imaging in Min6 cells. Scale bars = 10 μm. C. Quantification of pHrodo live staining signal intensity in Min6 cells pretreated with loperamide/FoxOi for 48h. D. Representative images of lysotracker staining in Min6 cells. Scale bars = 10 μm.
Supplementary Figure 8.
A. Chloroquine inhibits FoxOi-induced glucagon transcription in Min6 cells, as measured by RT-qPCR. B. Chloroquine dose response on glucagon transcription in Min6 cells treated with FoxO inhibitor (i.e. dedifferentiated Min6 cells). C. Quantification of pHrodo live staining signal intensity in Min6 cells pretreated with chloroquine/FoxOi for 48h.
Supplementary Figure 9.
A-C. Top: RNAseq expression levels after 48h treatment; Bottom: RT-qPCR time course of mRNA levels for A. P4hb. B. Atp2a2. C. Atp2a3. D. Quantification of SERCA2 immunofluorescence intensity in Min6 cells treated with loperamide and FoxOi for 48h. E. Representative immunofluorescence images of Min6 cells treated with loperamide and thapsigargin for 2 h. Scale bar = 10 μm. F. Quantification of proinsulin and SERCA2 immunofluorescence intensity in Min6 cells treated with loperamide and thapsigargin for 2 h. G-H. Protein levels of mature insulin or proinsulin in Min6 cells after 24h treatment with varying concentrations of thapsigargin +/− loperamide, as measured by ELISA. I. Ratio of insulin to proinsulin protein levels.
Supplementary Figure 10.
A-B. Intracellular insulin and proinsulin protein levels after 30 h loperamide treatment +/− 10 μM Golgicide A for the final 6 h, as measured by ELISA. C. Ratio of insulin to proinsulin protein levels. D. Western blot for glucagon and insulin protein levels in Min6 cells after 48 h FoxOi/loperamide/PCi treatment. E. Quantification of insulin and proinsulin immunofluorescence intensities at the cell plasma membrane after 48 h loperamide and FoxOi treatment. Plasma membrane defined using Harmony 4 (PerkinElmer) software. F. Quantification of insulin granule size distribution from electron microscopy images. N = 5 cells per treatment condition. G. Gene ontology enrichment terms for RNAseq dataset of genes significantly upregulated by loperamide after 48 h treatment in Min6 cells. H. Significantly upregulated genes involved in autophagosome maturation following 48 loperamide treatment in Min6 cells.
Supplementary Figure 11.
Additional representative electron microscopy images of Min6 cells at two different magnifications. Scale bar = 1 μM.
Supplementary Figure 12.
A-D. Effects of loperamide treatment on FoxO1 and glucagon levels in alpha cells. A. Western blot showing an increase of FoxO1 protein with loperamide treatment in alpha and HEK cells. B. FoxO1 transcription at different concentrations of loperamide in alpha cells, measured via RT-qPCR. C. Western blot showing decrease of glucagon protein with different concentrations of loperamide in alpha cells. D. Suppression of glucagon transcription by increasing doses of loperamide in alpha cells. E-G. Effects of chloroquine treatment on glucagon in alpha cells. E. Western blot showing changes to glucagon protein expression in alpha cells following 24h chloroquine treatment. F. Chloroquine suppresses glucagon transcription in alpha cells in a dose dependent manner. G. Glucagon secretion from alpha cells in low glucose medium pretreated with chloroquine (5 μM) for 24 h, concentration measured by ELISA. H. Western blot of glucagon gene protein products in alpha cells treated with different doses of bafilomycin (V-ATPase inhibitor) overnight. I. Increase in Atp2a2 and Atp2a3 mRNA levels in alpha cells after 24 h loperamide treatment. J. Western blot highlighting increase in SERCA2 protein in alpha cells after 24 h loperamide treatment.
Supplementary Figure 13.
A. Representative images of insulin positive cells in zebrafish larvae. B. Quantification of insulin or glucagon positive cells following different doses of loperamide treatment in zebrafish larvae. N = 8–10. C. Intracellular insulin protein in diabetic human islets pre-treated with loperamide for 48h, as measured by ELISA. D. RT-qPCR mRNA levels of CACNA1C, CACNA1D, FOXO1, ATP2A2 and ATP2A3 in diabetic human islets pre-treated with loperamide for 48 h. E. GSEA results show a downregulation of alpha cell specific genes in loperamide-treated diabetic human islets vs. DMSO. F. Intracellular insulin protein in db/db murine islets pre-treated with loperamide for 48h. G. Concentration of secreted insulin from db/db murine islets following 48h loperamide pre-treatment.
