Abstract
Gynostemma pentaphyllum (Thunb.) Makino is an economically valuable medicinal plant belonging to the Cucurbitaceae family that produces the bioactive compound gypenoside. Despite several transcriptomes having been generated for G. pentaphyllum, a reference genome is still unavailable, which has limited the understanding of the gypenoside biosynthesis and regulatory mechanism. Here, we report a high-quality G. pentaphyllum genome with a total length of 582 Mb comprising 1,232 contigs and a scaffold N50 of 50.78 Mb. The G. pentaphyllum genome comprised 59.14% repetitive sequences and 25,285 protein-coding genes. Comparative genome analysis revealed that G. pentaphyllum was related to Siraitia grosvenorii, with an estimated divergence time dating to the Paleogene (∼48 million years ago). By combining transcriptome data from seven tissues, we reconstructed the gypenoside biosynthetic pathway and potential regulatory network using tissue-specific gene co-expression network analysis. Four UDP-glucuronosyltransferases (UGTs), belonging to the UGT85 subfamily and forming a gene cluster, were involved in catalyzing glycosylation in leaf-specific gypenoside biosynthesis. Furthermore, candidate biosynthetic genes and transcription factors involved in the gypenoside regulatory network were identified. The genetic information obtained in this study provides insights into gypenoside biosynthesis and lays the foundation for further exploration of the gypenoside regulatory mechanism.
Keywords: Gynostemma pentaphyllum, genome assembly, gypenoside biosynthesis, co-expression, regulatory network
1. Introduction
Gynostemma pentaphyllum (Thunb.) Makino (2n = 2× = 22), a traditional Chinese medicinal herb of the Cucurbitaceae family, is widely distributed in Southeast Asia, including China, Korea, and Japan.1Gynostemmapentaphyllum was first recorded in Jiuhuang Bencao (Materia Medica for the Relief of Famine), which was compiled by Zhu Su in AD 1406 to prepare for natural disasters and to extend traditional Chinese medicine (Supplementary Fig. S1). Gypenosides, a class of triterpenoid saponin compounds with dammarane-type structure, are effective chemical ingredients in G. pentaphyllum.2 At present, more than 170 unique gypenosides have been isolated, with gypenosides III, IV, VIII, and XII homologous to ginsenosides Rb1, Rb3, Rd, and F2, and the structures of the remaining gypenosides similar to ginsenosides.3,4 Recent pharmacological studies reveal that gypenosides exhibit beneficial effects against cardiovascular disease, lower blood pressure, improve arteriosclerosis, and enhance anticancer activities.5–7 In plants, triterpenoids are regarded as defensive compounds against pathogens and phytophagous insects.8 Therefore, elucidating the genes responsible for gypenoside biosynthesis could provide essential clues for probing the gypenoside biosynthetic pathway and regulatory network as well as open up opportunities for improving the triterpenoid saponin content in medicinal plants using biotechnology.
Gypenoside biosynthesis is a branch of the triterpenoid pathway, and gypenoside enzyme-encoding genes can be divided into two groups—early and late. The early biosynthesis genes (EBGs) encode enzymes necessary for producing 2,3-oxidosqualene, such as farnesyl pyrophosphate synthase (FPS), squalene synthase (SS), and squalene epoxidase (SE); these enzymes have been well characterized in plants such as Panax ginseng from the Araliaceae family.9 The late biosynthesis genes (LBGs) encode enzymes involved in gypenoside production, such as 2,3-oxidosqualene cyclases (OSCs), cytochrome P450s (CYP450s), and UDP-glucuronosyltransferases (UGTs). Cyclization of 2,3-oxidosqualene catalysed by OSCs is the first committed step in gypenoside biosynthesis, and all characterized CYP450s involved in triterpenoid saponin hydroxylation belong to the CYP716 family, whereas UGT genes belonging to the UGT71, 73, 74, 85, 91, and 94 subfamilies are responsible for catalysing triterpene saponin glycosylation.8,10,11 However, because of the lack of genomic information, LBGs involved in gypenoside biosynthesis remain to be identified. In addition, little is known about the transcriptional regulation of the gypenoside biosynthetic pathway, which limits the improvement in gypenoside production using metabolic engineering.
Here, we present a chromosome-level reference genome for diploid G. pentaphyllum using Illumina, PacBio, and Hi-C data. We identified the repeat sequences and annotated the functions of protein-coding genes. By combining genomic and transcriptomic approaches, we systematically screened and identified a series of candidate genes responsible for the diversity, hydroxylation, and glycosylation of the gypenoside skeleton. Furthermore, we screened a series of transcription factors (TFs) involved in gypenoside regulation, thus opening up the possibility of producing and increasing the yield of gypenosides or their derivatives through metabolic engineering.
