Abstract
Major evolutionary transitions were always accompanied by genetic remodelling of phenotypic traits. For example, the vertebrate transition from water to land was accompanied by rapid evolution of olfactory receptors and by the expansion of genes encoding lipocalins, which – due to their transporting functions – represent an important interface between the external and internal organic world of an individual and also within an individual. Similarly, some lipocalin genes were lost along other genes when this transition went in the opposite direction leading, for example, to cetaceans. In terrestrial vertebrates, lipocalins are involved in the transport of lipophilic substances, chemical signalling, odour reception, antimicrobial defence and background odour clearance during ventilation. Many ancestral lipocalins have clear physiological functions across the vertebrate taxa while many other have – due to pleiotropic effects of their genes – multiple or complementary functions within the body homeostasis and development. The aim of this review is to deconstruct the physiological functions of lipocalins in light of current OMICs techniques. We concentrated on major findings in the house mouse in comparison to other model taxa (e.g., voles, humans, and birds) in which all or most coding genes within their genomes were repeatedly sequenced and their annotations are sufficiently informative.
Keywords: lipocalins, odorant, mouse, major urinary protein, odorant-binding protein, retinol-binding protein, LCN, microbiota
Introduction
Initial mouse genome sequencing (Abril et al., 2002), re-sequencing of wild derived mice and other laboratory strains (Chang et al., 2017; Lilue et al., 2018) and now widely used OMICs techniques helped to link genotypes and phenotypes and provided new pieces of evidence that lipocalins are essential for life for their capacity to bind and transport biologically active as well as the toxic organic compounds in many fitness-related contexts. The lipocalins are small soluble proteins (typically 20kDa) with an eight-stranded antiparallel β-barrel often with two α-helices (N-terminal, C-terminal) on both ends of the protein, reviewed in (Stopková et al., 2009, 2014; Phelan et al., 2014b). The structure of the lipocalin β-barrel is open at one end allowing for the binding of various hydrophobic substances. There is yet another group of highly similar proteins that have β-barrels, though 10-stranded, and they include cellular retinol-binding proteins (CRBPs, depicted in Figure 1A as RBP1,2,5,7) and fatty acid binding proteins. The exception is extracellular RBP4 which, similarly as other lipocalins, have eight-stranded β-barrel. All together these proteins and lipocalins form the calycin superfamily (Sanchez et al., 2003; Álvarez et al., 2005). The lipocalin family is diverse accounting for at least 55 genes in the house mouse (Stopková et al., 2014) with little DNA sequence homology but with conserved tertiary structure thus forming β-barrel in all the mouse lipocalins but also in all metazoans over the evolutionary history (Ganfornina et al., 2000, 2006; Sanchez et al., 2003, 2006). Another interesting feature of lipocalins is their ability to bind and transport a wide spectrum of compounds including potentially toxic metabolic end-products such as ROS (reactive oxygen species) and also xenobiotics in many bacterial, plant, insect, avian, and mammalian species (Charron and Sarhan, 2006; Kwak et al., 2011, 2016; Bhatia et al., 2012).
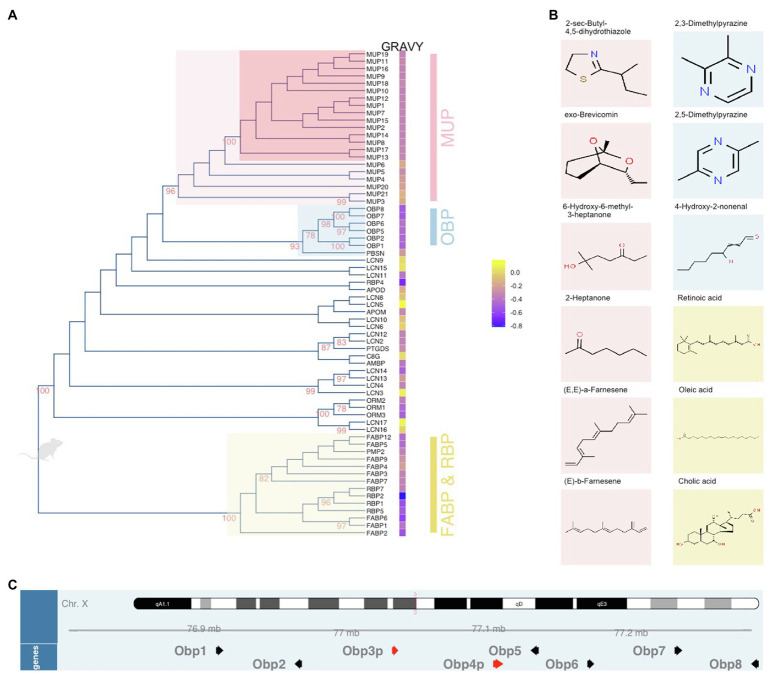
Figure 1.
Unrooted dendrogram of mouse lipocalins within the Calycin superfamily. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The tree (A) with the highest log likelihood (−17,639.2427) is shown. The percentage of trees in which the associated proteins clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 66 amino acid sequences. There were a total of 406 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 while the final figure was plotted with ggtree. Next to the tree, Gravy index is scaled from blue (hydrophilic) to yellow (hydrophobic). The colour code of highlighted subtrees is also used (B) to highlight the most common ligands with high affinities to particular protein groups. The position of Obp genes (black arrows) and pseudogenes (red arrows) on the X chromosome is visualized in (C) using Gviz Bioconductor package.
From the evolutionary point of view, ancestral lipocalins can be found in many animal taxa including bacteria. The closest homologues of bacterial lipocalins are lazarillo in insects or vertebrate apolipoproteinD – APOD (Ganfornina et al., 1995, 2008; Bishop et al., 2006). Concerning the vertebrate calycins (incl. lipocalins), some members were found through all the taxa (fish, amphibians, birds, and mammals), for example, APOD, FABPs, C8G, AMBP, LCNs (Bilkova et al., 2018), while others are highly specific for mammalian species – major urinary protein (MUP), odorant-binding protein (OBP). Although many OBPs were detected in insect species, where they are involved in chemical communication, being highly abundant in antennae and having specific affinities to insect pheromones, there is no true homology between the vertebrate and insect OBPs (Pelosi et al., 2006; Vieira and Rozas, 2011). This supports the view that the radiation of OBPs happened independently during the evolutionary history.
The aim of this review is to further explore what features make the lipocalins essential for life from the mouse perspective but also from other vertebrates which makes the view on lipocalin functions more robust. Thus, in this review, we aim to extract biologically relevant features typical for different lipocalin proteins in the mouse based on amino-acid sequence similarities using a dendrogram (Figure 1A) that was generated from public data1 and using other characteristics and expression sites of particular lipocalins. In addition, we also re-opened and explored proteomic datasets from our previous experiments that are also publicly available to discuss them in light of other studies published to date. We discuss the importance of lipocalins in three major biological areas in which lipocalins play crucial roles, namely in chemical communication, reproduction and development, and in the regulation of pathogens and natural microbiota.
Chemical Communication
Chemical communication is facilitated by behaviour-guiding olfactory signals, which in mice are abundant in body secretions such as tears, saliva, vaginal secretions, and urine (Thoß et al., 2015, 2016; Stopka et al., 2016; Cerna et al., 2017; Stopkova et al., 2017). These signals are often complex and provide information about multiple states of an individual including (sub-)species, kin, sex, health, and food sources (Hurst et al., 1997, 1998; Zala et al., 2004, 2015; Cheetham et al., 2007; Stopková et al., 2007; Bímová et al., 2009). Different types of molecules manifest the complexity of such signals which are detected via chemosensory G-protein coupled receptors of the main olfactory epithelia (MOE) and of the vomeronasal organ (VNO; Moss et al., 1997; Leinders-Zufall et al., 2000; Spehr et al., 2006; Wynn et al., 2012; Ibarra-Soria et al., 2014a; Nagel et al., 2018; Santoro and Jakob, 2018; Van Der Linden et al., 2018; Miller et al., 2020; Bansal et al., 2021). These receptors are tuned to particular signalling molecules including the volatile organic compounds – VOCs (Berghard and Buck, 1996; Malnic et al., 1999; Kwak et al., 2012, 2013), short peptides (Leinders-Zufall et al., 2004; Sturm et al., 2013) and non-volatile lipocalins, in mice, dominated by the male-biased MUPs (Chamero et al., 2007; Roberts et al., 2012). MUP like signals even have interspecific effects via the parallel mechanisms for kairomone detection. For example, lipocalins released from predators (cats, rats) are detected by mice and this induces aversive responses (Papes et al., 2010; Carvalho et al., 2015; Pérez-Gómez et al., 2015). Together, these secretory signals form signature mixtures, which generate stable representations in the accessory olfactory bulb (AOB) and, in other parts of the brain, these representations are linked to relevant behaviours (Bansal et al., 2021).
Major urinary proteins are essential for proper delivery of various volatile signals out of the body and for protecting and slowly releasing them from the urine marks (Hurst et al., 1998; Drickamer, 2001; Sharrow et al., 2002, 2003). Many of the ligands released from MUPs or just from the drying urine are directly detected by VNO neurons (Leinders-Zufall et al., 2000). MUPs were also demonstrated as signals themselves that are detected by basal, V2R-expressing sensory neurons in the VNO (Chamero et al., 2007) which due to small but detectable differences between different Mup genes and MUP protein structures (Phelan et al., 2014a,b) provide the basis for combinatorial coding of MUP signalling (Chamero et al., 2007; Kaur et al., 2014) which are then responsible for the display of specific mouse behaviours (Roberts et al., 2010, 2012; Dey et al., 2015; Demir et al., 2020). Importantly, these neurons are different from those that detect small hydrophobic compounds, which are apical V1R-expressing VNO neurons (Leinders-Zufall et al., 2000). In Figure 1A, MUPs are depicted in dendrogram as monophyletic group in which the associated proteins highly cluster together (bootstrap, BT=96). This group can be further separated onto sub-recently duplicated and highly similar “central” MUPs (BT=100) and the ancestral “outlier” MUPs 3, 4, 5, 6, 20, and 21 (Logan et al., 2008; Stopková et al., 2009, 2014; Phelan et al., 2014b; Sheehan et al., 2019). In Figure 1A, the Gravy index2 is provided in heatmap next to the protein names indicating that both groups of MUPs have different numbers of hydrophobic residua and thus the affinity to bind different VOCs; this difference is significant while their isoelectric points are similarly acidic pI=~4.9 (Stopková et al., 2016). This evolutionary differentiation of mouse MUPs has widened the spectrum of hydrophobic compounds that they protect, transport and release. This step was presumably crucial for the evolution of the house mouse as it enabled to “utilize” a wider and more informative odour space in individual recognition. Examples of typical MUP ligands (VOCs) previously studied in mice (Cavaggioni et al., 1987; Novotny et al., 1999; Zidek et al., 1999; Sharrow et al., 2003; Bingham et al., 2004) are presented in Figure 1B. For their complexity (volatiles, non-volatile proteins, and peptides), body secretions are now seen as signature mixtures rather than pheromones (Brennan and Kendrick, 2006; Nagel et al., 2018; Roberts et al., 2018; Bansal et al., 2021).