Supplementary Figure 14.
A-C. Results of 4-week loperamide treatment in wild-type non-diabetic mice. N = 4 mice/treatment. A. Serum glucose concentrations after overnight fast. B. Body weight. C. Serum insulin concentration measured by ELISA. D. Representative immunofluorescence panel of db/db mouse pancreas sections 4-weeks post loperamide treatment, stained for insulin and FoxO1. Scale bar = 20 μm. E. Quantification of immunofluorescence images. FoxO1 intensity was only calculated within insulin-positive cells. POC = percent of control. NDMSO = 32 islets and NLoperamide = 26 islets from five different mice each.
References
- 1.Gupta D., Leahy A.A., Monga N., Peshavaria M., Jetton T.L., Leahy J.L. Peroxisome proliferator-activated receptor γ (PPARγ) and its target genes are downstream effectors of FoxO1 protein in islet β-cells: mechanism of β-cell compensation and failure. The Journal of Biological Chemistry. 2013;288(35):25440–25449. doi: 10.1074/jbc.M113.486852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kluth O., Mirhashemi F., Scherneck S., Kaiser D., Kluge R., Neschen S. Dissociation of lipotoxicity and glucotoxicity in a mouse model of obesity associated diabetes: role of forkhead box O1 (FOXO1) in glucose-induced beta cell failure. Diabetologia. 2011;54(3):605–616. doi: 10.1007/s00125-010-1973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim-Muller J.Y., Zhao S., Srivastava S., Mugabo Y., Noh H.-L., Kim Y.R. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metabolism. 2014;20(4):593–602. doi: 10.1016/j.cmet.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cinti F., Bouchi R., Kim-Muller J.Y., Ohmura Y., Sandoval P.R., Masini M. Evidence of β-cell dedifferentiation in human type 2 diabetes. Journal of Clinical Endocrinology & Metabolism. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Efrat S. Beta-cell dedifferentiation in type 2 diabetes: concise review. Stem Cells. 2019;37(10):1267–1272. doi: 10.1002/stem.3059. [DOI] [PubMed] [Google Scholar]
- 7.Zeng H., Guo M., Zhou T., Tan L., Chong C.N., Zhang T. An isogenic human ESC platform for functional evaluation of genome-wide-association-study-identified diabetes genes and drug discovery. Cell Stem Cell. 2016;19(3):326–340. doi: 10.1016/j.stem.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diedisheim M., Oshima M., Albagli O., Huldt C.W., Ahlstedt I., Clausen M. Modeling human pancreatic beta cell dedifferentiation. Mol Metab. 2018;10:74–86. doi: 10.1016/j.molmet.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagashima T., Shigematsu N., Maruki R., Urano Y., Tanaka H., Shimaya A. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Molecular Pharmacology. 2010;78(5):961–970. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- 10.Kim-Muller J.Y., Fan J., Kim Y.J., Lee S.A., Ishida E., Blaner W.S. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic beta cells in diabetic mice. Nature Communications. 2016;7:12631. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo S., Dai C., Guo M., Taylor B., Harmon J.S., Sander M. Inactivation of specific beta cell transcription factors in type 2 diabetes. Journal of Clinical Investigation. 2013;123(8):3305–3316. doi: 10.1172/jci65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Licciardello M.P., Ringler A., Markt P., Klepsch F., Lardeau C.H., Sdelci S. A combinatorial screen of the CLOUD uncovers a synergy targeting the androgen receptor. Nature Chemical Biology. 2017;13(7):771–778. doi: 10.1038/nchembio.2382. [DOI] [PubMed] [Google Scholar]
- 13.Giagnoni G., Casiraghi L., Senini R., Revel L., Parolaro D., Sala M. Loperamide: evidence of interaction with mu and delta opioid receptors. Life Sciences. 1983;33(Suppl 1):315–318. doi: 10.1016/0024-3205(83)90506-4. [DOI] [PubMed] [Google Scholar]