2. Materials and methods
2.1. DNA extraction and genome sequencing
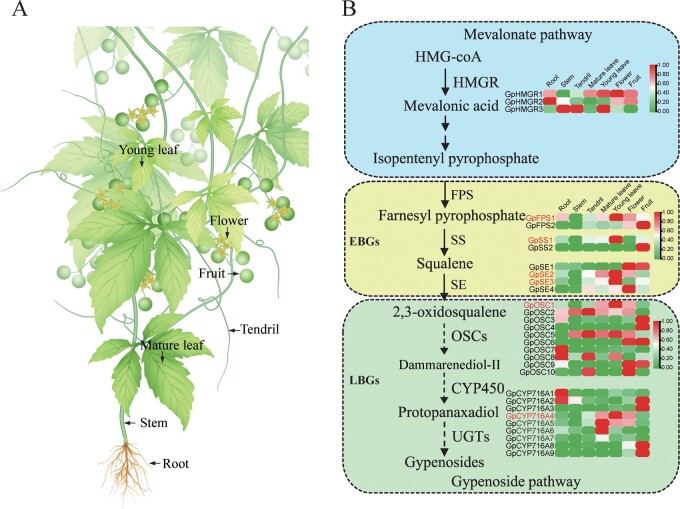
Gynostemma pentaphyllum were grown in an experimental medicinal botanical garden at Guangxi University of Chinese Medicine (Nanning, China). Root, stem, tendril, young leaf, mature leaf, flower, and fruit of G. pentaphyllum were collected as shown in Fig. 3B, and immediately frozen in liquid nitrogen and stored at −80°C until use. Tissues from three plants of G. pentaphyllum were used as three biological repetitions.
Figure 3.
Comparative transcriptome analysis of genes involved in the gypenoside biosynthetic pathway. (A) Different plant parts of G. pentaphyllum. (B) A simplified representation of the gypenoside biosynthetic pathway. Gypenoside enzyme-encoding genes are divided into two groups—early and late. Early biosynthesis genes (EBGs) encode enzymes leading to 2,3-oxidosqualene production, whereas late biosynthesis genes (LBGs) encode enzymes leading to gypenoside production. The expression value of each gene is coloured in log10 (FPKM) in seven tissues: root, stem, tendril, young leaf, mature leaf, flower, and fruit. Low to high expression is indicated by a change in colour from green to red. Genes showing young leaf-specific expression are indicated in red. Dashed arrows indicate that the gene function needs to be verified.
Genomic DNA was extracted from young leaves of G. pentaphyllum and used to construct Illumina DNA libraries whose fragment size was ∼350 bp, according to the standard protocols provided by the Illumina company. The paired-end libraries were sequenced on an BGISEQ-500 system for error correction and K-mer analysis12 and generated a total of ∼82 Gb of clean data.
A PacBio library was constructed using a SMRTbell Template Prep Kit 1.0 (PacBio, Menlo Park, CA, USA) and sequenced on a PacBio Sequel II system. We obtained one SMRT cells with ∼256 Gb of sequencing clean data (coverage of 403×) from the PacBio Sequel II platform a with an N50 read length of 27.91 kb. We denovo assembled contigs by NextDenovo v2.0-beta.1 (https://github.com/Nextomics/NextDenovo) with PacBio reads. Then the contigs were polished with NextPolish v1.0.413 using Illumina paired-end reads. Purgehaplotigs were further used to reduced redundant sequences. The detailed pipeline of genome assembly and polish was described in GitHub (https://github.com/hdh016/Gynostemma-pentaphyllum).
We also constructed Hi-C fragment libraries (insert size of 350 bp) and sequenced them using the BGISEQ-500 platform. Finally, we obtained a total of ∼33 Gb of clean reads for Hi-C analyses. Then, 3D-DNA version 18011414 were used to cluster, order, and orient the contigs with default parameters. The oriented contigs were used to build the interaction matrices with juicer,15 and were manually corrected with Juicebox assembly tools v2.1.0. We evaluated the completeness and quality of the final assembled genome through benchmarking universal single-copy ortholog (BUSCO) v5.1.216 tests using gene content from the Embryophyta_odb10 database.
2.2. Gene prediction and functional annotation
For repeat elements annotation, a curated TE library was first built by a combination of homology- and structure-based approaches using EDTA,17 RepeatMasker (version 4.0.7, rmblast-2.2.28) was then used to mask the whole genome.
We integrated ab initio gene predictions, homology searches, and RNA sequencing (RNA-seq) analysis to predict gene models. Ab initio gene prediction and annotation were performed by Augustus v3.318 and GlimmerHMM.19 Then published protein sequences of Cucurbitaceae family (Siraitia grosvenorii, Cucurbita moschata, Lagenaria siceraria, Citrullus lanatus, Benincasa hispida, Cucumis sativus, and Cucumis melo) were used to perform homologous searches by Genomethreader v1.7.1.20 For RNA analysis, we first obtained transcriptomes by sequencing high-quality RNA from root, stem, tendril, young leaf, mature leaf, flower, and fruit tissues and sequenced them by the BGISEQ-500 platform. We removed adapters and discarded reads with >10% N bases or reads having more than 20% bases of low quality (below 5) using NGS QC Toolkit21 v.2.3.372 and finally generated 144 Gb of clean data. RNA-seq reads were mapped to the assembled genome to obtain the mapping rate through HISAT2 v2.1.0.22 Then we de novo and genome-guided assembled the transcriptomes by Trinity v.2.8.5.23 The assemblies were further refined by using PASA pipeline. All the predicted gene structures above were integrated by EVM v1.1.1.24 Finally, we used PASA v2.3.325 to annotate the UTR and alternative splicing isoforms using for two rounds and obtain the final gene models.