The system of signal detection has also diversified within the vertebrates thus evolving clear morphological structures (VNO, MOE) in tetrapods (except birds) while genetic components (V1R, V2R vomeronasal receptors) have already been found in teleost fish and sharks (Grus and Zhang, 2008). The transition from water to land was accompanied by a change in the ratio of V1R and V2R receptors such that the terrestrial vertebrates have more V1R receptors detecting smaller molecules including volatiles, whilst aquatic vertebrates have more V2R receptors for soluble and larger molecules including peptides and proteins. For example, amphibians (xenopus) have 21 V1Rs and 330 V2Rs, reptiles (python) have 4 V1Rs and 216 V2Rs, while the mouse system of olfaction is highly specialized for terrestrial life with 239 V1Rs (Tirindelli, 2021) with distinct evolutionary trajectories across mouse species (Miller et al., 2020), and 122 V2Rs (Tirindelli, 2021). Humans have lost functional VNO and the genes with remaining five intact V1Rs (Shi and Zhang, 2007). Profound differentiation of VNO and MOE in mice resulted in a highly specialized system being able to detect a wide spectrum of compounds including soluble (V2R) and volatile (V1R) compounds with VNO and general odorants with MOE. The signals detected by VNO are then processed in the AOB which maintain stereotypic sensory representations for broad types of stimuli, providing a substrate for relevant behaviours (Bansal et al., 2021). The responses of VNO and MOE are fast because odours are transported in turbulent plumes from the sites of origin and for example mice are able to detect these dynamically changing odours with a frequency of up to 40Hz (Ackels et al., 2021). The main message from this study is that volatiles, released from MUPs in the urine marks, must directly interact with chemosensory receptors and not via nasal volatile-binding proteins that would slow down this interaction. This is also suggested in a study on elephants, who produce the OBPs along the whole trunks making it unlikely to fast transport the volatile signals to the vicinity of chemosensory receptors from distant trunk parts (Lazar et al., 2002).
The most interesting aspect of olfactory diversification is a co-evolutionary process during which the receptor diversification was accompanied by the duplication and neo-functionalization of lipocalins. Particularly, there is a group of highly similar lipocalins (LCN3, LCN4, LCN13, and LCN14) which cluster together (BT=99, Figure 1A) and which are predominantly expressed in the mouse VNO thus forming of up to 36% of all transcripts in males and almost 29% in females with Lcn14 being the most abundant transcript in VNO (Kuntova et al., 2018). Transcriptomic analysis of MOE provided similar results as what we have detected with proteomics of the nostrils with prevailing OBPs (Obp2, 1, 5, 8, and Mup4 transcripts). Interestingly, a total of 17 lipocalins were detected on the proteomic level in the mouse saliva including LCNs that were detected mainly in VNO (Stopka et al., 2016). Many of these proteins are presumably transported to the oral cavity by a system of tiny tunnels called naso-palatinal ducts which are required for proper pheromone signalling and functioning of VNO which is why they are concentrated near the VNO opening (Levy et al., 2020). It is likely that after the signals are detected in VNO, they are chelated by the VNO-specific LCNs with larger barrels and consequently pumped out and “released” to the oral cavity where they have been detected (Stopka et al., 2016) and where the digestion begins.
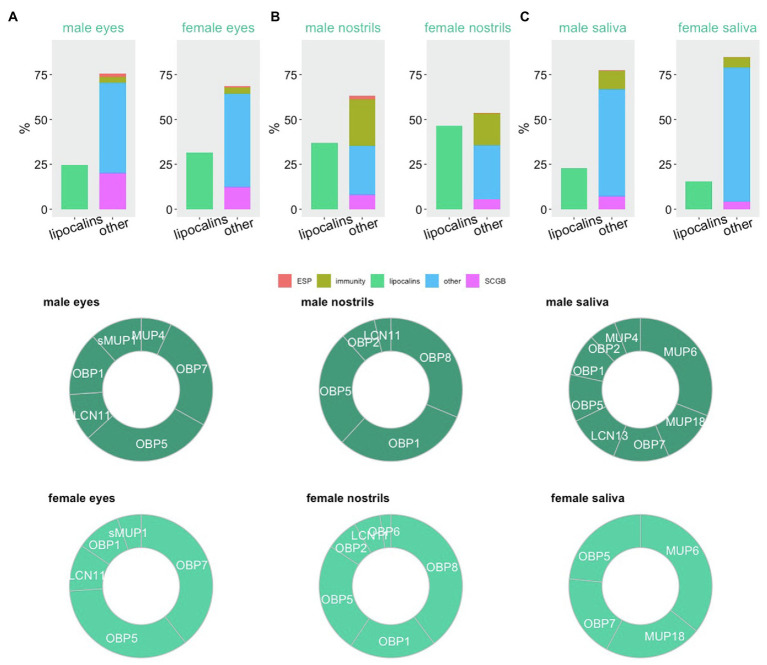
In mice, the neo-functionalization of lipocalins can be demonstrated by specific proteomic signatures, which in our case are presented as particular differences in the relative contribution to total protein content across different secretions. In Figures 2A–C,3 we present the data from our previous experiments where we generated the whole proteomes from lavages of eyes (Stopkova et al., 2017), nostrils (Kuntova et al., 2018), and oral cavity (Stopka et al., 2016) using label-free proteomics. We used just a few proxies to see relevant signatures, namely lipocalins, proteins involved in immunity and antimicrobial defence, secretoglobins [SCGB or ABP – androgen-binding proteins, formerly suggested as putative semiochemicals (Bimova et al., 2011)], exocrine-gland secreted peptides (ESP, putative pheromones; Haga et al., 2010; Abe and Touhara, 2014), and many other proteins with structural, homeostatic or cellular functions depicted as ‘other’. The most interesting result of this simple comparison is that the nostrils contain excessive amounts of lipocalins. In comparison with the urine which contains as much as 85% of lipocalins, mainly MUPs (Enk et al., 2016), the nostrils contain mainly the odorant binding proteins OBP8, OBP1, OBP5, OBP2 and a lipocalin named LCN11, Figure 2B.
Figure 2.
Proteomic signatures in major orofacial secretions of the house mouse. Estimation of protein abundances is represented as a fraction (%) of all proteins in male and female eyes (A), nostrils (B), and oral cavity (C). We have extracted this data from our previously published studies with publically available data. Proxy for functions (exocrine-gland secreted peptides – ESP, immunity, lipocalins, secretoglobins – SCGB) is based on public gene ontology tool.
The family of OBPs is coded by genes on the mouse chromosome X. To extend the knowledge on wild-living mice, we have sequenced all Obp transcripts and provided unique Obp sequences for feral M. m. domesticus (Obp1 – KJ605385, Obp2 – KJ605386, Obp5 – KJ605387, Obp6 – KJ605388, and Obp7 – KJ605389), and Mus musculus musculus (Obp1 – KJ605390, Obp2 – KJ605391, Obp5 – KJ605392, Obp6 – KJ605393, and Obp7 – KJ605394) now available in GenBank (Stopková et al., 2016). All the novel OBPs have a feature typical for the entire Obp cluster – a specific disulfide bond (Cys38–Cys42), which represents a strong OBP-diagnostic motif CXXXC (Cys-Xaa-Xaa-Xaa-Cys). OBPs are phylogenetically close to MUPs, Figure 1A. However, their biochemical properties are different from those of acidic MUPs in that their isoelectric points are closer to neutral and their hydropathy (Gravy) index is lower (Stopková et al., 2016). Thus, their affinity for less hydrophobic compounds, their prevalence in the proteomic profiles of eyes and nostrils and their close-to-neutral pI suggests that these proteins are important in removing the background organic molecules including odorants that are different from volatiles presented by MUPs in the urine. From the point of view of the “odour space” a fraction of odours and other compounds is being instantly removed from the nostrils and this presumably facilitates that the receptors preferentially bind relevant signals.
There is yet another perspective in the understanding of potential roles of OBPs in the mouse nostrils. They are sexually dimorphic similarly as the semiochemical responsive olfactory neurons (Vihani et al., 2020). This means that for some reason, being most likely a sharper detection of the ligands produced by males, females produce more OBPs (almost 50% of all soluble proteins) in the nostrils, which is likely to remove those VOCs that do not have the chemical properties of MUP ligands that males use for signalling. Similar pattern was detected in the eyes with similar levels of sexual dimorphism, Figure 2A. Because, eyes, nostrils and oral cavity are the inter-connected structures, with eyes being connected with nostrils via naso-lacrimal ducts, nostrils (VNO) with the oral cavity by the naso-palatinal ducts (Levy et al., 2020) and through naso-pharyngeal upper airways, OBPs may help removing the given spectrum of selected compounds from the majority of oro-facial mucosa. The selective forces that shaped the evolution of OBPs probably included the ability to remove organic toxins from nasal epithelia and upper airways. This has been shown in porcine OBPs, which bind with high affinity HNE (4-hydroxy-2-nonenal) depicted in Figure 1B, a toxic compound derived from lipid peroxidation. OBPs therefore protect living cells from damage caused by oxidative stress (Grolli et al., 2006). In non-rodent taxa, such as Artiodactyla (Ruminantia), the olfactory secretome becomes more complex during the reproductive period. For example in goats and sheeps, more OBP variants are expressed and the ligand reception by OBPs is often facilitated by protein modifications including O-GlcNAcylation (Cann et al., 2019), which was previously described also in pigs (Nagnan-Le Meillour et al., 2014).
In many mammals, Obp genes were selected for diverse functions across various taxa. Arvicolids (Cricetidae) are a parallel lineage to Murid rodents (Muridae) within the superfamily Muroidea and they seem to lack the genes for MUPs. However, they produce excessive amounts of OBPs in their urine and several studies provided evidence that they are produced by the liver. For example, the bank voles (Clethionomys glareolus) produce at least three OBPs coded by Obp1, Obp2, and Obp3 genes encoding the predominant lipocalins present in their urine and saliva (Stopkova et al., 2010), see also (Loxley et al., 2017). Similarly, the European water voles (Arvicola terrestris) produce one sexually dimorphic OBP named arvicolin with transcripts being highly abundant in the liver and proteins present in the urine. The reception of pheromones by this particular OBP may be facilitated via protein modifications (e.g., phosphorylation and O-GlcNAcylation; Nagnan-Le Meillour et al., 2019) while the hamster OBP (aphrodisin) is N-glycosilated (Singer et al., 1986). All these arvicolid and also mouse OBPs are highly similar to hamster aphrodisin, which is vaginal OBP that was named after its effect upon male-mounting behavior via its natural OBP ligands (Singer et al., 1986; Briand et al., 2004) and is also expressed by other hamster species (Turton et al., 2010). When looking further from the muroid rodents (Myomorpha), OBPs have also been detected in the urine of highly social African mole rats (Hystricomorpha; Hagemeyer et al., 2011) thus providing evidence that the involvement of OBPs in chemical signaling was likely the ancestral mode of chemical communication in rodents. Taken together, the selective pressures that drove the evolution of chemical communication in rodents acted upon the function of a range of lipocalins and not on particular lipocalin gene. This is evidenced by the fact that those species that lack the genes for MUPs evolved other lipocalins for this function so the function of MUPs as carriers of chemical signals was supplanted by OBPs in many rodent taxa.