- 14.Groer C.E., Tidgewell K., Moyer R.A., Harding W.W., Rothman R.B., Prisinzano T.E. An opioid agonist that does not induce mu-opioid receptor--arrestin interactions or receptor internalization. Molecular Pharmacology. 2007;71(2):549–557. doi: 10.1124/mol.106.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H., Partilla J.S., Wang X., Rutherford J.M., Tidgewell K., Prisinzano T.E. A comparison of noninternalizing (herkinorin) and internalizing (DAMGO) mu-opioid agonists on cellular markers related to opioid tolerance and dependence. Synapse (New York, N.Y.) 2007;61(3):166–175. doi: 10.1002/syn.20356. [DOI] [PubMed] [Google Scholar]
- 16.Kuo T., Kraakman M.J., Damle M., Gill R., Lazar M.A., Accili D. Identification of C2CD4A as a human diabetes susceptibility gene with a role in β cell insulin secretion. Proceedings of the National Academy of Sciences. 2019;116(40):20033 LP–20042. doi: 10.1073/pnas.1904311116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilon P., Chae H.-Y., Rutter G.A., Ravier M.A. Calcium signaling in pancreatic β-cells in health and in Type 2 diabetes. Cell Calcium. 2014;56(5):340–361. doi: 10.1016/j.ceca.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Gandasi N.R., Yin P., Riz M., Chibalina M.V., Cortese G., Lund P.-E. Ca2+ channel clustering with insulin-containing granules is disturbed in type 2 diabetes. The Journal of Clinical Investigation. 2017;127(6):2353–2364. doi: 10.1172/JCI88491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozcan L., Wong C.C.L., Li G., Xu T., Pajvani U., Park S.K.R. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metabolism. 2012;15(5):739–751. doi: 10.1016/j.cmet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barg S. Mechanisms of exocytosis in insulin-secreting B-cells and glucagon-secreting A-cells. Pharmacology & Toxicology. 2003;92(1):3–13. doi: 10.1034/j.1600-0773.2003.920102.x. [DOI] [PubMed] [Google Scholar]
- 21.Bensellam M., Jonas J.C., Laybutt D.R. Mechanisms of beta-cell dedifferentiation in diabetes: recent findings and future research directions. Journal of Endocrinology. 2018;236(2):R109–R143. doi: 10.1530/joe-17-0516. [DOI] [PubMed] [Google Scholar]
- 22.Weiss M., Steiner D.F., Philipson L.H. In: Feingold K.R., Anawalt B., Boyce A., Chrousos G., de Herder W.W., Dungan K., editors. 2000. Insulin biosynthesis, secretion, structure, and structure-activity relationships. South Dartmouth (MA) [PubMed] [Google Scholar]
- 23.Leibiger I.B., Leibiger B., Berggren P.O. Insulin signaling in the pancreatic beta-cell. Annual Review of Nutrition. 2008;28:233–251. doi: 10.1146/annurev.nutr.28.061807.155530. [DOI] [PubMed] [Google Scholar]
- 24.Leibiger B., Moede T., Muhandiramlage T.P., Kaiser D., Vaca Sanchez P., Leibiger I.B. Glucagon regulates its own synthesis by autocrine signaling. Proc Natl Acad Sci U S A. 2012;109(51):20925–20930. doi: 10.1073/pnas.1212870110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohkuma S., Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proceedings of the National Academy of Sciences of the U S A. 1978;75(7):3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajpal G., Schuiki I., Liu M., Volchuk A., Arvan P. Action of protein disulfide isomerase on proinsulin exit from endoplasmic reticulum of pancreatic β-cells. The Journal of Biological Chemistry. 2012;287(1):43–47. doi: 10.1074/jbc.C111.279927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang W., Liao W., Luo C.T., Yin N., Huse M., Kim M.V. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491(7425):554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalvisa A., Siersbæk M.S., Præstholm S.M., Christensen L.J.L., Nielsen R., Stohr O. Insulin signaling and reduced glucocorticoid receptor activity attenuate postprandial gene expression in liver. PLoS Biology. 2018;16(12) doi: 10.1371/journal.pbio.2006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuch B.E., Szymanska B., Yao M., Tabiin M.T., Gross D.J., Holman S. Function of a genetically modified human liver cell line that stores, processes and secretes insulin. Gene Therapy. 2003;10(6):490–503. doi: 10.1038/sj.gt.3301911. [DOI] [PubMed] [Google Scholar]