For functional annotation of protein-coding genes, nucleotide sequences of high-confidence genes were searched against SwissProt and TrEMBL databases. We further used Mercator sequence annotation website26 and eggNOG software27 to perform function prediction. All these function predictions were integrated.
2.3. Comparative genomic analysis
An all-vs-all blastp was first performed using corresponding protein sequences of G. pentaphyllum and other species in Cucurbitaceae family (mentioned above). Then the gene family clustering was conducted by OrthoMCL.28 Then single-copy orthologous genes were retrieved and aligned by Muscle v3.8.1551.29 The maximum likelihood tree was produced with grapefruit as the outgroup using the substitution model GTRGAMMA of the RAxML v8.2.12 software.30 A total of 1,000 rapid bootstrap inferences were performed. The bootstrap values for each node that were > 60 were labelled in the tree. The tree was visually validated using iTOL.31 For the estimation of divergence time, calibration times between Vitis vinifera and Cucurbitaceae family (mentioned above), C. sativus and C. melo were selected as calibration time32 and the estimated divergence time was confirmed using r8s v1.8.1.33
2.4. Whole-genome duplication analysis and collinearity analysis
To examine whole-genome duplication (WGD) in of G. pentaphyllum, we extracted all homologous proteins between within G. pentaphyllum using an all-to-all search in BLASTP with an e value cut-off of 1e−6. We then used MCScanX34 with default parameters to identify collinear blocks, each containing at least five collinear gene pairs. The Ks between the syntenic homologous gene pairs was calculated by KaKs_calculator v2.035 using the YN model.
2.5. Transcriptome sequencing and data analysis
Total RNA of all the tissues was extracted by using TRIzol reagent (Takara). The clean reads were mapped to the reference genome by HISAT v2.1.0.22 The expression level was evaluated by normalization to fragments per kilobase of exon model per million mapped reads (FPKM) value calculated from the number of aligned reads for each gene. Differential expression analyses were performed by using Cufflinks v2.2.1.36 Genes with adjusted P-values < 0.05 and at least 2-fold expression changes were defined as differentially expressed genes. The correlation between replicates was analysed by using R package. Gene Ontology (GO) enrichment analysis was performed by the web-based agriGO37 with the annotation data of assembled genome as the statistical background. Weighted gene co-expression network analysis (WGCNA) package38 was used for co-expression analysis, for which genes expressed (FPKM ≥ 2) at least in one tissue were used as the input data.
2.6. Promoter cloning and analysis
For the promoter cloning, the promoter sequences of GpFPS1, GpSS1, GpSE2, GpOSC1, and GpCYP716A4 genes were amplified and inserted into a pTOPO-Blunt vector (Aidlab, China). Primer information were presented in Supplementary Table S1. The DNA binding sites were predicted in about 2,000 bp region upstream of the start codon of GpFPS1, GpSS1, GpSE2, GpOSC1 and GpCYP716A4 genes by using PLACE (http://www.dna.affrc.go.jp/PLACE) and plantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
3. Results and discussion
3.1. Genome sequencing, assembly, and annotation
To acquire a high-quality reference genome sequence of G. pentaphyllum, a hierarchical approach was applied for the chromosome-level genome assembly. Before deep sequencing, ∼82.36 Gb (∼130× genome coverage) short reads of Illumina sequencing were obtained, and a genomic survey was performed. The estimated genome size of G. pentaphyllum was ∼635 Mb, and the estimated heterozygosity rate was ∼0.90% using K-mer analysis (Supplementary Fig. S2). The genome was de novo assembled based on ∼256 Gb (∼403× genome coverage) PacBio long-read sequencing data and improved by Illumina paired-end short reads to finally generate 11 chromosome-scale scaffolds (pseudochromosomes) using 33.02 Gb of clean Hi-C data (∼52× genome coverage). The total length of the final chromosome-level genome was 582 Mb, ∼91.65% of the estimated genome size, which is similar to that of Sechium edule (i.e. 606 Mb)39 but larger than those of other members of the Cucurbitaceae family, such as C.sativus,40C. melo,41 and S.grosvenorii.42 The genome assembly comprised 1,232 contigs and had a scaffold N50 of 50.78 Mb. Among the obtained sequences, 97.13% were anchored to the 11 chromosomes (Fig. 1A), with the pseudochromosome length ranging between 37.28 and 64.79 Mb (Supplementary Table S2). An overview of the genome assembly is shown in Fig. 1B and Supplementary Table S3.
Figure 1.
Overview of the G. pentaphyllum genome assembly. (A) Genome-wide Hi-C heatmap of G. pentaphyllum. Interaction frequency distribution of Hi-C links among chromosomes; the change in colour from white to red indicates the frequency of Hi-C links from low to high (0–10). (B) Synteny and distribution of genomic features. a, Gene density (genes per Mb); b, TE coverage; c, GC content; d, position of the AP2 gene family (green), ERF gene family (red), and RAV gene family (blue); e, collinearity regions.