An interesting aspect of the systems of signalling and detection is that they are socially (environmentally) modulated. To our knowledge, we have been the first laboratory that provided evidence that the abundance of MUPs in the urine of female mice correlates with the estrous cycle, reaching the highest levels od MUP abundance in the urine (Stopka et al., 2007) and vaginal secretions (Cerna et al., 2017) near estrus. There is also a strong influence of social environment, whereby males of wild M. m. musculus increase their production of MUPs in the urine when presented with a female behind metal grid (Janotova and Stopka, 2011). Socially induced MUP variation has also been demonstrated in seminatural enclosures, where males doubled the excretion of MUPs after acquiring a territory and became socially dominant (Thoss et al., 2019). Higher concentrations of MUPs may then yield various behavioural responses in the receiver due to MUP detection by VNO neurons via progesterone signalling (Dey et al., 2015). Recent development of transcriptomic techniques helped to reveal that the fitness-related social dynamics of protein expression was also demonstrated in olfactory tissues. The expression of many receptor genes was altered when presented with different stimuli in several strains of the laboratory mice (Ibarra-Soria et al., 2017) and similarly when mice are separated, the isolation induces sex-specific differences in the olfactory sensory receptor repertoires (Santoro and Jakob, 2018; Van Der Linden et al., 2018). These social and reproductive effects are relevant examples of the expression dynamics that is regulated by benefits and costs of lipocalin and receptor production driven by social stimuli and the potential to mate. Until this point, we concentrated on lipocalins that function as an interface between external and internal worlds of an individual. However, many lipocalins have important functions within an individual homeostasis, reproduction and development.
Roles of Lipocalins in Reproduction and Development
The production of fully mature and motile sperm cells is essential for successful reproduction. The first key event happens in the seminiferous tubules where, through a complex process of spermatogenesis, germ stem cells differentiate into highly polarized spermatozoa. Along testosterone, vitamin A in the form of retinoic acid (RA) is an important signalling molecule that initiates spermatogonial differentiation and meiotic entry (Zhou et al., 2008). After the conversion from retinol, which is synthetized in Sertoli cells, RA specifically binds to intracellular heterodimeric receptors RARs and RXRs and regulates gene expression, e.g., of Stra8 gene inducing the synthesis of downstream markers of meiosis (Livera et al., 2002; Zhou et al., 2008). Signalization mediated by RA and its receptors was shown to be important also for the normal function of steroidogenic Leydig cells producing testosterone (Jauregui et al., 2018). However, as retinoids are not soluble in an aqueous environment, they require both extracellular and intracellular binding proteins. Interestingly, lipocalin-type prostaglandin D2 synthase (L-PGDS or PTGDS) is expressed in many tissues, also including the mouse testis (Gerena et al., 2000). Since PTGDS has been reported to have a high affinity to RA and retinaldehyde, it was proposed that it might serve as a retinoid carrier supplying developing germ cells (Urade and Hayaishi, 2000; Rossitto et al., 2015). Moreover, Kido et al. (2005) showed the specific expression in transgenic mouse testis of fatty acid binding protein 9 (FABP9). Later study specified the FABP9 localization to advanced stages of spermiogenesis – from elongating spermatids on (Selvaraj et al., 2010). FABP9 shows high homology (58%) to FABP4, which binds long-chain fatty acids like stearate, palmitate, and oleate, but also RA, though with lower affinity (Oko and Morales, 1994; Richieri et al., 2000). Because spermatogenesis is a process demanding high levels of fatty acids (e.g., for membrane biogenesis), FABP9 likely contributes to providing them to the germ cells.
Fully developed testicular spermatozoa further undergo several maturation steps during their transit through epididymis, which is divided into three segments, each with unique luminal environment determined by proteins secreted from various epithelial cell types (Turner et al., 2003). The modification of sperm protein profile in region-specific manner is one of the proposed features of epididymal maturation. Support for this was provided by proteomic analysis of M. musculus whole sperm isolated from the caput, corpus and cauda regions (Skerget et al., 2015). The study showed that, among other proteins, lipocalins LCN2, LCN6, and LCN8 were downregulated in the caput sperm, whereas LCN12 was abundant in the corpus. On the contrary, LCN5 and FABP9 were common in sperm from all the segments. Moreover, Guyonnet et al. (2012) specified in CD 1 mice the FABP9, LCN5 and LCN12 localization to the matrix of sperm acrosome, while LCN2 and LCN6 signals were observed using fluorescent techniques to be associated with the mouse and human spermatozoa isolated from cauda epididymis (Chu et al., 2000; Hamil et al., 2003).
Despite the pieces of evidence, biological functions of many lipocalins in reproduction still remain unclear. LCN8 is hypothesized to bind and transfer retinoids (RA) similarly as LCN5 and thus might drive the development and maintenance of epididymal epithelium (Rankin et al., 1992; Costa et al., 1997; Lareyre et al., 2001). Based on an increased uptake of ferric ion by sperm mediated by LCN2, lipocalins are also suggested, by yet unknown mechanism, to undergo internalization into the sperm cell (Elangovan et al., 2004). In order to elucidate the role of lipocalins and cytosolic calycins in male reproduction, several knockout studies have been performed. Deletions of either of Lcn6, Lcn8, Lcn9, or Fabp9 did not cause abnormalities in the morphology of testis or epididymis nor in sperm production and fertility. However, the sperm from Lcn8−/− and Fabp9−/− double-knockout male mice showed abnormal head and tail morphology. An enhanced frequency of spontaneous acrosome reaction occurred in Lcn6 and Lcn8 deficient sperm (Selvaraj et al., 2010; Yin et al., 2018; Wen et al., 2021). All this evidence indicates that particular lipocalins are not essential for sperm development and its fertilizing ability because they can, to some extent, supplant each other. Very likely, it might be due to functional compensatory mechanism originating from their high structural and hence also binding homology. Therefore, double or triple knockout studies should be performed to undoubtedly elucidate this mechanism. The major significance of lipocalins in male reproduction thus remains in epididymal sperm maturation and events linked to capacitation and acrosome reaction occurring in the female reproductive tract.
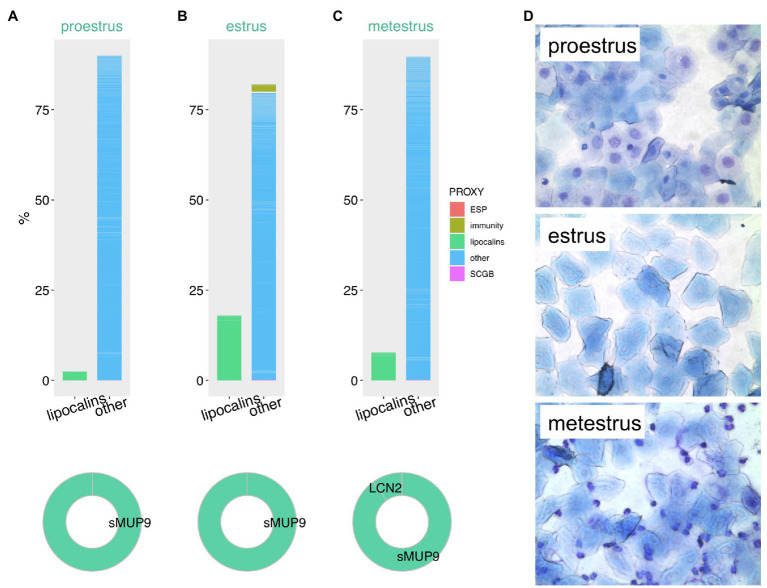
The estrous cycle along with the oogenesis, which is linked to folliculogenesis are the most important fitness-related processes in female reproduction. In mice and other rodents, the cycle is generally divided into the four phases known as proestrus, estrus, metestrus, and diestrus. Interestingly, in some rodents such as the wood mice (Apodemus sylvaticus) these phases dynamically change as a reaction to the presence or absence of male mating partners (Stopka and Macdonald, 1998). Particular phases are characterized by the differential gene expression and by varying cellular types of the utero-vaginal epithelia (Figure 3D) reflecting the maturation of ovarian follicles (Byers et al., 2012; Yip et al., 2013; Cora et al., 2015). In detail (see Figure 3D), the proestrus phase is typical with predominant nucleated epithelial cells, whereas estrus phase with keratinized epithelial cells on which bacteria feed. The metestrus is overpopulated with neutrophils. Hormonal imbalance or pathogen exposure may lead to disruption of cycling. As shown in Figures 3A–C using the vaginal fluid proteomic data from M. m. musculus, lipocalins are significantly up-regulated in estrus and metestrus compared to proestrus (Cerna et al., 2017). This stage-specific elevation was proportionally most obvious in sMUP9 (i.e., group of highly similar MUPs: MUP6, MUP9, MUP16, and MUP19) while LCN2 only mildly varied, Figure 3B, and was upregulated only in metestrus (Figure 3C). Except these, MUP20, sMUP17, LCN11, and OBP5 were significantly up-regulated in estrus but their proportion to total protein content was lower. On the contrary, retinol-binding protein 1 (RBP1) had the lowest abundances during estrus. Given their proposed ability to internalize and transport various ligands, highly abundant lipocalins in these stages possibly detoxify the mouse vaginal environment while some of these ligands might have become the signals by which males recognize female receptivity (Stopková et al., 2009).
Figure 3.
Proteomic variation in vaginal secretions throughout the mouse estrous cycle. Estimation of protein abundances is represented as a fraction (%) of all proteins in vaginal lavages during proestrus (A), estrus (B), and metestrus (C). We have extracted this data from our previously published study with publically available data. Proxy for functions (ESP, immunity, lipocalins, secretoglobins – SCGB) is based on public gene ontology tool. Representative microphotographs of the vaginal cytology were taken at magnification 100× after May-Gruendewald and Giemsa staining (D).
High abundances of LCN2 limits the bacterial growth during estrus by iron-chelating bacterial siderophores (Flo et al., 2004). Similarly to male reproduction, retinoids are essential also for the female genital system. Vitamin A sufficiency is important for the differentiation of meiotic germ cells, embryonic implantation and normal embryonic development. The significance of the uterine expression of retinoid binding proteins and their transcripts, e.g., for RBP1, RBP4, CRABP2 thus supports this view (Clagett-Dame and Knutson, 2011; Yip et al., 2013). Additionally to the LCN2 protective roles for female genital-tract epithelia, this lipocalin also functions as a sperm capacitation-promoting factor in vitro (Watanabe et al., 2014). Isolated sperm from the oviduct of Lcn2−/− knockout females showed a significant decrease in membrane lipid raft movement, which was retrieved after LCN2 addition. The association of LCN2 with sperm surface also enhances the progressive motility (Lee et al., 2003). The interaction between mammalian spermatozoa with seminal fluid, and female genital tract is known to be reciprocal. For instance, spermadhesins from ejaculate, apart from other functions, modulate the uterine immune responses. As lipocalins are contained in male genital fluid, they very likely co-participate on this direct interaction (Rodriguez-Martinez et al., 2010). To add, lipocalins are essential for various stages of reproduction both in males and females and similar lipocalins were detected also in avian ova (Bilkova et al., 2018), thus suggesting that the roles of lipocalins are conserved across distant vertebrate taxa.