- 30.Orci L., Ravazzola M., Amherdt M., Yanaihara C., Yanaihara N., Halban P. Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. The Journal of Cell Biology. 1984;98(1):222–228. doi: 10.1083/jcb.98.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh B.J., Soden C., Alarcón C., Wicksteed B.L., Yaekura K., Costin A.J. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Molecular Endocrinology (Baltimore, Md. 2007;21(9):2255–2269. doi: 10.1210/me.2007-0077. [DOI] [PubMed] [Google Scholar]
- 32.Jung H.S., Chung K.W., Won Kim J., Kim J., Komatsu M., Tanaka K. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metabolism. 2008;8(4):318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Fielenbach N., Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes & Development. 2008;22(16):2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fajardo R.J., Karim L., Calley V.I., Bouxsein M.L. A review of rodent models of type 2 diabetic skeletal fragility. Journal of Bone and Mineral Research. 2014;29(5):1025–1040. doi: 10.1002/jbmr.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spijker H.S., Song H., Ellenbroek J.H., Roefs M.M., Engelse M.A., Bos E. Loss of β-cell identity occurs in type 2 diabetes and is associated with islet amyloid deposits. Diabetes. 2015;64(8):2928. doi: 10.2337/db14-1752. [DOI] [PubMed] [Google Scholar]
- 36.White M.G., Marshall H.L., Rigby R., Huang G.C., Amer A., Booth T. Expression of mesenchymal and α-cell phenotypic markers in islet β-cells in recently diagnosed diabetes. Diabetes Care. 2013;36(11):3818. doi: 10.2337/dc13-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J., Ni Q., Xie J., Xu M., Zhang J., Kuang J. β-Cell dedifferentiation in patients with T2D with adequate glucose control and nondiabetic chronic pancreatitis. The Journal of Clinical Endocrinology & Metabolism. 2018;104(1):83–94. doi: 10.1210/jc.2018-00968. [DOI] [PubMed] [Google Scholar]
- 38.Marselli L., Suleiman M., Masini M., Campani D., Bugliani M., Syed F. Are we overestimating the loss of beta cells in type 2 diabetes? Diabetologia. 2014;57(2):362–365. doi: 10.1007/s00125-013-3098-3. [DOI] [PubMed] [Google Scholar]
- 39.Lu T.T.-H., Heyne S., Dror E., Casas E., Leonhardt L., Boenke T. The polycomb-dependent epigenome controls β cell dysfunction, dedifferentiation, and diabetes. Cell Metabolism. 2018;27(6):1294–1308. doi: 10.1016/j.cmet.2018.04.013. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fred R.G., Bang-Berthelsen C.H., Mandrup-Poulsen T., Grunnet L.G., Welsh N. High glucose suppresses human islet insulin biosynthesis by inducing miR-133a leading to decreased polypyrimidine tract binding protein-expression. PloS One. 2010;5(5) doi: 10.1371/journal.pone.0010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J.W., You Y.H., Jung S., Suh-Kim H., Lee I.K., Cho J.H. miRNA-30a-5p-mediated silencing of Beta2/NeuroD expression is an important initial event of glucotoxicity-induced beta cell dysfunction in rodent models. Diabetologia. 2013;56(4):847–855. doi: 10.1007/s00125-012-2812-x. [DOI] [PubMed] [Google Scholar]
- 42.Xu G., Chen J., Jing G., Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nature Medicine. 2013;19(9):1141–1146. doi: 10.1038/nm.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sebastiani G., Po A., Miele E., Ventriglia G., Ceccarelli E., Bugliani M. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetologica. 2015;52(3):523–530. doi: 10.1007/s00592-014-0675-y. [DOI] [PubMed] [Google Scholar]
- 44.Marquina-Sanchez B., Fortelny N., Farlik M., Vieira A., Collombat P., Bock C. Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets. Genome Biology. 2020;21(1):106. doi: 10.1186/s13059-020-02006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Idevall-Hagren O., Tengholm A. Metabolic regulation of calcium signaling in beta cells. Seminars in Cell & Developmental Biology. 2020;103:20–30. doi: 10.1016/j.semcdb.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Li S., Hao B., Lu Y., Yu P., Lee H.C., Yue J. Intracellular alkalinization induces cytosolic Ca2+ increases by inhibiting sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) PloS One. 2012;7(2) doi: 10.1371/journal.pone.0031905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orci L., Ravazzola M., Storch M.J., Anderson R.G., Vassalli J.D., Perrelet A. Proteolytic maturation of insulin is a post-Golgi event which occurs in acidifying clathrin-coated secretory vesicles. Cell. 1987;49(6):865–868. doi: 10.1016/0092-8674(87)90624-6. [DOI] [PubMed] [Google Scholar]