To test the high fidelity of the genome, Illumina short reads were mapped to the reference genome, and 99.38% of the Illumina sequencing data can be mapped back to the assembly with 95.25% overall coverage. In addition, the base error rate of the G. pentaphyllum genome assembly was < 0.001%, indicating its high accuracy. To assess the completeness of the genome assembly, BUSCO analysis was performed and indicated 94.90% completeness (Supplementary Table S4). Besides, the long terminal repeat (LTR) assembly index score was 14.60, suggesting that the G. pentaphyllum genome achieved reference level quality. Taken together, these results supported the high quality of the assembled G. pentaphyllum genome in both genic and intergenic regions.
To screen the repeat sequences in the assembled G. pentaphyllum genome, structure prediction and de novo prediction approaches were adopted. A total of 59.14% of the genome sequence was identified as transposable elements (TEs). LTR retrotransposons were the major class, accounting for 39.16% of the whole genome. Among the LTRs, LTR/Gypsy elements were the most abundant, occupying 19.32% of the whole genome, followed by LTR/Copia (9.12%; Supplementary Table S5). TEs were unevenly distributed across the chromosomes and were particularly enriched in the centromeric regions. In addition, we annotated 25,282 protein-coding genes in G. pentaphyllum using a comprehensive analysis method based on ab initio prediction and homology alignment from transcriptome data of seven tissues (Supplementary Table S6), including root, stem, tendril, young leaf, mature leaf, flower, and fruit. The number of protein-coding genes detected in the G. pentaphyllum genome was similar to that annotated for the C. sativus genome (i.e. 24,317 protein-coding genes)40 but less than that annotated for the C. moschata genome (i.e. 32,205 protein-coding genes).43 The average length of transcripts, coding sequences, and exons was 2,110, 1,238, and 335 bp, respectively (Supplementary Table S7).
3.2. Phylogenetic relationships and WGD analyses
To investigate the evolution of the G. pentaphyllum genome, we performed a comparative analysis with eight representative plant species, including monk fruit (S. grosvenorii), pumpkin (C. moschata), bottle gourd (L.siceraria), watermelon (C.lanatus), wax gourd (B.hispida), cucumber (C. sativus), melon (C. melo), and grape (V.vinifera). We identified 24,925 gene families, of which 2,600 gene families were considered single-copy orthologs. A phylogenetic tree was constructed based on these single-copy orthologs. According to the phylogenetic tree, G. pentaphyllum is a primitive species in the Cucurbitaceae family and diverged from it 48 million years ago (Fig. 2A). Among all gene families, 805 gene families accounting for 3,370 genes were specific to G. pentaphyllum. Gene ontology enrichment analysis of these specific genes revealed functional categories associated with biosynthetic processes such as defence response, collagen catabolic process, and multicellular organismal macromolecule metabolic process, which reflected their particular biological characteristics (Supplementary Table S8).
Figure 2.
Genome evolutionary history of G. pentaphyllum. (A) Phylogenetic analysis and divergence time estimation in G. pentaphyllum and eight other species. The phylogenetic tree was constructed based on 2,600 single-copy orthologs and rooted with V. vinifera as the outgroup. Red represents expanded gene families, and blue represents contracted gene families. Scale bar, 1 cm. (B) Ks distribution of G. pentaphyllum and other plant species. Lines show Ks distribution within (continuous) and between (dashed) genomes. (C) Gene copy number distribution in G. pentaphyllum and other sequenced plant genomes of the Cucurbitaceae family.
WGD has been considered a major driving force in plant evolution. To investigate WGD events of G. pentaphyllum, we first identified syntenic blocks within the G. pentaphyllum genome and extracted paralogous gene pairs in these syntenic blocks. The Ks distribution of orthologs suggested the absence of a recent WGD event in G. pentaphyllum as in other species of the Cucurbitaceae family (Fig. 2B), which is consistent with the results of a recent study.44 Ortholog cluster analysis was performed for each species, revealing that G. pentaphyllum contains 6.67% multiple-copy genes, which is similar to other cucurbit species, including L. siceraria, C. lanatus, B. hispida, C. sativus, and C. melo, in which no recent WGD events occurred, but remarkably lower than C. moschata, whose genome underwent a recent WGD event43 (Fig. 2C). In addition, we performed a genome collinearity analysis between G. pentaphyllum, C. sativus, C. melo, and L. siceraria, which showed that a large number of interchromosomal rearrangement events occurred between these species (Supplementary Fig. S3).
3.3. Gypenoside biosynthetic pathway genes in G. pentaphyllum
In gypenoside biosynthesis, EBGs comprising FPS, SS, and SE have been elucidated. However, despite studies on gypenoside biosynthesis using transcriptome data, LBGs comprising OSC, CYP450, and UGT have remained elusive since completeness and accuracy of RNA-seq limit their mining and identification. Our high-quality genome assembly allowed reconstruction of the gypenoside biosynthetic pathway, specifically capturing the implicated enzyme genes in the late stage. Comparative transcriptome analysis was performed on seven tissues, and samples for transcriptome sequencing were divided into root, stem, tendril, young leaf, mature leaf, flower, and fruit (Fig. 3A).