Regulation of Pathogens and Natural Microbiota
Lipocalins are expressed in all body parts with the complex microbial communities (Seo et al., 2006; Stopka et al., 2016; Cerna et al., 2017; Xiao et al., 2017) and their typical β-barrel structure with a central cavity for binding of various hydrophobic molecules (Flower, 1996) provides several strategies for constant sensing and controlling both pathogens and commensal bacteria. There is growing evidence that each microbiome-populated body site hosts unique bacterial ecosystems (Lee and Mazmanian, 2010; Matějková et al., 2020; Moudra et al., 2021) and that those tissues permanently face the challenge of discriminating between commensal and pathogenic bacteria. A rapid destruction of potential pathogens is especially important in the sites where pathogen burden could have detrimental effects upon the host. Such places include the oral cavity representing the main route to the digestive tract, urogenital openings, and eyes and nostrils as a gate to the brain via the optic and olfactory nerves (Stopková et al., 2014).
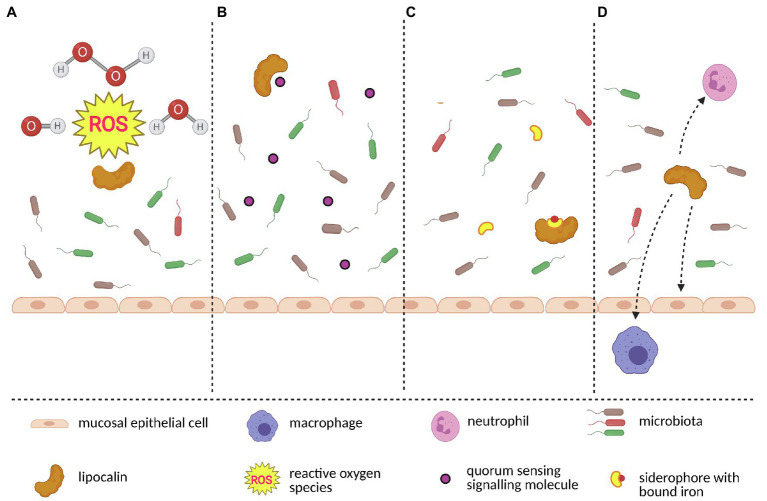
Communication between the host and its microbiome is facilitated by molecules secreted by bacteria. Microorganisms (and especially bacteria) belonging to the same species are able to synchronize their activities via so-called quorum sensing, Figure 4.4 Each quorum sensing is characterized by a signalling compound, usually a small molecule (QSM – quorum sensing molecule) which is released into the environment and recognized by other co-specific microorganisms (Rutherford and Bassler, 2012). When the concentration of QSM reaches a certain threshold, the whole population of microorganisms synchronizes its biosynthetic activity. QSMs can be inactivated by quorum quenching mechanism (QQ), which is usually mediated by specific enzymes (Uroz et al., 2009). However, the alternative strategy for QQ is the scavenging and removal of QSMs provided by the proteins with a QSM binding capacity. Ligands associated with bacterial infections and those from defeated bacteria during regulation of microbiota are also detected with MOE and VNO via the microorganism-associated molecular patterns (MAMPs), and they are sensed in many places in the body including specific chemosensory neurons in the mammalian nose (Bufe and Zufall, 2016). They also include the formyl peptide receptor-like proteins in VNO, which provide sensitivity to disease/inflammation-related ligands (Riviere et al., 2009) and presumably they are responsible for the activation of bactericidal proteins. Bactericidal proteins (i.e., such as BPI proteins) were previously detected in the olfactory transcriptomes of the mouse (Ibarra-Soria et al., 2014b) in tears (Stopkova et al., 2017) and saliva (Stopka et al., 2016).
Figure 4.
A model of interactions between lipocalins and microbiota. Lipocalins protect host’s epithelial cells and also commensal microbiota by scavenging toxic molecules such as reactive oxygen species which originate during oxidative stress (A). Keeping microbiome homeostasis requires blocking the origin of bacterial monoculture. Lipocalins contribute to diversified microbiome by scavenging bacteria-produced quorum sensing signalling molecules (B). Similarly, certain bacterial species use siderophores to gain iron and subseuently outcompete siderophore-lacking bacteria. By binding to siderophore-iron complex, lipocalins further contribute to stable microbiome (C). Finally, lipocalins closely interact with other members of the immune system network (i.e., neutrophils, macrophages and epithelial cells) and together create a complex microbiota-surveillace system (D). The figure was created in Biorender.
Lipocalins have the ability to bind a broad spectrum of ligands, therefore it is likely that they are involved in the regulation of microbiota and of the function of olfactory neurons (Bryche et al., 2021). For example, a recent study provided the evidence that bovine (bOBP) and porcine (pOBP) odorant binding proteins can effectively bind farnesol, which is a terpenoid produced by Candida albicans as QSM and which is able to affect transformation from the mycelial to the yeast state (Bianchi et al., 2019). Moreover, both OBPs have the capability to almost completely remove bacterial toxin pyocyanin produced by Pseudomonas aeruginosa. To some extent, both OBPs have also the affinity to various types of N-acyl-homoserine lactones produced as QSMs by P. aeruginosa and some other microbial organisms. Despite the lower affinity of OBPs to Pseudomonas signalling molecules, OBPs might scavenge QSMs when they reach potentially dangerous concentrations that activate quorum sensing. Interestingly, assays testing the direct antimicrobial or antifungal activity revealed different efficiencies of the two OBPs. While pOBP has higher inhibitory activity on fungi, bOBP can better reduce the growth of various bacterial species (Bianchi et al., 2019). It is the first evidence that vertebrate odorant binding proteins exert antimicrobial activity through their scavenging capacity. Furthermore, OBPs also shape the microbiomes by scavenging toxic molecules (i.e., aldehyde 4-hydroxy-2-nonenal) that are produced during oxidative stress (Lacazette et al., 2000; Grolli et al., 2006) and whose higher concentrations cause damage not only to epithelial cells but also to commensal microbiota (Macedo-Marquez et al., 2014) which play a crucial role in keeping epithelial homeostasis (Koskinen et al., 2018).
Although the members of microbial communities are often described as commensals, the relationship between the host and a particular bacterial species can simply change to other forms of relationship as mutualistic or parasitic depending on the host’s genetic background, nutritional status, or co-infection of the host (Belkaid and Harrison, 2017). One of the main aspects influencing the bacterial switch from commensal to pathogen is the availability of iron, which is strongly regulated by lipocalins (Lechner et al., 2001; Goetz et al., 2002; Fluckinger et al., 2004; Julien et al., 2019). Iron is one of the essential nutrients for almost all aerobic organisms. In mammals, the majority of iron is bound either in hemoglobin or in ferritin and transferrin. Therefore, the concentration of extracellular free iron is maintained below 10−24M (Correnti and Strong, 2012). This low concentration of available iron serves as a protection of the host against the reactivity of free iron and also against the potentially pathogenic bacteria that would thrive in an iron-rich environment. To “steal” iron from host proteins (i.e., ferritin, transferrin, and lactoferrin), bacteria synthesize and secrete low-molecular-weight (<1.0kDa) iron chelators called siderophores (Nairz et al., 2010). One of the most studied siderophores is enterochelin (Ent) whose affinity for iron allows it to effectively outcompete the majority of the host’s iron-binding proteins (Fischbach et al., 2006). Typical lipocalin involved in iron homeostasis is LCN2 (alias 24p3, SIP24, siderocalin, NGAL), which is constantly present in low amounts in mucosa where it chelates Ent in order to inhibit overgrowing of Ent-producing bacteria. To prevent Ent from delivering iron back to the potential pathogens (e.g., to Escherichia coli, one of the major pathogens of gut-origin sepsis), LCN2 chelates the Ent-iron complex and forms a structure which is not capable of transferring iron back to bacteria (Goetz et al., 2002; Johnson et al., 2010; Wu et al., 2010; Mori et al., 2016). While LCN2-siderophore binding capacity is limited towards one group of siderophores – catecholate-type (Xiao et al., 2017), human tear lipocalin (LCN1) is capable of binding a broader array of siderophores, including the bacterial catecholate and hydroxamate-type, and all major classes of fungal siderophores (Fluckinger et al., 2004). This physiological function of LCN2 and human LCN1 is widespread in vertebrates and for example, in birds, four proteins highly homologous to LCN2 (CALβ, CALγ, Ggal-C8GC and Ex-FABP) exerted almost identical roles in the protection against E. coli (Garénaux et al., 2013).
Protective function of LCN2 is documented in experiments with Lcn2-deficient mice, which are highly prone to bacterial infection and sepsis. In detail, the lack of LCN2 leads to significant changes in gut microbiota composition followed by microbiota dysbiosis with a disproportionate growth of gram-negative bacteria (Moschen et al., 2016; Singh et al., 2016, 2020). Reducing effects on bacteria were observed in a study focused on in vitro growth of Aeromonas hydrophila and E. coli in the presence of fish LCN2 (ortholog – 3nLcn2; Zhou et al., 2020). Similarly to in vitro experiments, there were lower bacterial loads in tissues of 3nLcn2-administered fish infected by A. hydrophila (Zhou et al., 2020). Homeostatic relationships with microbiota can be kept only when the contact between bacteria and the host’s epithelial cells surface is minimized. This segregation of microbiomes from the host’s cells is accomplished by combined action of epithelial cells, mucus and the involvement of both innate and adaptive immune systems (Shi et al., 2017). The observation that Lcn2-deficient animals are also susceptible to the siderophore-independent pathogens suggests that LCN2 protects the microbiome-host barrier not only as a siderophore scavenger but also plays an important role in a broader immune system network.
Conclusion
From the evolutionary point of view, lipocalins represent a fascinating protein family because, in vertebrates, many of their members underwent rapid evolution and neo-functionalization during or after the transition from water to land and this is well documented in rodents. At the same time, there is a group of lipocalins and other calycins that seem to be essential for life, because they are stereotypically expressed in almost all vertebrates and play important roles in spermatogenesis and embryonic development (FABP, RBP), regulation of oxidative stress (AMBP), and antimicrobial defence (e.g., LCN2). The transition from water to land was the major driving force that has driven the reorganization of sensory systems of detection (nose, eyes) of fitness-related cues such as food, predators and other individuals of the same species and particular sex. Mice are primarily dependent on olfactory cues. They use MUPs to protect and transport volatile pheromones that trigger many physiological, behavioural and reproductive responses via the vomeronasal and major olfactory systems. Various tissues in the oro-facial region, including lymphoid tissues, epithelia and lacrimal glands produce excessive amounts of OBPs which have different binding properties than MUPs (e.g., lower hydrophobicity) and thus they represent the major system of chelators of signals that are different from those presented by MUPs. In VNO these chelators include specific lipocalins LCN3, LCN4, LCN13, and LCN14, while MOE is overpopulated by OBPs. Interestingly, many lipocalins have similar barrels across the vertebrate taxa and thus they may have overlapping roles in tissue detoxification and in the regulation of bacterial growth. To conclude, for their capacity to bind various lipophilic and other organic molecules, lipocalins are essential for life, and this is documented by parallel evolution of their functions across animal taxa that inhabited the terrestrial ecosystems.