- 48.Sun-Wada G.H., Toyomura T., Murata Y., Yamamoto A., Futai M., Wada Y. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. Journal of Cell Science. 2006;119(Pt 21):4531–4540. doi: 10.1242/jcs.03234. [DOI] [PubMed] [Google Scholar]
- 49.Dechant R., Binda M., Lee S.S., Pelet S., Winderickx J., Peter M. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. The EMBO Journal. 2010;29(15):2515–2526. doi: 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu I.M., Chi T.C., Chen Y.C., Lu F.H., Cheng J.T. Activation of opioid mu-receptor by loperamide to lower plasma glucose in streptozotocin-induced diabetic rats. Neuroscience Letters. 1999;265(3):183–186. doi: 10.1016/s0304-3940(99)00226-8. [DOI] [PubMed] [Google Scholar]
- 51.Cheng J.T., Liu I.M., Chi T.C., Tzeng T.F. Increase of opioid mu-receptor gene expression in streptozotocin-induced diabetic rats. Hormone and Metabolic Research = Hormon- Und Stoffwechselforschung = Hormones et Metabolisme. 2001;33(8):467–471. doi: 10.1055/s-2001-16939. [DOI] [PubMed] [Google Scholar]
- 52.Tzeng T.F., Liu I.M., Lai T.Y., Tsai C.C., Chang W.C., Cheng J.T. Loperamide increases glucose ultilization in streptozotocin-induced diabetic rats. Clinical and Experimental Pharmacology and Physiology. 2003;30(10):734–738. doi: 10.1046/j.1440-1681.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 53.Tzeng T.-F., Lo C.-Y., Cheng J.-T., Liu I.-M. Activation of mu-opioid receptors improves insulin sensitivity in obese Zucker rats. Life Sciences. 2007;80(16):1508–1516. doi: 10.1016/j.lfs.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Wang S., Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics (Oxford, England) 2012;28(16):2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 56.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Research. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubicek S., Gilbert J.C., Fomina-Yadlin D., Gitlin A.D., Yuan Y., Wagner F.F. Chromatin-targeting small molecules cause class-specific transcriptional changes in pancreatic endocrine cells. Proceedings of the National Academy of Sciences of the U S A. 2012;109(14):5364–5369. doi: 10.1073/pnas.1201079109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xin Y., Kim J., Ni M., Wei Y., Okamoto H., Lee J. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proceedings of the National Academy of Sciences of the U S A. 2016;113(12):3293–3298. doi: 10.1073/pnas.1602306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fomina-Yadlin D., Kubicek S., Walpita D., Dancik V., Hecksher-Sorensen J., Bittker J.A. Small-molecule inducers of insulin expression in pancreatic alpha-cells. Proceedings of the National Academy of Sciences of the U S A. 2010;107(34):15099–15104. doi: 10.1073/pnas.1010018107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Casteels T., Huber K.V.M., Lardeau C.-H., Klughammer J., Farlik M. Artemisinins target GABAA receptor signaling and impair α cell identity. Cell. 2017;168(1–2) doi: 10.1016/j.cell.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harper J., Shin Y., Daly J. Loperamide: a positive modulator for store-operated calcium channels? Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14912–14917. doi: 10.1073/pnas.94.26.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He L.-P., Mears D., Atwater I., Rojas E., Cleemann L. Loperamide mobilizes intracellular Ca2+ stores in insulin-secreting HIT-T15 cells. British Journal of Pharmacology. 2003;139(2):351–361. doi: 10.1038/sj.bjp.0705263. [DOI] [PMC free article] [PubMed] [Google Scholar]