We discovered that eight genes belonging to EBGs functioned in the gypenoside biosynthetic pathway. According to their expression levels in the seven tissue types, at least one of each EBG was found to be highly expressed in young leaf tissue. In detail, EBGs GpFPS1, GpSS1, GpSE2, and GpSE3 showed young leaf-specific expression (Fig. 3B; Supplementary Table S9), indicating that gypenosides are mainly synthesized in the young leaves of G. pentaphyllum, which is consistent with previous studies.45 By further screening LBGs in the G. pentaphyllum genome, 10 enzyme-encoding genes functioning as OSC (Supplementary Fig. S4), 216 enzyme genes with predicted function as CYP450 (of which nine belonged to the CYP716 family; Supplementary Fig. S5), and 151 unigenes annotated as UGTs (Supplementary Fig. S6) were identified. Usually, the structural genes within the same pathway are co-expressed in a specific tissue. By screening young leaf-specific expression genes, 23 LBG genes of the gypenoside biosynthetic pathway, including an OSC gene, 10 CYP450 genes (with 1 CYP450 gene from the CYP716 family), and 12 UGT genes (with 10 UGT genes from UGT71, 73, 74, 85, and 94 subfamilies) were found. These genes were considered candidate genes responsible for gypenoside biosynthesis (Fig. 3B; Supplementary Table S9). Interestingly, among the 10 candidate UGT genes, Gp2g_007730, Gp2g_007750, Gp2g_007760, and Gp2g_007780, which belonged to the UGT85 subfamily and formed a gene cluster, were neighbouring each other by an intergenic region of 107.4 kb on chromosome 2 in the reference G. pentaphyllum genome. This finding provides a unique perspective on gypenoside biosynthesis innovation and diversification.
Subtle differences in the structures of specific metabolites can lead to changes in their pharmacological and biological activities. Glycosylation, a critical step determining the structure and pharmacological effects of triterpenoid compounds as well as the final step of the triterpenoid biosynthetic pathway, is catalysed by a series of UGTs. Interestingly, we discovered a gene cluster of four UGTs from the UGT85 subfamily on chromosome 2, which were regarded as important candidate genes involved in catalyzing glycosylation in tissue-specific gypenoside biosynthesis. In Catharanthus roseus, the ORCA gene cluster consists of five AP2/ERF TFs, which are known to regulate the biosynthesis of monoterpenoid indole alkaloids.46,47 Because functional metabolic gene clusters have been reported in C. roseus, identifying and analysing gene clusters are promising means to identify candidate genes involved in the biosynthesis of specialized metabolites.48 On the one hand, compared with non-cluster pathways, UGTs forming a gene cluster for potential triterpene scaffold-modifying enzymes is beneficial for plants to coordinate gene expression and regulation and improve the efficiency of product synthesis. On the other hand, because triterpenoid saponins were mainly involved in the defence against pathogens and pests, the formation of a gene cluster may be the result of natural selection.
3.4. Construction of the gypenoside biosynthesis regulatory network
Secondary metabolite biosynthesis often shows a tissue-specific expression pattern, as exemplified by cucurbitacins.49 Therefore, to construct the regulatory network of the gypenoside biosynthetic pathway, transcriptome data of seven tissues were selected to first generate a tissue-specific gene co-expression network using WGCNA (Fig. 4A). As shown in Fig. 4B, the seven-tissue network incorporated 11 clusters of co-expression modules, each of which represented the most notable component genes. Among the 11 distinct modules, six modules showed an expression pattern closely linked to a specific tissue (r > 0.9), such as MEdarkred for young leaf, MEmidnightblue for stem, MEyellow for fruit, MElightgreen for root, MEblack for tendril, and MEbrown for flower, indicating that these modules dominate various tissue-specific biological processes. In our study, gypenosides were mainly synthesized in the young leaves, and the MEdarkred module, which showed significant correlation with the young leaf-specific expression pattern (r > 0.9), was considered a key module closely related with gypenoside biosynthesis. A total of 910 genes, including the key candidate EBGs and LBGs, were included in the MEdarkred module. In addition, 64 genes (including 9 basic helix-loop-helix (bHLH), 5 MYB, 3 AP2/ERF, 2 bZIP, and 1 WRKY genes) from the MEdarkred module were identified as TF-encoding genes using iTAK50 (Supplementary Table S10).
Figure 4.
Tissue-specific gene co-expression network in G. pentaphyllum. (A) Dendrogram with colour annotation. (B) Module–tissue association matrix classifying the genes expressed in seven tissues into 11 co-expression modules; each row corresponds to a module and each column corresponds to a specific tissue. The colour of each cell at the row–column intersection indicates the correlation coefficient between the module and the tissue. A low to high degree of correlation between a specific module and tissue type is indicated by a change in colour from blue to red.