Author Contributions
RS and PS wrote the first draft of the manuscript. TO prepared the involvement of lipocalins in reproduction while TM worked on the microbiota section. BK helped with discussions and analysis of literature searches. All authors contributed to the article and approved the submitted version.
Funding
This research team was established as an output of MICOBION teaming project funded from EU H2020 (No. 810224). PS and TO were funded by the Czech Science Foundation (GAČR) project No: 19-22538S. TM was funded by the Grant Agency of the Charles University, Prague (GAUK; No. 1191419).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
- Abe T., Touhara K. (2014). Structure and function of a peptide pheromone family that stimulate the vomeronasal sensory system in mice. Biochem. Soc. Trans. 42, 873–877. doi: 10.1042/BST20140051, PMID: [DOI] [PubMed] [Google Scholar]
- Abril J. F., Agarwal P., Alexandersson M., Antonarakis S. E., Baertsch R., Berry E., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. doi: 10.1038/nature01262, PMID: [DOI] [PubMed] [Google Scholar]
- Ackels T., Erskine A., Dasgupta D., Marin A. C., Warner T. P. A., Tootoonian S., et al. (2021). Fast odour dynamics are encoded in the olfactory system and guide behaviour. Nature 593, 558–563. doi: 10.1038/s41586-021-03514-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez M. D. G., Romero D. S., Greene L. H., Flower D. R. (2005). The lipocalin protein family: protein sequence, structure and relationship to the calycin superfamily. 17–27.
- Bansal R., Nagel M., Stopkova R., Sofer Y., Kimchi T., Stopka P., et al. (2021). Do all mice smell the same? Chemosensory cues from inbred and wild mouse strains elicit stereotypic sensory representations in the accessory olfactory bulb. BMC Biol. 19:133. doi: 10.1186/s12915-021-01064-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Harrison O. (2017). Homeostatic immunity and the microbiota. Immunity 46, 562–576. doi: 10.1016/j.immuni.2017.04.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghard A., Buck L. B. (1996). Sensory transduction in vomeronasal neurons: evidence for G alpha o, G alpha i2, and adenylyl cyclase II as major components of a pheromone signaling cascade. J. Neurosci. 16, 909–918. doi: 10.1523/JNEUROSCI.16-03-00909.1996, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S., Knoch B., Wong J., Kim W. S., Else P. L., Oakley A. J., et al. (2012). Selective reduction of hydroperoxyeicosatetraenoic acids to their hydroxy derivatives by apolipoprotein D: implications for lipid antioxidant activity and Alzheimer’s disease. Biochem. J. 442, 713–721. doi: 10.1042/BJ20111166, PMID: [DOI] [PubMed] [Google Scholar]
- Bianchi F., Flisi S., Careri M., Riboni N., Resimini S., Sala A., et al. (2019). Vertebrate odorant binding proteins as antimicrobial humoral components of innate immunity for pathogenic microorganisms. PLoS One 14:e0213545. doi: 10.1371/journal.pone.0213545, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilkova B., Swiderska Z., Zita L., Laloe D., Charles M., Benes V., et al. (2018). Domestic fowl breed variation in egg white protein expression: application of proteomics and transcriptomics. J. Agric. Food Chem. 66, 11854–11863. doi: 10.1021/acs.jafc.8b03099, PMID: [DOI] [PubMed] [Google Scholar]
- Bímová B., Albrecht T., Macholán M., Piálek J. (2009). Signalling components of mate recognition system in the house mouse. Behav. Process. 80, 20–27. doi: 10.1016/j.beproc.2008.08.004, PMID: [DOI] [PubMed] [Google Scholar]
- Bimova B. V., Macholan M., Baird S. J., Munclinger P., Dufkova P., Laukaitis C. M., et al. (2011). Reinforcement selection acting on the European house mouse hybrid zone. Mol. Ecol. 20, 2403–2424. doi: 10.1111/j.1365-294X.2011.05106.x, PMID: [DOI] [PubMed] [Google Scholar]
- Bingham R. J., Findlay J. B. C., Hsieh S. Y., Kalverda A. P., Kjellberg A., Chiara P., et al. (2004). Thermodynamics of binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the major urinary protein. J. Am. Chem. Soc. 126, 1675–1681. doi: 10.1021/ja038461i, PMID: [DOI] [PubMed] [Google Scholar]
- Bishop R. E., Cambillau C., Prive G. G., Hsi D., Tillo D., Tillier E. R. M. (2006). “Bacterial lipocalins: origin, structure, and function,” in Lipocalins. eds. Akerstrom B., Borregaard N., Flower D. R., Salier J. P. (Georgetown, TX: Landes Bioscience; ), 28–40. [Google Scholar]
- Brennan P. A., Kendrick K. M. (2006). Mammalian social odours: attraction and individual recognition. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 361, 2061–2078. doi: 10.1098/rstb.2006.1931, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand L., Blon F., Trotier D., Pernollet J.-C. (2004). Natural ligands of hamster aphrodisin. Chem. Senses 29, 425–430. doi: 10.1093/chemse/bjh044, PMID: [DOI] [PubMed] [Google Scholar]
- Bryche B., Baly C., Meunier N. (2021). Modulation of olfactory signal detection in the olfactory epithelium: focus on the internal and external environment, and the emerging role of the immune system. Cell Tissue Res. 384, 589–605. doi: 10.1007/s00441-021-03467-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B., Zufall F. (2016). The sensing of bacteria: emerging principles for the detection of signal sequences by formyl peptide receptors. Biomol. Concepts 7, 205–214. doi: 10.1515/bmc-2016-0013, PMID: [DOI] [PubMed] [Google Scholar]
- Byers S. L., Wiles M. V., Dunn S. L., Taft R. A. (2012). Mouse estrous cycle identification tool and images. PLoS One 7:e35538. doi: 10.1371/journal.pone.0035538, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann P., Chabi M., Delsart A., Le Danvic C., Saliou J.-M., Chasles M., et al. (2019). The olfactory secretome varies according to season in female sheep and goat. BMC Genomics 20:794. doi: 10.1186/s12864-019-6194-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho V. M. A., Nakahara T. S., Cardozo L. M., Souza M. A. A., Camargo A. P., Trintinalia G. Z., et al. (2015). Lack of spatial segregation in the representation of pheromones and kairomones in the mouse medial amygdala. Front. Neurosci. 9:283. doi: 10.3389/fnins.2015.00283, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaggioni A., Sorbi R. T., Keen J. N., Pappin D. J. C., Findlay J. B. C. (1987). Homology between the pyrazine-binding protein form nasal mucosa and major urinary proteins. FEBS Lett. 212, 225–228. doi: 10.1016/0014-5793(87)81349-2, PMID: [DOI] [PubMed] [Google Scholar]
- Cerna M., Kuntova B., Talacko P., Stopkova R., Stopka P. (2017). Differential regulation of vaginal lipocalins (OBP, MUP) during the estrous cycle of the house mouse. Sci. Rep. 7:11674. doi: 10.1038/s41598-017-12021-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamero P., Marton T. F., Logan D. W., Flanagan K., Cruz J. R., Saghatelian A., et al. (2007). Identification of protein pheromones that promote aggressive behaviour. Nature 450, 899–903. doi: 10.1038/nature05997, PMID: [DOI] [PubMed] [Google Scholar]
- Chang P. L., Kopania E., Keeble S., Sarver B. A. J., Larson E., Orth A., et al. (2017). Whole exome sequencing of wild-derived inbred strains of mice improves power to link phenotype and genotype. Mamm. Genome 28, 416–425. doi: 10.1007/s00335-017-9704-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J. B. F., Sarhan F. (2006). “Plant lipocalins,” in Lipocalins. eds. Akerstrom B., Borregaard N., Flower D. R., Salier J. P. (Georgetown, TX: Landes Bioscience; ), 41–48. [Google Scholar]
- Cheetham S. A., Thom M. D., Jury F., Ollier W. E. R., Beynon R. J., Hurst J. L. (2007). The genetic basis of individual-recognition signals in the mouse. Curr. Biol. 17, 1771–1777. doi: 10.1016/j.cub.2007.10.007, PMID: [DOI] [PubMed] [Google Scholar]
- Chu S. T., Lee Y. C., Nein K. M., Chen Y. H. (2000). Expression, immunolocalization and sperm-association of a protein derived from 24p3 gene in mouse epididymis. Mol. Reprod. Dev. 57, 26–36. doi: , PMID: [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M., Knutson D. (2011). Vitamin A in reproduction and development. Nutrients 3, 385–428. doi: 10.3390/nu3040385, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cora M. C., Kooistra L., Travlos G. (2015). Vaginal cytology of the laboratory rat and mouse: review and criteria for the staging of the estrous cycle using stained vaginal smears. Toxicol. Pathol. 43, 776–793. doi: 10.1177/0192623315570339, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correnti C., Strong R. (2012). Mammalian siderophores, siderophore-binding lipocalins, and the labile iron pool. J. Biol. Chem. 287, 13524–13531. doi: 10.1074/jbc.R111.311829, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S. L., Boekelheide K., Vanderhyden B. C., Seth R., Mcburney M. W. (1997). Male infertility caused by epididymal dysfunction in transgenic mice expressing a dominant negative mutation of retinoic acid receptor alpha 1. Biol. Reprod. 56, 985–990. doi: 10.1095/biolreprod56.4.985, PMID: [DOI] [PubMed] [Google Scholar]
- Demir E., Li K., Bobrowski-Khoury N., Sanders J. I., Beynon R. J., Hurst J. L., et al. (2020). The pheromone darcin drives a circuit for innate and reinforced behaviours. Nature 578, 137–141. doi: 10.1038/s41586-020-1967-8, PMID: [DOI] [PubMed] [Google Scholar]
- Dey S., Chamero P., Pru J. K., Chien M.-S., Ibarra-Soria X., Spencer K. R., et al. (2015). Cyclic regulation of sensory perception by a female hormone alters behavior. Cell 161, 1334–1344. doi: 10.1016/j.cell.2015.04.052, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer L. C. (2001). Urine marking and social dominance in male house mice (Mus musculus domesticus). Behav. Process. 53, 113–120. doi: 10.1016/S0376-6357(00)00152-2, PMID: [DOI] [PubMed] [Google Scholar]
- Elangovan N., Lee Y. C., Tzeng W. F., Chu S. T. (2004). Delivery of ferric ion to mouse spermatozoa is mediated by lipocalin internalization. Biochem. Biophys. Res. Commun. 319, 1096–1104. doi: 10.1016/j.bbrc.2004.05.091, PMID: [DOI] [PubMed] [Google Scholar]