Overexpression or interference in FPS, SS, SE, OSC, and CYP716 gene expression can increase or decrease triterpenoid saponin biosynthesis,51–55 indicating that these genes are essential for regulating triterpenoid saponin biosynthesis. In this study, GpFPS1, GpSS1, GpSE2, GpOSC1, and GpCYP716A4 promoters were cloned and their DNA-binding sites were predicted. Among these promoters, bHLH-recognizing element was the most, followed by MYB-recognizing element, GCC-box and dehydration-responsive element/C-repeat (DRE/CRT) motif for AP2/ERF, ABA-responsive element for bZIP, and W-box for WRKY, indicating that bHLH, MYB, AP2/ERF, bZIP, and WRKY are candidate TFs for regulating gypenoside biosynthesis (Fig. 5A). Finally, the candidate genes identified from EBGs, LBGs, and TFs in the MEdarkred module were selected to generate a gene co-expression regulatory network involved in the gypenoside biosynthetic pathway (Fig. 5B). For gypenoside biosynthesis, we predicted that 17 enzyme genes expressed at high levels in young leaves were directly regulated by 64 potential upstream regulators, most of which were bHLH family members.
Figure 5.
Gene co-expression regulatory network for gypenoside biosynthesis. (A) DNA binding site prediction of gypenoside biosynthesis gene promoters. Promoter sequences 2 kb upstream of the initiation codon were analysed for each gene. DNA binding sites were identified based on a previous study. (B) Predicted regulatory network and the connections among TFs and structural genes involved in the gypenoside biosynthetic pathway. Structural genes are represented by octagons and TFs are represented by circles. Lines indicate correlation, and red lines indicate a positive correlation; the darker the colour, the higher the correlation.
Regulating the gene expression of secondary metabolic pathway enzymes through TFs as well as activating related secondary metabolic biosynthetic pathways is an effective way to achieve efficient synthesis and targeted accumulation of active ingredients in medicinal plants. In this study, the reconstructed gypenoside biosynthetic pathway unraveled that MYB, bHLH, AP2/ERF, bZIP, and WRKY TFs collectively regulate gypenoside biosynthesis in the young leaf tissue. When DNA-binding site predictions were incorporated, binding sites for bHLH TFs were the most, indicating that bHLH TFs are at the centre of the gene co-expression regulatory network. We speculate that these bHLH TF family members, particularly those belonging to Groups Ia and IIId, are known to be responsible for stress responses56,57 and play vital roles in gypenoside biosynthesis. Besides, 5 MYBs, 3 AP2/ERFs, 2 bZIPs, and 1 WRKY were identified from the gene co-expression regulatory network, implying that gypenoside biosynthesis regulation is a fine and complex biological process regulated by multiple TFs of different gene families.
4. Conclusion
In conclusion, the chromosome-level G. pentaphyllum reference genome presented in this study was obtained through evolutionary and phylogenetic analyses of plants of the Cucurbitaceae family. Although it is difficult to reveal the biosynthetic pathways of complex secondary metabolism in non-model plants using only transcriptome data, we have demonstrated that multi-omics data and co-expression analysis can contribute to reconstructing the metabolic biosynthetic pathway and regulatory mechanism. We successfully constructed a gene co-expression regulatory network for gypenoside biosynthesis and inferred the potential contributions of individual TF gene family members and structural genes involved in gypenoside biosynthesis. Our study provides insights for further investigations exploring the biological activities of triterpenoid saponins for applications in medicine and agriculture.
Supplementary data
Supplementary data are available at DNARES online.
Supplementary Material
Acknowledgements
We thank J.H. Liu (Huazhong Agricultural University) for providing improvement for the manuscript. This project was supported by the Natural Science Foundation of Guangxi Zhuang Autonomous Region (2020GXNSFBA297025, GuiKe AA18118015), the Guangxi Middle-aged and Young Teachers' Basic Ability Promotion Project (2020KY07039), the Guangxi University of Chinese Medicine Scientific Research fund (2019BS007), the Guangxi Key Laboratory of Zhuang and Yao Ethnic Medicine Open Project Fund (20-065-14) and the Youth Innovation Research Team Project of Guangxi University of Chinese Medicine (2018QT001). We would like to thank TopEdit (www.topeditsci.com) for linguistic assistance during the preparation of this manuscript.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
D.H. and Y.T. conceived this project. D.H. designed the experiments. D.H. and S.X. prepared the samples. D.H., R.H.M. and S.X. analysed the bioinformatics data. D.H. and R.H. wrote the article. J.W., S.Y., L.L., R.H. and Y.T provided valuable suggestions on the research design and the improvement of the manuscript.
Data availability
The raw genome and transcriptome sequencing data have been deposited in NCBI under accession codes PRJNA720501 and PRJNA631355.
References
- 1. Zhang X., Su H., Yang J., Feng L., Li Z., Zhao G.. 2019, Population genetic structure, migration, and polyploidy origin of a medicinal species Gynostemma pentaphyllum (Cucurbitaceae), Ecol. Evol., 9, 11145–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ji H.K., Yong N.H.. 2011, Dammarane-type saponins from Gynostemma pentaphyllum, Phytochemistry, 72, 1453–9. [DOI] [PubMed] [Google Scholar]
- 3. Lundqvist L.C.E., Rattigan D., Ehtesham E., Demmou C., Ostenson C.G., Sandstrom C.. 2019, Profiling and activity screening of Dammarane-type triterpen saponins from Gynostemma pentaphyllum with glucose-dependent insulin secretory activity, Sci. Rep., 9, 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Razmovski-Naumovski V., Huang T.H.-W., Tran V.H., Li G.Q., Duke C.C., Roufogalis B.D.. 2005, Chemistry and pharmacology of Gynostemma pentaphyllum, Phytochem. Rev., 4, 197–219. [Google Scholar]
- 5. Li Y., Lin W., Huang J., Xie Y., Ma W.. 2016, Anti-cancer effects of Gynostemma pentaphyllum (Thunb.) Makino (Jiaogulan), Chin. Med., 11, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang J., Ha T.K.Q., Shi Y.P., Oh W.K., Yang J.L.. 2018, Hypoglycemic triterpenes from Gynostemma pentaphyllum, Phytochemistry, 155, 171–81. [DOI] [PubMed] [Google Scholar]
- 7. Lee H.S., Lim S.M., Jung J.I., et al. 2019, Gynostemma pentaphyllum extract ameliorates high-fat diet-induced obesity in C57BL/6N mice by upregulating SIRT1, Nutrients, 11, 2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thimmappa R., Geisler K., Louveau T., O’Maille P., Osbourn A.. 2014, Triterpene biosynthesis in plants, Annu. Rev. Plant Biol., 65, 225–57. [DOI] [PubMed] [Google Scholar]
- 9. Xu J., Chu Y., Liao B., et al. 2017, Panax ginseng genome examination for ginsenoside biosynthesis, Gigascience, 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seki H., Tamura K., Muranaka T.. 2015, P450s and UGTs: key players in the structural diversity of triterpenoid saponins, Plant Cell Physiol., 56, 1463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rahimi S., Kim J., Mijakovic I., et al. 2019, Triterpenoid-biosynthetic UDP-glycosyltransferases from plants, Biotechnol. Adv., 37, 107394. [DOI] [PubMed] [Google Scholar]
- 12. Liu B., Shi Y., Yuan J., et al. 2013, Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects, Quant. Biol., 35, 62–7. [Google Scholar]
- 13. Hu J., Fan J., Sun Z., Liu S.. 2020, NextPolish: a fast and efficient genome polishing tool for long-read assembly, Bioinformatics, 36, 2253–5. [DOI] [PubMed] [Google Scholar]
- 14. Dudchenko O., Batra S.S., Omer A.D., et al. 2017, De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds, Science., 356, 92–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durand N.C., Shamim M.S., Machol I., et al. 2016, Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments, Cell Syst., 3, 95–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seppey M., Manni M., Zdobnov E.M.. 2019, BUSCO: assessing genome assembly and annotation completeness, Methods Mol. Biol., 1962, 227–45. [DOI] [PubMed] [Google Scholar]
- 17. Ou S., Su W., Liao Y., et al. 2019, Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline, Genome Biol., 20, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mario S., Oliver K., Irfan G., Alec H., Stephan W., Burkhard M.. 2006, AUGUSTUS: ab initio prediction of alternative transcripts, Nucleic Acids Res., 34, W435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Majoros W., Pertea M., Salzberg S.. 2004, TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders, Bioinformatics, 20, 2878–9. [DOI] [PubMed] [Google Scholar]
- 20. Gremme G., Brendel V., Sparks M.E., Kurtz S.. 2005, Engineering a software tool for gene structure prediction in higher organisms, Inform Software Tech., 47, 965–78. [Google Scholar]
- 21. Patel R.K., Mukesh J., Liu Z.. 2012, NGS QC toolkit: a toolkit for quality control of next generation sequencing data, PLoS One., 7, e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D., Langmead B., Salzberg S.L.. 2015, HISAT: a fast spliced aligner with low memory requirements, Nat. Methods., 12, 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grabherr M.G., Haas B.J., Yassour M., et al. 2011, Full-length transcriptome assembly from RNA-Seq data without a reference genome, Nat. Biotechnol., 29, 644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haas B.J., Salzberg S.L., Zhu W., et al. 2008, Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments, Genome Biol., 9, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haas B.J., Delcher A.L., Mount S.M., et al. 2003, Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies, Nucleic Acids Res., 31, 5654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lohse M., Nagel A., Herter T., et al. 2014, Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data, Plant. Cell Environ., 37, 1250–8. [DOI] [PubMed] [Google Scholar]
- 27. Huerta-Cepas J., Szklarczyk D., Heller D., et al. 2019, eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses, Nucleic Acids Res., 47, D309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L., Stoeckert C.J. Jr, Roos D.S.. 2003, OrthoMCL: identification of ortholog groups for eukaryotic genomes, Genome Res., 13, 2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edgar R.C. 2004, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 32, 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexandros S. 2014, RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies, Bioinformatics., 30, 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Letunic I., Bork P.. 2019, Interactive Tree Of Life (iTOL) v4: recent updates and new developments, Nucleic Acids Res., 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sudhir K., Glen S., Michael S., Blair H.S.. 2017, TimeTree: a resource for timelines, Timetrees, and divergence times, Mol Biol Evol., 7, 1812. [DOI] [PubMed] [Google Scholar]