- Enk V. M., Baumann C., Thoß M., Luzynski K. C., Razzazi-Fazeli E., Penn D. J. (2016). Regulation of highly homologous major urinary proteins in house mice quantified with label-free proteomic methods. Mol. BioSyst. 12, 3005–3016. doi: 10.1039/C6MB00278A, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M., Lin H., Zhou L., Yu Y., Abergel R., Liu D., et al. (2006). The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U. S. A. 103, 16502–16507. doi: 10.1073/pnas.0604636103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., et al. (2004). Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921. doi: 10.1038/nature03104, PMID: [DOI] [PubMed] [Google Scholar]
- Flower D. (1996). The lipocalin protein family: structure and function. Biochem. J. 318, 1–14. doi: 10.1042/bj3180001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckinger M., Haas H., Merschak P., Glasgow B., Redl B. (2004). Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob. Agents Chemother. 48, 3367–3372. doi: 10.1128/AAC.48.9.3367-3372.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganfornina M. D., Do Carmo S., Lora J. M., Torres-Schumann S., Vogel M., Allhorn M., et al. (2008). Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 7, 506–515. doi: 10.1111/j.1474-9726.2008.00395.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganfornina M. D., Gutierrez G., Bastiani M., Sanchez D. (2000). A phylogenetic analysis of the lipocalin protein family. Mol. Biol. Evol. 17, 114–126. doi: 10.1093/oxfordjournals.molbev.a026224, PMID: [DOI] [PubMed] [Google Scholar]
- Ganfornina M. D., Sánchez D., Bastiani M. J. (1995). Lazarillo, a new GPI-linked surface lipocalin, is restricted to a subset of neurons in the grasshopper embryo. Dev. Dent. 121, 123–134. [DOI] [PubMed] [Google Scholar]
- Ganfornina M. D., Sanchez D., Greene L. H., Flower D. R. (2006). “The lipocalin protein family: protein sequence, structure and relationship to the calycin superfamily,” in Lipocalins. eds. Akerstrom B., Borregaard N., Flower A. C. T., Salier J. P. (Georgetowns, TX: Landes Bioscience; ), 17–27. [Google Scholar]
- Garénaux A., Houle S., Folch B., Dallaire G., Truesdell M., Lépine F., et al. (2013). Avian lipocalin expression in chickens following Escherichia coli infection and inhibition of avian pathogenic Escherichia coli growth by ex-FABP. Vet. Immunol. Immunopathol. 152, 156–167. doi: 10.1016/j.vetimm.2012.09.018, PMID: [DOI] [PubMed] [Google Scholar]
- Gerena R. L., Eguchi N., Urade Y., Killian G. J. (2000). Stage and region-specific localization of lipocalin-type prostaglandin D synthase in the adult murine testis and epididymis. J. Androl. 21, 848–854. PMID: [PubMed] [Google Scholar]
- Goetz D., Holmes M., Borregaard N., Bluhm M., Raymond K., Strong R. (2002). The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10, 1033–1043. doi: 10.1016/S1097-2765(02)00708-6, PMID: [DOI] [PubMed] [Google Scholar]
- Grolli S., Merli E., Conti V., Scaltriti E., Ramoni R. (2006). Odorant binding protein has the biochemical properties of a scavenger for 4-hydroxy-2-nonenal in mammalian nasal mucosa. FEBS J. 273, 5131–5514. doi: 10.1111/j.1742-4658.2006.05510.x, PMID: [DOI] [PubMed] [Google Scholar]
- Grus W. E., Zhang J. (2008). Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol. Biol. Evol. 26, 407–419. doi: 10.1093/molbev/msn262, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyonnet B., Zabet-Moghaddam M., Sanfrancisco S., Cornwall G. A. (2012). Isolation and proteomic characterization of the mouse sperm acrosomal matrix. Mol. Cell. Proteomics 11, 758–774. doi: 10.1074/mcp.M112.020339, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S., Hattori T., Sato T., Sato K., Matsuda S., Kobayakawa R., et al. (2010). The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature 466, 118–122. doi: 10.1038/nature09142, PMID: [DOI] [PubMed] [Google Scholar]
- Hagemeyer P., Begall S., Janotova K., Todrank J., Heth G., Jedelsky P. L., et al. (2011). Searching for major urinary proteins (MUPs) as chemosignals in urine of subterranean rodents. J. Chem. Ecol. 37, 687–694. doi: 10.1007/s10886-011-9971-y, PMID: [DOI] [PubMed] [Google Scholar]
- Hamil K. G., Liu Q., Sivashanmugam P., Anbalagan M., Yenugu S., Soundararajan R., et al. (2003). LCN6, a novel human epididymal lipocalin. Reprod. Biol. Endocrinol. 1:112. doi: 10.1186/1477-7827-1-112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst J. L., Gray S. J., Davey P., Young D., Corbishley J., Dawson C. (1997). Social interaction alters attraction to competitor's odour in the mouse Mus spretus Lataste. Anim. Behav. 54, 941–953. doi: 10.1006/anbe.1997.0515, PMID: [DOI] [PubMed] [Google Scholar]
- Hurst J. L., Robertson D. H. L., Tolladay U., Beynon R. J. (1998). Proteins in urine scent marks of male house mice extend the longevity of olfactory signals. Anim. Behav. 55, 1289–1297. [DOI] [PubMed] [Google Scholar]
- Ibarra-Soria X., Levitin M. O., Logan D. W. (2014a). The genomic basis of vomeronasal-mediated behaviour. Mamm. Genome 25, 75–86. doi: 10.1007/s00335-013-9463-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Soria X., Levitin M. O., Saraiva L. R., Logan D. W. (2014b). The olfactory transcriptomes of mice. PLoS Genet. 10:e1004593. doi: 10.1371/journal.pgen.1004593, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Soria X., Nakahara T. S., Lilue J., Jiang Y., Trimmer C., Souza M. A., et al. (2017). Variation in olfactory neuron repertoires is genetically controlled and environmentally modulated. elife 6:e21476. doi: 10.7554/eLife.21476, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janotova K., Stopka P. (2011). The level of major urinary proteins is socially regulated in wild Mus musculus musculus. J. Chem. Ecol. 37, 647–656. doi: 10.1007/s10886-011-9966-8, PMID: [DOI] [PubMed] [Google Scholar]
- Jauregui E. J., Mitchell D., Topping T., Hogarth C. A., Griswold M. D. (2018). Retinoic acid receptor signaling is necessary in steroidogenic cells for normal spermatogenesis and epididymal function. Development 145:dev160465. doi: 10.1242/dev.160465, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E., Srikanth C., Sandgren A., Harrington L., Trebicka E., Wang L., et al. (2010). Siderocalin inhibits the intracellular replication of mycobacterium tuberculosis in macrophages. FEMS Immunol. Med. Microbiol. 58, 138–145. doi: 10.1111/j.1574-695X.2009.00622.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien L., Baron F., Bonnassie S., Nau F., Guerin C., Jan S., et al. (2019). The anti-bacterial iron-restriction defence mechanisms of egg white; the potential role of three lipocalin-like proteins in resistance against salmonella. Biometals 32, 453–467. doi: 10.1007/s10534-019-00180-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A. W., Ackels T., Kuo T. H., Cichy A., Dey S., Hays C., et al. (2014). Murine pheromone proteins constitute a context-dependent combinatorial code governing multiple social behaviors. Cell 157, 676–688. doi: 10.1016/j.cell.2014.02.025, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido T., Arata S., Suzuki R., Hosono T., Nakanishi Y., Miyazaki J., et al. (2005). The testicular fatty acid binding protein PERF15 regulates the fate of germ cells in PERF15 transgenic mice. Develop. Growth Differ. 47, 15–24. doi: 10.1111/j.1440-169x.2004.00775.x, PMID: [DOI] [PubMed] [Google Scholar]
- Koskinen K., Reichert J., Hoier S., Schachenreiter J., Duller S., Moissl-Eichinger C., et al. (2018). The nasal microbiome mirrors and potentially shapes olfactory function. Sci. Rep. 8:1296. doi: 10.1038/s41598-018-19438-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntova B., Stopkova R., Stopka P. (2018). Transcriptomic and proteomic profiling revealed high proportions of odorant binding and antimicrobial defense proteins in olfactory tissues of the house mouse. Front. Genet. 9:26. doi: 10.3389/fgene.2018.00026, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J., Grigsby C. C., Preti G., Rizki M. M., Yamazaki K., Beauchamp G. K. (2013). Changes in volatile compounds of mouse urine as it ages: their interactions with water and urinary proteins. Physiol. Behav. 120, 211–219. doi: 10.1016/j.physbeh.2013.08.011, PMID: [DOI] [PubMed] [Google Scholar]
- Kwak J., Grigsby C. C., Rizki M. M., Preti G., Koksal M., Josue J., et al. (2012). Differential binding between volatile ligands and major urinary proteins due to genetic variation in mice. Physiol. Behav. 107, 112–120. doi: 10.1016/j.physbeh.2012.06.008, PMID: [DOI] [PubMed] [Google Scholar]
- Kwak J., Josue J., Faranda A., Opiekun M. C., Preti G., Osada K., et al. (2011). Butylated hydroxytoluene is a ligand of urinary proteins derived from female mice. Chem. Senses 36, 443–452. doi: 10.1093/chemse/bjr015, PMID: [DOI] [PubMed] [Google Scholar]
- Kwak J., Strasser E., Luzynski K., Thoß M., Penn D. J. (2016). Are MUPs a toxic waste disposal system? PLoS One 11:e0151474. doi: 10.1371/journal.pone.0151474, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacazette E., Gachon A., Pitiot G. (2000). A novel human odorant-binding protein gene family resulting from genomic duplicons at 9q34: differential expression in the oral and genital spheres. Hum. Mol. Genet. 9, 289–301. doi: 10.1093/hmg/9.2.289, PMID: [DOI] [PubMed] [Google Scholar]
- Lareyre J. J., Winfrey V. P., Kasper S., Ong D. E., Matusik R. J., Olson G. E., et al. (2001). Gene duplication gives rise to a new 17-kilodalton lipocalin that shows epididymal region-specific expression and testicular factor(s) regulation. Endocrinology 142, 1296–1308. doi: 10.1210/endo.142.3.8045, PMID: [DOI] [PubMed] [Google Scholar]
- Lazar J., Greenwood D. R., Rasmussen L. E. L., Prestwich G. D. (2002). Molecular and functional characterization of an odorant binding protein of the Asian elephant, elephas maximus: implications for the role of lipocalins in mammalian olfaction. Biochemistry 41, 11786–11794. doi: 10.1021/bi0256734, PMID: [DOI] [PubMed] [Google Scholar]
- Lechner M., Wojnar P., Redl B. (2001). Human tear lipocalin acts as an oxidative-stress-induced scavenger of potentially harmful lipid peroxidation products in a cell culture system. Biochem. J. 356, 129–135. doi: 10.1042/0264-6021:3560129, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C., Liao C., Li P. T., Tzeng W. F., Chu S. T. (2003). Mouse lipocalin as an enhancer of spermatozoa motility. Mol. Biol. Rep. 30, 165–172. doi: 10.1023/A:1024985024661, PMID: [DOI] [PubMed] [Google Scholar]