- 33. Sanderson M.J. 2003, R8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock, Bioinformatics., 19, 301–2. [DOI] [PubMed] [Google Scholar]
- 34. Wang Y., Tang H., DeBarry J.D., et al. 2012, MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity, Nucleic Acids Res., 40, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang D., Zhang Y., Zhang Z., Zhu J., Yu J.. 2010, KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies, Genomics. Proteomics Bioinformatics., 8, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C., Williams B.A., Pertea G., et al. 2010, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotechnol., 28, 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tian T., Liu Y., Yan H., et al. 2017, agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update, Nucleic Acids Res., 45, W122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langfelder P., Horvath S.. 2008, WGCNA: an R package for weighted correlation network analysis, BMC Bioinformatics., 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fu A., Wang Q., Mu J., et al. 2021, Combined genomic, transcriptomic, and metabolomic analyses provide insights into chayote (Sechium edule) evolution and fruit development, Hortic. Res., 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Q., Li H., Huang W., et al. 2019, A chromosome-scale genome assembly of cucumber (Cucumis sativus L.), GigaScience., 8, giz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H., Li X., Yu H., et al. 2019, A high-quality melon genome assembly provides insights into genetic basis of fruit trait improvement, iScience., 22, 16–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia M., Xue H., Hang H., et al. 2018, Improved de novo genome assembly and analysis of the Chinese cucurbit Siraitia grosvenorii, also known as monk fruit or luo-han-guo, GigaScience., 7, giy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun H., Wu S., Zhang G., et al. 2017, Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes, Mol. Plant., 10, 1293–306. [DOI] [PubMed] [Google Scholar]
- 44. Guo J., Xu W., Hu Y., et al. 2020, Phylotranscriptomics in Cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations, Mol. Plant., 13, 1117–33. [DOI] [PubMed] [Google Scholar]
- 45. Xu S., Yao S., Huang R., Tan Y., Huang D.. 2020, Transcriptome-wide analysis of the AP2/ERF transcription factor gene family involved in the regulation of gypenoside biosynthesis in Gynostemma pentaphyllum, Plant Physiol. Biochem., 154, 238–47. [DOI] [PubMed] [Google Scholar]
- 46. Paul P., Singh S.K., Patra B., Sui X., Pattanaik S., Yuan L.. 2017, A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus, New Phytol., 213, 1107–23. [DOI] [PubMed] [Google Scholar]
- 47. Paul P., Singh S.K., Patra B., Liu X., Pattanaik S., Yuan L.. 2020, Mutually regulated AP2/ERF gene clusters modulate biosynthesis of specialized metabolites in plants, Plant Physiol., 182, 840–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh S.K., Patra B., Paul P., Liu Y., Pattanaik S., Yuan L.. 2020, Revisiting the ORCA gene cluster that regulates terpenoid indole alkaloid biosynthesis in Catharanthus roseus, Plant Sci., 293, 110408. [DOI] [PubMed] [Google Scholar]
- 49. Zhou Y., Ma Y., Zeng J., et al. 2016, Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae, Nat. Plants., 2, 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng Y., Jiao C., Sun H., et al. 2016, iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases, Mol. Plant., 9, 1667–70. [DOI] [PubMed] [Google Scholar]
- 51. Kim Y.K., Kim Y.B., Uddin M.R., Lee S., Kim S.U., Park S.U.. 2014, Enhanced triterpene accumulation in Panax ginseng hairy roots overexpressing mevalonate-5-pyrophosphate decarboxylase and farnesyl pyrophosphate synthase, ACS Synth. Biol., 3, 773–9. [DOI] [PubMed] [Google Scholar]
- 52. Lee M.H., Jeong J.H., Seo J.W., et al. 2004, Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene, Plant Cell Physiol., 45, 976–84. [DOI] [PubMed] [Google Scholar]
- 53. Han J.Y., In J.G., Kwon Y.S., Choi Y.E.. 2010, Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng, Phytochemistry., 71, 36–46. [DOI] [PubMed] [Google Scholar]
- 54. Han J.Y., Kwon Y.S., Yang D.C., Jung Y.R., Choi Y.E.. 2006, Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng, Plant Cell Physiol., 47, 1653–62. [DOI] [PubMed] [Google Scholar]
- 55. Chun J.H., Adhikari P.B., Park S.B., Han J.Y., Choi Y.E.. 2015, Production of the dammarene sapogenin (protopanaxadiol) in transgenic tobacco plants and cultured cells by heterologous expression of PgDDS and CYP716A47, Plant Cell Rep., 34, 1551–60. [DOI] [PubMed] [Google Scholar]
- 56. Ran J.H., Shen T.T., Liu W.J., Wang X.Q.. 2013, Evolution of the bHLH genes involved in stomatal development: implications for the expansion of developmental complexity of stomata in land plants, PloS One., 8, e78997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Qi T., Wang J., Huang H., et al. 2015, Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis, Plant Cell., 27, 1634–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw genome and transcriptome sequencing data have been deposited in NCBI under accession codes PRJNA720501 and PRJNA631355.