- Lee Y., Mazmanian S. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773. doi: 10.1126/science.1195568, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T., Brennan P., Widmayer P., Chandramani P. S., Maul-Pavicic A., Jager M., et al. (2004). MHC class I peptides as chemosensory signals in the vomeronasal organ. Science 306, 1003–1037. doi: 10.1126/science.1102818, PMID: [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T., Lane A. P., Puche A. C., Ma W., Novotny M. V., Shipley M. T., et al. (2000). Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature 405, 792–796. doi: 10.1038/35015572, PMID: [DOI] [PubMed] [Google Scholar]
- Levy D. R., Sofer Y., Brumfeld V., Zilkha N., Kimchi T. (2020). The nasopalatine ducts are required for proper pheromone signaling in mice. Front. Neurosci. 14:585323. doi: 10.3389/fnins.2020.585323, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilue J., Doran A. G., Fiddes I. T., Abrudan M., Armstrong J., Bennett R., et al. (2018). Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat. Genet. 50, 1574–1583. doi: 10.1038/s41588-018-0223-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livera G., Rouiller-Fabre V., Pairault C., Levacher C., Habert R. (2002). Regulation and perturbation of testicular functions by vitamin A. Reproduction 124, 173–180. PMID: [PubMed] [Google Scholar]
- Logan D. W., Marton T. F., Stowers L. (2008). Species specificity in major urinary proteins by parallel evolution. PLoS One 3:e3280. doi: 10.1371/journal.pone.0003280, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loxley G. M., Unsworth J., Turton M. J., Jebb A., Lilley K. S., Simpson D. M., et al. (2017). Glareosin: a novel sexually dimorphic urinary lipocalin in the bank vole, Myodes glareolus. Open Biol. 7:170135. doi: 10.1098/rsob.170135, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo-Marquez A., Vazquez-Acevedo M., Ongay-Larios L., Miranda-Astudillo H., Hernandez-Munoz R., Gonzalez-Halphen D., et al. (2014). Overexpression of a monomeric form of the bovine odorant-binding protein protects Escherichia coli from chemical-induced oxidative stress. Free Radic. Res. 48, 814–822. doi: 10.3109/10715762.2014.910867, PMID: [DOI] [PubMed] [Google Scholar]
- Malnic B., Hirono J., Sato T., Buck L. B. (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. doi: 10.1016/S0092-8674(00)80581-4, PMID: [DOI] [PubMed] [Google Scholar]
- Matějková T., Hájková P., Stopková R., Stanko M., Martin J.-F., Kreisinger J., et al. (2020). Oral and vaginal microbiota in selected field mice of the genus Apodemus: a wild population study. Sci. Rep. 10:13246. doi: 10.1038/s41598-020-70249-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. H., Campbell P., Sheehan M. J. (2020). Distinct evolutionary trajectories of V1R clades across mouse species. BMC Evol. Biol. 20:99. doi: 10.1186/s12862-020-01662-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Suzuki T., Minamishima S., Igarashi T., Inoue K., Nishimura D., et al. (2016). Neutrophil gelatinase-associated lipocalin regulates gut microbiota of mice. J. Gastroenterol. Hepatol. 31, 145–154. doi: 10.1111/jgh.13042, PMID: [DOI] [PubMed] [Google Scholar]
- Moschen A., Gerner R., Wang J., Klepsch V., Adolph T., Reider S., et al. (2016). Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe 19, 455–469. doi: 10.1016/j.chom.2016.03.007, PMID: [DOI] [PubMed] [Google Scholar]
- Moss R. L., Flynn R. E., Shen X., Dudley C., Shi J., Novotny M. (1997). Urine-derived compound evokes membrane responses in mouse vomeronasal receptor neurons. J. Neurophysiol. 77, 2856–2862. doi: 10.1152/jn.1997.77.5.2856, PMID: [DOI] [PubMed] [Google Scholar]
- Moudra A., Niederlova V., Novotny J., Schmiedova L., Kubovciak J., Matejkova T., et al. (2021). Phenotypic and clonal stability of antigen-inexperienced memory-like t cells across the genetic background, hygienic status, and aging. J. Immunol. 206, 2109–2121. doi: 10.4049/jimmunol.2001028, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel M., Bansal R., Stopka P., Kimchi T., Ben-Shaul Y., Spehr M. (2018). A systematic comparison of semiochemical signaling in the accessory olfactory system of wild and lab strain mice. Chem. Senses 43:E31. [Google Scholar]
- Nagnan-Le Meillour P., Descamps A., Le Danvic C., Grandmougin M., Saliou J.-M., Klopp C., et al. (2019). Identification of potential chemosignals in the European water vole Arvicola terrestris. Sci. Rep. 9:18378. doi: 10.1038/s41598-019-54935-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagnan-Le Meillour P., Vercoutter-Edouart A. S., Hilliou F., Le Danvic C., Levy F. (2014). Proteomic analysis of pig (Sus scrofa) olfactory soluble proteome reveals O-linked-N-acetylglucosaminylation of secreted odorant-binding proteins. Front. Endocrinol. 5:202. doi: 10.3389/fendo.2014.00202, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M., Schroll A., Sonnweber T., Weiss G. (2010). The struggle for iron – a metal at the host-pathogen interface. Cell. Microbiol. 12, 1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x, PMID: [DOI] [PubMed] [Google Scholar]
- Novotny M. V., Jemiolo B., Wiesler D., Ma W., Harvey S., Xu F., et al. (1999). A unique urinary constituent, 6-hydroxy-6-methyl-3-heptanone, is a pheromone that accelerates puberty in female mice. Chem. Biol. 6, 377–383. doi: 10.1016/S1074-5521(99)80049-0, PMID: [DOI] [PubMed] [Google Scholar]
- Oko R., Morales C. R. (1994). A novel testicular protein, with sequence similarities to a family of lipid binding proteins, is a major component of the rat sperm perinuclear theca. Dev. Biol. 166, 235–245. doi: 10.1006/dbio.1994.1310, PMID: [DOI] [PubMed] [Google Scholar]
- Papes F., Logan D. W., Stowers L. (2010). The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell 141, 692–703. doi: 10.1016/j.cell.2010.03.037, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P., Zhou J. J., Ban L. P., Calvello M. (2006). Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Gómez A., Bleymehl K., Stein B., Pyrski M., Birnbaumer L., Munger S. D., et al. (2015). Innate predator odor aversion driven by parallel olfactory subsystems that converge in the ventromedial hypothalamus. Curr. Biol. 25, 1340–1346. doi: 10.1016/j.cub.2015.03.026, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan M. M., Mclean L., Armstrong S. D., Hurst J. L., Beynon R. J., Lian L. Y. (2014a). The structure, stability and pheromone binding of the male mouse protein sex pheromone darcin. PLoS One 9:e108415. doi: 10.1371/journal.pone.0108415, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan M. M., Mclean L., Hurst J. L., Beynon R. J., Lian L. Y. (2014b). Comparative study of the molecular variation between ‘central’ and ‘peripheral’ MUPs and significance for behavioural signalling. Biochem. Soc. Trans. 42, 866–872. doi: 10.1042/BST20140082, PMID: [DOI] [PubMed] [Google Scholar]
- Rankin T. L., Ong D. E., Orgebin-Crist M. C. (1992). The 18-kDa mouse epididymal protein (MEP 10) binds retinoic acid. Biol. Reprod. 46, 767–771. doi: 10.1095/biolreprod46.5.767, PMID: [DOI] [PubMed] [Google Scholar]
- Richieri G. V., Ogata R. T., Zimmerman A. W., Veerkamp J. H., Kleinfeld A. M. (2000). Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry 39, 7197–7204. doi: 10.1021/bi000314z, PMID: [DOI] [PubMed] [Google Scholar]
- Riviere S., Challet L., Fluegge D., Spehr M., Rodriguez I. (2009). Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature 459, 574–577. doi: 10.1038/nature08029, PMID: [DOI] [PubMed] [Google Scholar]
- Roberts S. A., Davidson A. J., Mclean L., Beynon R. J., Hurst J. L. (2012). Pheromonal induction of spatial learning in mice. Science 338, 1462–1465. doi: 10.1126/science.1225638, PMID: [DOI] [PubMed] [Google Scholar]
- Roberts S. A., Prescott M. C., Davidson A. J., Mclean L., Beynon R. J., Hurst J. L. (2018). Individual odour signatures that mice learn are shaped by involatile major urinary proteins (MUPs). BMC Biol. 16:48. doi: 10.1186/s12915-018-0512-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. A., Simpson D. M., Armstrong S. D., Davidson A. J., Robertson D. H., Mclean L., et al. (2010). Darcin: a male pheromone that stimulates female memory and sexual attraction to an individual male’s odour. BMC Biol. 8:75. doi: 10.1186/1741-7007-8-75, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martinez H., Saravia F., Wallgren M., Martinez E. A., Sanz L., Roca J., et al. (2010). Spermadhesin PSP-I/PSP-II heterodimer induces migration of polymorphonuclear neutrophils into the uterine cavity of the sow. J. Reprod. Immunol. 84, 57–65. doi: 10.1016/j.jri.2009.10.007, PMID: [DOI] [PubMed] [Google Scholar]
- Rossitto M., Ujjan S., Poulat F., Boizet-Bonhoure B. (2015). Multiple roles of the prostaglandin D2 signaling pathway in reproduction. Reproduction 149, R49–R58. doi: 10.1530/REP-14-0381, PMID: [DOI] [PubMed] [Google Scholar]
- Rutherford S., Bassler B. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D., Ganfornina M. D., Gutierrez G., Gauthier-Jauneau A. C., Risler J. L., Salier J. P. (2006). “Lipocalin genes and their evolutionary history,” in Lipocalins. eds. Akerstrom B., Borregaard N., Flower A. C. T., Salier J. P. (Georgetown, TX: Landes Bioscience; ), 5–16. [Google Scholar]
- Sanchez D., Ganfornina M. D., Gutierrez G., Marin A. (2003). Exon-intron structure and evolution of the lipocalin gene family. Mol. Biol. Evol. 20, 775–783. doi: 10.1093/molbev/msg079, PMID: [DOI] [PubMed] [Google Scholar]
- Santoro S. W., Jakob S. (2018). Gene expression profiling of the olfactory tissues of sex-separated and sex-combined female and male mice. Sci. Data 5:180260. doi: 10.1038/sdata.2018.260, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj V., Asano A., Page J. L., Nelson J. L., Kothapalli K. S., Foster J. A., et al. (2010). Mice lacking FABP9/PERF15 develop sperm head abnormalities but are fertile. Dev. Biol. 348, 177–189. doi: 10.1016/j.ydbio.2010.09.019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Ahn J., Hong C., Seo E., Kye K., Lee W., et al. (2006). Expression of neutrophil gelatinase-associated lipocalin in skin epidermis. J. Investig. Dermatol. 126, 510–512. doi: 10.1038/sj.jid.5700035, PMID: [DOI] [PubMed] [Google Scholar]
- Sharrow S. D., Novotny M. V., Stone M. J. (2003). Thermodynamic analysis of binding between mouse major urinary protein-I and the pheromone 2-sec-butyl-4,5-dihydrothiazole. Biochemistry 42, 6302–6309. doi: 10.1021/bi026423q, PMID: [DOI] [PubMed] [Google Scholar]
- Sharrow S. D., Vaughn J. L., Žídek L., Novotny M. V., Stone M. J. (2002). Pheromone binding by polymorphic mouse major urinary proteins. Protein Sci. 11, 2247–2256. doi: 10.1110/ps.0204202, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan M. J., Campbell P., Miller C. H. (2019). Evolutionary patterns of major urinary protein scent signals in house mice and relatives. Mol. Ecol. 28, 3587–3601. doi: 10.1111/mec.15155, PMID: [DOI] [PubMed] [Google Scholar]
- Shi N., Li N., Duan X., Niu H. (2017). Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 4:14. doi: 10.1186/s40779-017-0122-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P., Zhang J. (2007). Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 17, 166–174. doi: 10.1101/gr.6040007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A., Macrides F., Clancy A. N., Agosta W. C. (1986). Purification and analysis of a proteinaceous aphrodisiac pheromone from hamster vaginal discharge. J. Biol. Chem. 261, 13323–13326. doi: 10.1016/S0021-9258(18)69307-X [DOI] [PubMed] [Google Scholar]
- Singh V., Galla S., Golonka R., Patterson A., Chassaing B., Joe B., et al. (2020). Lipocalin 2 deficiency-induced gut microbiota dysbiosis evokes metabolic syndrome in aged mice. Physiol. Genomics 52, 314–321. doi: 10.1152/physiolgenomics.00118.2019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Yeoh B., Chassaing B., Zhang B., Saha P., Xiao X., et al. (2016). Microbiota-inducible innate immune, siderophore binding protein lipocalin 2 is critical for intestinal homeostasis. Cell. Mol. Gastroenterol. Hepatol. 4, 482–498.e6. doi: 10.1016/j.jcmgh.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerget S., Rosenow M. A., Petritis K., Karr T. L. (2015). Sperm proteome maturation in the mouse epididymis. PLoS One 10:e0140650. doi: 10.1371/journal.pone.0140650, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M., Spehr J., Ukhanov K., Kelliher K. R., Leinders-Zufall T., Zufall F. (2006). Parallel processing of social signals by the mammalian main and accessory olfactory systems. Cell. Mol. Life Sci. 63, 1476–1484. doi: 10.1007/s00018-006-6109-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka P., Janotova K., Heyrovsky D. (2007). The advertisement role of major urinary proteins in mice. Physiol. Behav. 91, 667–670. doi: 10.1016/j.physbeh.2007.03.030, PMID: [DOI] [PubMed] [Google Scholar]
- Stopka P., Kuntova B., Klempt P., Havrdova L., Cerna M., Stopkova R. (2016). On the saliva proteome of the eastern European house mouse (Mus musculus musculus) focusing on sexual signalling and immunity. Sci. Rep. 6:32481. doi: 10.1038/srep32481, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka P., Macdonald D. W. (1998). Signal interchange during mating in the wood mouse (Apodemus sylvaticus): the concept of active and passive signalling. Behaviour 135, 231–249. [Google Scholar]
- Stopková R., Dudkova B., Hajkova P., Stopka P. (2014). Complementary roles of mouse lipocalins in chemical communication and immunity. Biochem. Soc. Trans. 42, 893–898. doi: 10.1042/BST20140053, PMID: [DOI] [PubMed] [Google Scholar]
- Stopková R., Hladovcová D. J. K., Vyoral D., Stopka P. (2009). Multiple roles of secretory lipocalins (MUP, OBP) in mice. Folia Zool. 58, 29–40. [Google Scholar]
- Stopkova R., Klempt P., Kuntova B., Stopka P. (2017). On the tear proteome of the house mouse (Mus musculus musculus) in relation to chemical signalling. PeerJ 5:e3541. doi: 10.7717/peerj.3541, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopková R., Stopka P., Janotová K., Jedelsky P. L. (2007). Species-specific expression of major urinary proteins in the house mice (Mus musculus musculus and Mus musculus domesticus). J. Chem. Ecol. 33, 861–869. doi: 10.1007/s10886-007-9262-9, PMID: [DOI] [PubMed] [Google Scholar]
- Stopková R., Vinkler D., Kuntová B., Sedo O., Albrecht T., Suchan J., et al. (2016). Mouse lipocalins (MUP, OBP, LCN) are co-expressed in tissues involved in chemical communication. Front. Ecol. Evol. 4:47. doi: 10.3389/fevo.2016.00047 [DOI] [Google Scholar]
- Stopkova R., Zdrahal Z., Ryba S., Sedo O., Sandera M., Stopka P. (2010). Novel OBP genes similar to hamster Aphrodisin in the bank vole, Myodes glareolus. BMC Genomics 11:45. doi: 10.1186/1471-2164-11-45, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm T., Leinders-Zufall T., Macek B., Walzer M., Jung S., Pommerl B., et al. (2013). Mouse urinary peptides provide a molecular basis for genotype discrimination by nasal sensory neurons. Nat. Commun. 4:1616. doi: 10.1038/ncomms2610, PMID: [DOI] [PubMed] [Google Scholar]
- Thoß M., Enk V., Yu H., Miller I., Luzynski K. C., Balint B., et al. (2016). Diversity of major urinary proteins (MUPs) in wild house mice. Sci. Rep. 6:38378. doi: 10.1038/srep38378, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoß M., Luzynski K., Ante M., Miller I., Penn D. J. (2015). Major urinary protein (MUP) profiles show dynamic changes rather than individual ‘barcode’ signatures. Front. Ecol. Evol. 3:71. doi: 10.3389/fevo.2015.00071, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoss M., Luzynski K. C., Enk V. M., Razzazi-Fazeli E., Kwak J., Ortner I., et al. (2019). Regulation of volatile and non-volatile pheromone attractants depends upon male social status. Sci. Rep. 9:489. doi: 10.1038/s41598-019-41666-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirindelli R. (2021). Coding of pheromones by vomeronasal receptors. Cell Tissue Res. 383, 367–386. doi: 10.1007/s00441-020-03376-6, PMID: [DOI] [PubMed] [Google Scholar]
- Turner T. T., Bomgardner D., Jacobs J. P., Nguyen Q. A. (2003). Association of segmentation of the epididymal interstitium with segmented tubule function in rats and mice. Reproduction 125, 871–878. doi: 10.1530/rep.0.1250871, PMID: [DOI] [PubMed] [Google Scholar]
- Turton M. J., Robertson D. H. L., Smith J. R., Hurst J. L., Beynon R. J. (2010). Roborovskin, a lipocalin in the urine of the Roborovski hamster, Phodopus roborovskii. Chem. Senses 35, 675–684. doi: 10.1093/chemse/bjq060, PMID: [DOI] [PubMed] [Google Scholar]
- Urade Y., Hayaishi O. (2000). Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Biochim. Biophys. Acta 1482, 259–271. doi: 10.1016/S0167-4838(00)00161-8, PMID: [DOI] [PubMed] [Google Scholar]
- Uroz S., Dessaux Y., Oger P. (2009). Quorum sensing and quorum quenching: the yin and yang of bacterial communication. Chembiochem 10, 205–216. doi: 10.1002/cbic.200800521, PMID: [DOI] [PubMed] [Google Scholar]
- Van Der Linden C., Jakob S., Gupta P., Dulac C., Santoro S. W. (2018). Sex separation induces differences in the olfactory sensory receptor repertoires of male and female mice. Nat. Commun. 9:5081. doi: 10.1038/s41467-018-07120-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira F. G., Rozas J. (2011). Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 3, 476–490. doi: 10.1093/gbe/evr033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihani A., Hu X. S., Gundala S., Koyama S., Block E., Matsunami H. (2020). Semiochemical responsive olfactory sensory neurons are sexually dimorphic and plastic. elife 9:e54501. doi: 10.7554/eLife.54501, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Takeo T., Tojo H., Sakoh K., Berger T., Nakagata N., et al. (2014). Lipocalin 2 binds to membrane phosphatidylethanolamine to induce lipid raft movement in a PKA-dependent manner and modulates sperm maturation. Development 141, 2157–2164. doi: 10.1242/dev.105148, PMID: [DOI] [PubMed] [Google Scholar]
- Wen Z., Liu D., Zhu H., Sun X., Xiao Y., Lin Z., et al. (2021). Deficiency for Lcn8 causes epididymal sperm maturation defects in mice. Biochem. Biophys. Res. Commun. 548, 7–13. doi: 10.1016/j.bbrc.2021.02.052, PMID: [DOI] [PubMed] [Google Scholar]
- Wu H., Santoni-Rugiu E., Ralfkiaer E., Porse B., Moser C., Hoiby N., et al. (2010). Lipocalin 2 is protective against E. coli pneumonia. Respir. Res. 11:96. doi: 10.1186/1465-9921-11-96, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn E. H., Sánchez-Andrade G., Carss K. J., Logan D. W. (2012). Genomic variation in the vomeronasal receptor gene repertoires of inbred mice. BMC Genomics 13:415. doi: 10.1186/1471-2164-13-415, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Yeoh B., Vijay-Kumar M., Stover P., Balling R. (2017). Lipocalin 2: an emerging player in iron homeostasis and inflammation. Annu. Rev. Nutr. 37, 103–130. doi: 10.1146/annurev-nutr-071816-064559, PMID: [DOI] [PubMed] [Google Scholar]
- Yin Q., Shen J., Wan X., Liu Q., Zhou Y., Zhang Y. (2018). Impaired sperm maturation in conditional Lcn6 knockout mice. Biol. Reprod. 98, 28–41. doi: 10.1093/biolre/iox128, PMID: [DOI] [PubMed] [Google Scholar]
- Yip K. S., Suvorov A., Connerney J., Lodato N. J., Waxman D. J. (2013). Changes in mouse uterine transcriptome in estrus and proestrus. Biol. Reprod. 89:13. doi: 10.1095/biolreprod.112.107334, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zala S. M., Bilak A., Perkins M., Potts W. K., Penn D. J. (2015). Female house mice initially shun infected males, but do not avoid mating with them. Behav. Ecol. Sociobiol. 69, 715–722. doi: 10.1007/s00265-015-1884-2 [DOI] [Google Scholar]
- Zala S. M., Potts W. K., Penn D. J. (2004). Scent-marking displays provide honest signals of health and infection. Behav. Ecol. 15, 338–344. doi: 10.1093/beheco/arh022 [DOI] [Google Scholar]
- Zhou Z., Feng C., Liu X., Liu S. (2020). 3nLcn2, a teleost lipocalin 2 that possesses antimicrobial activity and inhibits bacterial infection in triploid crucian carp. Fish Shellfish Immunol. 102, 47–55. doi: 10.1016/j.fsi.2020.04.015, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou Q., Li Y., Nie R., Friel P., Mitchell D., Evanoff R. M., et al. (2008). Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol. Reprod. 78, 537–545. doi: 10.1095/biolreprod.107.064337, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidek L., Stone M. J., Lato S. M., Pagel M. D., Miao Z., Ellington A. D., et al. (1999). NMR mapping of the recombinant mouse major urinary protein I binding site occupied by the pheromone 2-sec-butyl-4,5-dihydrothiazole. Biochemistry 38, 9850–9861. doi: 10.1021/bi990497t, PMID: [DOI] [PubMed] [Google Scholar]